Background: Type III polyketide synthases (PKSs) from fungi produce a wide variety of polyketides.
Results: CsyB catalyzes the formation of 3-acetyl-4-hydroxy-6-alkyl-α-pyrone (AcAP) from a fatty acyl-CoA, a malonyl-CoA, and an acetoacetyl-CoA.
Conclusion: CsyB is a novel type III PKS that possesses two β-ketoacyl-CoA coupling activities.
Significance: Characterization of CsyB expands the use of type III PKSs for polyketide production.
Keywords: Aspergillus, Enzyme Catalysis, Enzyme Mechanism, Fatty Acid Metabolism, Natural Product Biosynthesis, Polyketide, α-Pyrone, Type III Polyketide Synthase
Abstract
The type III polyketide synthases from fungi produce a variety of secondary metabolites including pyrones, resorcinols, and resorcylic acids. We previously reported that CsyB from Aspergillus oryzae forms α-pyrone csypyrone B compounds when expressed in A. oryzae. Feeding experiments of labeled acetates indicated that a fatty acyl starter is involved in the reaction catalyzed by CsyB. Here we report the in vivo and in vitro reconstitution analysis of CsyB. When CsyB was expressed in Escherichia coli, we observed the production of 3-acetyl-4-hydroxy-α-pyrones with saturated or unsaturated straight aliphatic chains of C9–C17 in length at the 6 position. Subsequent in vitro analysis using recombinant CsyB revealed that CsyB could accept butyryl-CoA as a starter substrate and malonyl-CoA and acetoacetyl-CoA as extender substrates to form 3-acetyl-4-hydroxy-6-propyl-α-pyrone. CsyB also afforded dehydroacetic acid from two molecules of acetoacetyl-CoA. Furthermore, synthetic N-acetylcysteamine thioester of β-ketohexanoic acid was converted to 3-butanoyl-4-hydroxy-6-propyl-α-pyrone by CsyB. These results therefore confirmed that CsyB catalyzed the synthesis of β-ketoacyl-CoA from the reaction of the starter fatty acyl CoA thioesters with malonyl-CoA as the extender through decarboxylative condensation and further coupling with acetoacetyl-CoA to form 3-acetyl-4-hydroxy-6-alkyl-α-pyrone. CsyB is the first type III polyketide synthase that synthesizes 3-acetyl-4-hydroxy-6-alkyl-α-pyrone by catalyzed the coupling of two β-ketoacyl-CoAs.
Introduction
Polyketides are secondary metabolites produced by various organisms including bacteria, fungi, and plants. Their basic carbon skeletons are constructed by polyketide synthases (PKSs),2 which catalyze condensation of acetate units. PKSs are categorized into three classes: types I, II, and III. Type I includes multidomain enzymes, and type II includes complex enzyme systems of monofunctional subunits. Type III PKSs are relatively small homodimeric enzymes of ketosynthases of ∼42 kDa. Distributed in diverse organisms from plants, bacteria, and fungi, type III PKSs are responsible for production of a wide variety of polyketides including chalcones, stilbenes, phloroglucinols, resorcinols, benzophenones, biphenyls, bibenzyls, chromones, acridones, curcuminoids, and pyrones (1–3).
Most plant type III PKSs accept bulky aromatic starters such as 4-coumaroyl-CoA, benzoyl-CoA, and N-methylanthraniloyl-CoA. Conversely, microbial type III PKSs use aliphatic acyl-CoAs and even malonyl-CoA as a starter. For example, RppA from Streptomyces griseus, the first microbial type III PKS characterized, normally synthesizes 1,3,6,8-tetrahydroxynaphthalene from 5 units of malonyl-CoA (4). Additionally, RppA can accept alkanoyl- or alkenoyl-CoA as the starter substrate to form α-pyrones and phloroglucinols (5, 6).
Although it was believed that type III PKSs existed exclusively in plants and bacteria, recent genome projects have revealed the presence of type III PKS genes in filamentous fungi. Following our first report in 2005 on the discovery of four type III PKS genes, csyA, csyB, csyC, and csyD, from Aspergillus oryzae (7), some type III PKS genes were reported to synthesize 6-alkyl-α-pyrones, resorcinols, and resorcylic acids (8–14). Neurospora crassa 2′-oxoalkylresorcylic acid synthase accepts the CoA ester of C2 to C20 straight chain fatty acids as a starter substrate to synthesize α-pyrones, resorcinols, and resorcylic acids (8).
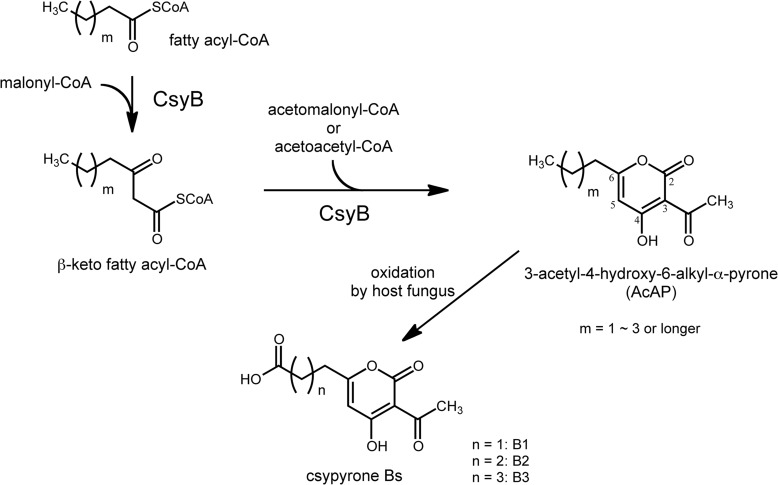
We previously reported that CsyB expression in A. oryzae resulted in the production of csypyrone Bs, which are 3-acetyl-α-pyrone compounds bearing carboxylic acid side chains (9, 11) (Fig. 1). Feeding experiments with 13C-labeled acetate indicated that fatty acyl-CoA serves as the starter for the biosynthesis of csypyrone Bs to form 3-acetyl-4-hydroxy-6-alkyl-α-pyrone (AcAP), a putative csypyrone B precursor (15).
