Background: The cell type-specific induction of Bmp2 expression by Wnt3a indicates that Bmp2 is controlled by epigenetic mechanisms.
Results: Epigenetic modification activates Bmp2 and Alp expression by Wnt3a in nonosteogenic cells.
Conclusion: Epigenetic induction of Bmp2 production by Wnt3a reveals a new mechanistic dimension in morphogen-mediated control of osteogenesis.
Significance: Epigenetic modifications/canonical Wnt3a signaling may provide a new method for trans-differentiation of nonosteogenic cells to osteoblasts.
Keywords: Bone Morphogenetic Protein (BMP), Cell Differentiation, Epigenetics, Osteoblast, Wnt Signaling
Abstract
Mesenchymal cells alter and retain their phenotype during skeletal development through activation or suppression of signaling pathways. For example, we have shown that Wnt3a only stimulates osteoblast differentiation in cells with intrinsic osteogenic potential (e.g. MC3T3-E1 pre-osteoblasts) and not in fat cell precursors or fibroblasts (3T3-L1 pre-adipocytes or NIH3T3 fibroblasts, respectively). Wnt3a promotes osteogenesis in part by stimulating autocrine production of the osteoinductive ligand Bmp2. Here, we show that the promoter regions of the genes for Bmp2 and the osteoblast marker Alp are epigenetically locked to prevent their expression in nonosteogenic cells. Both genes have conserved CpG islands that exhibit increased CpG methylation, as well as decreased acetylation and increased methylation of histone H3 lysine 9 (H3-K9) specifically in nonosteogenic cells. Treatment of pre-adipocytes or fibroblasts with the CpG-demethylating agent 5′-aza-2′-deoxycytidine or the histone deacetylase inhibitor trichostatin-A renders Bmp2 and Alp responsive to Wnt3a. Hence, drug-induced epigenetic activation of Bmp2 gene expression contributes to Wnt3a-mediated direct trans-differentiation of pre-adipocytes or fibroblasts into osteoblasts. We propose that direct conversion of nonosteogenic cells into osteoblastic cell types without inducing pluripotency may improve prospects for novel epigenetic therapies to treat skeletal afflictions.
Introduction
Skeletal development and osteoblast differentiation are controlled by ligands of the wingless/int-1 class (WNTs)3 and bone morphogenetic proteins (BMPs), each of which activates essential signaling pathways (1–4). WNTs are a family of secreted glycoproteins that control cell proliferation, differentiation, and migration (5) by both canonical and noncanonical pathways. The canonical pathway controls bone formation (6, 7) and is mediated by WNT ligand binding to two distinct receptor molecules, frizzled receptor and low density lipoprotein receptor-related proteins 5 or 6 (LRP 5/6). WNTs transduce signals through Dishevelled protein. The latter protein inhibits the intracellular activity of glycogen synthase kinase 3β and prevents glycogen synthase kinase 3β-mediated phosphorylation of β-catenin. The resulting stabilization of β-catenin promotes its translocation to the nucleus where it acts as a co-activator of T cell factor/lymphoid enhancer factor to induce WNT target genes (4, 8). BMPs also play crucial roles in osteoblast differentiation (2). BMP signaling is initiated by ligand binding to BMPR-II/BMPR-I receptors that represent serine/threonine kinases. BMPR-I phosphorylates receptor-regulated proteins Smad1 or -5 (R-Smads) to stimulate mRNA expression of two critical transcription factors, Runx2 and Osterix, that are genetically required for bone formation (9, 10). Notwithstanding this detailed knowledge of how Wnt3a and BMP2 signals are transduced to the nucleus, the epigenetic barriers that prevent promiscuous expression of osteogenic genes outside the osteoblast lineage remain to be explored.
Recently, we showed that Wnt3a stimulates Bmp2 expression to generate a positive autocrine loop in MC3T3-E1 pre-osteoblasts (11). This study also showed that matrix extracellular phosphoglycoprotein (MEPE), which is a specific marker gene of mineralizing osteoblasts and osteocytes (12), is regulated directly by canonical WNT signaling and indirectly by BMP2 (11). Similarly, the classical osteoblast-related marker alkaline phosphatase (Alp), which is stimulated by BMP2 (13) or Wnt3a (14), is regulated directly by BMP2 or Wnt3a and indirectly by Wnt3a through a BMP2-mediated autocrine loop (11). However, these feed-forward mechanisms are limited to osteogenic cells that are primed to differentiate into mature osteoblasts in response to exogenous BMP2 (e.g. MC3T3-E1 pre-osteoblast, C3H10T1/2 mesenchymal progenitor cells, ST2 bone marrow stromal cells, and C2C12 pre-myoblasts). Nonosteogenic cells that have entered alternative mesenchymal cell lineages (e.g. 3T3-L1 pre-adipocytes and NIH3T3 fibroblasts) do not show Bmp2 expression in response to Wnt3a. These qualitative differences in the effects of Wnt3a signaling prompted us to consider the idea that an epigenetic mechanism may block Wnt3a-responsive targets.
Mature mesenchymal cells do not typically change their phenotype, but there are many examples in which a (pre-)committed somatic cell type can transform into another (pre-)committed somatic cell type through trans-differentiation (15). For example, morphogenetic ligands like BMP2 are capable of trans-differentiating pre-myoblasts into osteoblastic cells. Such direct trans-differentiation of pre-committed somatic cells offers a more efficient route for cell type conversion than other reprogramming strategies. The latter strategies require the initial regression of cells into a more immature state (de-differentiation) and the subsequent induction of an alternative cell lineage. Natural trans-differentiation during newt lens regeneration provides insights into strategies for artificial trans-differentiation by direct programming (16). A number of mammalian programming strategies have recently emerged. For example, C/EBPα and -β efficiently convert differentiated B cells into macrophages (17); PDX-1 drives trans-differentiation of adult hepatocytes into pancreatic cells (18), and fibroblasts convert into cardiomyocytes using cardiac-related transcription factors and epigenetic remodeling proteins (19–21). Trans-differentiation among mesenchymal cell types may be achieved by direct programming using approaches involving overexpression of transcription factors, as well as epigenetic strategies in which chromatin structure is altered through changes in DNA methylation and/or histone modifications.
Epigenetic mechanisms that support transcriptional control of gene expression account for the phenotypic diversity that emerges as a result of cellular differentiation during fetal development. 5′-Cytosine methylation of 5′-CG-3′ dinucleotides (“CpG doublets”) is a prominent epigenetic modification of genomic DNA that occurs at clusters of CpG doublets (“CpG islands”) in transcriptional regulatory regions (22). DNA methyltransferases maintain the methylation status of CpG islands after DNA replication in a cell- or tissue-specific manner (23). Generally, DNA hypermethylation of CpG islands in promoter regions is associated with chromatin condensation through repressive histone modifications that support gene silencing (24). Both the state of DNA methylation and histone modifications play essential roles in gene expression (25, 26). In this study, we demonstrate that Wnt3a responsiveness of nonosteogenic cells is normally inhibited by CpG methylation in the Bmp2 and Alp promoters thus precluding induction of Bmp2 and Alp expression. However, 5′-aza-2′-deoxycytidine (5′-aza-dC) treatment, which indirectly demethylates these promoters, renders the Bmp2 and Alp genes responsive to Wnt3a. These findings indicate that trans-differentiation of nonosteogenic mesenchymal cells into the osteoblast can be provoked by interfering with epigenetic inhibitory mechanisms.
