Background: PsbP and PsbQ are extrinsic subunits of photosystem II (PSII).
Results: Using chemical cross-linking, multiple interactions of PsbP and PsbQ with PSII intrinsic subunits, as well as a light-harvesting protein, were detected.
Conclusion: Binding of PsbP and PsbQ affects the quaternary structure of the PSII supercomplex.
Significance: Our data provide new insights into the organization of PSII extrinsic subunits in higher plants.
Keywords: Mass Spectrometry (MS), Photosystem II, Plant Biochemistry, Protein Cross-linking, Western Blot, Extrinsic Protein, Oxygen-evolving Complex
Abstract
The extrinsic subunits of membrane-bound photosystem II (PSII) maintain an essential role in optimizing the water-splitting reaction of the oxygen-evolving complex (OEC), even though they have undergone drastic change during the evolution of oxyphototrophs from symbiotic cyanobacteria to chloroplasts. Two specific extrinsic proteins, PsbP and PsbQ, bind to the lumenal surface of PSII in green plants and maintain OEC conformation and stabilize overall enzymatic function; however, their precise location has not been fully resolved. In this study, PSII-enriched membranes, isolated from spinach, were subjected to chemical cross-linking combined with release-reconstitution experiments. We observed direct interactions between PsbP and PsbE, as well as with PsbR. Intriguingly, PsbP and PsbQ were further linked to the CP26 and CP43 light-harvesting proteins. In addition, two cross-linked sites, between PsbP and PsbR, and that of PsbP and CP26, were identified by tandem mass spectrometry. These data were used to estimate the binding topology and location of PsbP, and the putative positioning of PsbQ and PsbR on the lumenal surface of the PSII. Our model gives new insights into the organization of PSII extrinsic subunits in higher plants and their function in stabilizing the OEC of the PSII supercomplex.
Introduction
Photosystem II (PSII)3 is a water-plastoquinone oxidoreductase, driven by light energy, consisting of both membrane-intrinsic and membrane-extrinsic subunits (1). On the thylakoid lumenal side of PSII, the Mn4CaO5 cluster catalyzes water oxidation, leading to the production of molecular oxygen, essential for most living organisms on Earth (2). The membrane-intrinsic subunits of PSII are involved in pigment and/or cofactor binding for photochemical reactions, whereas the membrane-extrinsic subunits on the lumenal side play crucial roles in stabilizing the Mn4CaO5 cluster and in retaining Ca2+ and Cl−, required cofactors for water oxidation (3). X-ray structural analysis of the cyanobacterial PSII complex has revealed the organization of its subunits to a resolution of 1.9 Å (4). This crystallographic information, derived from prokaryotic cyanobacterial PSII, cannot necessarily be applied in the context of eukaryotes, due to differences in PSII subunit composition and light harvesting strategies.
It is known that the composition of the extrinsic subunits of PSII has undergone much evolutionary change. Green plant eukaryotes, such as higher plants, green algae, and euglena, have a set of three extrinsic proteins, PsbO, PsbP, and PsbQ, binding to the lumenal surface of PSII. Of note, cyanobacteria have PsbV and PsbU binding to this lumenal surface instead of PsbP and PsbQ, although none of these proteins share any sequence similarity with each other (5, 6). In cyanobacteria, there are also homologs of PsbP and PsbQ, termed CyanoP and CyanoQ, respectively (7, 8), but these have not yet been included in current structural models. Red algae and diatoms have their own specific lumenally bound extrinsic subunits, such as PsbQ′ and Psb31 (9, 10). Interestingly, the PsbP and PsbQ proteins in green plants seem to have evolved from CyanoP and CyanoQ in cyanobacteria, although considerable genetic and functional modifications have occurred to generate the forms of these proteins seen in eukaryotes today (11). The high resolution structures of isolated PsbP and PsbQ proteins have been reported (12–15). Furthermore, their locations and binding topologies in the green plant PSII complex have been proposed (16, 17), but these have not been confirmed experimentally.
The main purpose of PsbP and PsbQ is to optimize the availability of Ca2+ and Cl− cofactors for efficient water oxidation by PSII. It is reported that the N-terminal region of PsbP specifically interacts with PSII to induce conformational changes around the Mn4CaO5 cluster, required for Ca2+ and Cl− (18), and PsbQ supports this PsbP function (19). Other recent studies in higher plants suggest that the PsbP and PsbQ proteins are required for stable association of intramembranous light-harvesting proteins (LHCs) with PSII (20, 21); however, it remains unclear how the binding of extrinsic subunits of PSII affect the association of LHCs to PSII.
In our study, the interaction of PsbP and PsbQ within the PSII complex was investigated using a chemical zero-length cross-linker, a water-soluble carbodiimide, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). We found that PsbP can directly interact with the membrane-intrinsic protein, PsbE, and also with PsbR, a protein we suggest is mostly membrane-extrinsic; in addition, both PsbP and PsbQ interact with the CP26 and CP43 light-harvesting proteins of the LHCII-PSII supercomplex. It is proposed that the above, as a whole, provides a structural basis for the stabilization of the LHCII-PSII supercomplex in higher plants.