FIGURE 1.
Proposed biosynthesis of csypyrone Bs. The condensation of fatty acyl-CoA with malonyl-CoA to form β-keto acyl-CoA and then diketide coupling with acetomalonyl-CoA or acetoacetyl-CoA give the putative intermediate AcAP. Csypyrone Bs are formed by the oxidation of the side chains by the host fungus.
It was assumed that AcAP was formed via two step reactions: 1) condensation of a fatty acyl-CoA and a malonyl-CoA to yield a β-ketoacyl intermediate and 2) its further condensation with acetomalonyl-CoA or acetoacetyl-CoA and then pyrone cyclization. The alkyl side chain of the formed AcAP is likely oxidized to give csypyrone Bs by the host fungus (15) (Fig. 1). Some microbial type III PKSs are known to produce 6-alkyl-α-pyrone with alkyl substitution at the 3 position. Streptomyces coelicolor Gcs uses ethylmalonyl-CoA to yield 3-ethyl-4-hydroxy-6-alkyl-α-pyrone germicidins (16). However, csypyrone Bs are the first example of 3-acetyl-α-pyrones produced by type III PKS. In this study, we show that CsyB catalyzes condensation of two diketo acyl-CoAs, β-keto fatty acyl-CoA and acetoacetyl-CoA, to form AcAP.
EXPERIMENTAL PROCEDURES
Materials
Escherichia coli BL21(DE3) and pET22b were purchased from Novagen (Darmstadt, Germany). Restriction endonucleases and Mighty ligation mix were obtained from Takara Biochemicals (Shiga, Japan). The pColdTF vector for cold shock expression with a trigger factor chaperone was purchased from Takara Biochemicals. Phusion Hot Start high fidelity DNA polymerase was purchased from Finnzyme (Espoo, Finland). All fatty acyl-CoA reagents used in this study were purchased from Sigma. Dehydroacetic acid was obtained from Wako (Osaka, Japan).
Cloning of CsyB
For in vivo productions of AcAPs from E. coli fermentations, the coding region of csyB was amplified from pTA-csyB (9) by PCR using the forward primer 5′-GGAATTCCATATGATTGAGCCACTACCTACCGAAGAC-3′ (the bold letters indicate an NdeI site, and the italics indicate the translation start codon) and the reverse primer 5′-CCGGAATTCCGCATGAAGGTAGCTGGAAGAG-3′ (the bold letters indicated an EcoRI site). The amplified DNA fragment was digested with NdeI/EcoRI and ligated into a pET22b expression vector to give pET22-csyB. For in vitro experiments, the PCR fragment was obtained by the same method, except the reverse primer contained a stop codon. The PCR fragment was cloned into pColdTF to yield pColdTF-csyB.
In Vivo Production of AcAP
E. coli BL21(DE3) harboring plasmid pET22-csyB was inoculated into LB liquid medium containing 50 μg/ml carbenicillin and incubated at 37 °C until the A600 reached ∼0.6. After cooling on ice for 20 min, isopropyl 1-thio-β-galactopyranoside was added to the culture to a final concentration of 0.1 mm, and the culture was incubated at 18 °C for an additional 20 h. After removal of cells by centrifugation (9,400 × g), the supernatant was extracted twice with an equal volume of ethyl acetate. The ethyl acetate extract was concentrated in vacuo and dissolved in 50% methanol containing 0.1% formic acid. This solution was then subjected to reverse phase column chromatography (COSMOSIL 75C18-OPN; Nacalai Tesque, Kyoto, Japan). The column was washed with 70% methanol containing 0.1% formic acid and was eluted with 100% methanol containing 0.1% formic acid to give a mixture of AcAPs with different aliphatic chain lengths. These AcAP mixtures were further purified by octadecyl silica gel preparative HPLC. High resolution electrospray ionization (ESI)-MS) analysis was performed to determine the molecular formula of the products, and the structures of new metabolites were determined by chemical and spectroscopic methods (MS and 1H and 13C NMR spectra (supplemental Figs. S1–S8) can be found in the supplemental material).
Synthesis of N-Acetylcysteamine Thioester (SNAC) of β-Ketohexanoic Acid
A butyryl derivative of Meldrum's acid was synthesized according to the procedure described by Oikawa et al. (17). Briefly, a solution of Meldrum's acid (1.4 mmol) was treated with butyryl chloride (1.5 mmol, 1.1 eq) in the presence of pyridine (2.8 mmol, 2 eq). The butyryl derivative was then added to a benzene solution of N-acetylcysteamine (1.3 mmol), and the resulting mixture was heated at reflux under a nitrogen atmosphere for 7 h (18). The reaction mixture was then cooled to ambient temperature and purified by column chromatography over copper sulfate impregnated silica gel followed by normal silica gel. The β-ketohexanoyl-SNAC (1.1 mmol, 72%) gave an identical 1H NMR spectrum (supplemental Fig. S1) to that described previously in the literature (19).
Expression and Purification of Recombinant TF-CsyB
E. coli BL21(DE3) harboring plasmid pColdTF-csyB was incubated until the A600 reached ∼0.6 using the same method mentioned above. After cooling on ice for 30 min, isopropyl 1-thio-β-galactopyranoside was added to the culture to a final concentration of 0.1 mm, and the culture medium was incubated at 15 °C for an additional 20 h. The cells were harvested by centrifugation (9,400 × g); then resuspended in a wash buffer containing 50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 10 mm imidazole; and disrupted by sonication. The recombinant TF-CsyB was purified by using a nickel-nitrilotriacetic acid resin (Sigma) according to the manufacturer's instructions, and buffer was then exchanged by ultrafiltration with 50 mm sodium phosphate buffer (pH 7.5) to remove imidazole. The protein concentration was determined by the Bradford method (Bio-Rad) with bovine serum albumin as the standard.