EXPERIMENTAL PROCEDURES
Cell Culture
Mouse MC3T3-E1 pre-osteoblasts were cultured in α-minimal essential medium as described previously (12). Multipotent ST2 mesenchymal progenitor cells were maintained in RPMI 1640 medium, whereas C3H10T1/2 mesenchymal progenitor cells, mouse C2C12 pre-myoblasts, 3T3-L1 pre-adipocytes, and NIH3T3 fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM, Logan, UT). All media contain 10% fetal bovine serum or bovine calf serum with 1% penicillin/streptomycin. All cell lines were purchased from ATCC (Manassas, VA). Osteogenic medium includes 5 mm β-glycerophosphate and 50 μg/ml ascorbic acid.
Materials
Bioactive recombinant mouse Wnt3a and human BMP2 proteins were purchased from R&D Systems (Minneapolis, MN). Both 5′-aza-dC and trichostatin-A (TSA) were purchased from Sigma.
Reverse Transcription-PCR and Quantitative Real Time PCR
RNA was isolated using QIAzol lysis reagent (Qiagen, Valencia, CA). The PrimescriptTM RT-reagent kit for reverse transcription was purchased from TAKARA (Takara Bio, Japan). Quantitative real time PCR for murine Bmp2 and Alp were performed with primers we used previously (11). Quantitative real time PCR was performed using Takara SYBR premix Ex Taq (Takara Bio, JAPAN) on an Applied Biosystems 7500 Real Time PCR system (Foster city, CA). PCR primers were synthesized by Integrated DNA technology (IDT; Coralville, IA). All samples were run in duplicate, and the relative mRNA expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA. The primer sets for real time PCR were published previously (11).
In Silico Analysis of CpG Islands
The following programs were used to search for CpG islands: EMBOSS CpGPlot program, UCSC Genome Browser public database, and MethPrimer (27). These programs identified putative CpG islands that spans from −1500 to + 500 bp in the Bmp2 promoter (NCBI gene accession number 12156) and from −1000 to + 1000 bp in the Alp promoter (NCBI gene accession number 11647). CpG island prediction was performed using the following parameters: 100-bp window, observed/expected CpG ratio >0.6 and percentage of G plus C >50% (22).
Methylation-specific PCR
Genomic DNA was extracted from cells using LaboPass tissue miniprep kit (Cosmo Genetech, Korea). EpiTect Fast DNA bisulfite kit (Qiagen, Valencia, CA) was used for the bisulfite conversion. PCR was performed using LugenTM Sensi 5× PCR premix (Lugen Science Co., Korea) with primers listed in Table 1. The MethPrimer program was used for making methylation-specific PCR primer sets (27).
TABLE 1.
Primer lists
M and U represent amplification of methylated and unmethylated alleles, respectively.
| Name | Oligonucleotide sequence |
|---|---|
| Methylation PCR | |
| Bmp2, M (forward) | 5′-TATTCGGTTTGGTAATTCGAGAC-3′ |
| Bmp2, M (reverse) | 5′-TACATCCCTCAACCTAACTCTAACG-3′ |
| Bmp2, U (forward) | 5′-GGTTATTTGGTTTGGTAATTTGAGAT-3′ |
| Bmp2, U (reverse) | 5′-ACATCCCTCAACCTAACTCTAACAC-3′ |
| Alp, M (forward) | 5′-GAGGTTTAGAGGTGTATATGGTGAC-3′ |
| Alp, M (reverse) | 5′-TCTATAAAAAACTTTATCCCTCGAT-3′ |
| Alp, U (forward) | 5′-GGTTTAGAGGTGTATATGGTGATGG-3′ |
| Alp, U (reverse) | 5′-TCTATAAAAAACTTTATCCCTCAAT-3′ |
| Chromatin immunoprecipitation assay | |
| Bmp2, ChIP (forward) | 5′-CCGACGACAGCAGCAGCCTT-3′ |
| Bmp2, ChIP (reverse) | 5′-AAGACTGGATCCGCCGGGCG-3′ |
| Alp, ChIP (forward) | 5′-ACATGGTGACGGACAAGGAT-3′ |
| Alp, ChIP (reverse) | 5′-GCAGCTGGCTTGTCTTTAGG-3′ |
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP assays were performed as described previously (11). Protein G magnetic beads and single strand DNA were purchased from Upstate Biotechnology (Charlottesville, VA) and Sigma, respectively. Antibodies recognizing acetyl histone H3 (Lys-9), methyl histone H3 (Lys-9) or immunoglobulin G (IgG, control antibody) were purchased from Millipore (Temecula, CA). Anti-MeCP2 antibody was purchased from Abcam (Cambridge, MA), and anti-Lef-1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). 3T3-L1 or NIH3T3 cells were plated in 100-mm dishes and cultured up to 70% confluency; 5′-aza-dC (10 μm) and TSA (100 nm) were each administered for 24 h, followed by treatment with Wnt3a (50 ng/ml) 24 h later. PCR primer pairs used to detect DNA segments for ChIP assays are listed in Table 1.
Alkaline Phosphatase (ALP) Staining
A standardized kit for ALP staining was purchased from Sigma. Cells were washed twice with phosphate-buffered saline and stained for alkaline phosphatase according to the manufacturer's instructions.
DNA Construction
We used a Bmp2 promoter luciferase reporter vector spanning nucleotides from −1200 to 200 bp, including predicted CpG islands and previously validated Lef-1-binding elements (11). The Alp promoter region (−1000 to + 1000 bp) was generated by PCR amplification and cloned into to pGL3-basic vector (Promega, Madison, WI) using the In-Fusion HD cloning kit (Clontech) with primers forward 5′-TCT TAC GCG TGC TAG CCT TCA GGG TAG AAG TGA TCA-3′ and reverse 5′-CCG GAA TGC CAA GCT TAG CCA AAC GTT CTT TCA GGC-3′ using the in-fusion primer design tool according to the manufacturer's instruction. Expression of Lef-1 and Dlx5 proteins from their respective vectors were each confirmed by Western blot previously (11, 13). All plasmid DNAs were prepared using a DNA Maxi-prep kit (Genomed, Loehne, Germany).
Transient Transfection
C2C12 cells were plated in 100-mm plates and cultured up to 90–100% confluency. Cells were transfected by electroporation using a NeonTM transfection system (Invitrogen) with a 10-μl gold tip according to the manufacturer's instructions. Transfections were performed with 0.5 μg of Lef-1 or Dlx5 expression vector or pcDNA3.1 empty vector as a control and 0.15 μg of the Bmp2 or Alp promoter luciferase reporter vector.
In Vitro Methylation
Methylated reporter vectors were obtained by incubation with CpG methyltransferase M.SssI (New England Biolabs, Beverly, MA) in the presence of S-adenosylmethionine at 37 °C for 8 h according to the manufacturer's instruction. Completion of the methylation was confirmed by resistance to HpyCH4IV enzyme digestion (New England Biolabs).