EXPERIMENTAL PROCEDURES
Plasmid Construction, Recombinant Protein Expression, and Purification
The recombinant PsbP_A186C, in which Ala-186 is substituted with Cys, was produced as reported previously (22). To produce the expression plasmid for PsbQ_G149C, in which Gly-149 is substituted with Cys, cDNA fragments of spinach PsbQ were amplified by PCR with the primers (5′-CATATGAGGCCAGGCCCATCGTTGT-3′ and 5′-GAGCTCTTAACAGAGCTTGGCAAGAAC-3′) using 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 30 s. The amplified fragment was purified, digested with NdeI and SacI, and subcloned into the pET 41-a vector (Novagen). The recombinant PsbQ_G149C was expressed in the Escherichia coli strain BL21 (DE3) and purified as described previously (19). The N-terminal Glu-1 of mature PsbQ was truncated during the preparation process. PsbP_A186C and PsbQ_G149C were labeled with maleimide-PEG2-biotin (Pierce) according to the manufacturer's protocol. The presence of the desired mutation in each recombinant protein and in the maleimide-PEG2-biotin labeling was confirmed by MALDI-TOF mass spectrometry (Autoflex III, Bruker).
Reconstitution of the PsbP Protein to NaCl-washed PSII and Cross-linking Experiments
Oxygen-evolving PSII membranes of spinach were prepared as reported previously (23). The activity of isolated PSII membranes was ∼480 μmol of O2/mg of Chl/h in the activity measurement buffer (25 mm Mes-NaOH, pH 6.5, 2 m betaine, 20 mm NaCl, 0.4 mm 2,6-dichlorobenzoquinone). The PSII membranes were treated with the high salt buffer (25 mm Mes-NaOH, pH 6.5, 2 m NaCl) to remove the extrinsic PsbP and PsbQ proteins. After incubation for 30 min on ice in the dark, the membranes were sedimented, washed once with buffer (20 mm Mes-NaOH, pH 6.5, 10 mm NaCl), and then resuspended in cross-linking buffer (25 mm Hepes-NaOH, pH 7.2, 25 mm CaCl2). The NaCl-washed PSII membranes, at a concentration of 0.5 mg Chl/ml, were cross-linked with the recombinant biotin-labeled PsbP and/or PsbQ proteins in the cross-linking buffer, including 6.25 mm EDC and 5 mm N-hydroxysulfosuccinimide (sulfo-NHS). The solution was incubated for 2 h in darkness, and the reaction was terminated by adding ammonium acetate, to a final concentration of 0.2 m. The solution was centrifuged for 5 min at 20,400 × g at 4 °C, and the pellet was subjected to further affinity purification.
Purification of Cross-linked Products
PSII membranes reconstituted by biotin-labeled PsbP or PsbQ were solubilized in 100 mm Hepes-NaOH, pH 7.2, 150 mm NaCl, 2% n-octyl-β-d-glucoside and incubated with Strep-Tactin Sepharose (IBA) at 4 °C for 1 h. The resin was washed three times with 100 mm Hepes-NaOH, pH 7.2, 150 mm NaCl, and 1% SDS at room temperature for 10 min. The proteins bound to the resin were eluted by boiling the resin in SDS-PAGE sample buffer (62.5 mm Tris-HCl, pH 6.8, 10% (v/v) glycerol, 2.5% (w/v) SDS, 2.5% (v/v) 2-mercaptoethanol) at 90 °C for 10 min and used for subsequent analysis.
Immunoblot Analysis
Proteins separated by SDS-PAGE were transferred to PVDF membranes and analyzed by immunoblotting using specific antibodies. Rabbit antibodies against PsbP and D1 were prepared by the authors. The rabbit antibody against PsbQ was provided by the late Dr. A. Watanabe of Tokyo University. A rabbit antibody against CP43 was a gift from Dr. Y. Kashino of the University of Hyogo. Rabbit antibodies against PsbR, PsbE, and CP26 were purchased from Agrisera AB, Sweden.
Mass Spectrometric (MS) Analysis
The purified proteins were separated by SDS-PAGE and subjected to in-gel digestion with MS-grade modified trypsin (Promega). The peptides were extracted and loaded onto a column (100 μm internal diameter, 15 cm length; L-Column, CERI) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics). Buffers were as follows: A, 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in water and 0.1% (v/v) acetic acid; and B, 90% (v/v) acetonitrile in water. A linear gradient of buffer B from 5 to 45% was employed, and peptides eluted for 26 min from the column were introduced directly into an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) with a flow rate of 500 nl min−1 and a spray voltage of 2.0 kV. The range of the mass spectrometric scan was a mass-to-charge ratio of 450 to 1500, and the top three peaks were subjected to tandem mass spectrometry analysis.