Enzyme Assay and Product Characterization
A standard assay mixture contained 100 μm malonyl-CoA, 100 μm starter fatty acyl-CoA and 10 μg of purified enzyme in 50 mm sodium phosphate buffer (pH 7.5) in a total volume of 100 μl. After preincubation for 3 min at 37 °C, assays were started by adding 10 μl of 1 mm of acetoacetyl-CoA. Incubations were carried out at 37 °C for 1 h and stopped by adding 20 μl of 6 m HCl and then 80 μl of methanol. The assay samples were centrifuged at 20,400 × g for 5 min to remove the precipitates, and the products in the supernatant were analyzed using a TOSOH 8020 HPLC apparatus equipped with a COSMOSIL Cholester column (4.6 × 150 mm; Nacalai Tesque). The products were eluted with a linear methanol gradient containing 0.1% formic acid at a flow rate of 1.0 ml/min with detection at 310 nm. The products were also analyzed by LC-ESI-MS (Shimadzu, Kyoto, Japan) to determine the molecular formula of the PKS products.
Kinetic Parameters of CsyB
A standard reaction contained acetoacetyl-CoA (concentration varied between 2.5 and 300 μm) and 10 μg of purified TF-CsyB in 50 mm sodium phosphate buffer (pH 8.0) in a total volume of 100 μl. After preincubation in the absence of TF-CsyB at 30 °C for 3 min, the reactions were initiated by adding purified TF-CsyB and then further incubated at 30 °C for 3 min before quenching with 20 μl of 6 m HCl and 80 μl of methanol. The amounts of dehydroacetic acid formed were quantified by HPLC analysis using a COSMOSIL Cholester column (4.6 × 150 mm, Nacalai Tesque) eluted isocratically with 50% methanol containing 0.1% formic acid at a flow rate of 1.0 ml/min. Calibration of the peak area of the product was based on the area of an authentic dehydroacetic acid standard. Steady-state kinetic parameters were determined from Lineweaver-Burk plots.
RESULTS
In Vivo Function
The csyB gene was inserted into the pET22b vector between the NdeI and EcoRI site to construct pET22-csyB for expression of the C terminus His6-tagged CsyB protein in E. coli. When the E. coli BL21(DE3) transformant harboring pET22-csyB was cultured at 18 °C for 20 h in the induction medium (Fig. 2A), we noticed the production of compounds in the culture medium, which were not detected in the control transformant (Fig. 3A). These compounds found in the culture medium of the CsyB transformant were not csypyrone B1–B3 but were likely csypyrone B-related pyrone compounds because they had similar UV spectra to those of csypyrone Bs.
FIGURE 2.
SDS-PAGE analyses of CsyB expression and purification. A, His6-CsyB. Lane 1, empty vector total protein; lane 2, CsyB total protein; lane 3, empty vector soluble fraction; lane 4, CsyB soluble fraction; lane 5, empty vector insoluble fraction; lane 6, CsyB insoluble fraction. B, TF-CsyB. Lane 1, empty vector total protein; lane 2, CsyB total protein; lane 3, empty vector soluble fraction; lane 4, CsyB soluble fraction; lane 5, empty vector insoluble fraction; lane 6, CsyB insoluble fraction; lane 7, flow-through fraction; lane 8, wash fraction; lane 9, purified recombinant CsyB by nickel-nitrilotriacetic acid column.
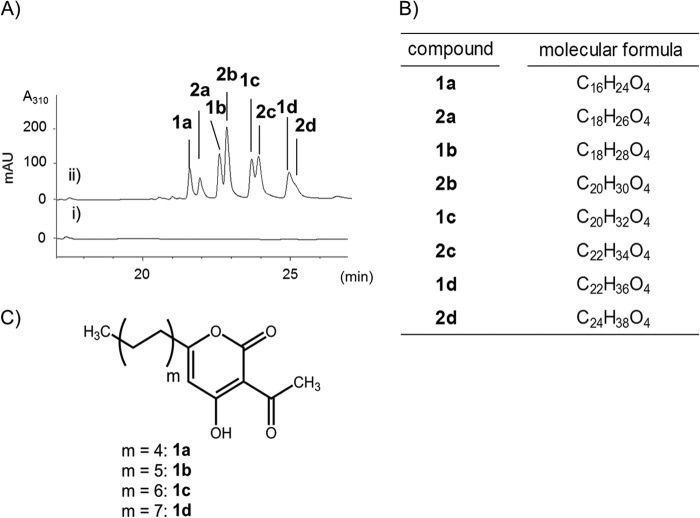
FIGURE 3.
AcAP production in the recombinant E. coli expressing CsyB. A, HPLC analysis of the ethyl acetate extract from E. coli BL21(DE3)/pET22b (panel i) or E. coli BL21(DE3)/pET22-csyB (panel ii). B, molecular formula of the products. C, structures of compounds 1a–1d.
These products were extracted with ethyl acetate and purified by reverse phase HPLC. LC-MS analysis indicated that these compounds were 3-acetyl-4-hydroxy-α-pyrones with saturated or unsaturated straight aliphatic chains of C9–C17 at the 6 position (compounds 1a–1d and 2a–2d in Fig. 3B). NMR analysis (supplemental Figs. S2–S7) confirmed that these compounds were AcAPs, csypyrone B derivatives with long alkyl or alkenyl side chains (Fig. 3C).