Luciferase Reporter Assay
Cells were lysed with passive lysis buffer, and luciferase activity was detected using a Bright-GloTM luciferase assay system with a GloMax-Multi Detection System machine (Promega, Madison, WI).
Statistical Analysis
Quantitative data are presented as the mean ± S.D. Each experiment was performed at least three times, and one representative experiment result is shown. Significant differences were analyzed using Student's t test. A value of p < 0.05 was considered statistically significant.
RESULTS
Wnt3a Stimulates Bmp2 and Alp Expression in Osteogenic Cells
Our group has previously shown that Wnt3a stimulates Bmp2 expression thus creating a Bmp2 autocrine loop in mouse MC3T3-E1 pre-osteoblastic cells (11). In this study, we examined Bmp2 expression upon Wnt3a administration in a number of mesenchymal cell lines that differ in their ability to differentiate into osteoblasts using the potent osteogenic ligands BMP2 or Wnt3a as evidenced by staining for the classical osteoblast marker alkaline phosphatase (Alp) (Fig. 1A). For brevity, we classified the various cell types in our study as either “osteogenic” (MC3T3-E1, C2C12, C3H10T1/2, and ST2 cells) or “nonosteogenic” (3T3-L1 and NIH3T3 cells) based on their intrinsic ability to differentiate to osteoblasts in the presence of either BMP2 and/or Wnt3a.
FIGURE 1.
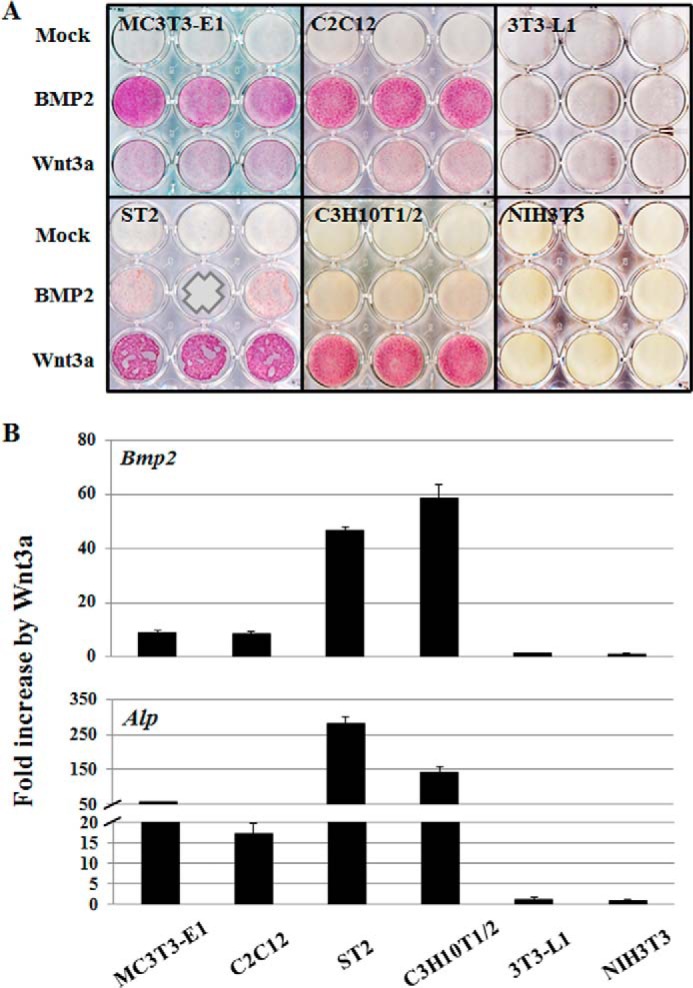
Wnt3a stimulates Bmp2 and Alp expression in the osteogenic cells. A, Alp staining. MC3T3-E1, C2C12, ST2, C3H10T1/2, 3T3-L1, and NIH3T3 cells were treated with BMP2 (50 ng/ml) or Wnt3a (50 ng/ml) and cultured for 3 days. Alp activity was determined by cytochemical staining. B, cells were treated with Wnt3a (50 ng/ml) for 24 h. Data are presented as fold increase by Wnt3a. Expression of Bmp2 and Alp mRNAs was determined by quantitative real time PCR and normalized to the Gapdh expression level. The expression levels of NIH3T3 cell were used as a comparison standard, which showed the lowest expression levels of Bmp2 and Alp. Data were confirmed by triplicate determinations from each mRNA and at least three biological replicates. Values are the means ± S.D. of a representative triplicate determination. A value of p < 0.05 was considered statistically significant.
The osteogenic cell types (i.e. MC3T3-E1, C2C12, C3H10T1/2, and ST2 cells), but not 3T3-L1 pre-adipocytes and NIH3T3 fibroblasts, exhibit robust Alp staining upon BMP2 administration. However, MC3T3-E1 pre-osteoblasts and C2C12 pre-myoblasts are more effectively induced by BMP2, whereas C3H10T1/2 mesenchymal progenitor cells and ST2 bone marrow stromal cells are more efficiently stimulated by Wnt3a (Fig. 1A). It seems that less differentiated cells (C3H10T1/2 and ST2) are more reactive to Wnt3a; however, differentiated cells (MC3T3-E1 and C2C12) are more to the BMP2 in the osteogenic cells. Strikingly, we observed that Bmp2 expression is induced by Wnt3a only in MC3T3-E1, C2C12, C3H10T1/2, and ST2 cells but not 3T3-L1 and NIH3T3 cells (Fig. 1B) (28). Interestingly, the four Wnt3a-responsive cell types that up-regulate Bmp2 expression also have in common the potential to differentiate into the osteoblasts in the presence of Wnt3a based on Alp staining (Fig. 1A). Notably, the Alp gene itself is also a downstream target of the Bmp2 signaling pathway (13), and Alp mRNA expression is indeed induced by Wnt3a in the same four cell types that display Wnt3a induction of Bmp2 expression (Fig. 1B). These findings indicate that the inability to initiate osteogenic differentiation in 3T3-L1 and NIH3T3 cells in response to Wnt3a may be directly linked to the inability of these two cell types to produce endogenous Bmp2.
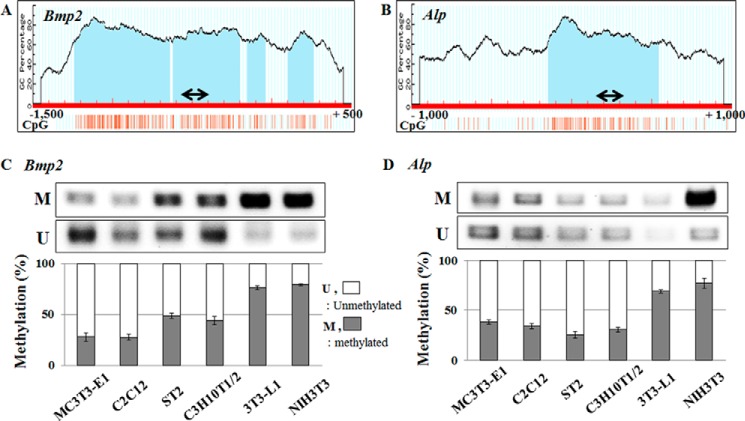
Differential CpG Methylation between Osteo-inducible and Noninducible Cell Lines
Because of qualitative differences between the mouse cell lines in their ability to react to Wnt3a and BMP2, we postulated that the cell type-specific enhancement of Bmp2 and Alp expression by these two ligands is epigenetically regulated. In silico analysis using three different bioinformatics programs (CpG plot tool, EMBOSS CpGPlot, and MethPrimer) (27) revealed CpG islands in the Bmp2 and Alp promoter regions within the initial 1.5-kb region upstream of the transcription start site (Fig. 2, A and B) (22). Methylation-specific PCR showed that CpG islands of Bmp2 and Alp promoter regions of osteo-inducible cells are highly under-methylated relative to noninducible cells (Fig. 2, C and D). Based on this differential methylation of CpG islands in the Bmp2 and Alp promoter regions, we reasoned that the cell type specificity of their induction is due to epigenetic regulation at the level of CpG methylation.