Search for Cross-linked Sites
Collected data were analyzed using Mass Matrix version 2.4.2 (24). Mass Matrix adopts a staged search strategy. In the first stage, the tandem mass spectrometric data set was searched against the UniProt database using the target-decoy search strategy for protein identification. Reversed sequences were used as a decoy, and the data were filtrated with a ≤1% false discovery rate. In the second stage, protein matches selected from the first stage were searched with the search algorithm that Mass Matrix employs to identify the cross-linked peptides (25). The quality of peptide match was evaluated by three validated statistical measures as follows: pp, probabilistic score based on number of matched peaks; pp2, probabilistic score based on ion intensity distribution of matched peaks; pptag, probabilistic score based on consecutiveness of matched peaks. A peptide match with maximum (pp and pp2) >4.1 and pptag >3 was considered to be significant with a p value of <0.001. The settings of Mass Matrix were as follows: enzyme, trypsin, no Pro rule; decoy database, reversed; missed cleavages, 4; variable modifications, oxidation of Met, His, and Trp, amidation of Asp and Glu, carbamidomethyl of Cys; precursor ion tolerance, 10 ppm; product ion tolerance, 0.8 Da; maximum number of post-translational modification per peptide, 2; mass type, monoisotopic; minimum peptide length, 5; maximum peptide length, 40; cross-link, EDC; cross-link mode, exploratory; cross-link site cleavability, noncleavable by enzyme or not applicable; maximum number of cross-links per peptide, 2.
Structural Modeling
Atomic coordinates from the Protein Data Bank (PDB) of the Research Collaboratory for Structural Bioinformatics were downloaded for the protein subunits or complexes of cyanobacterial PSII (PDB ID 3ARC), higher plant LHCII (PDB ID 2BHW), CP29 (PDB ID 3PL9), PsbP (PDB ID 1V2B), and PsbQ (PDB ID 1NZE). For subunits not yet present in the PDB, spinach sequences for PsbR, CP26, and CP24 were subjected to I-TASSER homology modeling analysis, and in each case the top-scoring model, model1.pdb, was preferred. The most highly ranked protein with a highly similar structure in the PDB, as identified by the TM-align algorithm within I-TASSER, was PDB ID 1V34_A2, PDB ID 2BHW_C, and PDB ID 2BHW_A, respectively, for PsbR, CP26, and CP24. UCSF Chimera (26) was the program used to form the modeling environment, and all subunits were fitted, as surface-rendered rigid bodies, by visual inspection. First, PDB files for the PSII dimer, LHCII trimers, and CP29 subunits were modeled within a molecular envelope derived from a transmission electron microscopy (TEM) as detailed previously (27). This was followed by the substitution of CP26 and CP24 homology models of I-TASSER, in place of previously modeled subunits based upon monomeric LHCII (27). Next, the PDB files for PsbP and PsbQ structures were overlaid across the lumenal surface, taking into account the data from cross-linked sites reported in this study, and finally, the I-TASSER homology model for PsbR was introduced.
RESULTS
Interaction of the PsbP and PsbQ Proteins with LHCII-PSII Supercomplex Membrane-intrinsic Subunits
Previous studies using EDC hardly detected the cross-linking between PsbP/Q and PSII intrinsic subunits in higher plants. In this study, we combined the conventional release-reconstitution experiment with a chemical cross-linking, followed by affinity chromatography. NaCl-washed PSII membranes were treated with EDC in the presence or absence of biotinylated PsbP or PsbQ. It was confirmed that treatment by 6.25 mm EDC and 5 mm sulfo-NHS did not affect the reconstituted activity of PSII. After purification using Strep-Tactin Sepharose, cross-linked peptides were analyzed by SDS-PAGE, and the bands specifically observed in the presence of biotinylated proteins were used for MS analysis.
As shown in Fig. 1, four cross-linked bands were reproducibly purified by affinity chromatography with biotinylated-PsbP or -PsbQ (P-1 to P-4 and Q-1 to Q-4). The corresponding bands in the SDS-polyacrylamide gels were excised and subjected to in-gel digestion with trypsin. The MS analyses indicated that the bands P-1 to P-4 contained CP43, CP26, PsbR, and PsbE in addition to PsbP, respectively (Table 1 and supplemental Tables S1–S4). The band P-4 corresponds to the cross-linked peptide between PsbP and PsbE that we reported recently (22). Molecular masses of P-1 to P-4 (about 60, 46, 33, and 30 kDa, respectively) indicate that those subunits were cross-linked in a one-to-one (1:1) manner. Similarly, the bands Q-3 (55 kDa) and Q-4 (41 kDa) contained CP43 and CP26 in addition to PsbQ, respectively (Table 1 and supplemental Tables S5 and S6). The bands Q-1 (77 kDa) and Q-2 (65 kDa) also contained CP26 in addition to PsbQ, although their molecular masses indicate that additional subunits should be included; however, they could not be identified due to low database search scores.
FIGURE 1.
