Background: REV-ERB activity is regulated by the endogenous porphyrin agonist, heme.
Results: REV-ERB binds other porphyrin ligands, including CoPP and ZnPP, which function as REV-ERB antagonists.
Conclusion: Differential regulation of REV-ERBs by porphyrins with different metal centers indicates that REV-ERB function is sensitive to the metal center.
Significance: Antagonist porphyrin ligands may be useful chemical tools for probing the function of REV-ERBs.
Keywords: Crystal Structure, Gene Expression, Heme, Ligand-binding Protein, Nuclear Magnetic Resonance (NMR), Nuclear Receptor, Porphyrin, Transcription
Abstract
REV-ERBα and REV-ERBβ are members of the nuclear receptor (NR) superfamily of ligand-regulated transcription factors that play important roles in the regulation of circadian physiology, metabolism, and immune function. Although the REV-ERBs were originally characterized as orphan receptors, recent studies have demonstrated that they function as receptors for heme. Here, we demonstrate that cobalt protoporphyrin IX (CoPP) and zinc protoporphyrin IX (ZnPP) are ligands that bind directly to the REV-ERBs. However, instead of mimicking the agonist action of heme, CoPP and ZnPP function as antagonists of REV-ERB function. This was unexpected because the only distinction between these ligands is the metal ion that is coordinated. To understand the structural basis by which REV-ERBβ can differentiate between a porphyrin agonist and antagonist, we characterized the interaction between REV-ERBβ with heme, CoPP, and ZnPP using biochemical and structural approaches, including x-ray crystallography and NMR. The crystal structure of CoPP-bound REV-ERBβ indicates only minor conformational changes induced by CoPP compared with heme, including the porphyrin ring of CoPP, which adopts a planar conformation as opposed to the puckered conformation observed in the heme-bound REV-ERBβ crystal structure. Thus, subtle changes in the porphyrin metal center and ring conformation may influence the agonist versus antagonist action of porphyrins and when considered with other studies suggest that gas binding to the iron metal center heme may drive alterations in REV-ERB activity.
Introduction
The nuclear receptors (NRs)6 REV-ERBα and REV-ERBβ regulate a number of physiological functions including circadian rhythm, lipid metabolism, cellular differentiation, and muscle endurance (1–5). The REV-ERBs were originally identified as orphan receptors due to the lack of an identifiable physiological ligand (6, 7). The REV-ERBs lack the activation function-2 region that is associated with the ability of NRs to recruit coactivators and activate target gene transcription (8, 9). Thus, REV-ERBs were thought to be constitutive repressors of transcription based on their ability to recruit corepressor proteins and suppress target gene transcription. Recently, we and others identified the porphyrin heme as a physiological agonist for both REV-ERBs (4, 10). Heme binds directly to the REV-ERB ligand-binding domain (LBD) and regulates the ability of REV-ERBs to recruit NR corepressors such as NCoR to target gene promoters and repress transcription of downstream genes (4, 10). A crystal structure of heme bound within the ligand-binding pocket of REV-ERBβ has also been described confirming these biological observations (11). The apparent constitutive repressor effect previously noted for the REV-ERBs is most likely due to the fact that all cells have some level of intracellular heme present, thus providing the ligand-dependent repression activity of these receptors.
The first synthetic REV-ERBα agonist (GSK4112, also known as SR6452) was soon after described and displayed activity in altering the circadian rhythm in a cell-based system, but unfortunately displayed unfavorable pharmacokinetic properties that limited further development (12). We further characterized GSK4112 as a dual REV-ERBα/REV-ERBβ agonist that activated REV-ERB activity in cell-based models of adipogenesis (13). We also described the first REV-ERB antagonist, SR8278, which was developed based on the GSK4112 scaffold (14). These studies suggested that the REV-ERBs are tractable targets for development of improved synthetic ligands that may have the ability to modulate REV-ERB-dependent physiological processes including circadian rhythm and metabolism. Indeed, we recently characterized the pharmacological effects in vivo of two newer synthetic REV-ERB agonists also derived from GSK4112 that modulate circadian behavior and a range of metabolic effects including increased energy expenditure and reduced plasma lipids (5, 15).
The physiological REV-ERB ligand, heme, is a prosthetic group that consists of a heterocyclic porphyrin ring with an iron ion metal center. Heme is an essential component of a range of proteins including oxygen transport proteins such as hemoglobin and myoglobin as well as the cytochrome P450 enzymes where the heme moiety carries out electron transport. Beyond heme, there is an array of additional porphyrin compounds that have been synthesized and/or are found naturally in cells depending on the physiological metal availability in tissues. Here we demonstrate that cobalt protoporphyrin IX (CoPP) and zinc protoporphyrin IX (ZnPP) also bind to REV-ERBs. CoPP and ZnPP are identical to heme except for the replacement of the iron metal center with a cobalt or zinc ion. We demonstrate that this subtle modification switches the activity of the porphyrin from a REV-ERB agonist (heme) to an antagonist (CoPP and ZnPP). We used structural, biochemical, and biophysical approaches to characterize the interaction of these porphyrins with REV-ERBβ. Because CoPP in particular has been shown to display in vivo efficacy including anti-obesity activity (16), this suggests that porphyrin REV-ERB antagonists may be useful chemical tools to probe REV-ERB function.
EXPERIMENTAL PROCEDURES
Recombinant Protein Expression and Purification
DNA encoding the human REV-ERBα-LBD (NR1D1; residues 281–614) and REV-ERBβ-LBD (NR1D2; residues 381–579) was amplified by PCR and cloned into the expression vector a pET-46 vector using the Ek/LIC system (EMD Chemicals/Novagen) as a tobacco etch virus protease-cleavable N-terminal His tag fusion protein. Protein was expressed in Escherichia coli BL21(DE3) grown in M9 minimum medium (iron-free to produce apoprotein) at 37 °C. When the cell density reached an A600 of 0.8–1.0, protein expression was induced with 1 mm isopropyl-1-thio-d-galactopyranoside and grown for 12 h at 16 °C and 250 rpm then harvested. Cells were resuspended and lysed, and protein was purified using His-Trap affinity chromatography followed His tag excision by tobacco etch virus and a final purification size exclusion step (S200 GE HealthCare) in buffer containing 50 mm HEPES, pH 7.5, 200 mm NaCl, and 0.5 mm TCEP. Protein was concentrated using 10 K Amicon-Ultra centrifugal filters and quantified measuring A280 in an Epoch BioTek system. For NMR experiments, REV-ERBβ-LBD was expressed in E. coli BL21(DE3) grown in M9 medium with either [15N]ammonium sulfate as the sole source of nitrogen, [13C]glucose as the sole carbon source, and medium was prepared with ∼99% deuterium oxide (D2O) to produce triple-labeled [2H13C15N]REV-ERBβ-LBD protein suitable for NMR experiments.
Spectroscopic Analysis of Porphyrin Binding to REV-ERB
Nine different porphyrin containing compounds were purchased from Frontier Scientific: protoporphyrin IX (PPIX), Co(III)PPIX, Cr(III)-mesoporphyryn IX, Cu(II)PPIX, Mg(II)PPIX, Mn(III)IX, N-methyl mesoporphyrin IX, Sn(IV)PPIX, and Zn(II)PPIX. Electronic absorption spectrum for the free form and in complex with REV-ERBβ-LBD(381–579) of these compounds was acquired in a Epoch BioTek system and analyzed with the Gen5 software version 1.11.5. Scans for every porphyrin (5 μm) in buffer (50 mm HEPES, pH 7.5, 200 mm NaCl, and 0.5 mm TCEP) were collected between 250 and 700 nm in 0.5-nm steps. REV-ERBβ-LBD(381–579) was added in a 1:3 molar excess to assure the completeness of the complex formation, and scans were collected again after a 2-min incubation at 25 °C in the dark.
Circular Dichroism
His-REV-ERBα-LBD was buffer exchanged (20 mm Tris, pH 8.0, 200 mm NaCl, 5% glycerol) and diluted 9-fold into assay buffer (45 mm Tris, 7.4, 17 mm NaCl, 5% glycerol) resulting in 9 μm REV-ERBα-LBD protein. Equal volumes of CoPP or Heme in 1 m NaOH or 1 m NaOH were added bringing the pH to 8.0 at room temperature. This mixture was continuously monitored at 222 nm via circular dichroism (JASCO J-815 CD Spectrometer) while raising the temperature 1 °C/min from 20 to 75–85 °C. The melting temperature (Tm) was calculated as the intersection of the melting curve with the midpoint between lines fit to the pre- and post-transition slopes. Tm and dH were fit using an in-house fitting algorithm written in Python.
