Background: There is a big controversy about how mitochondria dysfunction affects skeletal myogenesis and insulin signaling.
Results: Mitochondrial complex I deficiency inactivates SIRT1 by decreasing the NAD+/NADH ratio, leading to skeletal myogenesis enhancement and insulin resistance.
Conclusion: Mitochondrial dysfunction-elicited SIRT1 inactivation enhances skeletal myogenesis but impairs insulin signaling.
Significance: This work provides a precise molecular mechanism of how mitochondrial dysfunction induces skeletal myogenesis enhancement and insulin resistance.
Keywords: Insulin Resistance, Mitochondrial Disease, Mitochondrial Metabolism, Myogenesis, Sirtuin 1 (SIRT1)
Abstract
To address whether mitochondrial biogenesis is essential for skeletal myogenesis, C2C12 myogenesis was investigated after knockdown of NADH dehydrogenase (ubiquintone) flavoprotein 1 (NDUFV1), which is an oxidative phosphorylation complex I subunit that is the first subunit to accept electrons from NADH. The NDUFVI knockdown enhanced C2C12 myogenesis by decreasing the NAD+/NADH ratio and subsequently inactivating SIRT1 and SIRT1 activators (pyruvate, SRT1720, and resveratrol) abolished the NDUFV1 knockdown-induced myogenesis enhancement. However, the insulin-elicited activation of insulin receptor β (IRβ) and insulin receptor substrate-1 (IRS-1) was reduced with elevated levels of protein-tyrosine phosphatase 1B after NDUFV1 knockdown in C2C12 myotubes. The NDUFV1 knockdown-induced blockage of insulin signaling was released by protein-tyrosine phosphatase 1B knockdown in C2C12 myotubes, and we found that NDUFV1 or SIRT1 knockdown did not affect mitochondria biogenesis during C2C12 myogenesis. Based on these data, we can conclude that complex I dysfunction-induced SIRT1 inactivation leads to myogenesis enhancement but blocks insulin signaling without affecting mitochondria biogenesis.
Introduction
Skeletal myogenesis is frequently accompanied by mitochondrial biogenesis because the expression levels of mitochondrial oxidative phosphorylation (OXPHOS)3 complexes, mitochondrial DNA (mtDNA) content, and oxygen consumption rate (OCR) are dramatically increased during C2C12 myogenesis (1, 2). Myocyte enhancer factor 2 (Mef2), a myogenic transcription factor, up-regulates peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), which is a master switch of mitochondrial biogenesis (3). PGC-1α in turn binds to and activates Mef2 to drive the formation of slow-oxidative muscle fibers that contain high numbers of mitochondria (4, 5). Through this positive auto-regulatory loop, skeletal myogenesis progresses with mitochondrial biogenesis (5). Because mitochondrial function is highly increased during skeletal myogenesis, it is tempting to speculate that functional OXPHOS enzymes are required for skeletal myogenesis. For example, skeletal myogenesis is completely abrogated in the presence of OXPHOS inhibitors such as rotenone, antimycin A, and oligomycin (6–8). However, C2C12 myogenesis is not prevented after the knockdown of ATP synthase β (9), which challenges the requirement of mitochondrial biogenesis on skeletal myogenesis.
The mitochondrial OXPHOS system is composed of five complexes: complex I (CI, NADH:ubiquinone oxidoreductase), II (CII, succinate:ubiquinone oxidoreductase), III (CIII, ubiquinol:cytochrome c oxidoreductase), IV (CIV, cytochrome c oxidase), and V (CV, ATP synthase). Two electrons are transferred from NADH and FADH2 to CI and CII, respectively, and subsequently to ubiquinone, CIII, cytochrome c, and CIV, thus generating proton a gradient across the mitochondrial inner membrane that is utilized by CV for ATP synthesis. Nuclear or mitochondrial gene mutations in the mitochondrial OXPHOS complexes causes a chronic lack of ATP in neurons and striated muscles and lactic acidosis, which leads to Leigh's disease with symptoms of encephalomyopathy and cardiomyopathy (10, 11). In particular, many mutations have been found in the catalytic subunits of CI in Leigh's disease patients (12). For example, patients with an NADH dehydrogenase (ubiquinone) flavoprotein 1 (NDUFV1, a subunit of CI) mutation exhibit Leigh's disease with encephalomyopathy, leukoencephalopathy, and lethal infantile mitochondrial disease (13–17). CI deficiency is also found in patients with Parkinson disease, autism, and diabetes (18–20).
Mitochondrial dysfunction has been proposed to lead to type 2 diabetes and obesity, which causes insulin resistance in skeletal muscle and adipose tissue (21, 22). An inverse correlation between OXPHOS function and insulin resistance has been revealed in the skeletal muscle of diabetes patients, as determined by microarray, proteomics, oxygen consumption, and in vivo nuclear magnetic resonance analyses (23–27). However, this proposal has been challenged because the expression levels of the OXPHOS proteins are similar in the skeletal muscles of lean and db/db mice, and OXPHOS function is similar in diabetic and nondiabetic Asian Indians (28–30). In addition, disruption of mitochondrial transcription factor A (TFAM), apoptosis-inducing factor, or PGC-1α and PGC-1ββ improves glucose tolerance and increases insulin sensitivity despite severe CI activity loss and mitochondrial defects (31–33).
Silent information regulator 2 homologue 1 (SIRT1), an NAD+-dependent deacetylase enzyme, has been known to regulate myogenesis and mitochondrial biogenesis in skeletal muscle (34–36). SIRT1 expression level and its deacetylase activity are decreased due to the low ratio of NAD+ to NADH during skeletal myogenesis (37). Thus, acetylated and active MyoD accelerates skeletal myogenesis. Low glucose has been shown to prevent skeletal myogenesis by increasing the ratio of NAD+ to NADH and activating SIRT1, and low glucose-induced myogenesis inhibition is released by SIRT1 knockdown (38). Ironically, PGC-1α, a SIRT1 deacetylase substrate, is up-regulated and deacetylated despite SIRT1 inactivation during skeletal myogenesis (2, 39). These findings create a paradox for the SIRT1-PGC-1α pathway in mitochondrial biogenesis during skeletal myogenesis.
The present study aimed to investigate the role of mitochondrial function in skeletal myogenesis and insulin signaling after NDUFV1 knockdown. Here we demonstrate that NDUFV1 knockdown enhances skeletal myogenesis by lowering the ratio of NAD+ to NADH and then inactivating SIRT1. In addition, we show that NDUFV1 knockdown blunts the insulin-elicited activation of insulin receptor β (IRβ) through PTP1B up-regulation, which supports the notion that mitochondrial dysfunction is a causative factor of insulin resistance. In addition, we demonstrate that SIRT1 is not required for mitochondrial biogenesis during skeletal myogenesis.
EXPERIMENTAL PROCEDURES
Materials
Table 1 shows the information on the antibodies used for immunoblotting, immunoprecipitation, and immunofluorescence. Resveratrol and pyruvate were obtained from Sigma, and SRT1720 was obtained from Selleckchem.
TABLE 1.
List of primary antibodies for immunoblotting (IB), immunoprecipitation (IP), and immunofluorescence (IF)
| Name | Company | Host | Experiments (dilution factor) |
|---|---|---|---|
| NDUFV1 | Santa Cruz | Rabbit polyclonal | IB (1:500) |
| NDUFV2 | Santa Cruz | Rabbit polyclonal | IB (1:1,000) |
| NDUFS3 | Santa Cruz | Mouse monoclonal | IB (1:1,000) |
| NDUFS7 | Santa Cruz | Rabbit polyclonal | IB (1:500) |
| SDHB | Abcam | Mouse monoclonal | IB (1:1,000) |
| UQCRC1 | Abcam | Mouse monoclonal | IB (1:1,000) |
| COX Va | Invitrogen | Mouse monoclonal | IB (1:500) |
| COX Vb | Invitrogen | Mouse monoclonal | IB (1:1000) |
| ATP synthase α | Abcam | Mouse monoclonal | IB (1:2,000) |
| β-Actin | Santa Cruz | Mouse monoclonal | IB (1:2,000) |
| Tubulin | Lab Frontier | Rabbit polyclonal | IB (1:1,000) |
| p53 | Santa Cruz | Rabbit polyclonal | IB (1:1,000) |
| Ac-p53 | Cell Signaling | Rabbit polyclonal | IB (1:1,000) |
| MyHC | Sigma | Mouse monoclonal | IB (1:2,000), IF (1:100) |
| Myogenin | BD Biosciences | Mouse monoclonal | IB (1:1000), IF (1:100) |
| Mef2 | Santa Cruz | Goat polyclonal | IB (1:1,000), IF (1:100) |
| Caveolin-3 | BD Sciences | Mouse monoclonal | IB (1:2,000) |
| MG53 | Abcam | Mouse polyclonal | IB (1:1,000) |
| FLAG | Sigma | Mouse monoclonal | IF (1:100) |
| SIRT1 | Millipore | Rabbit polyclonal | IB (1:1,000) |
| PTP1B | Millipore | Rabbit polyclonal | IB (1:1,000) |
| IRβ | Millipore | Mouse monoclonal | IB (1:500), IP (2 μg/mg) |
| IRS-1 | Millipore | Mouse monoclonal | IB (1:1000), IP (1 μg/mg) |
| pY | BD Sciences | Mouse monoclonal | IB (1:1,000) |
| p-AKT | Cell Signaling | Rabbit polyclonal | IB (1:1,000) |
| AKT | Millipore | Mouse monoclonal | IB (1:1,000) |
| PGC-1α | Santa Cruz | Rabbit polyclonal | IB (1:1,000) |
| PGC-1β | Santa Cruz | Mouse monoclonal | IB (1:500) |
| HRP-conjugated anti-mouse IgG | Pierce | Goat polyclonal | IB (1:20,000∼40,000) |
| HRP-conjugated anti-goat IgG | Pierce | Goat polyclonal | IB (1:20,000) |
| HRP-conjugated anti-rabbit IgG | Pierce | Goat polyclonal | IB (1:20,000∼40,000) |
C2C12 Cell Culture
C2C12 cells were purchased from ATCC and grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% penicillin/streptomycin (Thermo Scientific) and 10% fetal bovine serum (Thermo Scientific) in a 5% CO2 incubator at 37 °C. Confluent C2C12 cells were differentiated into myotubes by incubating them with DMEM supplemented with 2% horse serum (Invitrogen) and refeeding them every 24 h.