Fatty acid biosynthesis in E. coli is catalyzed by an enzyme system consisting of nine distinct proteins: FabA, FabB, FabD, FabF, FabG, FabH, FabI, FabZ, and ACP. They convert acetyl-CoA and malonyl-CoA into various acyl-ACP species. Thus these fatty acyl-ACPs (or CoAs) could be accepted as starters of CsyB condensation reactions because of the broad starter flexibility of CsyB. E. coli possesses an anaerobic pathway to synthesize unsaturated fatty acids (20). The FabA β-hydroxydecanoyl-ACP dehydratase catalyzes the dehydration of β-hydroxydecanoyl-ACP to cis-3-decenoyl-ACP (C10:1, Δ3) and trans-2-decenoyl-ACP (C10:1, Δ2). The cis-isomer is then elongated by a β-ketoacyl-ACP synthase, FabB or FabF, to synthesize the unsaturated fatty acids (21). ACP thioesters of unsaturated fatty acids such as palmitoleate (C16:1, Δ9) and cis-vaccenate (C18:1, Δ11) are also thought to be used by CsyB to form 3-acetyl-4-hydroxy-6-alkenyl-α-pyrones (compounds 2a–2d), although the position of the double bond in the unsaturated aliphatic chain of compounds 2a–2d was not confirmed yet.
From these results together with the data obtained from the acetate feeding experiments (15), we assumed that CsyB catalyzes the condensation of fatty acyl-ACP (or CoA) starter with malonyl-CoA to form the β-ketoacyl intermediate followed by further condensation with acetomalonyl-CoA or acetoacetyl-CoA. CsyB then catalyzes cyclization to form 3-acetyl-4-hydroxy-6-alkyl (or alkenyl)-α-pyrone.
In Vitro Analysis of CsyB Activity
When E. coli BL21(DE3) harboring plasmid pET22-csyB was cultured in the induction medium, recombinant C terminus His6-tagged CsyB was obtained as a soluble protein at 44 kDa (Fig. 2A), but its purity was low even after nickel affinity column purification. Thus the csyB gene was inserted into the pColdTF vector between the NdeI and EcoRI sites to prepare TF-CsyB, CsyB with both a His6 tag and trigger factor, which catalyzes the proper in vivo folding of the protein (22) at its N terminus. The recombinant TF-CsyB protein was purified using nickel-nitrilotriacetic acid resin to almost homogeneity as shown by a single protein band at 95 kDa on SDS-PAGE (Fig. 2B). This TF-CsyB was used for the following in vitro experiment.
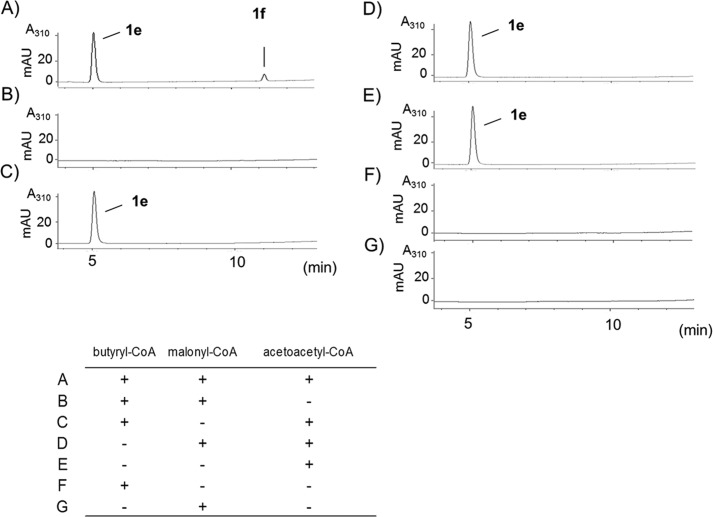
We first analyzed whether TF-CsyB could accept butyryl-CoA as a starter substrate and malonyl-CoA and acetoacetyl-CoA as extender substrates (Fig. 4). HPLC analysis of the incubation products revealed that two major products, compounds 1e and 1f, were formed by TF-CsyB (Fig. 4A). The UV spectra of these two compounds were identical to those of csypyrone Bs with absorption maxima at 311 nm, suggesting that they are AcAPs.
FIGURE 4.
In vitro assay of CsyB with butyryl-CoA, malonyl-CoA, and acetoacetyl-CoA. For reactions A–G, the CsyB assays were carried out in the presence or absence of the substrates, as indicated.
When butyryl-CoA or malonyl-CoA was absent in the assay mixture, compound 1e was detected, but compound 1f was not (Fig. 4, C and D). However, neither compound 1e nor 1f was formed when acetoacetyl-CoA was removed from the incubation mixture (Fig. 4, B, F, and G). Compound 1e was also obtained in the enzymatic reaction when only acetoacetyl-CoA was used as a substrate (Fig. 4E). These results revealed that all three substrates were required for the synthesis of compound 1f, whereas compound 1e was derived from acetoacetyl-CoA alone. High resolution ESI-MS analyses revealed that compounds 1e and 1f had molecular formulas of C8H8O4 and C10H12O4, respectively (supplemental data). Therefore, compound 1f was confirmed to be 3-acetyl-4-hydroxy-6-propyl-α-pyrone. Compound 1e was identified as dehydroacetic acid by direct comparisons with the authentic sample.