Isothermal Titration Calorimetry
Experiments were carried out on a MicroCal iTC200 calorimeter (GE/MicroCal, Northampton, MA) at 25 °C. CoPP powder was dissolved in 100% DMSO at a concentration of 10 mm, heme was prepared similarly, then each was diluted to 500 μm (5% DMSO final concentration) in the same buffer as apo-REV-ERBβ-LBD (50 mm HEPES, pH 7.5, 200 mm NaCl, and 0.5 mm TCEP). 5% DMSO was added to the apoprotein sample and the reference buffer, and the pH was evaluated for all (protein, ligand, and reference) before performing ITC. The reaction cell contained 50 μm protein (200 μl) and was titrated with 19 injections of 2 μl of 500 μm CoPP or heme. The binding isotherm was fit with a binding model employing a single set of independent sites to determine the thermodynamic binding constants and stoichiometry.
Biochemical Corepressor Peptide Interaction Assay
Thirty-three-residue coregulator peptides designated for NCoR-1 (Biotin-KGGVPRTHRLITLADHICQIITQDFARNQVSSQ)and SMRT-1 (Biotin-KGGVKGHQRVVTLAQHISEVITQDYTRHHPQQL) were synthesized by Anaspec, Inc. (San Jose, CA). The amino acid peptide sequence design was based on the known amphipathic helical core (CoRNR box) motif. Low capacity streptavidin beads were purchased from Radix Biosolutions (Georgetown, TX). To couple peptides to beads, 50 μg/ml working concentrations of peptides were prepared in distilled H2O and used to couple to streptavidin beads overnight at 4 °C. All bead-peptide conjugates were washed twice with PBS/BSA buffer (10 mm NaH2PO4, 150 mm NaCl, 0.1% w/v BSA, 2 mm dithiothreitol, pH 7.4) and resuspended in 600 μl of PBS/BSA buffer. All bead-peptide conjugates were mixed into a single homogeneous bead mix before addition to the nuclear receptor protein/PentaHis-Alexa Fluor 532 complex. PentaHis-Alexa Fluor 532 antibody (Qiagen) was diluted to a final concentration of 0.8 μg/ml in 1× Luminex buffer (25 mm HEPES, 100 mm NaCl, 0.1% BSA, 2 mm dithiothreitol, pH 7.4). For each 96-well Luminex reaction, 10 μl of PentaHis-Alexa Fluor 532 antibody was added to 10 μl of 25× His-tagged-REV-ERB-LBD proteins being studied. REV-ERB·antibody complexes were allowed to form for a minimum of 30 min before addition of 220 μl of peptide bead mix. Ten μl of 25× cobalt protoporphyrin at each respective concentration was added to the appropriate wells. REV-ERB/NCoR1 and REV-ERB/SMRT-1 peptide interactions were allowed to proceed for ∼3 h with shaking at room temperature. REV-ERB/NCoR1 and REV-ERB/SMRT-1 interaction data were obtained by xMAP technology using the Bio-Plex 200 system and suspension array platform (Bio-Rad Laboratories). Dose-dependent effects of cobalt protoporphyrin treatment on REV-ERB/NCoR1 and REV-ERB/SMRT-1 interaction data were plotted using GraphPad Prism (GraphPad Inc., La Jolla, CA).
Cell Culture, Cotransfection, RNA Isolation, and Quantitative RT-PCR
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. HepG2 cells were maintained and routinely propagated in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. Twenty-four hours prior to transfection, HepG2 cells were plated in 96-well plates at a density of 15 × 103 cells/well. The PAI-1::luc reporter was constructed by inserting the human PAI-1 promoter (−867 to +122) into the pTAL-Luc luciferase vector (Clontech). GAL4-REV-ERBα-LBD, GAL4-REV-ERBβ-LBD, FLAG tag-REV-ERBα, and FLAG-REV-ERBβ plasmids were described previously (10, 15). Transfections contained 100 ng of the pTAL-Luc or pG5 reporter, 50 ng of pGL4.73 (Renilla) reporter, and 50 ng of receptor expression plasmid (GAL4-LBD or FLAG tag-full-length). Transfections were performed using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, the cells were treated with vehicle or compound. Twenty-four hours after treatment, the luciferase activity was measured using the Dual-Glo luciferase assay system (Promega). The results were analyzed using GraphPad Prism software, and values indicated represent the means ± S.E. from four independently transfected wells. The experiments were repeated at least three times.
RNA was isolated using the RNeasy kit (Qiagen) and quantified. Gene expression was determined using an ABI PRISM 7700 sequence detector (Applied Biosystems) and a SYBR Green detection system (Roche Applied Science). A standard curve was generated using cDNA pooled from the experimental samples. Relative expression levels were determined by normalization to cyclophilin B and expressed as arbitrary units.
Crystallography
Crystallization conditions were screening for REV-ERBβ-LBD bound to CoPP by sitting drop vapor diffusion using previous reported reservoir solution for REV-ERBβ-LBD bound to heme crystal structure (11). Crystals of REV-ERBβ-LBD bound to CoPP (added at a 2 molar excess) were obtained by equilibrating 1 μl of the CoPP-bound protein (15 mg/ml) with 1 μl of the reservoir solution containing 1.7 m ammonium sulfate, 4% Jeffamine, and 0.1 m Tris, pH 8.5. Initial crystals grew with in 5 days at 20 °C and diffracted at 3.2 Å resolution. These crystals were further optimized by microseeding into reservoir solution containing 1.4 m ammonium sulfate, 4% Jeffamine, and 0.1 m Tris, pH 8.5, in a ratio of protein-CoPP:reservoir solution (1:0.7) to obtain final crystals diffracting at 1.8 Å resolution. For data collection, crystals were cryoprotected by addition of 20% (v/v) glycerol and flash-frozen by immersion in liquid nitrogen. Crystals of REV-ERBβ-LBD bound to CoPP belong to the primitive orthorhombic system, space group P21212, with one molecule/asymmetric unit corresponding to a solvent content of 54%. Diffraction data from a single crystal of the complex were collected in a Micromax007-HF x-ray generator equipped with the MAR345 detector installed on a MAR345dtb table at The Scripps Research Institute Florida. Data were integrated using iMOSFLM (17) and scaled using SCALA in the CCP4 suite of programs (18). The structure was determined by molecular replacement using Phaser (19) and previously reported REV-ERBβ-LBD-heme structure (PDB ID code 3CQV) (11) as search model. To avoid bias of our maps, all water molecules and the heme ligand were excluded. The initial model obtained was improved by several cycles of refinement using PHENIX (20), and model manual building was carried out in Coot (21). At the last stage of refinement, we also applied the translation-libration-screw (TLS). The coordinates and structure factors have been deposited in the RCSB Protein Data Bank as entry 4N73. Structure figures were produced using PyMOL and chimera software for the B-factor color surface (22).
NMR Spectroscopy
NMR data were collected at 298 K on a Bruker spectrometer at 700 MHz 1H frequencies equipped with a TXI probe. The proton carrier frequency was set coincident with the water resonance for all experiments. Backbone assignments for the apo-REV-ERBβ or bound to zinc(II) protoporphyrin IX (ZnPP, 1:2 ratio) were obtained using three-dimensional NMR experiments available in Bruker Topspin 3.0, including TROSY versions of the HNCO, HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, as well as 15N-NOESY-HSQC, using 2H,13C,15N-labeled REV-ERBβ-LBD (1.2 mm). Data were processed with Topspin and analyzed by NMRview (23). The NMR sample buffer contained 20 mm potassium phosphate, pH 7.4, 50 mm potassium chloride, and 0.5 mm TCEP. Ligands were added from DMSO stock solutions.
Statistical Analysis
All data are expressed as the mean ± S.E. (n = 3 or more). Statistical evaluation was performed using one-way analysis of variance followed by Dunnet's or Tukey's post hoc test.