Measurement of Myogenic Index
C2C12 myotubes were co-stained with an anti-MyHC antibody and DAPI and observed using a fluorescence microscope (Nikon). The myogenic index was determined as the average number of nuclei from the myosin heavy chain (MyHC)-positive myotubes in five separate images.
RNA Interference
siRNA oligomers targeting NDUFV1 (si-NDUFV1), NADH dehydrogenase (ubiquinone) iron-sulfur protein 7 (NDUFS7; si-NDUFS7), and a scrambled oligomer (si-control) were obtained from Invitrogen. siRNA oligomers targeting SIRT1 (si-SIRT1) and protein-tyrosine phosphatase 1B (si-PTP1B) were purchased from Ambion. C2C12 myoblasts were transfected with 50 nm siRNA by electroporation according to the protocol of electroporator MP-100 (Invitrogen). Table 2 shows the target sequence for each gene.
TABLE 2.
siRNA oligomer sequence for each gene
| Gene | Target sequence |
|---|---|
| NDUFV1 | 5′-CUACUCCACUGAUUCCCAAtt-3′ |
| NDUFV1 | 5′-CCUUCCAGCAGAUGUGGGAGUGUU-3 |
| NDUFV1 | 5′-CCGUGCUAAUGGACUUCGAUGCACU-3′ |
| NDUFV1 | 5′-GCUGUGAUUCCUGGUGGCUCAUCUA-3′ |
| NDUFS7 | 5′-CCACUACUCCUACUCGGUGUUCGU-3′ |
| SIRT1 | 5′-GCUUGGAAGAUGAUACGGATT-3′ |
| PTP1B | 5′-GCCUCUUACUGAUGGACAATT-3′ |
Antibody-based Assays
Cells were lysed with a radioimmune precipitation lysis buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2 mm EDTA, protease inhibitor mixture, and phosphatase inhibitor mixture (Roche Applied Science). After microcentrifugation at 14,000 rpm for 10 min at 4 °C, the whole cell lysates (supernatant) were separated in SDS-polyacrylamide gels and transferred to a PVDF membrane. Antigens were visualized by sequential treatment with specific antibodies, HRP-conjugated secondary antibodies, and an enhanced chemiluminescence substrate kit.
C2C12 cells were fixed with 4% paraformaldehyde in PBS for 10 min and then permeabilized with 0.2% Triton X-100 for 10 min. The cells were then incubated with blocking buffer (2% BSA in PBS) and primary antibodies, and the primary antibodies were detected with fluorescein-conjugated secondary antibodies. The cells were also incubated with DAPI for 10 min and then observed with a LSM 510 META confocal microscope (Carl ZEISS) or a fluorescence microscope (Nikon). For immunoprecipitation, the cells were lysed in a buffer containing 20 mm Tris-HCl, pH 7.4, 137 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, 20 mm NaF, 10 mm Na4P2O7, 1 mm Na3VO4, 1% Nonidet P-40, 1 mm PMSF, and a protease inhibitor mixture (Roche Applied Science). The whole cell lysates (1∼2 mg of total protein) were incubated with specific antibodies (1∼3 μg) for 1 h and then with 50 μl of a protein A-agarose bead (Roche Applied Science) slurry overnight. The immunoprecipitates were then analyzed using immunoblotting. Table 1 shows the information on the antibodies used for immunoblotting, immunoprecipitation, and immunofluorescence.
Plasmids, Transient Transfection, and Luciferase Assay
The FLAG-SIRT1 and FLAG-SIRT1 H355A genes were kindly provided by Professor Soo-Jong Um at Sejong University, and the myogenin- and creatine kinase promoter-luciferase reporter genes were provided by Professor Jeong-Ho Hong at Korea University. The mitsugumin 53 (MG53) promoter-luciferase reporter gene was previously generated (40). Gene transfection was performed using electroporation according to the protocol of electroporator MP-100 (Invitrogen) for C2C12 myoblasts. Luciferase activity was measured by using the luciferase assay system (Promega) using a Luminoskan Ascent (Thermo Labsystems). We normalized the luciferase activity to the activity of coexpressed β-galactosidase. To calculate the relative luciferase activity, we divided each normalized luciferase activity by that of the empty vector-transfected cells.
Oxygen Consumption Rate and Extracellular Acidification Rate
The mitochondrial OCR and the extracellular acidification rate (ECAR) were measured using a Seahorse XF24 Extracellular Flux Analyzer (Seahorse Bioscience) according to the manufacturer's instructions. C2C12 myoblasts and 4-day-differentiated myotubes were cultured on an XF assay plate at a density of 50,000 cells per well. After 24 h the cells were washed and incubated with DMEM without bicarbonate in a non-CO2 incubator for 1 h at 37 °C. For calibration, the sensor cartridge was loaded into an XF24 analyzer, and after calibration was complete, the assay plate was loaded into the XF24 analyzer. Oligomycin (5 mg/ml), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (5 mm), and rotenone (1 mm) were automatically injected according to priority. OCR and ECAR were automatically calculated and recorded by the Seahorse XF24 software.
Assays for Mitochondrial Respiratory Chain Activities
For mitochondria isolation, C2C12 myotubes were homogenized in 10 mm Tris-HCl buffer containing 0.25 m sucrose and centrifuged at 600 × g. The supernatant was centrifuged at 12,000 × g for 30 min at 4 °C; the resulting pellets were resuspended in 0.25 m sucrose containing 10 mm Tris-HCl buffer, pH 7.4, and loaded onto a discontinuous gradient consisting of 50% w/v and 40% w/v and 20% w/v sucrose containing 10 mm Tris-HCl buffer, pH 7.4, and centrifuged at 105,000 × g in an SW41 rotor for 1 h at 4 °C. Mitochondria were collected at the 40–50% sucrose interface. Mitochondria (80 μg of protein) were prepared from C2C12 myoblasts and analyzed for their OXPHOS I, II, and IV activities according to the manufacturer's protocol (Mitosciences). In addition, OXPHOS III activity was estimated by the reduction of oxidized cytochrome c at 550 nm as described previously (41).
Quantification of ATP Using Bioluminescent Luciferase Assay
Intracellular ATP was measured as previously described (42). After removal of the medium, the residual C2C12 cells were lysed in 0.2 ml of 0.1 n NaOH for 10 min at room temperature, and the lysates were neutralized by the addition of the same volume of 0.1 n HCl. Aliquots (50 μl) of the cell lysates were used to determine intracellular ATP content, and ATP levels were measured using the bioluminescence assay according to the protocol provided with the ATP determination kit (Sigma).
Pyruvate and Lactate Assay
C2C12 cells were lysed in 0.2 ml of 0.1 n NaOH for 10 min at room temperature, and the lysates were neutralized by the addition of the same volume of 0.1 n HCl. The lysates were then neutralized with a volume of 4 m KOH and centrifuged for 5 min at 10,000 × g, and the resulting supernatant (10 μl) was used to determine the intracellular lactate and pyruvate levels. The contents of pyruvate were detected using assay kits according to the manufacturer's protocol (BioVision).
To quantify the lactate level, C2C12 myoblasts were treated with trypsin-EDTA and washed with PBS. After sonication for 10 s, the lysates were concentrated using Centricon filters (Millipore) with a 10-kDa cutoff. The lactate contents were measured using a lactate assay kit (BioVision) according to manufacturer's protocol. The contents of pyruvate and lactate were normalized to the protein amount.
Reactive Oxygen Species Measurement
C2C12 cells were incubated with serum-free media containing 2′,7′-dichlorodihydrofluorescein diacetate (10 μm, Invitrogen) for 20 min, scraped off, and then transferred to 96-well plates. The cells were incubated for the indicated times with Krebs-HEPES buffer, and then fluorescence was monitored using a UV spectrofluorophotometer (excitation, 480 nm; emission, 530 nm).
Proliferation Assay
Approximately 5 × 105 C2C12 cells were transfected with siRNAs, and then 5,000 cells were seeded into 96-well plates and maintained in 10% FBS-containing DMEM for 24 h. Cell proliferation was measured using a non-radioactive cell proliferation assay with MTS (Promega) according to the manufacturer's instructions.
Determination of NAD+ and NADH Cellular Concentrations
Cellular NAD+ and NADH concentrations were determined according to the protocol of the NAD+/NADH assay kit (Bioassay ECND-100). The concentration of NAD+ (or NADH) was obtained by spectrophotometrically measuring the rate of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide reduction by alcohol dehydrogenase in the presence of phenazine ethosulfate at 595 nm.
Glucose Uptake Assay
Briefly, after serum starvation with low glucose DMEM for 5 h, C2C12 myotubes were incubated with or without 100 nm insulin in DMEM for 30 min and then washed 2 times with HEPES buffer (1.2 mm MgSO4, 0.33 mm CaCl2, 121 mm NaCl, 4.9 mm KCl, and 12 mm HEPES, pH 7.4. Glucose uptake was determined by adding 50 μCi/ml 2-deoxy-[3H]glucose (PerkinElmer Life Sciences). After 10 min of incubation, the reaction was stopped with ice-cold PBS, and the cells were washed three times with ice-cold PBS. The cells were then lysed with 0.1% SDS, and glucose uptake was assessed using scintillation counting. Nonspecific, 2-deoxy-[3H]glucose uptake was measured by including 100 μm cytochalasin B (Sigma), and the glucose uptake was determined by subtracting the nonspecific counts from the total counts and normalizing to the protein amount. The assay was performed three times.