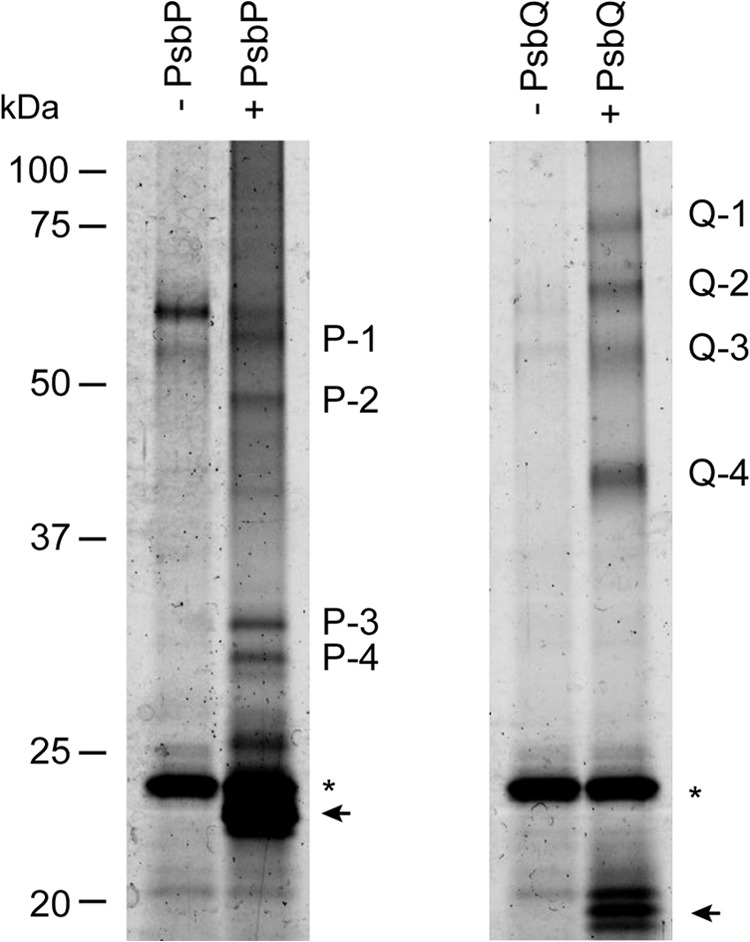
Fluorescence imaging of affinity-purified cross-linked products. NaCl-washed PSII membranes were incubated with 6.25 mm EDC and 5 mm sulfo-NHS in either the presence (+) or the absence (−) of biotinylated-PsbP (left) or-PsbQ (right). Cross-linked products were purified with Strep-Tactin Sepharose, and the purified peptides corresponding to 2 μg of Chl were separated by SDS-PAGE. The proteins were stained with Flamingo gel stain and visualized with a fluorescence gel imager. Arrows indicate the bands of noncross-linked proteins, and asterisks indicate the bands of unrelated proteins from the resin.
TABLE 1.
Summary of mass spectrometric analysis on the affinity-purified peptides
| Band | Identified protein | Score | No. of identified peptidesa | Sequence coverage |
|---|---|---|---|---|
| % | ||||
| P-1 | CP43 | 201 | 9 | 27 |
| P-2 | CP26 | 225 | 9 | 48 |
| P-3 | PsbR | 128 | 5 | 37 |
| P-4 | PsbE | 111 | 4 | 48 |
| Q-1 | ||||
| Q-2 | ||||
| Q-3 | CP43 | 233 | 10 | 25 |
| Q-4 | CP26 | 212 | 7 | 47 |
a Details of identified peptides are listed in supplemental Tables S1–S6.
The results of the MS analysis on the cross-linked peptides were supported by immunoblot analysis using specific antibodies (Fig. 2, A and B). Bands P-1 to P-4 reacted positively with anti-PsbP antibodies and also with the antibodies specific to CP43, CP26, PsbR, and PsbE. Bands Q-1, Q-2, and Q-4 clearly reacted with antibodies raised against CP26, whereas Q-3 specifically reacted with the anti-CP43 antibodies. It is reasonable to assume that PsbP has a direct interaction with PsbR, because PsbR is required for the stable binding of PsbP and PsbQ in Arabidopsis (28, 29). It is also likely that PsbP and PsbQ can interact with both CP43 and CP26, given the report that CP26 is located close to CP43 in the LHCII-PSII supercomplex (30). Our results are highly consistent with the report demonstrating that PsbQ binding requires the peripheral antenna system, at least the domain that is composed of CP26/LHCII-S (30).
FIGURE 2.