RESULTS
A Screen Identifies Porphyrin-Metal Compounds That Bind to REV-ERB
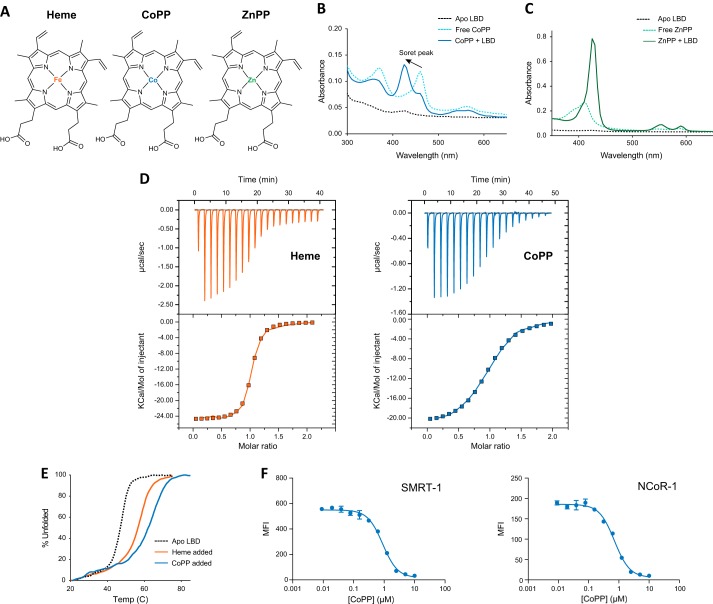
To determine whether porphyrins other than heme are capable of binding to REV-ERB, we performed a UV/visible electronic absorption assay to evaluate the ability of REV-ERB to interact with nine porphyrin metal derivatives. CoPP and ZnPP (Fig. 1A) specifically formed a complex with REV-ERBβ-LBD as observed by the shift of the soret peak from the free porphyrin form to the REV-ERB complex (Fig. 1, B and C, respectively). Other metal-porphyrin derivatives that did not significantly change the absorption spectrum include protoporphyrin IX, Cr(III) mesoporphyrin IX, Cu(II) protoporphyrin IX, Mg(II) protoporphyrin IX, Mn(III) protoporphyrin IX, N-methyl mesoporphyrin IX, and Sn(IV) protoporphyrin IX (data not shown). ITC also demonstrates binding of CoPP (KD = 2.56 μm) with and heme (KD = 353 nm) (Fig. 1D). Notably, the affinity of heme for REV-ERBβ that we determined is higher than the Kd we previously reported (2.07 μm) (10). As we explain below, the likely source of this difference is the shorter LBD fragment we used in our experiments here, which does not aggregate like the full-length REV-ERB-LBD used in our prior studies. We also observed that dialysis of the heme-bound REV-ERBβ-LBD fragment does not exchange away heme as easily as the full-length LBD, which is consistent with the higher heme affinity observed in our ITC data here. Of note, the ITC determined heme affinity for this shorter LBD fragment is lower than previously reported absorbance-based affinities (16–117 nm) for various redox forms of the full-length REV-ERBβ-LBD (24). This indicates there is likely an effect, although not dramatic, of the smaller LBD construct on binding affinity. Nuclear receptors typically display increased thermal stability when bound to a ligand. This can be detected by an increase in Tm detected in a thermal melt protein-unfolding assay monitored by CD spectroscopy. We previously demonstrated that heme binding causes an increase in the melting temperature of the REV-ERB-LBD (10). CoPP also causes an increase in melting temperature (Tm = 65 °C) relative to apo-LBD (Tm = 47.7 °C) (Fig. 1E).
FIGURE 1.
CoPP and ZnPP are REV-ERB ligands. A, structures of heme, CoPP, and ZnPP. B, absorption spectra of apo-REV-ERBβ-LBD(381–579; black dotted line), free cobalt(III)-protoporphyrin IX (CoPP, light blue dotted line) and the complex (dark blue line). The shift of the soret peak for the free porphyrin at approximately 460–425 nm in the presence of REV-ERBβ-LBD indicates complex formation. C, absorption spectra similarly show differences between free zinc(II)-protoporphyrin IX (ZnPP, light green dotted line) and the complex (dark green line). D, ITC analysis demonstrates direct binding of heme (KD = 353 nm; n = 0.988; ΔH = −2.488 E4 cal/mol; ΔS = −55.4 cal/mol/degree) and CoPP (KD = 2.56 μm; n = 0.967; ΔH = −2.132 E4 cal/mol; ΔS = −47.1 cal/mol/degree) to REV-ERBβ-LBD. E, circular dichroism analysis reveals CoPP (blue) and heme (orange) increase the melting temperature of apo-REV-ERBα-LBD (black dotted line). F, biochemical corepressor interaction assay illustrating the ability of REV-ERBα-LBD to recruit the NCoR CoRNR box peptide derived from either corepressor SMRT or NCoR with increasing concentrations of CoPP using Luminex technology. Error bars indicate mean ± S.E.; n = 3.
Based on these data indicating that CoPP binds to the REV-ERB-LBD, we tested the ability of CoPP to induce a conformational change in the receptor that might result in an alteration in the ability to recruit a corepressor protein. We performed a biochemical corepressor peptide recruitment assay using Luminex technology (10). CoPP dose-dependently inhibited the interaction of the REV-ERB-LBD with corepressor peptides (Fig. 1F). Thus, CoPP binds to the REV-ERB-LBD and induces a conformational change that alters the ability of the receptor to interact with biologically relevant factors. Interestingly, even though heme acts as an agonist in cells causing REV-ERB to recruit NCoR and suppression of target gene transcription, in biochemical assays such as Luminex (10) or TR-FRET (25) the natural porphyrin agonist, heme, causes a decrease in corepressor peptide recruitment. This appears to be a unique feature of heme as synthetic agonists that induce NCoR recruitment in a cellular context cause an increase in corepressor peptide binding in biochemical assays in contrast to heme (13). Thus, based on the effects of CoPP in this experiment and its structural similarity to heme, we could not predict whether this compound would function as a REV-ERB agonist or antagonist. Thus, we turned to cell-based assays to determine the functional effect of these porphyrins on REV-ERB activity.
Cobalt Protoporphyrin IX and Zinc Protoporphyrin IX Are REV-ERB Antagonists
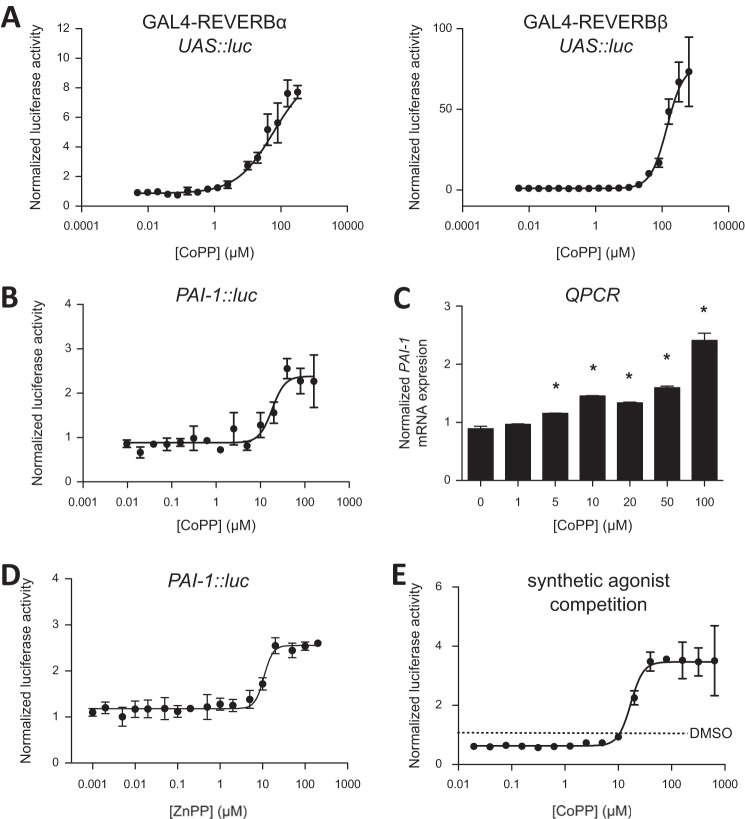
To clarify the biological action of CoPP and ZnPP on REV-ERB function, we performed a cell-based cotransfection assay using GAL4-REV-ERB chimeric receptors along with a luciferase reporter with a multimerized GAL4-binding element in the promoter (10). Addition of CoPP results in increased transcription of the luciferase reporter (Fig. 2A) consistent with REV-ERB antagonism. Because all cells have some level of heme present, REV-ERB is typically maintained at some level of constitutive activity resulting in suppression of target genes; thus, addition of a porphyrin antagonist, even without addition of an exogenous agonist, results in an increase in transcriptional activity.