Mitochondrial DNA Analysis
To determine the intracellular mtDNA levels, two primer sets, a set for ND2, which is found in mtDNA, and a set for 28 S RNA, which is found in nuclear DNA, were used for PCR analysis with total genomic DNA that was isolated using DNeasy blood and tissue kit (Qiagen). The final PCR products were electrophoresed on a 1.5% agarose gel, stained with 0.5 μg/ml ethidium bromide solution, and visualized using a UV transilluminator (Gel Doc 2000; Bio-Rad). The ratio of the ND2:28 S RNA was determined by comparing the band intensities using the Image Gauge software (Fusifilm). The sequences of the primer sets that were used in this study were as follows: for mitochondrial ND2, 5′-CAAGGGATCCCACTGCACATAGG-3′ and 5′-GTGCTTATGATAGCTAGGGTG-3′; for 28 S RNA, 5′-TAGCAGCCGACTTAGAACTGG-3′ and 5′-CTCCCACTTATTCTACACCTC-3′.
Determination of Citrate Synthase Activity
Citrate synthase activity was measured spectrophotometrically using 5,5′-dithiobis(nitrobenzoic acid) reduction at 412–360 nm (ϵ = 13.6 mm−1 × cm−1) as previously described (43). C2C12 myoblasts and 4 day-differentiated myotubes (1 × 106) were trypsinized, pelleted, and resuspended in 0.2 ml of medium containing 25 mm phosphate buffer, pH 7.4, 5 mm MgCl2, and 1 μl of protease inhibitor mixture. After freezing and thawing three times and centrifugation at 14,000 rpm for 10 min at 4 °C, the whole cell lysates (supernatant) were incubated with a reaction mixture (100 mm Tris-HCl, pH 8.0, 0.3 mm acetyl-coenzyme A, 0.2 mm 5,5′-dithiobis(nitrobenzoic acid), and 0.2% (v/v) Triton X-100) for 2 min at 25 °C. After the incubation, the base line was measured using a spectrophotometer set at 412 nm, and the citrate synthase enzymatic reaction was started by the addition of 0.5 mm oxaloacetate. After another 2 min of incubation, the citrate synthase activity was assessed at 412 nm, and the specific activity was determined by subtracting the absorbance value of the base line from the citrate synthase activity value.
MitoTracker Staining
C2C12 myoblasts were washed with PBS and incubated with 100 nm MitoTracker Green FM (100 nm) at 37 °C for 30 min. The cells were then detached using trypsin-EDTA and resuspended in PBS. The fluorescence intensity was detected using excitation at 490 nm and emission at 516 nm.
Mitochondrial Membrane Potential
C2C12 myoblasts were incubated with MitoTracker Red CMXRos (200 nm) for 20 min and washed 3 times with media. The fluorescence intensity was detected using excitation at 579 nm and emission at 599 nm.
Statistical Analysis
Statistical values are presented as the mean ± S.E. A two-tailed Student's t test and analysis of variance test were used to calculate the p values.
RESULTS
CI Dysfunction Enhances Skeletal Myogenesis
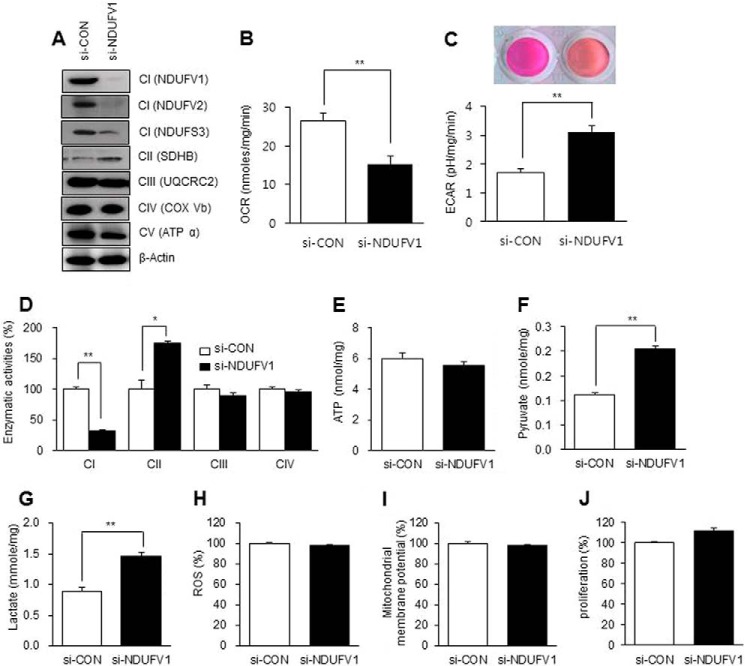
The mitochondrial biogenesis that accompanies with skeletal myogenesis raises a big question as to whether increased mitochondrial function is essential for myogenesis (1). To address the issue, we induced mitochondrial dysfunction in C2C12 myoblasts by knocking down NDUFV1, which is the first CI subunit to accept electrons from NADH (44). We then investigated the protein expression levels of CI subunits (NDUFV1, NDUFV2, and NDUFS3) in the NDUFV1-knockdown myoblasts. As shown in Fig. 3A, the expression levels of these CI subunits were dramatically reduced after NDUFV1 knockdown, suggesting that NDUFV1 knockdown induces CI deficiency in mitochondria.
FIGURE 3.
NDUFV1 knockdown increases glycolysis-dependent ATP production in C2C12 myoblasts. C2C12 myoblasts were transfected with 50 nm si-control (si-CON) or si-NDUFV1 for 24 h. A, the expression levels of CI (NDUFV1, NDUFV2, and NDUFS3), CII (SDHA), CIII (UQCRC2), CIV (COX Vb), and CV (ATPα) were assessed using immunoblotting with β-actin as a loading control. NDUFV1 and -2, NADH dehydrogenase (ubiquinone) flavoprotein 1 and 2; NDUFS3, NADH dehydrogenase (ubiquinone) iron-sulfur protein 3; SDHA, succinate dehydrogenase complex, subunit A, flavoprotein variant; UQCRC2, ubiquinol cytochrome c reductase complex III subunit 2; COX Vb, cytochrome c oxidase Vb; ATPα, ATP synthase α. B and C, the OCR and ECAR were determined and normalized by protein concentration. Media acidification was also assessed by phenol red color change (C, upper panel). D, the CI, CII, CIII, and CIV activities were measured as described under “Experimental Procedures.” E, the intracellular ATP content was measured using a luminescent luciferase assay, and the ATP content was normalized to the protein amount. F and G, the intracellular pyruvate and lactate contents were measured as described under “Experimental Procedures.” H, reactive oxygen species (ROS) were measured after staining C2C12 myoblasts with 2′-7′-dichlorofluorescein diacetate (10 μm) for 20 min. The fluorescent intensity was analyzed as described under “Experimental Procedures.” I, mitochondrial membrane potential was measured after staining C2C12 myoblasts with MitoTracker CMXRos (200 nm) for 20 min. J, the cellular proliferation of C2C12 myoblasts, as determined by an MTS assay. All data are presented as the means ± S.E.; t test. *, p < 0.05 and **, p < 0.01.