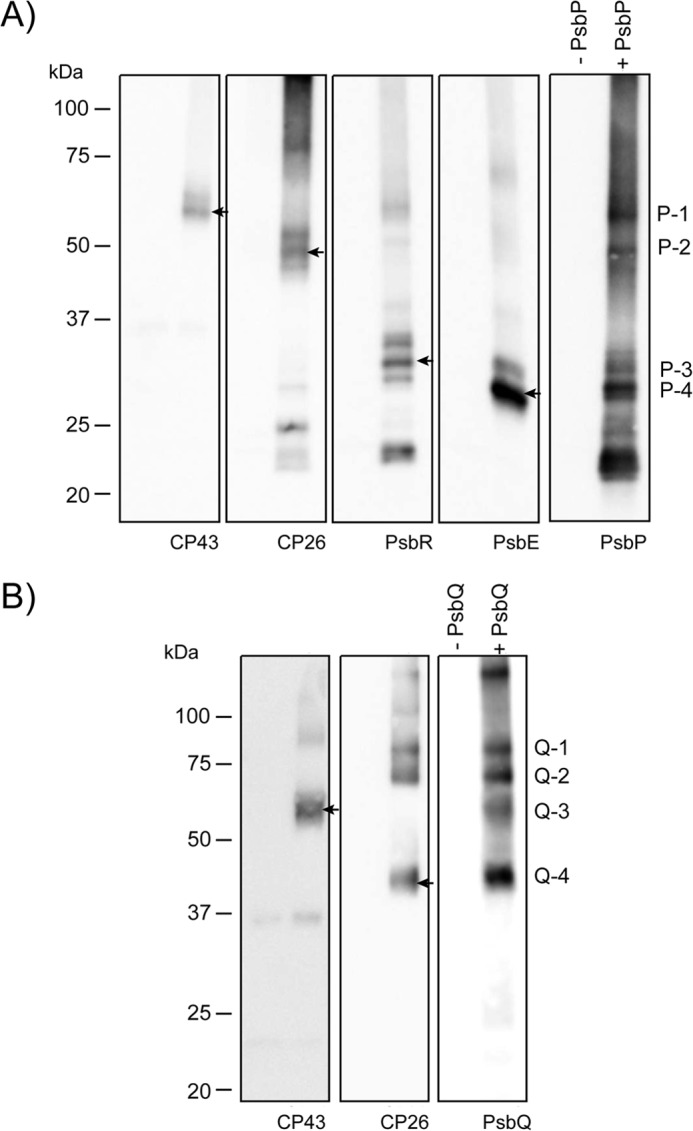
Immunodetection of affinity-purified cross-linked products. The affinity-purified cross-linked peptides using biotinylated PsbP (A) or PsbQ (B) corresponding to 2 μg of Chl were separated by SDS-PAGE and immunodetected with antisera against CP43, CP26, PsbP, PsbR, and PsbE, or PsbQ, as indicated on each lane. Arrows indicate the protein bands corresponding to P-1, P-2, P-3, P-4, Q-3 and Q-4, respectively.
Cross-linked Residues between PsbP and PsbR, and PsbP and CP26
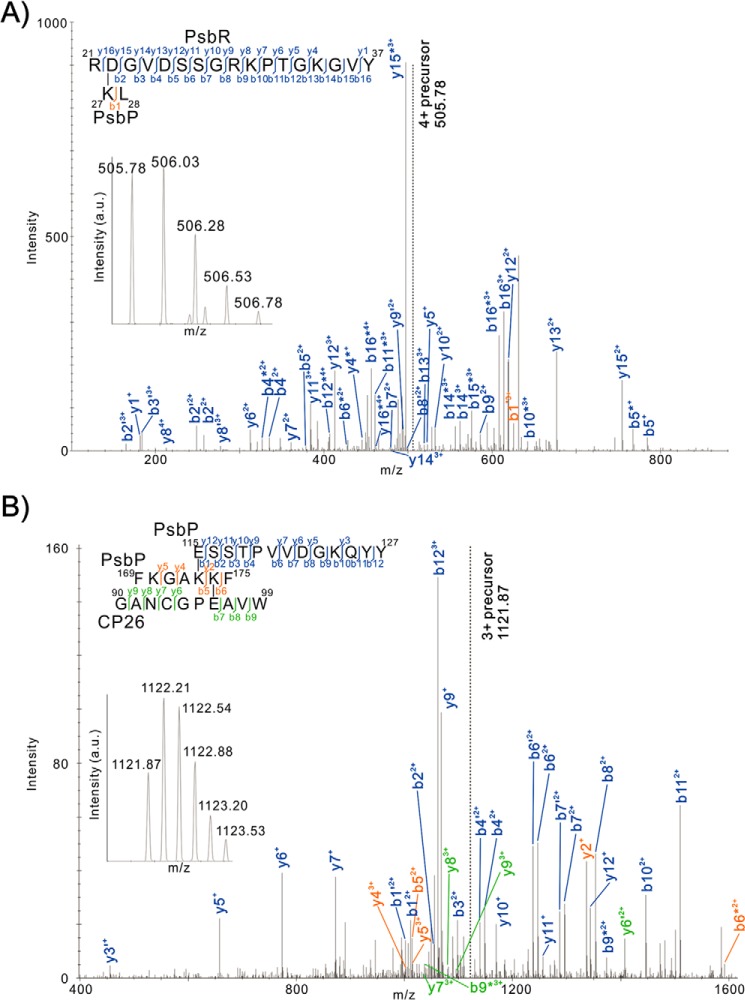
To identify the cross-linked sites within purified cross-linked peptides, tandem MS/MS spectra were analyzed by the Mass Matrix search engine (24). Possible cross-linking sites between the amino acids Asp to Lys, Glu to Lys, Asp to Ala-1, and Glu to Ala-1 were examined. As a result, cross-linked peptides were identified in the bands P-2 (PsbP and CP26), P-3 (PsbP and PsbR), and P-4 (PsbP and PsbE), although the cross-linked site within other purified peptides could not be identified in this analysis. Fig. 3A shows the tandem MS/MS spectrum of the cross-linked peptide, including RDGVDSSGRKPTGKGVY37 from PsbR and KL28 from PsbP, in which the Asp-22 residue in PsbR and Lys-27 in PsbP were cross-linked. Comparatively, Fig. 3B shows the spectrum of ESSTPVVDGKQYY127 and FKGAKKF175 from PsbP and GANCGPEAVW99 from CP26, in which the Glu-96 in CP26 and the Lys-174 in PsbP were cross-linked. The cross-linking between Ala-1 in PsbP and Glu-57 in PsbE was also detected in P-4, as reported previously (22). The scores of those peptides were statistically significant (p < 0.001, supplemental Tables S7 and S8), and the amino acid residues identified as cross-linked sites are highly conserved in their respective proteins in higher plants.