FIGURE 2.
CoPP functions as a REV-ERB antagonist. A, cotransfection assays in HEK293 cells demonstrating antagonist activity of CoPP for GAL4-REV-ERBα and GAL4-REV-ERBβ. B, cotransfection assay in HEK293 cells with full-length REV-ERBα and a luciferase reporter driven by the PAI-1 promoter. C, expression of REV-ERB target gene PAI-1 after treatment with various doses of CoPP for 24 h. D, ZnPP also functioning as a REV-ERB antagonist in a cell-based cotransfection assay using full-length REV-ERBα and a luciferase reporter driven by the PAI-1 promoter. *, p < 0.05. E, cotransfection assay in HEK293 cells performed with GAL4-REV-ERBα in the presence of the agonist, SR9011 (1 μm) and increasing amounts of the antagonist CoPP. SR9011 decreased transcription (agonist activity), and increasing amounts of CoPP blocked this activity dose-dependently (IC50 = 17 μm). This is cell-based, and there are physiological levels of heme present. Error bars indicate mean ± S.E. and n = 3.
REV-ERB regulates gene transcription after binding to specific DNA sequences in the promoter of REV-ERB target genes. To further characterize the activity of CoPP on REV-ERB function, we assessed the ability of CoPP to modulate the expression of target gene expression. PAI-1, also known as endothelial plasminogen activator inhibitor, is a REV-ERB target gene (26). We placed a fragment of the PAI-1 promoter upstream of a luciferase reporter and cotransfected this plasmid into HEK293 cells along with REV-ERBα; CoPP dose-dependently increased transcription of this PAI-1 promoter reporter (Fig. 2B), consistent with CoPP functioning as a REV-ERB antagonist. In HepG2 cells, which express REV-ERBα endogenously, endogenous levels of heme acting on REV-ERBα maintain a tonic suppression of target genes, and addition of CoPP resulted in an increase in expression of these genes due to blocking of the action of the endogenous agonist, heme (Fig. 2C). These data are consistent with our hypothesis that CoPP regulates REV-ERB function acting as an antagonist of REV-ERB activity.
CoPP might also be expected to have indirect effects on REV-ERB activity due to its effects on a key enzyme that regulates heme degradation, heme oxygenase (HMOX1). CoPP induces HMOX1 gene expression resulting in a decrease in intracellular heme levels (27), which would presumably result in loss of REV-ERB-mediated transcriptional repression. Two lines of evidence suggest that the CoPP effects we observed on REV-ERB function in our cotransfection assays are not indirect via HMOX1. First, our screen identified that ZnPP also binds to REV-ERB. ZnPP also functions as REV-ERB antagonists in a cell-based luciferase cotransfection assay (Fig. 2D). However, whereas CoPP is an inducer of HMOX1 activity, ZnPP is an inhibitor of HMOX1 activity (28). If HMOX1 plays an indirect role in regulating REV-ERB function in our assays, then CoPP and ZnPP would be expected to give different REV-ERB functional responses, but they display the same functional response (REV-ERB antagonism). Second, to help differentiate the potential effects of loss of heme-dependent repression versus direct antagonism of REV-ERB, we performed an additional cotransfection experiment as described above, but we added a synthetic REV-ERB agonist, SR9011 (15) and assessed the ability of CoPP to block the SR9011-dependent suppression of REV-ERB target gene expression (Fig. 2E). Addition of saturating amounts of SR9011 suppressed the basal transcriptional activity (normalized luciferase activity below a value of 1) as would be expected for an REV-ERB agonist, and titration of CoPP resulted in a dose-dependent loss of the suppression due to SR9011. Additionally, there was a further increase in transcription, presumably due to CoPP antagonizing the action of endogenous heme functioning.
It is noteworthy to mention that CoPP and ZnPP concentrations required to elicit a functional response in these cell-based assays are higher than the dissociation constant (Kd) for REV-ERB that we determined biophysically via ITC and biochemically via Luminex using purified protein. There are several possible explanations for this, including differences in redox environment in the cells versus the more controlled biophysical and biochemical experiments, which could change the binding affinity of the porphyrins for REV-ERB. In addition, all cells contain endogenous heme, and the binding affinity of heme for REV-ERB is much higher than that for CoPP or ZnPP. Thus, concentrations of CoPP and ZnPP that are higher than their Kd would be required to compete with heme and elicit a functional response.
Crystal Structure of CoPP-bound REV-ERBβ
To understand the function of CoPP as a REV-ERB antagonist, we determined a high resolution crystal structure of a C-terminal fragment REV-ERBβ-LBD (residues 381–576) bound to CoPP (Table 1). Importantly, we verified that CoPP inhibited CoRNR box recruitment for this C-terminal fragment using a Luminex biochemical assay (data not shown) in which we observed results similar to those obtained for the full-length LBD. The resolution of the CoPP-bound REV-ERBβ structure is similar to the resolution reported for heme-bound REV-ERBβ (PDB ID code 3CQV; solved to 1.9 Å) and comprises the same structural elements for ligand binding and recognition (11, 29), which provides confidence to compare the two structures directly.
TABLE 1.
Crystallography data collection and refinement statistics
| REV-ERBβ–LBD bound to CoPP | |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c, (Å) | 73.660, 48.680, and 69.750 |
| a, b, g (°) | 90.00, 90.00, and 90.00 |
| Resolution (Å) | 40.5–1.87 (1.92–1.87)a |
| Rmerge | 0.027 (0.349) |
| I/σI | 14.1 (2.8) |
| Completeness (%) | 99.8 (97.9) |
| Redundancy | 3.4 (3.5) |
| Refinement | |
| Resolution (Å) | 40.5-1.86 |
| No. reflections for refinement | 21161 |
| Rwork/Rfree | 0.215/0.264 |
| No. of atoms (B-factors) | |
| Protein | 3,087 (11.63) |
| Water | 220 (21.35) |
| Root mean square deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.35 |
| Ramachandran statistics (%) | |
| Residues in favored regions | 96.4 |
| Residues in additional allowed regions | 3.05 |
a Values in parentheses are for the highest-resolution shell.
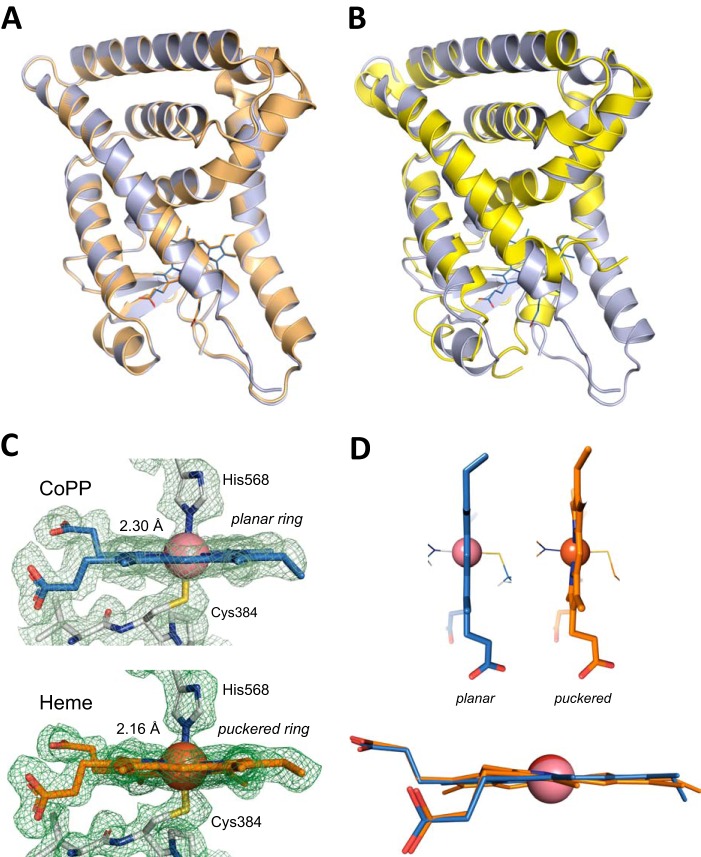
The overall domain structures of heme-bound and CoPP-bound REV-ERBβ are very similar with a root mean square deviation of 0.16 Å obtained from a structural backbone superposition (Fig. 3A). In contrast, similar to the heme-bound structure (11), significant differences are observed within the ligand-binding pocket between the apo and CoPP-bound forms of REV-ERBβ (Fig. 3B). One difference between these two structures involves residues Cys-384 and His-568, which are coordinated by the metal center of the porphyrin ring (Fig. 3C). The position and the distance of Cys-384 to the metal center (2.34 Å) in the holo structures (heme and CoPP) are conserved. However, the distance between His-568 and the cobalt metal of CoPP is 0.14 Å longer than that for iron in the heme complex. A similar difference was observed for another heme-binding protein, hemoglobin, between the coordinating histidine residue and heme versus CoPP (30–32). It was posited that these structural changes in the CoPP-hemoglobin complex affect oxygen binding to hemoglobin with affinities 50–100 times weaker compared with the iron-heme counterpart. It is possible that similar to hemoglobin, small structural arrangements in REV-ERB-LBD could affect corepressor recruitment and/or small molecule donor (e.g. NO or CO) binding (11, 24), providing a potential mechanism for the antagonist activity of CoPP.