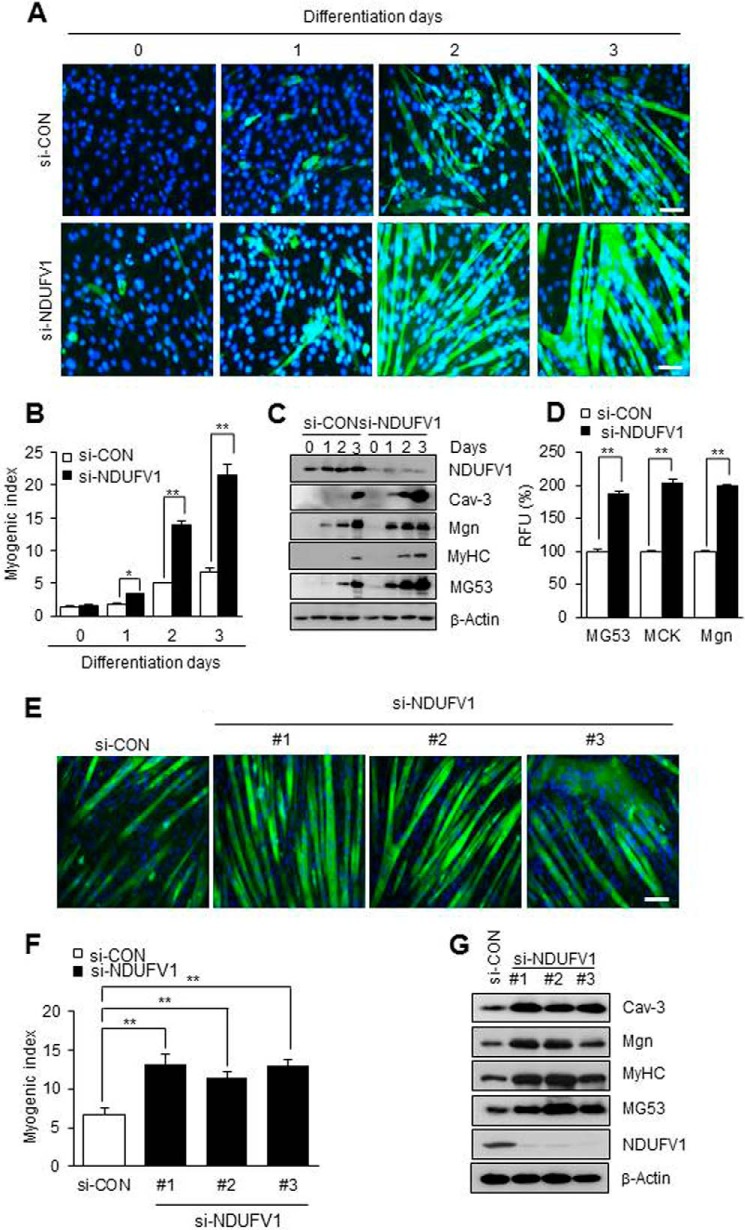
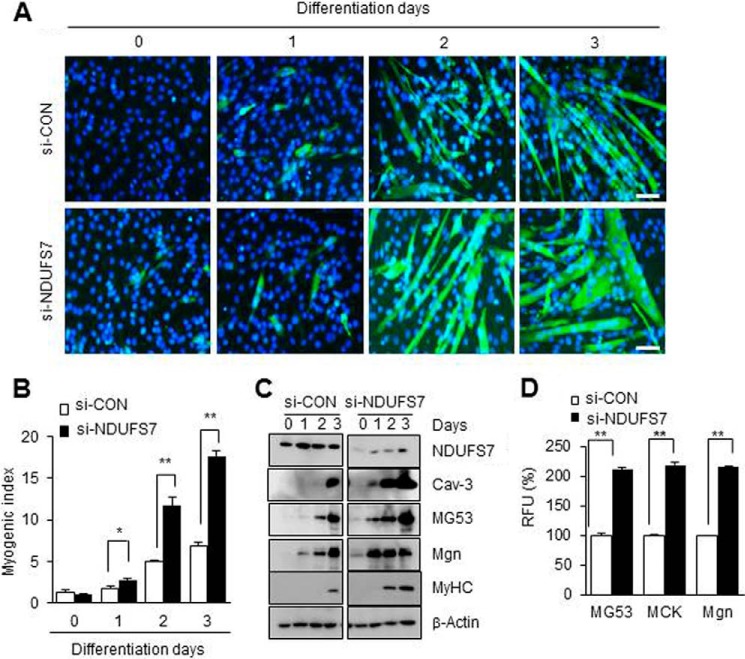
We differentiated the NDUFV1-knockdown C2C12 myoblasts to myotubes and monitored skeletal myogenesis using MyHC immunofluorescence, the myogenic index and immunoblotting for myogenic marker proteins such as MyHC, myogenin (Mgn), caveolin-3 (Cav-3), and MG53 (40, 45, 46). As shown in Fig. 1, A and B, the nuclei number of MyHC-positive cells was increased ∼4-fold in 3-day-differentiated myotubes after NDUFV1 knockdown. NDUFV1 knockdown also increased the protein levels of MyHC, Mgn, Cav-3, and MG53 in the differentiated myotubes (Fig. 1C). Additionally, we found that the promoter activities of MG53, muscle creatine kinase, and Mgn were increased more than 1.8-fold in NDUFV1 knockdown C2C12 myotubes compared with si-CON cells (Fig. 1D). To ensure that myogenesis enhancement was not an off-target effect of NDUFV1 knockdown, we treated C2C12 myoblasts with three other si-NDUFV1 oligomers and observed skeletal myogenesis. As shown in Fig. 1, E–G, all si-NDUFV1 oligomers enhanced C2C12 myogenesis. Knockdown of NDUFS7, which is another subunit of CI, also highly enhanced skeletal myogenesis (Fig. 2). All of these data indicate that NDUFV1- or NDUFS7-induced CI deficiency enhances rather than prevents skeletal myogenesis.
FIGURE 1.
NDUFV1 knockdown enhances skeletal myogenesis. A–C, C2C12 myoblasts were treated with 50 nm si-control (si-CON) or si-NDUFV1 for 24 h and further differentiated to myotubes for the indicated days. NDUFV1 indicates NADH dehydrogenase (ubiquinone) flavoprotein 1. Myogenesis was monitored using immunofluorescence for MyHC and DAPI staining (A). The myogenic index was determined by counting the nuclei number in MyHC-positive cells from five separate image fields (B). The expression levels of NDUFV1 and myogenic marker proteins such as Cav-3, Mgn, MyHC and MG53 were determined by immunoblotting with β-actin as a loading control (C). D, C2C12 myoblasts were transfected with MG53, muscle creatine kinase (MCK), or Mgn promoter-luciferase fusion genes along with si-CON or si-NDUFV1 for 24 h and further differentiated into myotubes. A luciferase activity assay was performed on the 1-day differentiated myotubes, and the relative luciferase units (RLU) from three independent experiments were calculated as follows: relative luciferase activity of si-NDUFV1 cells/relative luciferase activity of si-CON cells. E–G, C2C12 myoblasts were transfected with three different si-NDUFV1 oligomers and further differentiated into myotubes for 4 days. Skeletal myogenesis was determined using MyHC immunofluorescence (E), the myogenic index (F), and immunoblotting for myogenic marker proteins, such as Cav-3, Mgn, MyHC, and MG53, with β-actin as a loading control (G). All data are presented as the means ± S.E. t test and analysis of variance. *, p < 0.05 and **, p < 0.01.
FIGURE 2.
NDUFS7 knockdown enhances skeletal myogenesis. A–C, C2C12 myoblasts were treated with 50 nm si-control (si-CON) or si-NDUFS7 for 24 h and further differentiated into myotubes for the indicated days. NDUFS7 indicates NADH dehydrogenase (ubiquinone) iron-sulfur protein 7. Myogenesis was monitored using immunofluorescence for MyHC and DAPI staining (A). The myogenic index was determined by counting the number of nuclei in MyHC-positive cells from five separate image fields (B). The expression levels of NDUFS7 and myogenic marker proteins, such as Cav-3, Mgn, MyHC, and MG53, were determined using immunoblotting with β-Actin as a loading control (C). D, C2C12 myoblasts were transfected with the MG53, muscle creatine kinase (MCK), or Mgn promoter-luciferase fusion genes along with si-CON or si-NDUFS7 for 24 h and further differentiated into myotubes. Luciferase activity assays were performed on the 1-day differentiated myotubes. Relative luciferase units (RLU) from three independent experiments were calculated as follows: relative luciferase activity of si-NDUFS7 cells/relative luciferase activity of si-CON cells. All data are shown as the means ± S.E. t test. *, p < 0.05 and **, p < 0.01.
CI Deficiency Increases Glycolysis Activity in C2C12 Myoblasts
To determine if other intrinsic factors enhance skeletal myogenesis in NDUFV1 knockdown C2C12 cells, we investigated the effect of NDUFV1 knockdown on mitochondrial respiration, glycolysis, reactive oxygen species generation, and cellular proliferation in myoblasts. Despite the loss of CI subunits (NDUFV1, NDUFV2, and NDUFS3), the expression levels of UQCRC2 (ubiquinol-cytochrome c reductase complex core protein 2, a CIII subunit), COX Vb (cytochrome c oxidase Vb, a CIV subunit), and ATPα (ATP synthase α, a CV subunit) were not changed after NDUFV1 knockdown in C2C12 myoblasts (Fig. 3A).
The OCR was reduced by ∼40%, whereas the ECAR was increased to ∼150% after NDUFV1 knockdown (Fig. 3, B and C). The NDUFV1 knockdown changed the phenol red color to orange, indicating acidification of the culture medium (Fig. 3C, upper panel). Without changing the CIII and CVI enzymatic activity, the CI enzymatic activity was reduced to ∼30%, and CII enzymatic activity was increased to ∼170% after NDUFV1 knockdown (Fig. 3D). Taken together with data in Fig. 3, A–D, we conclude that NDUFV1 knockdown-induced CI dysfunction leads to a severe reduction in mitochondrial respiration despite the compensation of CII activity.
Next, we measured the intracellular ATP content in C2C12 myoblasts after NDUFV1 knockdown. The intracellular ATP level was not changed despite CI dysfunction (Fig. 3E). Because glycolysis is an alternative ATP source in mammalian cells with mitochondrial dysfunction (47), NDUFV1 knockdown myoblasts may have a strong activity of glycolysis. To exploit the possibility, we assessed the glycolysis activity by measuring pyruvate and lactate production. Fig. 3, F and G, showed that pyruvate and lactate production was highly increased by NDUFV1 knockdown in myoblasts, indicating that CI dysfunction induces glycolysis-dependent ATP production in myoblasts. We also measured reactive oxygen species generation and mitochondrial membrane potential after NDUFV1 knockdown by staining the myotubes with 2′-7′-dichlorofluorescein diacetate (DCF-DA) and MitoTracker CMXRos, respectively As shown in Fig. 3, H and I, CI dysfunction did not change the reactive oxygen species production and mitochondrial membrane potential in myoblasts. In addition, NDUFV1 knockdown did not change the cellular proliferation rate, as determined using an MTS assay (Fig. 3J). Taken together, we conclude that ATP level was maintained under the condition of CI dysfunction by activation of the glycolysis pathway.
CI Dysfunction Enhances Myogenesis by SIRT1 Inactivation
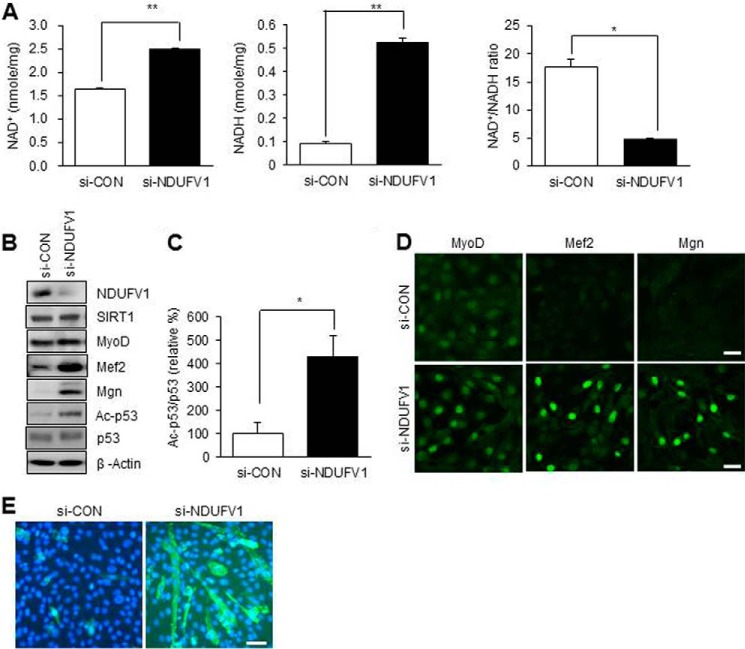
Because NDUFV1 transfers electrons from NADH to the mitochondrial oxidation-reduction chain, NDUFV1 knockdown may lead to a low NAD+/NADH ratio and thus inactivate SIRT1. Because the SIRT1 inactivation is essential for C2C12 myogenesis (37), NDUFV1 knockdown may enhance skeletal myogenesis by decreasing the NAD+/NADH ratio and inactivating SIRT1. The cellular levels of NAD+ and NADH were increased to 170 and 500%, respectively, but the NAD+/NADH ratio was reduced to 24% after NDUFV1 knockdown (Fig. 4A). To determine the SIRT1 activity in myoblasts, we measured the acetylation of p53, which is deacetylated by SIRT1. As shown in Fig. 4, B and C, the acetylated p53 protein level was increased after NDUFV1 knockdown, indicating that SIRT1 is inactivated after NDUFV1 knockdown.