FIGURE 2.
Bmp2 and Alp promoter regions are highly methylated in the nonosteogenic cell lines. A and B, diagram shows CpG islands in the Bmp2 (A, −1500 to + 500 bp) and Alp (B, −1000 to + 1000 bp) promoter regions. The detection region of methylation PCR is indicated using an arrow in the diagrams and sequences are shown in Table 1. C and D, MSP analysis of Bmp2 (C) and Alp (D) promoter regions shows amplification of methylated (M) and unmethylated (U) alleles, respectively. The bottom panel illustrates band density quantification of MSP. The percentage of unmethylated (open bars) and methylated DNA (closed bars) are indicated. Primer sets used for MSP are listed in Table 1. Values are the means ± S.D. of a representative triplicate determination.
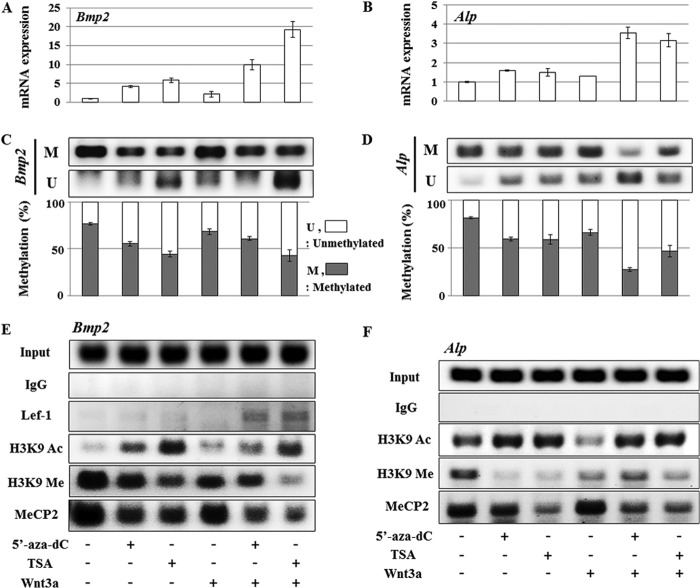
Epigenetic Modifications Enable Stimulation of Bmp2 or Alp Expression in the 3T3-L1 Pre-adipocytes
To modify the DNA methylation state, we administered the nonmethylatable nucleotide analog 5′-aza-dC, which prevents propagation of CpG methylation by DNA methyltransferase activity upon incorporation into DNA. Furthermore, we used the histone deacetylase inhibitor TSA to promote acetylation of histones H3 and H4 to generate a hyper-acetylated chromatin state that promotes gene expression (29). Single treatment with either 5′-aza-dC (10 μm) or TSA (100 nm) increases Bmp2 or Alp mRNA expression to a limited degree in 3T3-L1 cells (Fig. 3, A and B). However, administration of Wnt3a after treatment with 5′-aza-dC or TSA dramatically stimulates Bmp2 and Alp expression levels within 24 h in cells that are normally not competent to support induction of expression of these genes by Wnt3a (Fig. 3, A and B). We also examined the effects of LiCl and SB213763, which are glycogen synthase kinase 3β inhibitors that mimic activation of Wnt3a signaling by stabilizing β-catenin. These inhibitors also stimulate Bmp2 and Alp gene expression, but at levels lower than those observed after Wnt3a stimulation (data not shown). Sequential treatments with Wnt3a and either 5′-aza-dC or TSA are more effective in inducing expression of Bmp2 mRNA than Alp mRNA based on real time RT-quantitative PCR data (Fig. 3, A and B). We note that in 3T3-L1 cells, the combination treatment with 5′-aza-dC and Wnt3a is slightly more effective in increasing the expression level of Alp mRNA (Fig. 3B).
FIGURE 3.
Epigenetic regulation enables stimulation of Bmp2 and Alp expression in 3T3-L1 pre-adipocytes. 3T3-L1 cells were treated with 5′-aza-dC (10 μm) or TSA (100 nm) for 24 h and were then treated with Wnt3a (50 ng/ml) for an additional 24 h. Bmp2 (A) and Alp (B) mRNA expression was determined by quantitative real time PCR and normalized to the Gapdh expression level. Data were confirmed by triplicate determinations from each mRNA and by at least triplicate cultures. Values are the means ± S.D. of a representative triplicate determination. A value of p < 0.05 was considered statistically significant. All values are for fold induction relative to the respective control value. C and D, MSP analysis of Bmp2 (C) and Alp (D) genes are shown. M and U represent amplification of methylated and unmethylated alleles, respectively. The bottom panel illustrates band density quantification of MSP show as, respectively, percentage of unmethylated alleles (open bars) or methylated alleles (closed bars). Values are the means ± S.D. of a representative triplicate determination. Primer sets for MSP are listed in the Table 1. E and F, chromatin immunoprecipitation (ChIP) assays were performed using antibodies for acetyl-histone H3 (Lys-9), trimethyl histone H3 (Lys-9), MeCP2 and Lef-1 (E). The chromatin fragments were PCR-amplified with primers for Bmp2 (E) and Alp (F) as indicated in Table 1.
Methylation States of CpG Islands in the Bmp2 and Alp Promoters Determine Wnt3a Mediated Induction of Gene Expression
Our data show that epigenetic modifications enable stimulation of gene expression by Wnt3a in cell types that otherwise repress osteogenic genes. Therefore, we investigated DNA methylation in CpG islands within the Bmp2 and Alp promoters. Methylation-specific PCR (MSP) was applied to interrogate methylation and demethylation patterns in selected regulatory regions of the Bmp2 and Alp promoters. Bisulfite-sensitive primers flanking the CpG islands were selected using the “MethPrimer” program (27). We find that 5′-aza-dC or TSA treatment results in hypomethylation of 5′-cytosine in CpG islands reflected by decreased PCR amplification of methylated DNA fragments in the 3T3-L1 cells (Fig. 3, C and D). Furthermore, detection of unmethylated DNA correlates with Bmp2 and Alp gene expression levels (Fig. 3, A--D). The percentage of methylation at promoter fragments tightly correlates with gene expression levels (bottom panels, Fig. 3, C and D). Data obtained using chromatin immunoprecipitation (ChIP) assays demonstrate that DNA methylation status reflects genomic interactions of the methyl CpG-binding protein 2 (MeCP2) to the Bmp2 and Alp promoters (Fig. 3, E and F). Taken together, the MSP and ChIP assay results indicate that the DNA methylation states of the Bmp2 and Alp gene promoters are regulated by epigenetic modification and correlate inversely with gene expression levels.