FIGURE 3.
Crystal structure of CoPP bound to the REV-ERBβ-LBD. A, overlay of CoPP- (blue) and heme- (orange; PDB code 3CQV) bound structures of the REV-ERBβ-LBD. B, overlay of the CoPP-bound (blue) and apo (yellow; PDB 2V0V) structures of the REV-ERBβ-LBD. C, cobalt-His-568 coordination distance (2.30 Å). The distance is 0.14 Å larger than the one observed for the iron-His-568 holo structure (2.16 Å) (PDB code 3CQV (11)). D, CoPP adopts a planar conformation in the LBD of REV-ERBβ-LBD different from the non-planar, puckered distortion observed for the heme group.
Another structural difference between the complexes is the imidazole group of His-568, which deviates from the perpendicular plane of the metal center in the CoPP complex compared with the heme complex. In addition, the CoPP porphyrin ring adopts a planar conformation, in contrast to the planar, puckered conformation observed for the iron-based porphyrin ring (Fig. 3D), suggesting that the CoPP ligand adopts a subtle but different conformation within the REV-ERBβ ligand-binding pocket. This observation suggests that REV-ERBβ is very sensitive and responds differently to accommodate different porphyrin ligands into the ligand-binding pocket and more importantly indicates that His-568 plays a major role in this response.
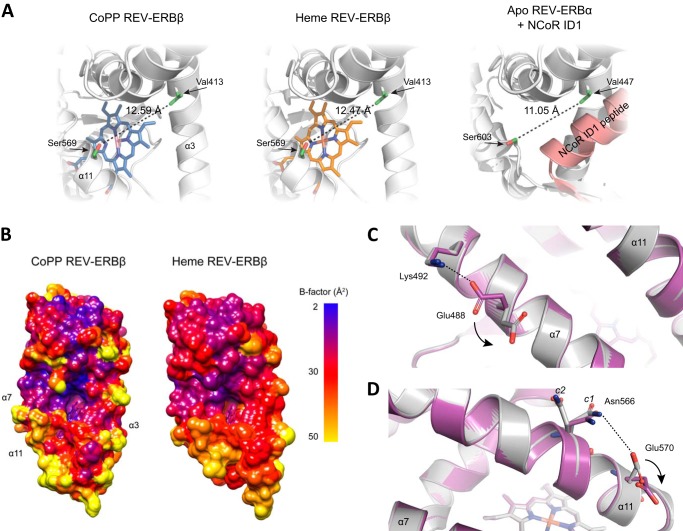
Helix 11 is one of the key structural elements for ligand binding and corepressor recruitment in REV-ERBs (11, 29, 33). The subtle movement of His-568, which is involved in metal-porphyrin binding and positioned on helix 11, causes a concomitant shift of proximal residues resulting in an expansion of the ligand-binding pocket when bound to CoPP (12.59 Å) compared with heme (12.47 Å) (Fig. 4A). Residues comprising this surface are involved in corepressor peptide binding, and in the crystal structure of REV-ERBα-LBD in complex with NCoR1 (29) the analogous surface distance is narrower (11.05 Å). This suggests that a compacted ligand-binding pocket is needed to assist corepressor recruitment. The enlarged CoPP/REV-ERBβ-LBD could weaken and negatively affect corepressor recruitment in agreement with our functional findings above. Thus ligands may control the open-closed conformation for corepressor recruitment of this surface.
FIGURE 4.
Additional structural comparisons of the CoPP and heme bound REV-ERBβ LBDs. A, CoPP binding increases the distance between helix 3 and 11 compared with the heme-bound REV-ERBβ (PDB ID Code 3CQV (11)) and NCoR-bound apo-REV-ERBα structures (PDB ID Code 3N00 (29)). B, B-factor colored surface represents CoPP and heme complexes. Helix 3, 7, and 11 display the highest B-factors in the CoPP structure and are increased compared with the heme complex. C and D, overview shows electrostatic interactions that might contribute to the antagonist function of CoPP. The disruption of the salt bridge interaction between Lys-492 and Glu-477 (C) in the CoPP/REV-ERBβ may confer less rigidity to helix 7. An ion pair is formed between Asn-566 and Glu-570 (D) on helix 11 in the CoPP/REV-ERBβ structure that is not present in the heme-bound complex. This electrostatic attraction exerts an arm moment in helix 11 that, considered with the increased distance between helix 3 and 11, may push 11 further away from the ligand-binding pocket.
Comparing the crystallographic B-factors in the CoPP and heme-bound structures also reveal flexible regions that could negatively affect corepressor recruitment. Because the CoPP and heme-bound structures were solved at a similar resolution (1.8 and 1.9 Å, respectively), this avoids uncertainty in determining the relative contributions from local dynamics and lattice disorder. Regions around helix 3, 7, and 11 display the highest B-factors for the CoPP complex compared with the heme complex (Fig. 4B). This is supported by a detailed inspection of the CoPP and heme-bound REV-ERBβ-LBD structures. In the heme-bound structure, there is a well defined salt bridge between residues Glu-488 and Lys-492 (Fig. 4C). This salt bridge interaction is absent in the CoPP-REV-ERBβ complex due to the different conformation that Glu-488 adopts in the presence of CoPP. This salt bridge disruption imposes flexibility to helix 7 in the CoPP complex. In addition, Asn-566 and Glu-570 on helix 11 are in close proximity to each other in the CoPP complex (4.1 Å) and weakly connected by an electrostatic interaction (Fig. 4D). In contrast, in the heme complex the side chain of Glu-570 flips, causing an increased distance between these two residues (5.9 Å) that disrupts this interaction. This ion pair interaction between Asn-566 and Glu-570 slightly moves helix 11 out of the ligand-binding pocket (open conformation) modulating the degree to which the corepressor-binding surface is expanded. Notably, in both the CoPP and heme complexes, there is clear electron density, indicating that Asn-566 can be modeled into two distinct conformations, c1 and c2 (Fig. 4D), which may have a role controlling the open-closed conformation of helix 11. It is important to notice that these two residues are exposed to the solvent; however, the asparagine side chain (amide group) is not dissociable, and the glutamic acid side chain is negatively charged below pH 7, suggesting that their chemical properties are similar for the two complex structures (heme pH 7.6 and CoPP pH 8.5).
Conformational Analysis of the REV-ERBβ-LBD by Solution NMR
We previously used NMR to demonstrate a dynamic mechanism by which ligands that bind to another nuclear receptor, PPARγ, partially stabilize the PPARγ activation function-2 surface to provide full versus partial agonist activity (34). Protein NMR analysis of apo-REV-ERBβ-LBD reveals that approximately half of the residues comprising the LBD are dynamic (flexible) on the intermediate NMR time scale and are not observed in the NMR experiments. Apo-PPARγ and RXRα also display a similar dynamic phenotype in the apo form (34, 35), suggesting generally that the ligand-binding pockets of apo-ligand-regulated NR LBDs are conformationally dynamic.
REV-ERBβ-LBD in complex with heme and CoPP, which contain a paramagnetic metal ion center, caused severe NMR line broadening (data not shown), preventing us from undertaking a detailed NMR analysis to determine the structural and dynamic effects of these ligands on the REV-ERBβ-LBD. Some notable chemical shift differences are observed, but in addition to structural changes these could be caused by differences in the pseudocontact chemical shift arising from the different paramagnetic centers of heme versus CoPP. However, another porphyrin derivative identified in our screen, ZnPP, also binds to REV-ERB and functions as an antagonist. Unlike heme and CoPP, the zinc center is diamagnetic. We therefore used ZnPP to study the effect of porphyrin binding in general to the REV-ERBβ-LBD using NMR.