FIGURE 3.
Product ion (tandem MS/MS) spectra of peptides that were predicted to be cross-linked between RDGVDSSGRKPTGKGVY37 from PsbR and KL28 from PsbP (A) and ESSTPVVDGKQYY127 and FKGAKKF175 from PsbP and GANCGPEAVW99 from CP26 (B). Prime and asterisk show unhydrated and deammoniated product ions, respectively. Note that for clarity not all product ions assigned by Mass Matrix are labeled. Complete sets of the assigned product ions are listed in supplemental Tables S7 and S8. Insets are the respective precursor ion spectra. a.u., arbitrary units.
The Lys-27 and Lys-174 residues are included in the six domains on the PsbP protein (Lys-11 to Lys-14, Lys-27 to Lys-38, Lys-40, Lys-90 to Lys-96, Lys-143 to Lys-152, and Lys-166 to Lys-174), the modification of which with N-succinimidyl propionate (NSP), which reacts with positively charged amino groups, decreases the binding of PsbP to PSII (31). Also, we recently found that the basic surface on PsbP consisting of conserved Arg-48, Lys-143, and Lys-160 is crucial for the electrostatic interaction with PSII (32). The above information allows us to estimate the binding topology of PsbP to PSII (Fig. 4). The Lys-27 and Lys-174 residues are located on the opposite side of the basic surface of the PsbP structure, indicating that PsbP has multiple interacting surfaces and may function as a link between membrane subunits. The fact that Lys-27 and Lys-174 interact with PsbR and CP26, both peripherally attached to the PSII core complex, suggests that a basic pocket, composed of Arg-48, Lys-143, and Lys-160, would interact with the inner surface of CP43, which is cross-linked with PsbP by EDC (Figs. 1 and 2). However, the residues closest to its N terminus of PsbP would extend toward cytochrome b559 (PsbE), around which PsbF, PsbJ, and most likely PsbR as well would make up a subdomain required for the stable binding of PsbP to PSII.
FIGURE 4.
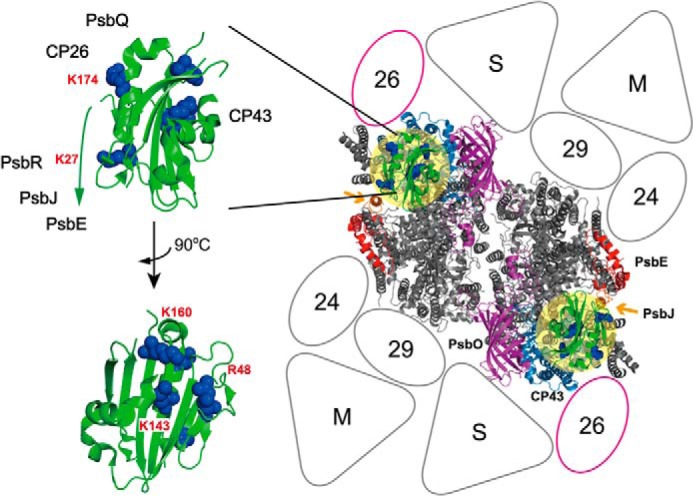
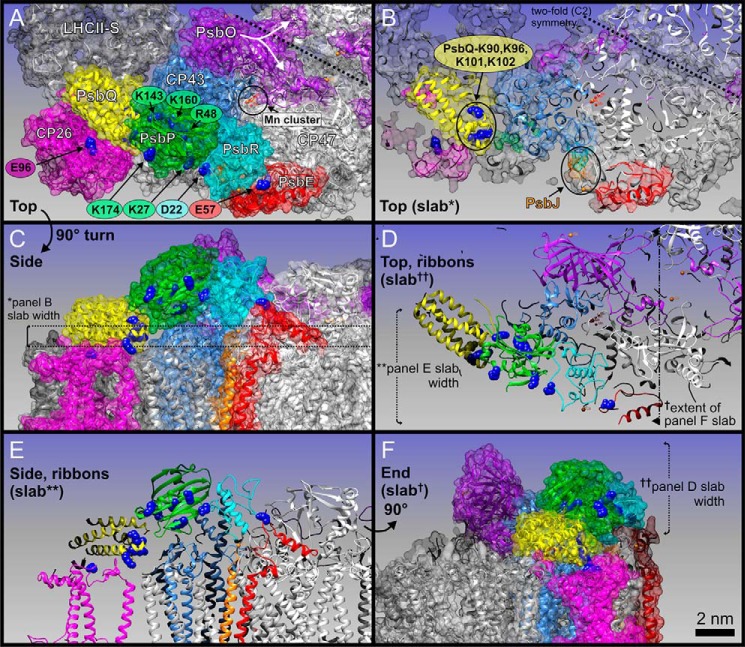
Proposed binding topology of PsbP in the LHCII-PSII supercomplex. Left, schematic of the PsbP structure from spinach (PDB ID 2VU4). The side chains of the residues Lys-27, Lys-K174, K160A, Lys-143, and Arg-48 involved in PsbP binding to PSII are shown as spherical models. Right, model of the LHCII-PSII supercomplex (C2S2M2 complex), viewed from the top onto the lumenal surface and utilizing the high resolution structure of the cyanobacterial PSII core (4), based upon comparisons with the literature (27, 30). Putative PsbP-binding sites are indicated by light yellow circular overlays. PsbV and PsbU, known to be absent in plant PSII, were excluded. CP43 and PsbE, which interact with PsbP, are shown in light blue and red, respectively. PsbJ and PsbO, required for the stable binding of PsbP and PsbQ, are shown in orange (indicated by arrows) and purple, respectively. Positioning of the strongly (S) or moderately (M) bound LHC trimers, as well as the minor LHC antennae CP24 (24), CP26 (26), and CP29 (29), are schematically depicted. CP26 is outlined in magenta to indicate its interaction with PsbP and PsbQ.