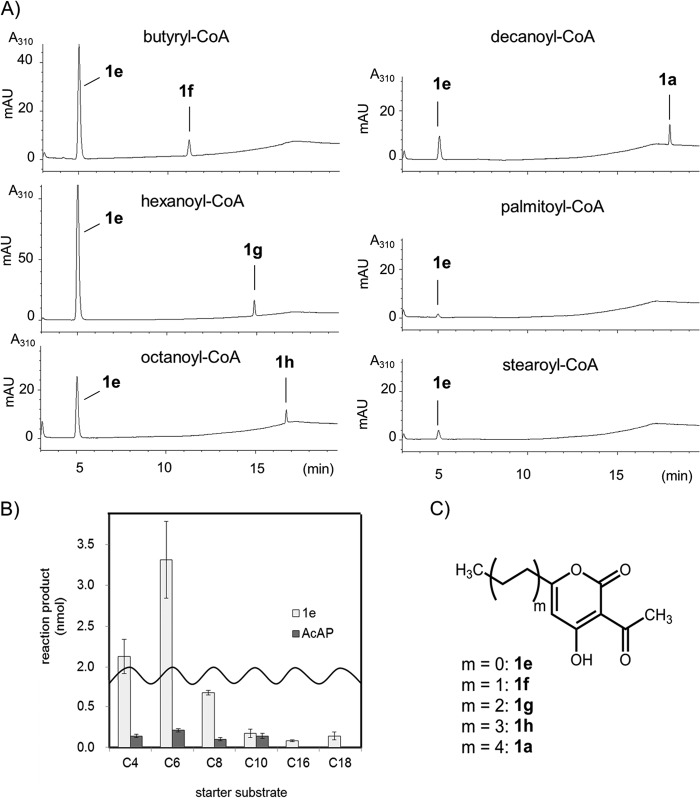
We then examined whether CsyB could accept longer straight chain aliphatic fatty acyl-CoAs as starter substrates (Fig. 5). When hexanoyl-, octanoyl-, or decanoyl-CoA was used as the starter substrate in place of butyryl-CoA, CsyB catalyzed the formation of the new products, compounds 1g, 1h, and 1a, respectively, with the same UV absorption profile as compound 1e. The molecular formulas of these products were confirmed to be C12H16O4 for compound 1g, C14H20O4 for compound 1h, and C16H24O4 for compound 1a, respectively, by high resolution ESI-MS analyses (supplemental data), indicating that CsyB could accept the aliphatic C4 to C10 fatty acyl-CoAs as starter substrates. In contrast, when palmitoyl-CoA or stearoyl-CoA was used as a substrate, no product except compound 1e was detected from the enzymatic reaction.
FIGURE 5.
AcAP formation by TF-CsyB from various fatty acyl-CoA starters. A, HPLC profiles of the reaction products catalyzed by TF-CsyB with various starter fatty acyl-CoAs. B, comparison of the products from different starter acyl-CoAs. The quantity of polyketide products was estimated from the intensity of its peak by HPLC analysis in comparison with authentic compound 1e. (means ± S.D., n = 3). C, structures of AcAPs obtained in vitro experiments.
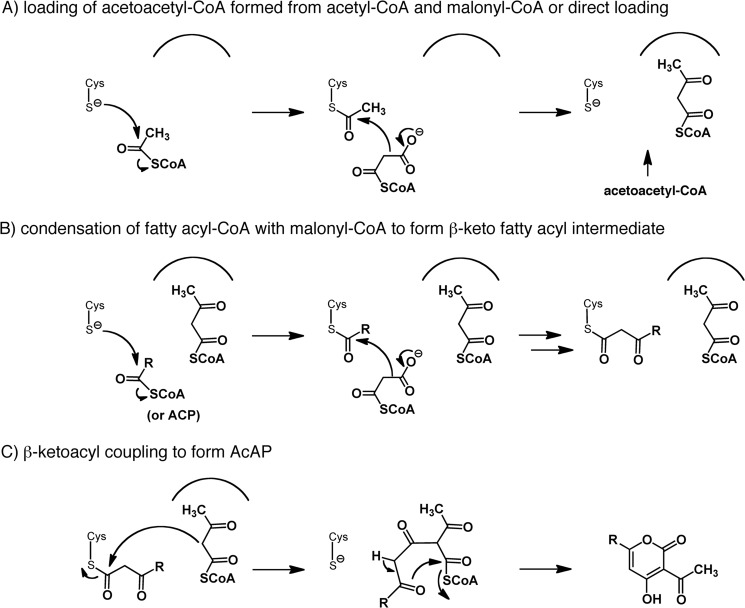
We further investigated whether CsyB could synthesize AcAP from acetyl-CoA and malonyl-CoA together with fatty acyl-CoA. Although the yield was very low, CsyB could synthesize compound 1f from acetyl-CoA, malonyl-CoA, and butyryl-CoA (Fig. 6). This thus indicated that CsyB could catalyze the condensation of acetyl-CoA with malonyl-CoA to form acetoacetyl-CoA. Overall, the results from these in vitro assays showed that CsyB uses C4 to C10 fatty acyl-CoAs as starter substrates to catalyze the condensation of two diketide CoA esters, β-keto fatty acyl-CoA and acetoacetyl-CoA, to form AcAP. In this reaction, compound 3 was also detected; it has an absorption maximum at 311 nm and the molecular formula C12H16O4 (Fig. 6A). This product was thought to be 3-butanoyl-4-hydroxy-6-propyl-α-pyrone formed by coupling of two β-ketohexanoyl-CoA intermediates, which resulted from condensation of butyryl-CoA and malonyl-CoA (Fig. 6B). With this in mind, we checked whether CsyB could synthesize compound 3 when β-ketohexanoyl-SNAC was used as a sole substrate. When 1 mm β-ketohexanoyl-SNAC was used in the CsyB reaction, the production of 3 was confirmed by HPLC and NMR analysis (supplemental Fig. S8).
FIGURE 6.
In vitro assay of CsyB. A, HPLC profiles of the reaction products from the reactions of butyryl-CoA and malonyl-CoA with acetoacetyl-CoA (panel i) or acetyl-CoA (panel ii). B, structure of compound 3.