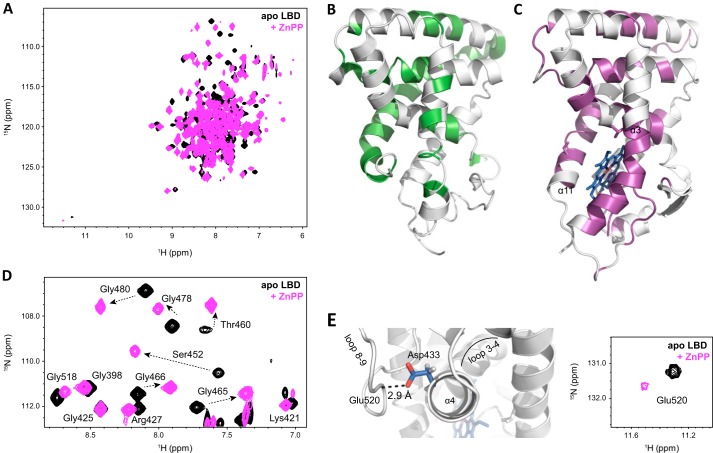
NMR data reveal significant NMR chemical shift changes for many residues in the REV-ERBβ-LBD between the apo and ZnPP-bound forms (Fig. 5A). This is consistent with the large structural changes observed in crystal structures of REV-ERBβ in the apo form compared with structures bound to porphyrin ligands. The large structural changes manifest as different chemical environments in the NMR data, likely due to the large surface area occupied by the porphyrin ring in the REV-ERBβ ligand-binding pocket. A comparison of the residues stabilized from intermediate exchange in the apo form would give insight into whether ZnPP binding significantly stabilizes the REV-ERBβ-LBD. Compared with resonances visible in the apo-REV-ERBβ NMR data (Fig. 5B), several new NMR resonances are observed in the ZnPP-bound complex that appear to be missing in the apo form (Fig. 5C). Still, not all resonances missing in the apo form are observed. Furthermore, most of the new NMR resonances in the ZnPP-bound complex have lower peak intensities, making it difficult to assign these new resonances confidently. The low peak intensities for these new resonances could be due to the low micromolar binding affinity of porphyrin derivatives for REV-ERB under the conditions of our study. Binding events that occur in this affinity regime show intermediate exchange or line broadening in the NMR data because the rate of interconversion between the free and bound states (kex) during the detection period of the NMR experiment is similar to the chemical shift difference (Δv) of the resonance frequencies of the two states (vfree and vbound).
FIGURE 5.
NMR analysis of the REV-ERBβ-LBD. A, overlay shows two-dimensional 1H,15N-TROSY-HSQC spectra of apo-REV-ERBβ-LBD (black) and ZnPP-bound complex (pink). B and C, unassigned residues in the apo-REV-ERBβ NMR data (B) and ZnPP-bound complex (C) reveal that ZnPP binding stabilizes some regions of REV-ERBβ. D, glycine fingerprint region of the NMR data demonstrates significant chemical shift perturbations resulting from porphyrin binding. E, NMR data detect a change in the structure for Glu-520 and more generally the loops between helix 3–4 and helix 8–9, upon porphyrin binding.
Although a comparison of the magnitude of the chemical shift change would indicate that in residues sensitive to ZnPP binding, ring current effects caused by the porphyrin ring scaffold would overestimate the analysis of a strict chemical shift perturbation analysis. However, significant NMR chemical shift perturbations between the apo and ZnPP-bound forms are quite apparent for residues with NMR chemical shifts within a well resolved “fingerprint” region in the NMR data comprised mostly of glycine residues (Fig. 5D). Residues near the porphyrin-binding pocket displaying significant chemical shift changes between apo and ZnPP-bound forms in this fingerprint region include Ser-452, Thr-460, Gly-465, Gly-478, and Gly-480. Another structural area that displays large chemical shift changes is the end of helix 11, which forms part of the porphyrin-binding site. These results are in agreement with the observations in the crystal structure of the CoPP-bound REV-ERBβ complex. In the apo-REV-ERBβ-LBD structure, the end of helix 11 is buried into the ligand-binding pocket. Binding of a porphyrin ligand displaces the end of helix 11 from the ligand-binding site. Residues on helix 11 N-terminal to Leu-572 and Leu-573 in the ZnPP and apo forms, respectively, are not observed in the NMR data. In the apo form, this is likely due to conformational exchange on the intermediate NMR time scale due to a dynamic ligand-binding pocket. However, in the case of the ZnPP complex, these missing resonances could be due to intermediate exchange caused by ligand exchange kinetics for a porphyrin with micromolar binding affinity.
In addition to the significant NMR chemical shift changes for residues within or near the ligand-binding pocket, a number of more modest chemical shift changes are observed from residues further away from the ligand-binding site. Interestingly, for some of the residues that display modest chemical shift changes, no major structural differences are observed in crystal structures of apo versus porphyrin-bound REV-ERBβ-LBD. This suggests that NMR is more sensitive to subtle structural perturbations that occur upon porphyrin binding than that observed in the crystal structures or detects structural states not present in the crystal structure. One surface remote porphyrin-binding site that is affected by porphyrin binding is the loop between helix 3 and 4 (loop 3–4), as well as the loop between helix 8 and 9 (loop 8–9) (Fig. 5E). The NMR resonance for Glu-520 is shifted downfield with a 1NH chemical shift of 11.3 ppm in the apo form. This unique chemical environment, far removed from the other backbone NH NMR resonances, is likely due to a strong hydrogen bond observed in the REV-ERBβ structure between the backbone amide of Glu-520 in loop 8–9 and the side chain carboxyl of Asp-433 at the beginning of helix 4. ZnPP binding causes a chemical shift perturbation of the Glu-520 and Asp-433 resonances, as well as significant line broadening of the Glu-520 resonance resulting in lower peak intensity that suggests a change in the dynamics of loop 8–9 in response to porphyrin binding.
DISCUSSION
REV-ERBs function as transcriptional repressors by recruiting nuclear receptor corepressor (NCoR)-HDAC3 complexes to REV-ERB response elements in enhancers and promoters of target genes. REV-ERB agonists, such as the endogenous porphyrin heme or synthetic agonists, function by increasing the transcriptional repression of REV-ERB target genes. Here, we determined that two porphyrin ligands, CoPP and ZnPP, function as antagonists of REV-ERB. We recently demonstrated that synthetic REV-ERB agonists reduce body weight in diet-induced obese mice (15), and it is therefore interesting that CoPP has also been shown to have an anti-obesity effect in vivo (36). However, CoPP appears to display complex pharmacology where it is also an inducer of HMOX1 expression (27), and the induction of HMOX1 has been suggested as the mechanism by which CoPP displays its anti-obesity activity (16). Given that CoPP induction of HMOX1 activity would also be associated with an alteration of heme levels and thus alter REV-ERB activity indirectly, we were careful to characterize the direct interaction of CoPP with REV-ERB as well as modulation of REV-ERB interaction with corepressor in our studies. One would expect induction of HMOX1 by CoPP to reduce intracellular heme levels leading to a decrease in REV-ERB-mediated transcriptional repression. This created potential issues with determining the REV-ERB antagonist activity of CoPP in cells, but we addressed this by providing a saturating amount of a synthetic agonist, SR9011, and demonstrated that CoPP could antagonize the action of the synthetic agonist. Furthermore, CoPP and ZnPP have opposite effects on HMOX1 activity, but we demonstrate that both porphyrins function as REV-ERB antagonists, further supporting that our observations are REV-ERB-dependent.
Particularly intriguing is that antagonists CoPP and ZnPP differ from the natural agonist heme only at the point of the metal ion that is coordinated by the porphyrin ring; and despite this fact REV-ERB function is affected differently when these different ligands are bound to it. Binding of CoPP to REV-ERBβ causes only subtle conformational changes within the crystal structure of the REV-ERBβ-LBD. Somehow, binding of CoPP leads to diminished affinity for corepressors, resulting in loss of the receptor transcriptional repressor activity. CoPP increased the B-factor of some residues compared with the heme-bound complex that likely contact a corepressor peptide if bound in the mode observed in a REV-ERBα crystal structure (29). Increased B-factors can indicate increased conformational dynamics, which could affect corepressor binding. What still remains a quandary is the discrepancy between cell-based assays and biochemical assays in assessing the function of heme. Heme acts as an endogenous agonist of REV-ERB, which means heme increases the ability of REV-ERB to repress target gene expression through increasing binding of corepressors such as NCoR to REV-ERB target genes (10, 25). However, biochemical and structural studies appear to contradict this, as heme and other porphyrins displace a NCoR peptide from REV-ERB, and the crystal structure of REV-ERBα bound to NCoR was determined in the absence of ligand.