Epigenetic Protein Modifications on Histone H3 Parallel Changes in DNA Methylation and Gene Expression
Changes in CpG methylation are functionally interrelated with histone modifications and control of promoter accessibility in chromatins that mediate silencing or activation of gene transcription (30). ChIP assay data indicate that inhibition of histone deacetylases using TSA promotes histone H3 acetylation of lysine 9 in H3 (H3-K9), a transcriptionally active mark, and simultaneously down-regulates its methylation, a transcriptionally repressive mark (Fig. 3, E and F). Increased acetylation of histone H3-K9 coincides with Bmp2 and Alp expression levels, whereas H3-K9 methylation patterns are inversely correlated in 3T3-L1 cells (Fig. 3, A, B, E, and F). Furthermore, ChIP assays for Lef-1 using the same genomic segments with increased H3-K9 acetylation (Fig. 3E) indicate that Lef-1 binding is definitely enhanced by serial treatment with 5′-aza-dC and Wnt3a, as well as with TSA-Wnt3a. Hence, the levels of CpG methylation, as well as post-translational modifications of histones that are linked to transcriptional repression or induction, together mediate epigenetic silencing or activation of the Bmp2 and Alp genes in mesenchymal cells.
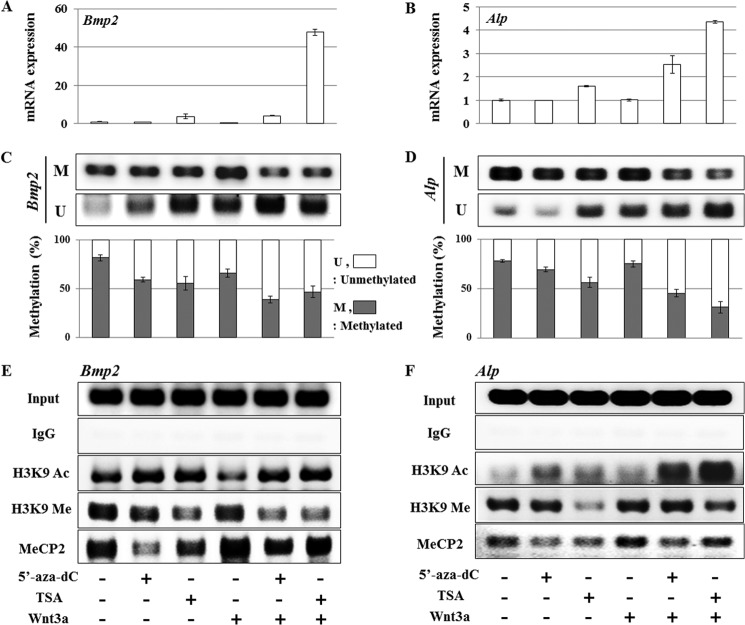
Validation That Epigenetic Modifications Change Gene Expression in Nonosteogenic Fibroblasts
For comparison with osteoinducible cell lines, we also checked noninducible NIH3T3 fibroblasts for changes in response to epigenetic drugs. Single treatment with either 5′-aza-dC (10 μm) or TSA (100 nm) increases Bmp2 or Alp expression to a limited degree, yet administration of Wnt3a after treatment with 5′-aza-dC or TSA dramatically stimulates Bmp2 and Alp expression levels within 24 h (Fig. 4, A and B). Similar to observations for 3T3-L1 cells, the effectiveness of Wnt3a treatments with either 5′-aza-dC or TSA in NIH3T3 cells is more dramatic for Bmp2 than Alp based on real time PCR data. Furthermore, Wnt3a synergizes more effectively with TSA than 5′-aza-dC in enhancing bone-related gene expression levels (Fig. 4, A and B). MSP (Fig. 4, C and D) and ChIP assays (Fig. 4, E and F) were performed using the same treatment conditions with 3T3-L1 cells, and the results are qualitatively similar. Collectively, initial epigenetic modifications with either 5′-aza-dC or TSA are required for Wnt3a stimulation of osteogenic cell commitment and concomitant activation of osteoblast-related genes in nonosteogenic cells.
FIGURE 4.
Epigenetic regulation enables stimulation of Bmp2 and Alp expression in NIH3T3 fibroblasts. NIH3T3 cells were treated with 5′-aza-dC (10 μm) or TSA (100 nm) for 24 h, then treated with Wnt3a (50 ng/ml) for 24 h. Bmp2 (A) and Alp (B) mRNA expression was determined by quantitative real time PCR and normalized to the Gapdh expression level. Expression data were confirmed by triplicate determinations from each mRNA and by at least triplicate cultures. Values are the means ± S.D. of a representative triplicate determination. A value of p < 0.05 was considered statistically significant. All values are for relative fold induction from the respective control value. C and D, MSP analysis of Bmp2 (C) and Alp (D) genes are shown. M and U represent amplification of methylated and unmethylated alleles, respectively. The bottom panel illustrates band density quantification of MSP for the percentage of unmethylated (open bar) or methylated (closed bar) DNA. Values are the means ± S.D. of a representative triplicate determination. Primer sets for MSP are listed in Table 1. E and F, ChIP assays were performed by immunoprecipitation of promoter fragments with antibodies for acetyl histone H3 (Lys-9), trimethyl histone H3 (Lys-9), or MeCP2. The chromatin fragments were PCR-amplified using primers for Bmp2 (E) and Alp (F) as indicated in Table 1.
Effect of CpG Methylation on Bmp2 and Alp Expression
To examine the functional effects of CpG methylation on expression of the Bmp2 and Alp genes, we performed luciferase reporter assays with in vitro methylated reporter constructs using SssI methyltransferase (31, 32). Bmp2 and Alp promoter regions generated by PCR were cloned into a luciferase reporter vector and then methylated in vitro on CpG doublets using SssI methyltransferase. CpG methylation of the constructs was confirmed by digestion with the methylation-sensitive endonuclease HpyCH4IV. Unmethylated DNA promoter fragments are clearly digested by the HpyCH4IV restriction enzyme, but fragments methylated by SssI are not (Fig. 5A), indicating that the two luciferase reporter vectors containing either the Bmp2 and Alp promoters were completely methylated in vitro. These methylated reporters were then co-transfected with expression vectors for the Wnt-dependent transcription factor Lef-1 or the BMP2-inducible transcription factor Dlx5 (Fig. 5B). Reporter constructs transfected without expression vectors exhibit reduced basal luciferase activity level when methylated. In addition, methylation of the Bmp2 and Alp promoters in each case suppresses luciferase activity upon co-expression of transcription factors Lef-1 or Dlx5 (Fig. 5B).
FIGURE 5.