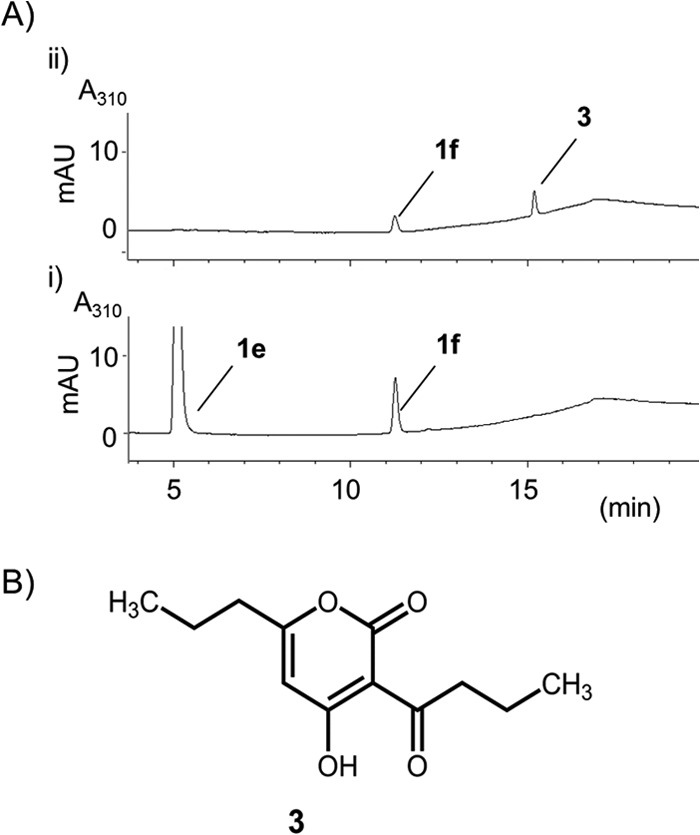
Enzymatic Properties of CsyB
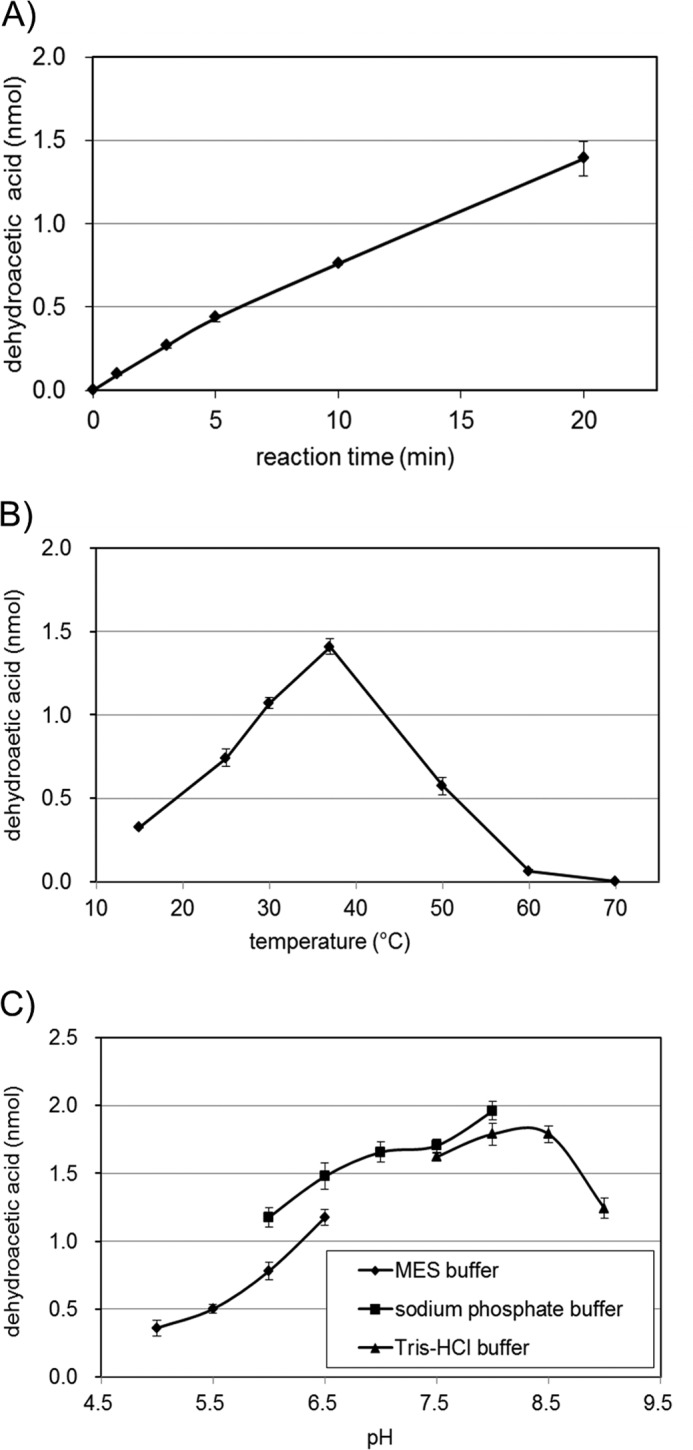
To further characterize the enzymatic properties of CsyB, an assay system to monitor the formation rate of compound 1e from acetoacetyl-CoA by CsyB was established. The optimum temperature was 37 °C, and the optimum pH was around 8.0 in 50 mm sodium phosphate buffer (Fig. 7). The Km value for acetoacetyl-CoA was determined to be 14.4 ± 0.5 μm, and kcat and kcat/Km were 20.1 ± 0.8 s−1 and 1.39 ± 0.02 × 105 s−1 m−1, respectively (Fig. 8).
FIGURE 7.
Enzymatic properties of CsyB. A–C, time (A), pH (B), and temperature (C) dependences for the formation of dehydroacetic acid (compound 1e) from acetoacetyl-CoA by CsyB. The quantity of compound 1e was estimated from the intensity of its peak by HPLC analysis at 310 nm through a comparison with authentic compound 1e (means ± S.D., n = 3).
FIGURE 8.
Steady-state kinetic analysis of dehydroacetic acid formation by CsyB. Km for acetoacetyl-CoA was 14.4 ± 0.5 μm. kcat and kcat/Km were 20.1 ± 0.8 s−1 and 1.39 ± 0.02 × 105 s−1 m−1, respectively. The quantity of compound 1e was estimated from the HPLC peak intensity at 310 nm by comparison with the authentic compound 1e (means ± S.D., n = 3).