FIGURE 4.
NDUFV1 knockdown inactivates SIRT1 via decreasing the NAD+/NADH ratio. C2C12 myoblasts were transfected with 50 nm si-CON or si-NDUFV1 for 24 h. A, the intracellular contents of NAD+ and NADH and the NAD+/NADH ratio were determined. All data are shown as the means ± S.E. t test. *, p < 0.05 and **, p < 0.01. B, the expression levels of SIRT1, MyoD, Mef2, Mgn, p53, acetylated p53 (Ac-p53) and NDUFV1 were determined using immunoblotting with β-actin as a loading control. C, the ratio of Ac-p53/p53 was statistically analyzed with three independent experiments. D, the cellular localization of MyoD, Mef2, and Mgn was monitored using immunofluorescence. E, C2C12 myoblasts at 100% confluence were further incubated with 10% FBS-containing growth medium for 3 days and immuno-stained with an anti-MyHC antibody.
SIRT1 inactivation is known to prematurely induce myogenesis (37). Thus, we investigated premature myogenesis in NDUFV1-knockdown C2C12 myoblasts by monitoring the protein expression levels and cellular localization of myogenic transcription factors such as MyoD, Mgn, and Mef2. As shown in Fig. 4B, the protein levels of Mgn and Mef2 were dramatically increased in NDUFV1-knocked-down C2C12 myoblasts. In addition, all of the myogenic transcription factors including MyoD, Mgn, and Mef2 were localized in the nucleus after NDUFV1 knockdown (Fig. 4D). These data indicate that myogenic transcription factors are ready to initiate skeletal myogenesis in myoblasts after NDUFV1 knockdown. In addition, MyHC immunofluorescence revealed that C2C12 myoblasts were differentiated to myotubes even in the presence of 10% FBS, which prevents myogenesis (Fig. 4E). These data indicate that NDUFV1 knockdown enhances skeletal myogenesis by decreasing the NAD+/NADH ratio and thus inactivating SIRT1.
SIRT1 Activation Abolishes CI Dysfunction-induced Myogenesis Enhancement
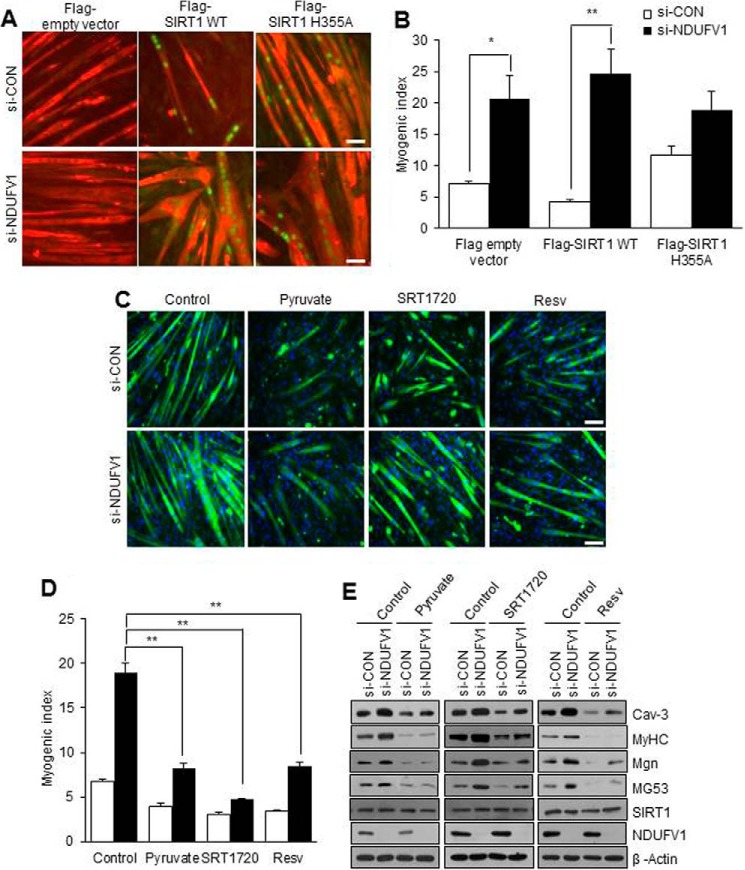
C2C12 myogenesis is prevented by SIRT1 overexpression and activation (37). Thus, we tested whether NDUFV1 knockdown-induced myogenesis enhancement was abolished by SIRT1 overexpression or activation. C2C12 myoblasts were transiently transfected with FLAG-tagged SIRT1 or an H355A mutant with si-CON or si-NDUFV1 and were further differentiated to myotubes. Myogenesis was analyzed by immunofluorescence using anti-MyHC and FLAG antibodies and the myogenic index. As shown in Fig. 5, A and B, myogenesis was prevented by SIRT1 overexpression but not by the SIRT1 H355A mutant in si-CON cells. However, the prevention of SIRT1 overexpression-induced myogenesis was released by NDUFV1 knockdown. These data indicate that NDUFV1 knockdown inactivates the overexpressed SIRT1 by decreasing the NAD+/NADH ratio and then enhancing myogenesis.
FIGURE 5.
NDUFV1 knockdown-induced myogenesis enhancement is completely abolished by SIRT1 activation. A and B, C2C12 myoblasts were transfected with different combinations of si-CON, si-NDUFV1, FLAG-empty vector, FLAG-SIRT1, and FLAG-SIRT1 H355A (H355A) for 24 h and then further differentiated into myotubes for 4 days. The cells were immuno-stained with anti-MyHC and anti-FLAG antibodies (A). The myogenic index was determined by counting the number of FLAG-positive nuclei in MyHC-positive cells. Because FLAG tag was not be monitored by immunofluorescence in FLAG empty vector-transfected cells, we measured the myogenic index from the FLAG-empty vector-transfected cells by counting the nuclei number in MyHC-positive cells (B). C–E, C2C12 myoblasts were transfected with si-CON and si-NDUFV1 for 24 h and then further differentiated into myotubes for 4 days in the absence or presence of pyruvate (25 mm), SRT1720 (2 μm), or resveratrol (Resv; 25 μm). Myogenesis was monitored using MyHC immunofluorescence and DAPI (C), the myogenic index (D), and immunoblotting for Cav-3, MyHC, Mgn, MG53, SIRT1, and NDUFV1, with β-actin as a loading control (E). The myogenic index was determined by counting the number of the nuclei in MyHC-positive cells. All data are shown as the means ± S.E.; t test and analysis of variance. **, p < 0.01.
We also tested whether the enhancement of NDUFV1 knockdown-induced myogenesis was abolished after activating SIRT1 by treating NDUFV1-knockdown myotubes with SIRT1 activators such as pyruvate, SRT1720, and resveratrol. It should be noted that pyruvate functions as a SIRT1 activator because it is converted to lactate and increases the NAD+/NADH ratio (37). We then monitored myogenesis using MyHC immunofluorescence, the myogenic index, and immunoblotting for myogenic marker proteins. As shown in Fig. 5, C–E, all of the SIRT1 activators abolished the enhancement of NDUFV1 knockdown-induced myogenesis. These data suggest that the enhancement of NDUFV1-induced myogenesis results from lowering the NAD+/NADH ratio and inactivating SIRT1.