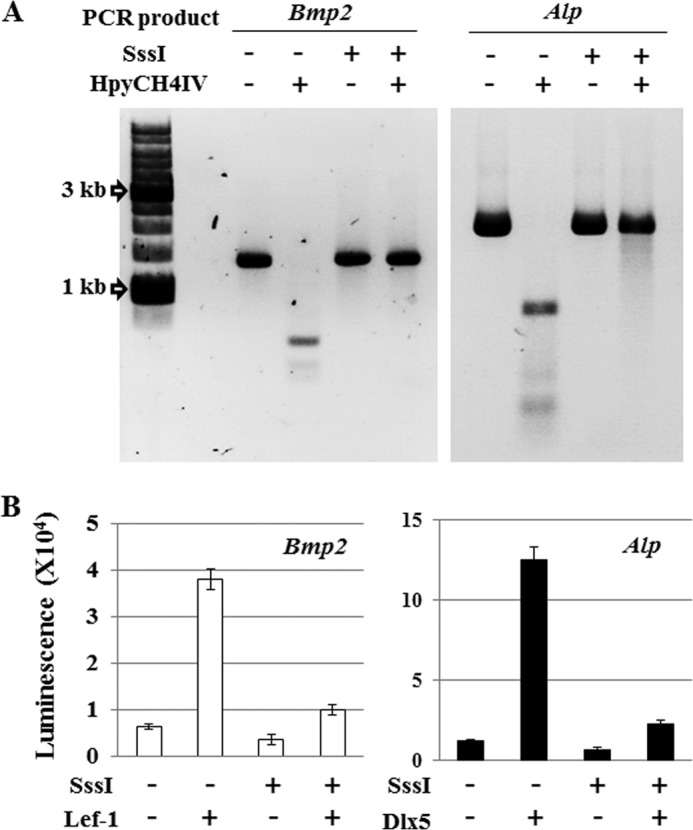
Methylation of the CpG islands in the promoter region regulates gene expression. A, PCR products are inserts of Bmp2 and Alp luciferase reporter vectors digested by HpyCH4IV enzyme for confirmation of in vitro methylation status. Methyl-sensitive endonuclease HpyCH4IV recognizes the sequence “ACGT”; however, its action is blocked by 5-methylcytosine. B, luciferase reporter activities from Bmp2 and Alp promoter reporter vectors with or without in vitro DNA methylation in the C2C12. C2C12 cells were co-transfected by electroporation with Bmp2 or Alp promoter reporter vectors and Lef-1 or Dlx5 expression vectors. Luciferase reporter activity was determined in triplicate for each experiment and three independent experiments per condition. Values are means ± S.D. of representative triplicate experiments. A value of p < 0.05 was considered statistically significant.
Epigenetic Modification Induces Trans-differentiation of Nonosteogenic Cells to Osteoblasts
Because both Bmp2 and Alp expressions are acutely stimulated in 3T3-L1 and NIH3T3 cells, which were treated 5′-aza-dC or TSA and Wnt3a, we examined staining of Alp as a biomarker for osteoblast differentiation in longer term cultures. Cells were treated with the epigenetic agents 5′-aza-dC or TSA for 24 h, followed by Wnt3a administration and culture in osteogenic media for 3 additional days (Fig. 6A). In 3T3-L1 cells, 5′-aza-dC is more effective in promoting Alp expression (Fig. 3B), whereas TSA is more effective in the NIH3T3 cells (Fig. 4B). Therefore, we selected 5′-aza-dC and TSA for induction of osteogenic differentiation in 3T3-L1 and NIH3T3 cells, respectively (Fig. 6, B and C). The Alp staining results clearly show osteoblast differentiation is promoted in pre-adipocytes and fibroblasts treated with Wnt3a and either 5′-aza-dC or TSA (Fig. 6, B and C). Therefore, epigenetic mechanisms account for the cell type specificity in repression of Wnt3a responsiveness in nonosteogenic cells.
FIGURE 6.
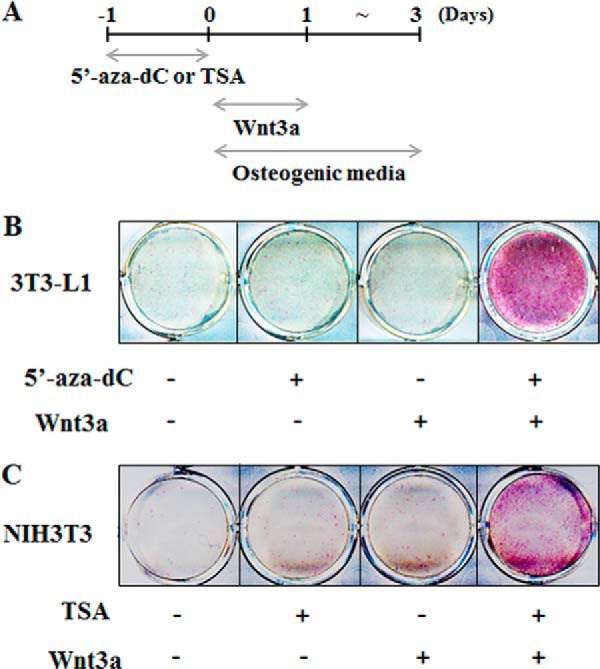
Epigenetic modification induces osteoblast differentiation in the nonosteogenic cells. A, cells were treated with 5′-aza-dC (10 μm) and TSA (100 nm) for 24 h individually, and then Wnt3a was added sequentially and cultured in the osteogenic media including β-glycerophosphate (5 mm) and ascorbic acid (50 μg/ml) for 3 days. Day 0 means initiation of differentiation. B and C, Alp staining. 3T3-L1 cells (B) were treated with 5′-aza-dC, and NIH3T3 cells (C) were treated with TSA as indicated in A. ALP activity was determined by cytochemical staining.
DISCUSSION
In this study, we validated the hypothesis that selective cell type-specific activation of the Bmp2 and Alp genes by Wnt3a and BMP2 in osteoinducible cells is controlled by epigenetic mechanisms at the level of CpG methylation. This renewed understanding of the cell type-dependent effects of Wnt3a and BMP2 resolves previous interpretative discrepancies in this field. Furthermore, our studies have yielded novel insights into epigenetic modifications that can be experimentally manipulated to convert nonosteogenic mesenchymal cells into osteoblasts by direct trans-differentiation.
Although many reports have examined relationships between WNT and BMP signaling in osteoblasts, these studies have not yet yielded an unequivocal model for their mechanistic roles in osteogenic differentiation. Several papers reported that BMP2 induces Wnt/β-catenin signaling. For example, it has been suggested that BMP2 controls the induction of ALP and mineralization by autocrine effects of WNTs in pre-osteoblastic cells (14). Consistent with this model, it has been shown that BMP2 induces expression of several WNT ligands and their cognate receptor that together enhance β-catenin activity during BMP2-induced ectopic bone formation (33). Similarly, other experiments have suggested that BMP2 promotes osteogenic differentiation by stimulation of LRP5 expression, which stabilizes β-catenin (34). Furthermore, conditional null mutation of the BMPR-IA receptor in osteoblasts increases bone mass and affects expression of the Wnt inhibitors, Dkk1 and Sost, that thus are genetically controlled by BMP signaling (35). Thus, these previous studies suggest that BMP2 signaling can occur upstream of the Wnt/β-catenin pathway.