DISCUSSION
In this study, we carried out in vivo and in vitro analysis of the reaction catalyzed by CsyB, a type III PKS from A. oryzae. CsyB was shown to catalyze the synthesis of a β-keto fatty acyl-CoA from a starter fatty acyl-CoA and malonyl-CoA through a decarboxylative condensation followed by its further condensation with acetoacetyl-CoA to form AcAP. CsyB is the first PKS that uses acetoacetyl-CoA as the extender substrate for a direct Claisen-type attack by an active methylene instead of a decarboxylative attack by a malonyl-type extender such as acetomalonyl-CoA. The proposed reaction mechanism catalyzed by CsyB is summarized in Fig. 9.
FIGURE 9.
Proposed mechanism for the formation of AcAP by CsyB. A, formation of acetoacetyl-CoA from acetyl-CoA and malonyl-CoA, which is loaded into a possible acetoacetyl-CoA pocket. When acetoacetyl-CoA was supplied as a substrate, acetoacetyl-CoA could be loaded directly into the pocket. B, decarboxylative condensation of the starting fatty acyl-CoA (or fatty acyl-ACP) and malonyl-CoA substrates to form a β-ketoacyl intermediate. C, coupling reaction of the β-keto fatty acyl intermediate with acetoacetyl-CoA followed by pyrone formation to yield AcAP. The arc shown in the figure represents a possible acetoacetyl-CoA pocket of CysB.
CsyB initially accepts acetoacetyl-CoA and loads it directly into the possible substrate pocket because CsyB can catalyze the condensation of two acetoacetyl-CoAs to form dehydroacetic acid. If acetoacetyl-CoA is not available, CsyB forms acetoacetyl-CoA from acetyl-CoA and malonyl-CoA, and loads it into the pocket (Fig. 9A). Then the fatty acyl-CoA starter is loaded onto the active site at the catalytic center Cys-155, and decarboxylative condensation with malonyl-CoA occurs to yield a β-keto fatty acyl intermediate loaded as a thioester on the active site (Fig. 9B). This β-keto fatty acyl intermediate is subsequently subjected to the coupling reaction with the acetoacetyl-CoA, and following pyrone cyclization, AcAP is released from CsyB (Fig. 9C).
CsyB is the first type III PKS that possesses two β-ketoacyl-CoA coupling activities to synthesize AcAP. AcAPs with longer alkyl or alkenyl chains were detected in the culture medium of E. coli BL21(DE3) harboring pET22-csyB. However, palmitoyl-CoA and stearoyl-CoA could not be substrates of CsyB in the in vitro assay condition used. Although CsyB favors short chain fatty acyl-CoAs as starter, the fatty acids in E. coli are usually comprised of C16 and C18 fatty acids (C16:0, 16:1, 18:1), which represent ∼80% of the total weight (23, 24). The number of short chain fatty acyl-CoAs available as CsyB substrates would therefore be very low, and AcAPs with long carbon chains would only be detectable by in vivo reconstitution analysis.
In the CsyB in vitro reaction with acetyl-CoA, butyryl-CoA, and malonyl-CoA, compound 3 (3-butanoyl-4-hydroxy-6-propyl-α-pyrone) was formed in addition to compound 1f (3-acetyl-4-hydroxy-6-propyl-α-pyrone). When synthetic β-ketohexanoyl-SNAC was used as a substrate, CsyB catalyzed the formation of compound 3, albeit in a low yield. These results therefore indicated that CsyB can catalyze the coupling of two molecules of β-keto fatty acyl-CoA to form 4-hydroxy-6-alkyl-α-pyrone with a longer acyl substitution at the 3 position. Although the size of the cavity for the acetoacetyl extender may restrict the carbon chain length of the extender β-keto fatty acyl-CoA, AcAP derivatives with a longer acyl group at the 3 position could be formed by CsyB, which could possess useful biological activities because some AcAP derivatives have been reported to inhibit RNA polymerase and HIV-1 integrase (25–27). Elucidation of the three-dimensional structure of CsyB will provide further information on the enzyme selectivity for the substrates and the reaction mechanism for this unique coupling reaction.
Supplementary Material
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and grant from Keiryokai Research Foundation, Japan.

This article contains supplemental information and Figs. S1–S8.
- PKS
- polyketide synthase
- AcAP
- 3-acetyl-4-hydroxy-6-alkyl-α-pyrone
- ESI
- electrospray ionization
- SNAC
- N-acetylcysteamine thioeste.