CI Dysfunction Blunts Insulin Signaling
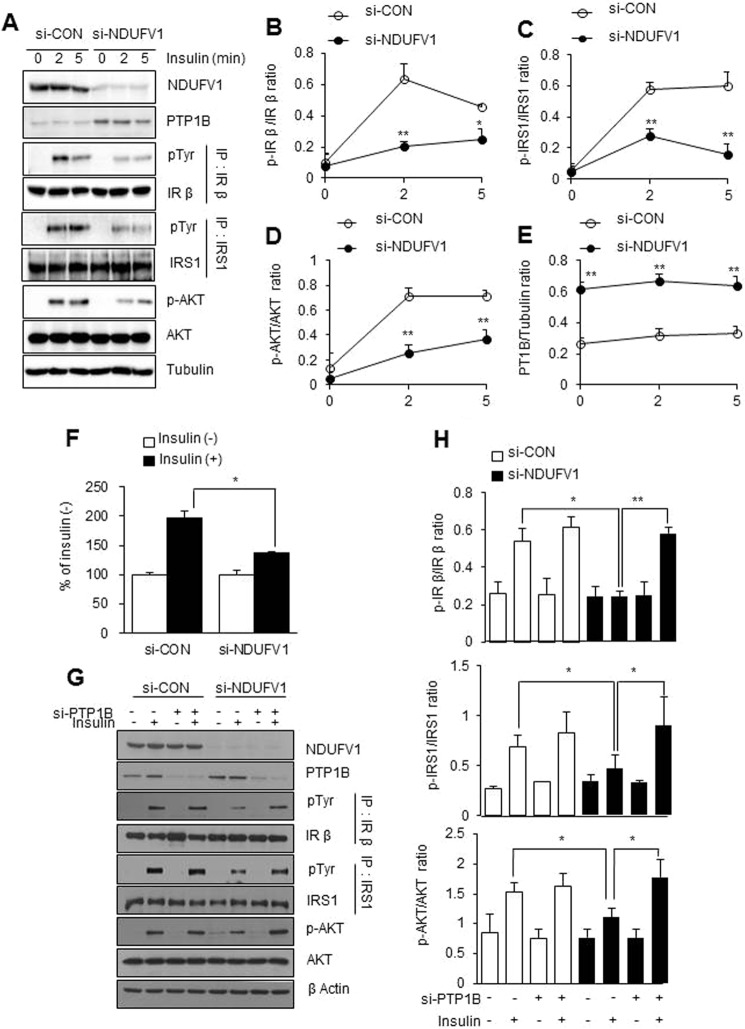
SIRT1 activation is known to improve insulin signaling by repressing the transcription of PTP1B, which dephosphorylates and inactivates IRβ and insulin receptor substrate-1 (IRS-1) (48). Because SIRT1 was inactivated by NDUFV1 knockdown (Fig. 4), insulin signaling may be blunted in NDUFV1-knockdown myotubes. Thus, we monitored insulin signaling after NDUFV1 knockdown in C2C12 myotubes. As shown in Fig. 6, A–E, insulin-elicited IR, IRS-1, and Akt phosphorylation was dramatically decreased along with the high expression level of the PTP1B protein after NDUFV1 knockdown. Furthermore, insulin-elicited glucose uptake was significantly reduced in NDUFV1-knockdown C2C12 myotubes (Fig. 6F). Next, we investigated insulin signaling after double knockdown of NDUFV1 and PTP1B in C2C12 myoblasts to determine whether the NDUFV1 knockdown-induced blockage of insulin signaling was released by PTP1B knockdown. As shown in Fig. 6, G and H, insulin-elicited IR phosphorylation was increased by double knockdown of NDUFV1 and PTP1B compared with the single knockdown of NDUFV1.
FIGURE 6.
NDUFV1 knockdown blunts insulin signaling via PTP1B up-regulation. A and E, C2C12 myoblasts were transfected with si-CON and si-NDUFV1 for 24 h and then further differentiated into myotubes for 4 days, serum-starved for 8 h, and stimulated with insulin (100 nm) for the indicated times. The expression levels of NDUFV1, SIRT1, PTP1B, AKT, and phospho-Akt (p-AKT) were determined using immunoblotting with tubulin as a loading control. Tyrosine phosphorylation of IRβ and IRS-1 was determined using immunoprecipitation. IP, immunoprecipitation; pTyr, phosphotyrosine (A). The ratios of p-IRβ/IRβ, p-IRS-1/IRS-1, p-Akt/Akt, and PTP1B/tubulin were assessed (n = 5 for each group) (B–E). F, the myotubes were serum-starved for 8 h, exposed to 100 nm insulin for 30 min, and incubated with 2-[3H]deoxyglucose for 10 min. The cell-associated radioactivity was measured and normalized to the protein amount, and insulin-stimulated 2-[3H]deoxyglucose uptake was presented as the percentage of the control (no treatment) in both cells. G, C2C12 myoblasts were treated with different combinations of si-CON, si-NDUFV1, and si-PTP1B for 24 h, serum-starved for 8 h, and stimulated with insulin (100 nm) for 5 min. The expression levels of NDUFV1, Akt, phospho-Akt, PTP1B, and β-actin were determined using immunoblotting. Tyrosine phosphorylation of IRβ and IRS-1 was determined using immunoprecipitation. H, the IRβ, IRS-1, and AKT phosphorylation and PTP1B/tubulin expression levels were statistically assessed based on the data shown in G (n = 3 for each condition). All data are shown as the means ± S.E. t test. *, p < 0.05 and **, p < 0.01 versus si-CON.
SIRT1 Is Not Required for Mitochondrial Biogenesis in Skeletal Myogenesis
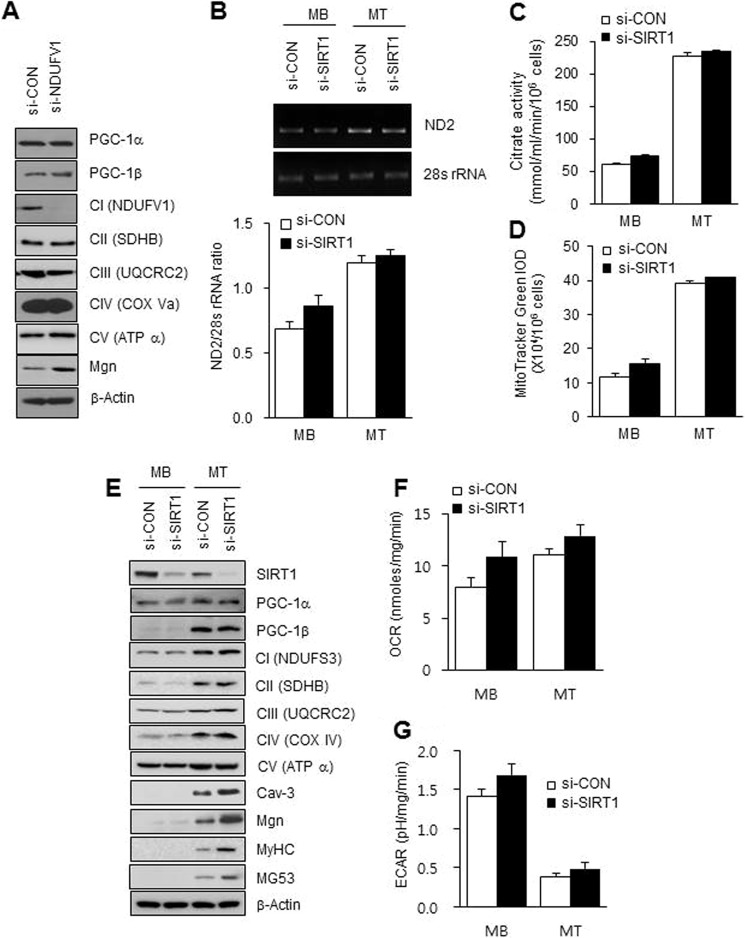
It has been known that PGC-1α is up-regulated, deacetylated, and activated by SIRT1 in C2C12 myotubes (49). However, SIRT1 is inactivated due to lowering of the NAD+/NADH ratio during myogenesis (37), which challenges the correlation between SIRT1 activation and mitochondrial biogenesis. To resolve this paradox, we monitored mitochondrial biogenesis during C2C12 myogenesis by immunoblotting for the OXPHOS proteins after NDUFV1 knockdown, which inactivated SIRT1. As shown in Fig. 7A, NDUFV1 knockdown did not change the OXPHOS (CII-CV) expression levels in myotubes, which indicated that NDUFV1 knockdown-induced SIRT1 inactivation does not affect mitochondrial biogenesis during myogenesis. To reconfirm the effect of SIRT1 on mitochondrial biogenesis that accompanies myogenesis, we investigated mitochondrial biogenesis in C2C12 myotubes after SIRT1 knockdown. The mitochondrial DNA content and number and citrate synthase activity were not altered by SIRT1 knockdown (Fig. 7, B–D), and the expression levels of the OXPHOS proteins (CI-V) and PGC-1α and PGC-1β were not changed after SIRT1 knockdown in myoblasts and myotubes (Fig. 7E). In addition, OCR and ECAR were not changed after SIRT1 knockdown (Fig. 7, F–G). All of these data indicate that NDUFV1 knockdown-induced SIRT1 inactivation does not affect mitochondrial biogenesis during skeletal myogenesis.
FIGURE 7.
SIRT1 is not required for mitochondrial biogenesis during myogenesis. A, C2C12 myoblasts were transfected with si-CON or si-NDUFV1 for 24 h and then further differentiated into myotubes for 4 days. The expression levels of PGC-1α and -1β, the mitochondrial OXPHOS proteins (NDUFV1, SDHB, UQCRC2, COX Va, and ATPα) and Mgn were determined using immunoblotting with β-actin as a loading control. B–G, C2C12 myoblasts were transfected with si-CON or si-SIRT1 for 24 h and then further differentiated into myotubes for 4 days. B, genomic DNA and the mtDNA contents of the C2C12 myoblasts and myotubes were assessed using PCR with primers designed to target the 28 S RNA and ND2, which encodes subunit 2 of OXPHOS I (upper panel). The ND2:28 S RNA ratio was obtained by measuring the intensity of each band (lower panel). C, the citrate synthase activity was measured for the C2C12 myoblasts and myotubes as described under “Experimental Procedures.” D, the fluorescent intensities of myoblasts and myotubes were measured, stained with MitoTracker Green for 20 min, and measured. E, the expression levels of the mitochondrial OXPHOS proteins (NDUFS3, SDHB, UQCRC2, COX IV, and ATPα), SIRT1, PGC-1α and -1β, Cav-3, Mgn, MyHC, and MG53 were determined using immunoblotting with β-actin as a loading control. F and G, OCR and ECAR were assessed for the myoblasts and myotubes and normalized by protein concentration. Data were obtained from three independent experiments. MB, myoblasts; MT, myotubes. All data are shown as the mean ± S.E. t test. *, p < 0.05.