Wnt3A Precedes BMP2 in the Osteogenic Cells
In this paper, we performed Alp staining after 3 days of Wnt3a or BMP2 treatment (Fig. 1A). In the case of ST2 and C3H10T1/2 cells, stimulation of Alp activity by BMP2 is lower than that achieved by Wnt3a. Wnt3a is more effective than BMP2 in stimulating osteogenic differentiation of ST2 or C3H10T1/2 cells, whereas BMP2 is more effective than Wnt3a in C2C12 and MC3T3-E1 cells. Because it is generally accepted that ST2 or 10T1/2 cells represent developmentally more immature cells than C2C12 or MC3T3 cells, we believe that Wnt3a is more effective than BMP2 at earlier stages of osteogenic differentiation. This concept is consistent with a model for osteogenic differentiation that was previously presented (6).
In our laboratory, we have obtained direct evidence showing that Wnt3a can act upstream of BMP2 by stimulating Bmp2 expression and generating a Bmp2 autocrine loop in MC3T3-E1 cells (11). This Wnt3a-dependent induction of Bmp2 expression is restricted to osteoinducible cells, which have the potential to differentiate into osteoblasts (28).
Our definition of a “nonosteogenic” to 3T3-L1 and NIH3T3 cells is somewhat arbitrary. Li et al. (36) reported NIH3T3 cells respond to BMP4 induction to differentiate into cartilage and bone. BMP4 first stimulates chondrogenic differentiation of NIH3T3 cells, and this stimulation is not mediated by Runx2, suggesting that BMP4 induction of NIH3T3 fibroblasts recapitulates stages of endochondral bone formation. Furthermore, NIH3T3 cells required 5-fold higher amounts of BMP4 or four times longer term treatment times compared with BMP4 treatment of C2C12 cells. These differences indicate that NIH3T3 cells tend to be far less responsive to BMP2s compared with C2C12 cells. This mechanism of BMP4 responsiveness in NIH3T3 cells is different from osteogenic differentiation of C2C12 cells, which directly induces osteogenic differentiation without evidence of a chondrogenic intermediate.
Our findings are corroborated by very recent studies showing that activation of the Wnt/β-catenin/T cell factor axis by Wnt3a results in β-catenin/TCF4 stimulation of Bmp2 gene expression at both promoter and mRNA levels (37). In addition, Wnt/β-catenin signaling stimulates osteoblast differentiation through effects on Bmp2-mediated signal transduction (38). Thus, the findings from our group and others clearly indicate that Bmp2 signaling is downstream of the Wnt/β-catenin pathway.
Consistent with the latter conclusion, WNT activation of BMPs is also reflected by WNT effects on BMP4 in different biological contexts. It has been shown that Wnt3a induces Bmp4 mRNA expression prior to the appearance of the osteoblast biomarker Alp in C3H10T1/2 mesenchymal progenitor cells (39). Kim et al. (40) showed that β-catenin specifically is absolutely required for BMP4 expression in human cancer cells, and Kuroda et al. (41) reported that canonical WNT signaling induces BMP4 to specify slow myofibrogenesis in fetal myoblasts. The collective body of evidence that has accumulated strongly suggests that the BMP and WNT pathways are intricately cross-regulated. The most salient feature of this proposed reciprocal cross-regulation is that both ligands are mutually stimulatory to generate two interlocked and self-reinforcing autocrine/paracrine pathways.
Epigenetic Mechanisms Are Essential for Cell Type-specific Cross-regulation of BMP2 by WNT3A
Epigenetic mechanisms are crucial for limiting or accommodating gene expression in different mesenchymal tissues. This paper shows that cell type-specific methylation of 5′-cytosines in CpG islands, which renders chromatin resistant to recognition by transcription factors (42, 43), creates an epigenetic barrier for the osteogenic activity of Wnt3a. Consequently, relief of this epigenetic inhibition by preceding 5′-aza-dC or TSA poises the Bmp2 and Alp genes for transcriptional stimulation by Wnt3a. We have performed co-treatment experiments with 5′-aza-dC and TSA to assess whether these compounds could have co-stimulatory effects. However, we observed that although these compounds are effective on their own, they do not synergize (data not shown). Our findings provide a novel epigenetic view for interpretation of previous studies. In a broader context, we show that manipulation of the epigenetic landscape of mesenchymal cell types at the level of CpG methylation and histone acetylation can be applied to induce trans-differentiation. This direct programming of otherwise nonosteogenic cells into osteoblastic cells is mechanistically linked to rendering Bmp2 expression responsive to Wnt3a. Trans-differentiation of cells into osteoblasts by modification of the epigenetic landscape in mesenchymal cells offers new opportunities for treatment of bone-related diseases.
In conclusion, studies from our laboratory and others in the past decade are converging toward the view that osteogenic differentiation requires synergistic cross-talk between both BMP2 and WNT3a. The mechanistic distinction between these ligands as being upstream or downstream of each other may be arbitrary and could depend on timing and the biological starting point of interlocked feedback circuits. The observation that induction of autocrine BMP2 production by WNT3a is epigenetically controlled affords a renewed understanding of regulatory mechanisms that control osteogenesis. This knowledge provides opportunities for identifying nontoxic epigenetic drugs that modify the state of CpG methylation and histone acetylation as the basis for novel bone anabolic strategies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AR049069. This work was also supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning Grant 2013R1A1A3011102.
- WNT
- wingless/int-1 class
- 5′-aza-dC
- 5′-aza-2′-deoxycytidine
- ALP
- alkaline phosphatase
- TSA
- trichostatin-A
- MSP
- methylation-specific PCR.