REFERENCES
- 1. Austin M. B., Noel J. P. (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110 [DOI] [PubMed] [Google Scholar]
- 2. Morita H., Abe I., Noguchi H. (2010) Comprehensive Natural Products II (Mander L., Liu H.-W., eds) pp. 171–255, Elsevier, Oxford, UK [Google Scholar]
- 3. Katsuyama Y., Horinouchi S. (2010) Comprehensive Natural Products II (Mander L., Liu H.-W., eds) pp. 147–170, Elsevier, Oxford, UK [Google Scholar]
- 4. Funa N., Ohnishi Y., Fujii I., Shibuya M., Ebizuka Y., Horinouchi S. (1999) A new pathway for polyketide synthase in microorganisms. Nature 400, 897–899 [DOI] [PubMed] [Google Scholar]
- 5. Funa N., Ohnishi Y., Ebizuka Y., Horinouchi S. (2002) Properties and substrates specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J. Biol. Chem. 277, 4628–4635 [DOI] [PubMed] [Google Scholar]
- 6. Jeong J.-C., Srinivasan A., Grüschow S., Bach H., Sherman D. H., Dordick J. S. (2005) Exploiting the reaction flexibility of a type III polyketide synthase through in vitro pathway manipulation. J. Am. Chem. Soc. 127, 64–65 [DOI] [PubMed] [Google Scholar]
- 7. Seshime Y., Juvvadi P. R., Fujii I., Kitamoto K. (2005) Discovery of novel superfamily of type III polyketide synthases in Aspergillus oryzae. Biochem. Biophys. Res. Commun. 331, 253–260 [DOI] [PubMed] [Google Scholar]
- 8. Funa N., Awakawa T., Horinouchi S. (2007) Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J. Biol. Chem. 282, 14476–14481 [DOI] [PubMed] [Google Scholar]
- 9. Seshime Y., Juvvadi P. R., Kitamoto K., Ebizuka Y., Fujii I. (2010) Identification of csypyrone B1 as the novel product of Aspergillus oryzae type III polyketide synthase CsyB. Bioorg. Med. Chem. 18, 4542–4546 [DOI] [PubMed] [Google Scholar]
- 10. Seshime Y., Juvvadi P. R., Kitamoto K., Ebizuka Y., Nonaka T., Fujii I. (2010) Aspergillus oryzae type III polyketide synthase CsyA is involved in the biosynthesis of 3,5-dihydroxybenzoic acid. Bioorg. Med. Chem. Lett. 20, 4785–4788 [DOI] [PubMed] [Google Scholar]
- 11. Hashimoto M., Seshime Y., Kitamoto K., Uchiyama N., Goda Y., Fujii I. (2013) Identification of csypyrone B2 and B3 as the minor products of Aspergillus oryzae type III polyketide synthase CsyB. Bioorg. Med. Chem. Lett. 23, 650–653 [DOI] [PubMed] [Google Scholar]
- 12. Yu D., Zeng J., Chen D., Zhan J. (2010) Characterization and reconstitution of a new fungal type III polyketide synthase from Aspergillus oryzae. Enzyme Microb. Technol. 46, 575–580 [Google Scholar]
- 13. Li J., Luo Y., Lee J.-K., Zhao H. (2011) Cloning and characterization of type III polyketide synthase from Aspergillus niger. Bioorg. Med. Chem. Lett. 21, 6085–6089 [DOI] [PubMed] [Google Scholar]
- 14. Jeya M., Kim T.-S., Tiwari M. K., Li J., Zhao H., Lee J.-K. (2012) The Botrytis cinerea type III polyketide synthase show unprecedented high catalytic efficiency toward long chain acyl-CoAs. Mol. BioSyst. 8, 2864–2867 [DOI] [PubMed] [Google Scholar]
- 15. Hashimoto M., Ishida S., Seshime Y., Kitamoto K., Fujii I. (2013) Aspergillus oryzae type III polyketide synthase CsyB uses a fatty acyl starter for the biosynthesis of csypyrone B compounds. Bioorg. Med. Chem. Lett. 23, 5637–5640 [DOI] [PubMed] [Google Scholar]
- 16. Song L., Barona-Gomez F., Corre C., Xiang L., Udwary D. W., Austin M. B., Noel J. P., Moore B. S., Challis G. L. (2006) Type III polyketide synthase β-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J. Am. Chem. Soc. 128, 14754–14755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oikawa Y., Sugano K., Yonemitsu O. (1978) Meldrum's acid in organic synthesis. 2. A general and versatile synthesis of β-keto esters. J. Org. Chem. 43, 2087–2088 [Google Scholar]
- 18. Gilbert I. H., Ginty M., O'Neill J. A., Simpson T. J., Staunton J., Willis C. L. (1995) Synthesis of β-keto and α,β-unsaturated N-acetylcysteamine thioesters. Bioorg. Med. Chem. Lett. 5, 1587–1590 [Google Scholar]
- 19. Piasecki S. K., Taylor C. A., Detelich J. F., Liu J., Zheng J., Komsoukaniants A., Siegel D. R., Keatinge-Clay A. T. (2011) Employing modular polyketide synthase ketoreductases as biocatalysts in the preparative chemoenzymatic synthases of diketide chiral building blocks. Chem. Biol. 18, 1331–1340 [DOI] [PubMed] [Google Scholar]
- 20. White S. W., Zheng J., Zhang Y.-M., Rock C. O. (2005) The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 [DOI] [PubMed] [Google Scholar]
- 21. Feng Y., Cronan J. E. (2009) Escherichia coli unsaturated fatty acid synthesis complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 284, 29526–29535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stoller G., Rücknagel K. P., Nierhaus K. H., Schmid F. X., Fischer G., Rahfeld J.-U. (1995) A ribsome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 14, 4939–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaneshiro T., Marr A. G. (1961) cis-9,10-Methylene hexadecanoic acid from the phospholipids of Escherichia coli. J. Biol. Chem. 236, 2615–2619 [PubMed] [Google Scholar]
- 24. Marr A. G., Ingraham J. L. (1962) Effect of temperature on the composition of fatty acids in Escherichia coli. J. Bacteriol. 84, 1260–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erol O., Schäberle T. F., Schmitz A., Rachid S., Gurgui C., El Omari M., Lohr F., Kehraus S., Piel J., Müller R., König G. M. (2010) Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem 11, 1253–1265 [DOI] [PubMed] [Google Scholar]
- 26. Sucipto H., Wenzel S. C., Müller R. (2013) Exploring chemical diversity of α-pyrone antibiotics: molecular basis of myxopyronin biosynthesis. ChemBioChem 14, 1581–1589 [DOI] [PubMed] [Google Scholar]
- 27. Ramkumar K., Tambov K. V., Gundla R., Manaev A. V., Yarovenko V., Traven V. F., Neamati N. (2008) Discovery of 3-acetyl-4-hydroxy-2-pyranone derivatives and their difluoridoborate complexes as a novel class of HIV-1 integrase inhibitors. Bioorg. Med. Chem. 16, 8988–8998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.