DISCUSSION
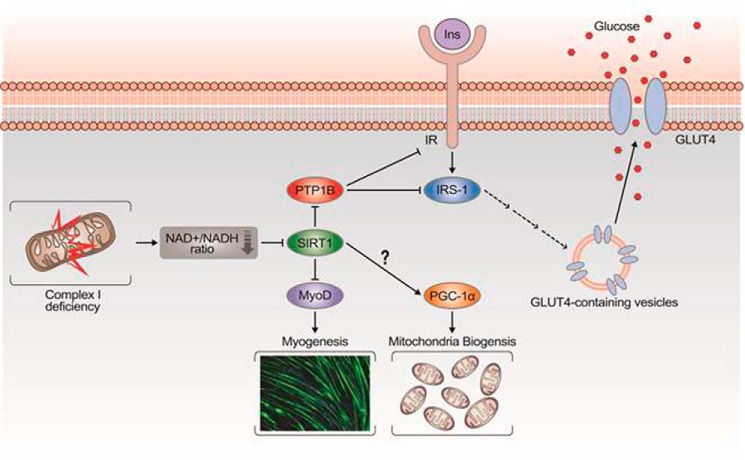
NDUFV1 knockdown disrupts CI assembly and results in CI deficiency because the expression levels of the other CI subunits (NDUFV2 and NDUFS3) were also dramatically decreased by NDUFV1 knockdown (Fig. 3A). Based on our experimental data, we propose for the first time a model explaining how CI dysfunction enhances skeletal myogenesis and blunts insulin signaling (Fig. 8). In the skeletal muscle with CI dysfunction, electrons are not transferred from NADH to the CI complex, which leads to a low NAD+/NADH ratio and subsequent SIRT1 inactivation. Because of inactive SIRT1 deacetylase, MyoD is acetylated and activated, which leads to myogenesis enhancement. CI dysfunction-induced SIRT1 inactivation also increases the expression level of PTP1B, a phosphatase that prevents the tyrosine phosphorylation and activation of IRβ and IRS-1 and leads to the blockage of insulin signaling. Thus, CI dysfunction-induced SIRT1 inactivation may be a causative factor for inducing insulin resistance.
FIGURE 8.
Model explaining how CI dysfunction induces myogenesis enhancement and insulin resistance. CI dysfunction decreases the NAD+/NADH ratio, which leads to SIRT1 inactivation. Thus, MyoD is acetylated and activated because of an inactive SIRT1 deacetylase, which enhances skeletal myogenesis. In addition, the PTP1B protein level is increased because of inactive SIRT1, which blunts insulin-elicited tyrosine phosphorylation of IR and IRS-1. However, mitochondrial biogenesis is not regulated by SIRT1 during skeletal myogenesis.
Because mitochondrial biogenesis accompanies skeletal myogenesis, the increased mitochondrial function may be essential for skeletal myogenesis (50). For example, C2C12 myoblasts lose their ability to be differentiated in the presence of chloramphenicol and ethidium bromide (inhibitors of mitochondrial DNA replication), rotenone (CI inhibitor), myxothiazol (CIII inhibitor), or oligomycin (CV inhibitor) (6, 8). Because these mitochondrial drugs may have other cellular targets that cause side effects such as myogenesis inhibition, genetic studies should be performed to understand the connection between mitochondrial biogenesis and skeletal myogenesis. Here, we re-assessed the role of mitochondrial function in skeletal myogenesis after the knockdown of NDUFV1, a subunit of CI. Unexpectedly, NDUFV1 knockdown did not affect C2C12 cell survival and proliferation but enhanced skeletal myogenesis (Fig. 1). Similar to our results, the systemic disruption of NDUFS4, another subunit of CI, did not show an abnormal muscular phenotype (51). The disruption of apoptosis-inducing factor with reduced CI activity also did not result in abnormal muscular development (32). Combined with our results and these previous findings, we conclude that mitochondrial function is not required for skeletal myogenesis.
SIRT1 may be essential for mitochondrial biogenesis in skeletal muscle because it up-regulates, deacetylates, and activates PGC-1α, which is a master transcriptional factor for mitochondrial biogenesis (49, 52). SIRT1 deacetylation of PGC-1α up-regulates genes encoding mitochondrial transcription factors and genes involved in oxidative phosphorylation and fatty acid utilization in C2C12 myotubes and mouse primary skeletal muscle cells, thus increasing glucose and fatty acid oxidation (53). Resveratrol-induced PGC-1α deacetylation and mitochondrial gene up-regulation is abolished by SIRT1 knockdown in mouse skeletal muscle (54). Paradoxically, SIRT1 is gradually inactivated during skeletal myogenesis because its expression level and the NAD+/NADH ratio are reduced (37). In addition, skeletal muscle-specific SIRT1 ablation does not reduce exercise-induced PGC-1α up-regulation, mitochondrial biogenesis, or cellular respiration (55). SIRT1 knockdown also does not reduce the mRNA levels of mitochondrial transcriptional factors and OXPHOS subunits in C2C12 myoblasts (56). Our data also challenge the hypothesis of SIRT1-mediated mitochondrial biogenesis in skeletal muscles and show that SIRT1 knockdown does not change the mtDNA content, OXPHOS protein expression levels, or cellular respiration in C2C12 myoblasts and myotubes (Fig. 7). Our results and those of previous reports suggest that SIRT1 activation is not required for mitochondrial biogenesis during C2C12 myogenesis.
The role of dysfunctional mitochondria in the development of type 2 diabetes has been highly controversial because mitochondrial dysfunction leads to the protection against obesity and insulin resistance in apoptosis-inducing factor 1 (AIF1)-, mitochondrial transcription factor A (TFAM)-, or PGC-1α/β-deficient mice (32, 33, 57). However, our data showed that CI dysfunction resulted in a decrease of the NAD+/NADH ratio, the deactivation of SIRT1, an increased expression level of PTP1B, the inactivation of IRβ and IRS-1, and insulin resistance (Fig. 8), which supports the hypothesis that mitochondrial dysfunction is a cause of insulin resistance. Thus, we can conclude that the activation of SIRT1 or inhibition of PTP1B could be used as a therapeutic strategy for the treatment of insulin resistance that results from CI dysfunction-induced SIRT1 inactivation and PTP1B up-regulation.
Acknowledgments
We thank Professor Soo-Jong Um, Sejong University, for the FLAG-SIRT1 and FLAG-SIRT1 H355A genes and Professor Jeong-Ho Hong, Korea University, for the myogenin and creatine kinase promoter-luciferase reporter genes.
This work was supported by Korea Research Foundation Grants 2011-0017562 and C00286 (to Y.-G. K.) and by the Korea University Research Fund.
- OXPHOS
- oxidative phosphorylation
- CI
- complex 1
- OCR
- oxygen consumption rate
- NDUFV1
- NADH dehydrogenase (ubiquinone) flavoprotein 1
- NDUFV1
- NADH dehydrogenase (ubiquintone) flavoprotein 1
- SIRT1
- sirtuin 1
- IR
- insulin receptor
- IRS-1
- insulin receptor substrate-1
- PTP1B
- protein-tyrosine phosphatase 1B
- Mef2
- myocyte enhancer factor 2
- ECAR
- extracellular acidification rate
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- MyHC
- myosin heavy chain
- Mgn
- myogenin
- Cav-3
- caveolin-3
- MG53
- mitsugumin 53
- si-CON
- si-control
- IP
- immunoprecipitation.
REFERENCES
- 1. Kim B. W., Lee J. W., Choo H. J., Lee C. S., Jung S. Y., Yi J. S., Ham Y. M., Lee J. H., Hong J., Kang M. J., Chi S. G., Hyung S. W., Lee S. W., Kim H. M., Cho B. R., Min D. S., Yoon G., Ko Y. G. (2010) Mitochondrial oxidative phosphorylation system is recruited to detergent-resistant lipid rafts during myogenesis. Proteomics 10, 2498–2515 [DOI] [PubMed] [Google Scholar]
- 2. Remels A. H., Langen R. C., Schrauwen P., Schaart G., Schols A. M., Gosker H. R. (2010) Regulation of mitochondrial biogenesis during myogenesis. Mol. Cell. Endocrinol. 315, 113–120 [DOI] [PubMed] [Google Scholar]
- 3. Gossett L. A., Kelvin D. J., Sternberg E. A., Olson E. N. (1989) A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 9, 5022–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 5. Handschin C., Rhee J., Lin J., Tarr P. T., Spiegelman B. M. (2003) An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. U.S.A. 100, 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pawlikowska P., Gajkowska B., Hocquette J. F., Orzechowski A. (2006) Not only insulin stimulates mitochondriogenesis in muscle cells, but mitochondria are also essential for insulin-mediated myogenesis. Cell Prolif. 39, 127–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biswas G., Adebanjo O. A., Freedman B. D., Anandatheerthavarada H. K., Vijayasarathy C., Zaidi M., Kotlikoff M., Avadhani N. G. (1999) Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle cross-talk. EMBO J. 18, 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lim J. H., Lee J. I., Suh Y. H., Kim W., Song J. H., Jung M. H. (2006) Mitochondrial dysfunction induces aberrant insulin signalling and glucose utilisation in murine C2C12 myotube cells. Diabetologia 49, 1924–1936 [DOI] [PubMed] [Google Scholar]
- 9. Choo H. J., Kim B. W., Kwon O. B., Lee C. S., Choi J. S., Ko Y. G. (2008) Secretion of adenylate kinase 1 is required for extracellular ATP synthesis in C2C12 myotubes. Exp. Mol. Med. 40, 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koopman W. J., Distelmaier F., Smeitink J. A., Willems P. H. (2013) OXPHOS mutations and neurodegeneration. EMBO J. 32, 9–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson S. C., Yanos M. E., Kayser E. B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V. Z., Gagnidze A., Oh K., Wasko B. M., Ramos F. J., Palmiter R. D., Rabinovitch P. S., Morgan P. G., Sedensky M. M., Kaeberlein M. (2013) mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeviani M., Tiranti V., Piantadosi C. (1998) Mitochondrial disorders. Medicine 77, 59–72 [DOI] [PubMed] [Google Scholar]
- 13. Schuelke M., Smeitink J., Mariman E., Loeffen J., Plecko B., Trijbels F., Stöckler-Ipsiroglu S., van den Heuvel L. (1999) Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat. Genet. 21, 260–261 [DOI] [PubMed] [Google Scholar]