Perhaps these structure-function differences are a result not of porphyrin binding solely, but rather subsequent modifications of porphyrin-bound REV-ERB via redox conditions and small molecule gases. Several models have been suggested for the regulation of REV-ERB activity, and a Drosophila ortholog of the human REV-ERB nuclear receptors, E75, by redox conditions and small molecule gases (11, 24, 37). In the Fe(III) state, a six-coordinate heme-receptor complex is formed involving Cys-384 and His-568 residues (referred to as heme “ligands”) and, upon reduction to Fe(II), the Cys-384 ligand is replaced by another neutral donor ligand to form a six-coordinate species or can exist as a five-coordinate species with only the His-568 ligand. It is to these reduced forms that NO and CO bind via the neutral, or non-His-568, ligand site. In cell-based assays, addition of NO or CO reduces the repressive transcriptional activity of REV-ERB.
This type of ligand switching mechanism has been observed in other gas sensor proteins that contain a prosthetic heme ligand, including cystathionine β-synthase (38, 39), soluble guanylate cyclase (40), H-NOX domain-containing proteins (41), and the CO sensing transcription factor CooA (42, 43). In general, several features are thought to coordinate structure-function responses in these heme sensors, including the coordination chemistry of the porphyrin metal center, the conformation of the porphyrin ring, and the dynamics of the ligand-binding domain, all of which allows these proteins to sense and react to different chemical environments. Given the chemical diversity of the metal center of heme, CoPP and ZnPP, it is feasible to suggest that differences in ability of these metal centers to respond to redox conditions and small molecule gas binding to REV-ERBs may impart different conformational changes that affect function. For example, in hemoglobin substitution of the heme iron metal center for other non-iron metals, including Mn and Zn, does not allow CO binding to hemoglobin (44) likely because these non-iron porphyrin analogs are unable to bind a sixth ligand and therefore do not readily interact with small molecule gases (45). Co-substituted porphyrin can bind O2, albeit with significantly lower affinity than heme (44), and can also bind NO (46) but not CO (32, 47). In another heme-regulatory sensor, the H-NOX protein from Thermoanaerobacter tengcongensis (Tt H-NOX), mutations that resulted in a more flattened heme porphyrin ring changed the activity of H-NOX, indicating an important role for porphyrin ring conformation in H-NOX function. Also related to this, in hemoglobin the heme porphyrin ring is non-planar (puckered) in the absence of O2, but upon O2 binding it forms a planar ring. The heme-bound REV-ERBβ structure also shows a non-planar, puckered ring conformation (11). If small molecule gas binding to heme-bound REV-ERB forms a planar ring, as does the non-gas bound form of CoPP, this would support a model by which the porphyrin ring conformation is involved in regulating REV-ERB activity. It is also possible that in the absence of gas the planar conformation of CoPP, compared with the puckered conformation of heme, may also play a role in negatively regulating the activity of REV-ERB.
Gas binding to a heme sensor transcription factor provides a means to coordinate gas levels to gene expression. For example, the transcriptional activity of the bacterial transcription factor CooA is affected both by redox conditions and CO levels. When bacteria are deprived of ligand and oxygen, CO levels increase, allowing CO to bind and activate heme-CooA, resulting in an increase in the expression of enzymes that oxidize CO (active on CooA) to CO2 (inactive on CooA) thus forming a negative feedback regulatory loop (48). Another example of how gas binding can affect transcription is in the bacterial two-component signal transduction system involving the heme sensor kinase FixL and the response regulator transcription factor FixJ. FixL is a sensor that binds O2 reversibly to affect the activation of FixJ via phosphorylation to regulate the expression of genes with roles in nitrogen fixation and respiration (49). Although FixL can bind a variety of diatomic gases, including CO and NO, FixL uses a specific structural mechanism involving an Arg side chain within the heme-binding pocket to discriminate among these ligands to functionally respond to O2 only (50). However, in contrast to gas binding activating CooA, O2 binding to FixL inhibits its activity. In both of these cases for CooA and FixL, though, it is not heme binding itself that regulates these functions, but rather gas binding events and redox conditions that control output activity.
Intriguingly, both phase and period of the circadian rhythm are controlled by NO (52), and due to the transcriptional roles REV-ERBs in regulating circadian rhythm (15) this further suggests a functional role for small molecule gas binding to REV-ERB and has been previously proposed (11). Our studies using CoPP and ZnPP, which likely respond differently to, or do not bind some, small molecule gases (44, 51) suggest that modifying the ability of the porphyrin-REV-ERB complex to bind small molecule gases may change REV-ERB function. The next structural mechanism to be tested should determine differences in how the bound metal-porphyrin responds to small molecule gases may play a role in the regulation of REV-ERB activity. Obtaining crystal structures of REV-ERB under different redox conditions and in the presence of small molecule gases is more technically challenging (11, 37) but something that we are pursuing. These future studies could provide insight into a potential structural mechanism by which redox conditions and small molecule gases play a role in the differing activity of heme and CoPP/ZnPP on REV-ERB function.
Acknowledgments
We thank Dr. Jerome Nwachukwu for helpful discussions and insights using the Phenix software and Dr. HaJeung Park at The Scripps Research Institute Florida for assistance with the Micromax007-HF x-ray generator and data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants DK080201 (to T. P. B.) and DK101871 (to D. J. K.). This work was also supported by the James and Esther King Biomedical Research Program from the Florida Department of Health Grant 1KN-09 (to D. J. K.) and start-up funds provided by The Scripps Research Institute (to D. J. K.).
The atomic coordinates and structure factors (code 4N73) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- NR
- nuclear receptor
- CoPP
- cobalt protoporphyrin IX
- DMSO
- dimethyl sulfoxide
- HMOX1
- heme oxygenase
- ITC
- isothermal titration calorimetry
- LBD
- ligand-binding domain
- Luc
- luciferase
- NCoR
- nuclear receptor corepressor
- PAI-1
- plasminogen activator inhibitor-1
- PDB
- Protein Data Bank
- Tm
- melting temperature
- ZnPP
- zinc protoporphyrin IX
- TCEP
- Tris(2-carboxyethyl)phosphine.