Localization of PsbP and PsbQ within a Higher Plant LHCII-PSII Supercomplex
To further investigate the location of PsbP in the complex, its structure (PDB ID 1V2B) was fitted (Fig. 5) into a TEM-based structural model of negatively stained C2S2M2 LHCII-PSII supercomplex samples (27). PsbP was also tentatively positioned close to the lumenal surface of CP43, consistent with a previous report of a three-dimensional reconstruction of a PSII supercomplex from spinach, derived from unstained cryo-TEM samples, calculated by angular reconstitution single-particle analysis (16). Note that the structure of the cyanobacterial PSII core dimer (PDB ID 3ARC) was used, as no eukaryotic PSII structure has been resolved to date; however, the extrinsic subunits PsbU and PsbV, not present in green plants, were removed. Also tentatively placed in our model are PsbQ and PsbR, although their exact binding topology has yet to be directly visualized by microscopy or crystallographic techniques. Furthermore, three-dimensional homology models of spinach PsbR (UniProt/SwissProt accession number P10690), CP26 (GenBankTM accession number AB908271), and CP24 (UniProt/SwissProt accession number P36494) were calculated using the program I-TASSER, and its model1.pdb output was used in all cases (see under “Experimental Procedures”). Of interest, it was found that the structures predicted for CP26 and CP24 were more closely aligned to that of pea LHCII (PDB ID 2BHW) and its chains C and A, respectively, rather than the recent structure of spinach CP29 (PDB ID 3PL9).
FIGURE 5.
Localization of PsbP, PsbQ, and PsbR within a pseudo-atomic C2S2M2 LHCII-PSII supercomplex three-dimensional model for higher plants. Protein coordinates (see text for PDB IDs) are shown either as schematic ribbons or transparent space-filling spheres as follows: PsbP (green), PsbQ (yellow), CP43 (light blue), PsbO (purple), PsbR (cyan), PsbE (red), CP26 (magenta), LHCII-S (light gray), and CP47 (white). A, top view, looking down onto the membrane plane from the lumen. Subunit labels are in white. Key amino acid residues of cross-linked sites are shown as blue spheres, and labels are color-coded as per associated proteins. Position of the Mn4CaO5 cluster (Mn cluster) is circled in black; manganese atoms are small red spheres and calcium atoms are orange. The PsbO protein of each PSII core monomer has a distinct feature that extends over to the adjacent PSII monomer, here noted by a white star and emphasized with a split arrow. B, slab (cut-away) view of A. PsbJ is highlighted in orange, due to it being mostly hidden in A. The 2-fold symmetry of the overall PSII dimeric core is emphasized by dashed line. C, side view of A. The thickness of the slab in B (∼1.2 nm) is marked with * and shown with dashed lines. D, slab of the entire lumenally protruding protein domain shown in schematic ribbon form, emphasizing the cross-links between key residues of PsbP, PsbQ, PsbR, and PsbE; note this is also in the same orientation as A and B. The extent of E and F slabs is also shown. E, slabbed side view of C, emphasizing all cross-link site key residues. The depth of this slab is shown in D. F, end view of E and C, slabbed from the tip of LHCII-S to PsbE. The depth (∼5 nm) of D slab is also shown. Scale bar, 2 nm (for all panels).