- 14. Bénit P., Chretien D., Kadhom N., de Lonlay-Debeney P., Cormier-Daire V., Cabral A., Peudenier S., Rustin P., Munnich A., Rötig A. (2001) Large-scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am. J. Hum. Genet. 68, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogilvie I., Kennaway N. G., Shoubridge E. A. (2005) A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Investig. 115, 2784–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calvo S. E., Tucker E. J., Compton A. G., Kirby D. M., Crawford G., Burtt N. P., Rivas M., Guiducci C., Bruno D. L., Goldberger O. A., Redman M. C., Wiltshire E., Wilson C. J., Altshuler D., Gabriel S. B., Daly M. J., Thorburn D. R., Mootha V. K. (2010) High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 42, 851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swalwell H., Kirby D. M., Blakely E. L., Mitchell A., Salemi R., Sugiana C., Compton A. G., Tucker E. J., Ke B. X., Lamont P. J., Turnbull D. M., McFarland R., Taylor R. W., Thorburn D. R. (2011) Respiratory chain complex I deficiency caused by mitochondrial DNA mutations. Eur. J. Hum. Genet 19, 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schapira A. H. (2012) Mitochondrial diseases. Lancet 379, 1825–1834 [DOI] [PubMed] [Google Scholar]
- 19. Weissman J. R., Kelley R. I., Bauman M. L., Cohen B. H., Murray K. F., Mitchell R. L., Kern R. L., Natowicz M. R. (2008) Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PloS ONE 3, e3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lefort N., Glancy B., Bowen B., Willis W. T., Bailowitz Z., De Filippis E. A., Brophy C., Meyer C., Højlund K., Yi Z., Mandarino L. J. (2010) Increased reactive oxygen species production and lower abundance of complex I subunits and carnitine palmitoyltransferase 1B protein despite normal mitochondrial respiration in insulin-resistant human skeletal muscle. Diabetes 59, 2444–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lowell B. B., Shulman G. I. (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 22. Rabøl R., Boushel R., Dela F. (2006) Mitochondrial oxidative function and type 2 diabetes. Appl. Physiol. Nutr. Metab. 31, 675–683 [DOI] [PubMed] [Google Scholar]
- 23. Petersen K. F., Befroy D., Dufour S., Dziura J., Ariyan C., Rothman D. L., DiPietro L., Cline G. W., Shulman G. I. (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300, 1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 25. Petersen K. F., Dufour S., Befroy D., Garcia R., Shulman G. I. (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 350, 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phielix E., Schrauwen-Hinderling V. B., Mensink M., Lenaers E., Meex R., Hoeks J., Kooi M. E., Moonen-Kornips E., Sels J. P., Hesselink M. K., Schrauwen P. (2008) Lower intrinsic ADP-stimulated mitochondrial respiration underlies in vivo mitochondrial dysfunction in muscle of male type 2 diabetic patients. Diabetes 57, 2943–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hwang H., Bowen B. P., Lefort N., Flynn C. R., De Filippis E. A., Roberts C., Smoke C. C., Meyer C., Højlund K., Yi Z., Mandarino L. J. (2010) Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes 59, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holloszy J. O. (2013) “Deficiency” of mitochondria in muscle does not cause insulin resistance. Diabetes 62, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choo H. J., Kim J. H., Kwon O. B., Lee C. S., Mun J. Y., Han S. S., Yoon Y. S., Yoon G., Choi K. M., Ko Y. G. (2006) Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia 49, 784–791 [DOI] [PubMed] [Google Scholar]
- 30. Nair K. S., Bigelow M. L., Asmann Y. W., Chow L. S., Coenen-Schimke J. M., Klaus K. A., Guo Z. K., Sreekumar R., Irving B. A. (2008) Asian Indians have enhanced skeletal muscle mitochondrial capacity to produce ATP in association with severe insulin resistance. Diabetes 57, 1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wredenberg A., Freyer C., Sandström M. E., Katz A., Wibom R., Westerblad H., Larsson N. G. (2006) Respiratory chain dysfunction in skeletal muscle does not cause insulin resistance. Biochem. Biophys. Res. Commun. 350, 202–207 [DOI] [PubMed] [Google Scholar]
- 32. Pospisilik J. A., Knauf C., Joza N., Benit P., Orthofer M., Cani P. D., Ebersberger I., Nakashima T., Sarao R., Neely G., Esterbauer H., Kozlov A., Kahn C. R., Kroemer G., Rustin P., Burcelin R., Penninger J. M. (2007) Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131, 476–491 [DOI] [PubMed] [Google Scholar]
- 33. Zechner C., Lai L., Zechner J. F., Geng T., Yan Z., Rumsey J. W., Collia D., Chen Z., Wozniak D. F., Leone T. C., Kelly D. P. (2010) Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 12, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chalkiadaki A., Guarente L. (2012) Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296 [DOI] [PubMed] [Google Scholar]
- 35. Menzies K. J., Hood D. A. (2012) The role of SirT1 in muscle mitochondrial turnover. Mitochondrion 12, 5–13 [DOI] [PubMed] [Google Scholar]
- 36. Krag T. O., Hauerslev S., Jeppesen T. D., Duno M., Vissing J. (2013) Muscle regeneration in mitochondrial myopathies. Mitochondrion 13, 63–70 [DOI] [PubMed] [Google Scholar]
- 37. Fulco M., Schiltz R. L., Iezzi S., King M. T., Zhao P., Kashiwaya Y., Hoffman E., Veech R. L., Sartorelli V. (2003) Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12, 51–62 [DOI] [PubMed] [Google Scholar]
- 38. Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. (2008) Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barbieri E., Battistelli M., Casadei L., Vallorani L., Piccoli G., Guescini M., Gioacchini A. M., Polidori E., Zeppa S., Ceccaroli P., Stocchi L., Stocchi V., Falcieri E. (2011) Morphofunctional and biochemical approaches for studying mitochondrial changes during myoblasts differentiation. J. Aging Res. 2011, 845379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee C. S., Yi J. S., Jung S. Y., Kim B. W., Lee N. R., Choo H. J., Jang S. Y., Han J., Chi S. G., Park M., Lee J. H., Ko Y. G. (2010) TRIM72 negatively regulates myogenesis via targeting insulin receptor substrate-1. Cell Death Diff. 17, 1254–1265 [DOI] [PubMed] [Google Scholar]
- 41. Spinazzi M., Casarin A., Pertegato V., Salviati L., Angelini C. (2012) Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 7, 1235–1246 [DOI] [PubMed] [Google Scholar]
- 42. Kim B. W., Choo H. J., Lee J. W., Kim J. H., Ko Y. G. (2004) Extracellular ATP is generated by ATP synthase complex in adipocyte lipid rafts. Exp. Mol. Med. 36, 476–485 [DOI] [PubMed] [Google Scholar]
- 43. Del Prete A., Zaccagnino P., Di Paola M., Saltarella M., Oliveros Celis C., Nico B., Santoro G., Lorusso M. (2008) Role of mitochondria and reactive oxygen species in dendritic cell differentiation and functions. Free Radic. Biol. Med. 44, 1443–1451 [DOI] [PubMed] [Google Scholar]
- 44. Ugalde C., Janssen R. J., van den Heuvel L. P., Smeitink J. A., Nijtmans L. G. (2004) Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 13, 659–667 [DOI] [PubMed] [Google Scholar]
- 45. Jung S. Y., Ko Y. G. (2010) TRIM72, a novel negative feedback regulator of myogenesis, is transcriptionally activated by the synergism of MyoD (or myogenin) and MEF2. Biochem. Biophys. Res. Commun. 396, 238–245 [DOI] [PubMed] [Google Scholar]
- 46. Yi J. S., Park J. S., Ham Y. M., Nguyen N., Lee N. R., Hong J., Kim B. W., Lee H., Lee C. S., Jeong B. C., Song H. K., Cho H., Kim Y. K., Lee J. S., Park K. S., Shin H., Choi I., Lee S. H., Park W. J., Park S. Y., Choi C. S., Lin P., Karunasiri M., Tan T., Duann P., Zhu H., Ma J., Ko Y. G. (2013) MG53-induced IRS-1 ubiquitination negatively regulates skeletal myogenesis and insulin signalling. Nat. Commun. 4, 2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jafri M. S., Dudycha S. J., O'Rourke B. (2001) Cardiac energy metabolism: models of cellular respiration. Annu. Rev. Biomed. Eng. 3, 57–81 [DOI] [PubMed] [Google Scholar]
- 48. Sun C., Zhang F., Ge X., Yan T., Chen X., Shi X., Zhai Q. (2007) SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 6, 307–319 [DOI] [PubMed] [Google Scholar]
- 49. Amat R., Planavila A., Chen S. L., Iglesias R., Giralt M., Villarroya F. (2009) SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-γ co-activator-1α (PGC-1α) gene in skeletal muscle through the PGC-1α autoregulatory loop and interaction with MyoD. J. Biol. Chem. 284, 21872–21880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wagatsuma A., Sakuma K. (2013) Mitochondria as a potential regulator of myogenesis. ScientificWorldJournal 2013, 593267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kruse S. E., Watt W. C., Marcinek D. J., Kapur R. P., Schenkman K. A., Palmiter R. D. (2008) Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 7, 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodgers J. T., Lerin C., Gerhart-Hines Z., Puigserver P. (2008) Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 582, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 55. Philp A., Chen A., Lan D., Meyer G. A., Murphy A. N., Knapp A. E., Olfert I. M., McCurdy C. E., Marcotte G. R., Hogan M. C., Baar K., Schenk S. (2011) Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) deacetylation following endurance exercise. J. Biol. Chem. 286, 30561–30570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Price N. L., Gomes A. P., Ling A. J., Duarte F. V., Martin-Montalvo A., North B. J., Agarwal B., Ye L., Ramadori G., Teodoro J. S., Hubbard B. P., Varela A. T., Davis J. G., Varamini B., Hafner A., Moaddel R., Rolo A. P., Coppari R., Palmeira C. M., de Cabo R., Baur J. A., Sinclair D. A. (2012) SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 15, 675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vernochet C., Mourier A., Bezy O., Macotela Y., Boucher J., Rardin M. J., An D., Lee K. Y., Ilkayeva O. R., Zingaretti C. M., Emanuelli B., Smyth G., Cinti S., Newgard C. B., Gibson B. W., Larsson N. G., Kahn C. R. (2012) Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 16, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]