REFERENCES
- 1. Guillaumond F., Dardente H., Giguère V., Cermakian N. (2005) Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403 [DOI] [PubMed] [Google Scholar]
- 2. Burris T. P. (2008) Nuclear hormone receptors for heme: REV-ERBα and REV-ERBβ are ligand-regulated components of the mammalian clock. Mol. Endocrinol. 22, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Solt L. A., Kojetin D. J., Burris T. P. (2011) The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med. Chem. 3, 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yin L., Wu N., Lazar M. A. (2010) Nuclear receptor Rev-erbα: a heme receptor that coordinates circadian rhythm and metabolism. Nucl. Recept. Signal. 8, e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woldt E., Sebti Y., Solt L. A., Duhem C., Lancel S., Eeckhoute J., Hesselink M. K., Paquet C., Delhaye S., Shin Y., Kamenecka T. M., Schaart G., Lefebvre P., Nevière R., Burris T. P., Schrauwen P., Staels B., Duez H. (2013) Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lazar M. A., Hodin R. A., Darling D. S., Chin W. W. (1989) A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbAα transcriptional unit. Mol. Cell. Biol. 9, 1128–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dumas B., Harding H. P., Choi H. S., Lehmann K. A., Chung M., Lazar M. A., Moore D. D. (1994) A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol. Endocrinol. 8, 996–1005 [DOI] [PubMed] [Google Scholar]
- 8. Savkur R. S., Bramlett K. S., Clawson D., Burris T. P. (2004) Pharmacology of nuclear receptor-coregulator recognition. Vitamins Hormones 68, 145–183 [DOI] [PubMed] [Google Scholar]
- 9. Savkur R. S., Burris T. P. (2004) The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63, 207–212 [DOI] [PubMed] [Google Scholar]
- 10. Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. (2007) Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol. 14, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pardee K. I., Xu X., Reinking J., Schuetz A., Dong A., Liu S., Zhang R., Tiefenbach J., Lajoie G., Plotnikov A. N., Botchkarev A., Krause H. M., Edwards A. (2009) The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. PLoS Biol. 7, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meng Q. J., McMaster A., Beesley S., Lu W. Q., Gibbs J., Parks D., Collins J., Farrow S., Donn R., Ray D., Loudon A. (2008) Ligand modulation of REV-ERBα function resets the peripheral circadian clock in a phasic manner. J. Cell Sci. 121, 3629–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar N., Solt L. A., Wang Y., Rogers P. M., Bhattacharyya G., Kamenecka T. M., Stayrook K. R., Crumbley C., Floyd Z. E., Gimble J. M., Griffin P. R., Burris T. P. (2010) Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology 151, 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kojetin D., Wang Y., Kamenecka T. M., Burris T. P. (2011) Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem. Biol. 6, 131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Solt L. A., Wang Y., Banerjee S., Hughes T., Kojetin D. J., Lundasen T., Shin Y., Liu J., Cameron M. D., Noel R., Yoo S. H., Takahashi J. S., Butler A. A., Kamenecka T. M., Burris T. P. (2012) Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Csongradi E., Docarmo J. M., Dubinion J. H., Vera T., Stec D. E. (2012) Chronic HO-1 induction with cobalt protoporphyrin (CoPP) treatment increases oxygen consumption, activity, heat production and lowers body weight in obese melanocortin-4 receptor-deficient mice. Int. J. Obesity 36, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 19. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Afonine P. V., Mustyakimov M., Grosse-Kunstleve R. W., Moriarty N. W., Langan P., Adams P. D. (2010) Joint x-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 24. Gupta N., Ragsdale S. W. (2011) Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erbβ. J. Biol. Chem. 286, 4392–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 26. Wang J., Yin L., Lazar M. A. (2006) The orphan nuclear receptor Rev-erbα regulates circadian expression of plasminogen activator inhibitor type 1. J. Biol. Chem. 281, 33842–33848 [DOI] [PubMed] [Google Scholar]
- 27. Smith A., Alam J., Escriba P. V., Morgan W. T. (1993) Regulation of heme oxygenase and metallothionein gene expression by the heme analogs, cobalt-, and tin-protoporphyrin. J. Biol. Chem. 268, 7365–7371 [PubMed] [Google Scholar]
- 28. Maines M. D. (1981) Zinc protoporphyrin is a selective inhibitor of heme oxygenase activity in the neonatal rat. Biochim. Biophys. Acta 673, 339–350 [DOI] [PubMed] [Google Scholar]
- 29. Phelan C. A., Gampe R. T., Jr., Lambert M. H., Parks D. J., Montana V., Bynum J., Broderick T. M., Hu X., Williams S. P., Nolte R. T., Lazar M. A. (2010) Structure of Rev-erbα bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat. Struct. Mol. Biol. 17, 808–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brucker E. A., Olson J. S., Phillips G. N., Jr., Dou Y., Ikeda-Saito M. (1996) High resolution crystal structures of the deoxy, oxy, and aquomet forms of cobalt myoglobin. J. Biol. Chem. 271, 25419–25422 [DOI] [PubMed] [Google Scholar]
- 31. Degtyarenko I., Nieminen R. M., Rovira C. (2006) Structure and dynamics of dioxygen bound to cobalt and iron heme. Biophys. J. 91, 2024–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffman B. M., Petering D. H. (1970) Coboglobins: oxygen-carrying cobalt-reconstituted hemoglobin and myoglobin. Proc. Natl. Acad. Sci. U.S.A. 67, 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renaud J. P., Harris J. M., Downes M., Burke L. J., Muscat G. E. (2000) Structure-function analysis of the Rev-erbA and RVR ligand-binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Mol. Endocrinol. 14, 700–717 [DOI] [PubMed] [Google Scholar]
- 34. Hughes T. S., Chalmers M. J., Novick S., Kuruvilla D. S., Chang M. R., Kamenecka T. M., Rance M., Johnson B. A., Burris T. P., Griffin P. R., Kojetin D. J. (2012) Ligand and receptor dynamics contribute to the mechanism of graded PPARγ agonism. Structure 20, 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu J., Cistola D. P., Li E. (2006) Analysis of ligand binding and protein dynamics of human retinoid X receptor α ligand-binding domain by nuclear magnetic resonance. Biochemistry 45, 1629–1639 [DOI] [PubMed] [Google Scholar]
- 36. Galbraith R. A., Kappas A. (1990) Cobalt-protoporphyrin suppresses expression of genetic obesity in homozygous fa/fa Zucker rats. Pharmacology 41, 292–298 [DOI] [PubMed] [Google Scholar]
- 37. Marvin K. A., Reinking J. L., Lee A. J., Pardee K., Krause H. M., Burstyn J. N. (2009) Nuclear receptors homo sapiens Rev-erbβ and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO. Biochemistry 48, 7056–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh S., Madzelan P., Stasser J., Weeks C. L., Becker D., Spiro T. G., Penner-Hahn J., Banerjee R. (2009) Modulation of the heme electronic structure and cystathionine β-synthase activity by second coordination sphere ligands: the role of heme ligand switching in redox regulation. J. Inorg. Biochem. 103, 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weeks C. L., Singh S., Madzelan P., Banerjee R., Spiro T. G. (2009) Heme regulation of human cystathionine β-synthase activity: insights from fluorescence and Raman spectroscopy. J. Am. Chem. Soc. 131, 12809–12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsai A. L., Berka V., Sharina I., Martin E. (2011) Dynamic ligand exchange in soluble guanylyl cyclase (sGC): implications for sGC regulation and desensitization. J. Biol. Chem. 286, 43182–43192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olea C., Boon E. M., Pellicena P., Kuriyan J., Marletta M. A. (2008) Probing the function of heme distortion in the H-NOX family. ACS Chem. Biol. 3, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shelver D., Kerby R. L., He Y., Roberts G. P. (1997) CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. U.S.A. 94, 11216–11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Puranik M., Nielsen S. B., Youn H., Hvitved A. N., Bourassa J. L., Case M. A., Tengroth C., Balakrishnan G., Thorsteinsson M. V., Groves J. T., McLendon G. L., Roberts G. P., Olson J. S., Spiro T. G. (2004) Dynamics of carbon monoxide binding to CooA. J. Biol. Chem. 279, 21096–21108 [DOI] [PubMed] [Google Scholar]
- 44. Blough N. V., Zemel H., Hoffman B. M., Lee T. C. K., Gibson Q. H. (1980) Kinetics of carbon monoxide binding to manganese, zinc, and cobalt hybrid hemoglobins. J. Am. Chem. Soc. 102, 5683–5685 [Google Scholar]
- 45. Zenke-Kawasaki Y., Dohi Y., Katoh Y., Ikura T., Ikura M., Asahara T., Tokunaga F., Iwai K., Igarashi K. (2007) Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 27, 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yonetani T., Yamamoto H., Erman J. E., Leigh J. S., Jr., Reed G. H. (1972) Electromagnetic properties of hemoproteins. V. Optical and electron paramagnetic resonance characteristics of nitric oxide derivatives of metalloporphyrin-apohemoprotein complexes. J. Biol. Chem. 247, 2447–2455 [PubMed] [Google Scholar]
- 47. Marden M. C., Kiger L., Poyart C., Rashid A. K., Kister J., Stetzkowski-Marden F., Caron G., Haque M., Moens L. (2000) Modulation of the oxygen affinity of cobalt-porphyrin by globin. FEBS Lett. 472, 221–224 [DOI] [PubMed] [Google Scholar]
- 48. Gilles-Gonzalez M. A., Gonzalez G. (2005) Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99, 1–22 [DOI] [PubMed] [Google Scholar]
- 49. Rodgers K. R., Lukat-Rodgers G. S. (2005) Insights into heme-based O2 sensing from structure-function relationships in the FixL proteins. J. Inorg. Biochem. 99, 963–977 [DOI] [PubMed] [Google Scholar]
- 50. Hao B., Isaza C., Arndt J., Soltis M., Chan M. K. (2002) Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 41, 12952–12958 [DOI] [PubMed] [Google Scholar]
- 51. Labbé R. F., Vreman H. J., Stevenson D. K. (1999) Zinc protoporphyrin: A metabolite with a mission. Clin. Chem. 45, 2060–2072 [PubMed] [Google Scholar]
- 52. Ding J. M., Chen D., Weber E. T., Faiman L. E., Rea M. A., Gillette M. U. (1994) Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science 266, 1713–1717 [DOI] [PubMed] [Google Scholar]