REFERENCES
- 1. Chen D., Zhao M., Mundy G. R. (2004) Bone morphogenetic proteins. Growth Factors 22, 233–241 [DOI] [PubMed] [Google Scholar]
- 2. Ryoo H. M., Lee M. H., Kim Y. J. (2006) Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene 366, 51–57 [DOI] [PubMed] [Google Scholar]
- 3. Yavropoulou M. P., Yovos J. G. (2007) The role of the Wnt signaling pathway in osteoblast commitment and differentiation. Hormones 6, 279–294 [DOI] [PubMed] [Google Scholar]
- 4. Baron R., Kneissel M. (2013) WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med. 19, 179–192 [DOI] [PubMed] [Google Scholar]
- 5. Nusse R., Varmus H. (2012) Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J. 31, 2670–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaur T., Lengner C. J., Hovhannisyan H., Bhat R. A., Bodine P. V., Komm B. S., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2005) Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 280, 33132–33140 [DOI] [PubMed] [Google Scholar]
- 7. Krishnan V., Bryant H. U., Macdougald O. A. (2006) Regulation of bone mass by Wnt signaling. J. Clin. Invest. 116, 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Logan C. Y., Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 9. Lee M. H., Javed A., Kim H. J., Shin H. I., Gutierrez S., Choi J. Y., Rosen V., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B., Ryoo H. M. (1999) Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor β1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J. Cell. Biochem. 73, 114–125 [PubMed] [Google Scholar]
- 10. Lee K. S., Hong S. H., Bae S. C. (2002) Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-β and bone morphogenetic protein. Oncogene 21, 7156–7163 [DOI] [PubMed] [Google Scholar]
- 11. Cho Y. D., Kim W. J., Yoon W. J., Woo K. M., Baek J. H., Lee G., Kim G. S., Ryoo H. M. (2012) Wnt3a stimulates Mepe, matrix extracellular phosphoglycoprotein, expression directly by the activation of the canonical Wnt signaling pathway and indirectly through the stimulation of autocrine Bmp-2 expression. J. Cell. Physiol. 227, 2287–2296 [DOI] [PubMed] [Google Scholar]
- 12. Cho Y. D., Yoon W. J., Woo K. M., Baek J. H., Lee G., Cho J. Y., Ryoo H. M. (2009) Molecular regulation of matrix extracellular phosphoglycoprotein expression by bone morphogenetic protein-2. J. Biol. Chem. 284, 25230–25240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim Y. J., Lee M. H., Wozney J. M., Cho J. Y., Ryoo H. M. (2004) Bone morphogenetic protein-2-induced alkaline phosphatase expression is stimulated by Dlx5 and repressed by Msx2. J. Biol. Chem. 279, 50773–50780 [DOI] [PubMed] [Google Scholar]
- 14. Rawadi G., Vayssière B., Dunn F., Baron R., Roman-Roman S. (2003) BMP-2 controls alkaline phosphatase expression and osteoblast mineralization by a Wnt autocrine loop. J. Bone Miner. Res. 18, 1842–1853 [DOI] [PubMed] [Google Scholar]
- 15. Graf T., Enver T. (2009) Forcing cells to change lineages. Nature 462, 587–594 [DOI] [PubMed] [Google Scholar]
- 16. Jopling C., Boue S., Izpisua Belmonte J. C. (2011) Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 12, 79–89 [DOI] [PubMed] [Google Scholar]
- 17. Xie H., Ye M., Feng R., Graf T. (2004) Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 [DOI] [PubMed] [Google Scholar]
- 18. Meivar-Levy I., Sapir T., Gefen-Halevi S., Aviv V., Barshack I., Onaca N., Mor E., Ferber S. (2007) Pancreatic and duodenal homeobox gene 1 induces hepatic dedifferentiation by suppressing the expression of CCAAT/enhancer-binding protein β. Hepatology 46, 898–905 [DOI] [PubMed] [Google Scholar]
- 19. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garg V., Kathiriya I. S., Barnes R., Schluterman M. K., King I. N., Butler C. A., Rothrock C. R., Eapen R. S., Hirayama-Yamada K., Joo K., Matsuoka R., Cohen J. C., Srivastava D. (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 [DOI] [PubMed] [Google Scholar]
- 21. Lin Q., Schwarz J., Bucana C., Olson E. N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276, 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardiner-Garden M., Frommer M. (1987) CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261–282 [DOI] [PubMed] [Google Scholar]
- 23. Bestor T., Laudano A., Mattaliano R., Ingram V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983 [DOI] [PubMed] [Google Scholar]
- 24. Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. (1992) Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 6, 705–714 [DOI] [PubMed] [Google Scholar]
- 25. Shiota K. (2004) DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet. Genome Res. 105, 325–334 [DOI] [PubMed] [Google Scholar]
- 26. Ohgane J., Yagi S., Shiota K. (2008) Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta 29, S29–S35 [DOI] [PubMed] [Google Scholar]
- 27. Li L. C., Dahiya R. (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 [DOI] [PubMed] [Google Scholar]
- 28. Cho Y.-D., Kim W.-J., Yoon W.-J., Woo K. M., Baek J.-H., Ryoo H.-M. (2011) Wnt3a stimulates Bmp2 expression in osteogenic cells that are epigenetically ready. J. Bone Miner. Res. 26, Suppl. 1, 161 [Google Scholar]
- 29. Moon C., Kim S. H., Park K. S., Choi B. K., Lee H. S., Park J. B., Choi G. S., Kwan J. H., Joh J. W., Kim S. J. (2009) Use of epigenetic modification to induce FOXP3 expression in nalive T cells. Transplant. Proc. 41, 1848–1854 [DOI] [PubMed] [Google Scholar]
- 30. Cedar H., Bergman Y. (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304 [DOI] [PubMed] [Google Scholar]
- 31. DiNardo D. N., Butcher D. T., Robinson D. P., Archer T. K., Rodenhiser D. I. (2001) Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene 20, 5331–5340 [DOI] [PubMed] [Google Scholar]
- 32. Fujita N., Takebayashi S., Okumura K., Kudo S., Chiba T., Saya H., Nakao M. (1999) Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol. Cell. Biol. 19, 6415–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Y., Whetstone H. C., Youn A., Nadesan P., Chow E. C., Lin A. C., Alman B. A. (2007) β-Catenin signaling pathway is crucial for bone morphogenetic protein 2 to induce new bone formation. J. Biol. Chem. 282, 526–533 [DOI] [PubMed] [Google Scholar]
- 34. Zhang M., Yan Y., Lim Y. B., Tang D., Xie R., Chen A., Tai P., Harris S. E., Xing L., Qin Y. X., Chen D. (2009) BMP-2 modulates β-catenin signaling through stimulation of Lrp5 expression and inhibition of β-TrCP expression in osteoblasts. J. Cell. Biochem. 108, 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamiya N., Kobayashi T., Mochida Y., Yu P. B., Yamauchi M., Kronenberg H. M., Mishina Y. (2010) Wnt inhibitors Dkk1 and sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J. Bone Miner. Res. 25, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li G., Peng H., Corsi K., Usas A., Olshanski A., Huard J. (2005) Differential effect of BMP4 on NIH/3T3 and C2C12 cells: implications for endochondral bone formation. J. Bone Miner. Res. 20, 1611–1623 [DOI] [PubMed] [Google Scholar]
- 37. Zhang R., Oyajobi B. O., Harris S. E., Chen D., Tsao C., Deng H. W., Zhao M. (2013) Wnt/β-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone 52, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bain G., Müller T., Wang X., Papkoff J. (2003) Activated β-catenin induces osteoblast differentiation of C3H10T1/2 cells and participates in BMP2 mediated signal transduction. Biochem. Biophys. Res. Commun. 301, 84–91 [DOI] [PubMed] [Google Scholar]
- 39. Winkler D. G., Sutherland M. S., Ojala E., Turcott E., Geoghegan J. C., Shpektor D., Skonier J. E., Yu C., Latham J. A. (2005) Sclerostin inhibition of Wnt-3a-induced C3H10T1/2 cell differentiation is indirect and mediated by bone morphogenetic proteins. J. Biol. Chem. 280, 2498–2502 [DOI] [PubMed] [Google Scholar]
- 40. Kim J. S., Crooks H., Dracheva T., Nishanian T. G., Singh B., Jen J., Waldman T. (2002) Oncogenic β-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 62, 2744–2748 [PubMed] [Google Scholar]
- 41. Kuroda K., Kuang S., Taketo M. M., Rudnicki M. A. (2013) Canonical Wnt signaling induces BMP-4 to specify slow myofibrogenesis of fetal myoblasts. Skelet. Muscle 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuks F., Hurd P. J., Wolf D., Nan X., Bird A. P., Kouzarides T. (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278, 4035–4040 [DOI] [PubMed] [Google Scholar]
- 43. Khavinson V., Solov'ev AIu., Zhilinskiĭ D. V., Shataeva L. K., Vaniushin B. F. (2012) Epigenetic aspects of peptide regulation of aging. Adv. Gerontol. 25, 11–22 [PubMed] [Google Scholar]