As discussed above, the main structure of PsbP was localized as straddling over the lumenally exposed surface of CP43, where a basic surface consisting of residues Arg-48, Lys-143, and Lys-160 faces toward the extrinsic loop of CP43, although another domain of PsbP, whose surface includes Lys-174, faces outward to CP26 (Fig. 5A). The cross-link between PsbP and CP26 is particularly novel, whereas direct interaction with major LHCs was not suggested by our analysis. In our model, PsbP together with PsbO stabilize the extrinsic loop of CP43 on either side, leading to the optimization of the microenvironment required for the oxygen-evolving center and efficient water-splitting activity (Fig. 5, A, D, and F). Without PsbO, the extrinsic loop of CP43 was likely to be highly flexible. This may explain why PsbP requires PsbO to bind to PSII, although no direct interaction has been detected between them (33), with the extrinsic loop of CP43 acting as a central pillar between them.
PsbP was observed to interact with PsbR, a polypeptide specific to PSII in green plants. The location of PsbR in PSII is predicted to be close to both PsbE and PsbJ (28). The structural model of PsbR predicted by I-TASSER shows a pronounced hydrophobic wedge shape that has been proposed to anchor it to the thylakoid membrane (34). This wedge shape in our study fits well into the gap formed between PsbP and the extrinsic portion of PsbE (Fig. 5B). In this way, the region, including the Lys-27 residue of PsbP, faces toward Asp-22 in PsbR, and the N-terminal 15-amino acid extension of PsbP, unfortunately missing in known x-ray structures, could still reach over to the contact point on the PsbE C terminus (Fig. 5A), an assumption consistent with our cross-linking results.
PsbQ was fitted within a pronounced pocket, observed between PsbP, CP43, and the lumenal membrane surface of the strongly bound LHCII-S trimer (Fig. 5, A and B) (PDB ID 2BHW), oriented to accommodate those previous studies that have suggested its multiple lysine residues (Lys-90, -96, -101, and -102) to be involved in interactions with other PSII components (35). Our results suggest that these particular residues of PsbQ interact with CP43 or CP26, or with PsbP, to stabilize the functional association of PsbP with PSII (19). Such a binding site of PsbQ is surprisingly different from that proposed for CyanoQ in PSII from Synechocystis 6803, as suggested recently by a separate chemical cross-linking study (36). It is therefore likely that the binding site(s) of PsbQ homologs have undergone significant changes during evolution, perhaps manifested in the limited sequence identity between the PsbQ and CyanoQ protein families.
DISCUSSION
In this study, we suggest the direct interaction of PsbP and PsbQ with the membrane-intrinsic PSII subunits, providing additional data for modifying current models of the organization of lumenally bound extrinsic subunits in higher plant PSII. It has been widely believed that once PsbO directly binds to the PSII core, PsbP binds to PsbO, and then PsbQ binds to PsbP or PsbO (33). However, several reports suggest that PsbO can be extracted without affecting PsbP and PsbQ binding, thereby indicating that PsbP and PsbQ directly associate with the PSII intrinsic subunits (37–39). Indeed, in our MS analysis, none of the bands P-1 to P-4 includes PsbO peptides nor Q-1 to Q-4 PsbO and PsbP. We do not exclude the possibility of there being a direct interaction between extrinsic subunits in higher plants PSII. In fact, it is reported that an EDC cross-linked product formed between PsbP and PsbQ in the green alga Chlamydomonas (40).
A direct interaction of PsbP with both PsbE and CP43 indicates that PsbP occupies a position roughly similar to that occupied by PsbV in the cyanobacterial crystal structure, in which the N terminus of PsbV (Ala-1) forms a charge-pair interaction with Asp-53 in PsbE, and the C-terminal main structure is associated with the CP43 protein (4). PsbV is suggested to have a function similar to PsbP regarding ion retention; hence, it may be reasonable to assume that its structural interactions are also similar. Given genetic studies using Arabidopsis report that knockdown of PsbP expression correlates with a loss in PsbQ, D2, and CP47 accumulation (41), PsbP being localized next to CP43 would also affect the stability of the further D2/CP47 side of each PSII monomer, probably through interaction with PsbR and also with PsbE via its N-terminal sequence.
It is probable that PsbP, PsbQ, and PsbR in higher plant PSII may all play significant roles, most likely via a defined temporal sequence, in providing enhanced structural rigidity for CP43 against PsbO. This rigidity could then be transmitted down into the membrane, locking in the protein scaffold and the ligands surrounding the Mn4CaO5 cluster of the oxygen-evolving center, thereby optimizing enzymatic activity. A further interesting observation to be noted is that a substantial part of the PsbO subunit, of one PSII monomer, reaches over to the CP47 extrinsic loop of the second monomer (Fig. 5A, white arrows) in the overall PSII dimer (PDB ID 3ARC). The binding of PsbP, PsbQ, and PsbR, via CP43 and its extrinsic loop, may make PsbO rigid enough to even stabilize the CP47 extrinsic loop of the second monomer, across the dimeric symmetry, further stabilizing the whole dimeric complex.
Our results also indicate that PsbP, together with PsbQ, can be structurally linked in stabilizing the association of a minor antenna protein, CP26, to the inner membrane-facing domains of CP43. This, in turn, will be relevant to the significance of these extrinsic subunits for the overall supramolecular organization of PSII (20, 30, 42) and their influence on the rate of state transitions (21). A major problem for our model is Lys-174 of PsbP. This is located about 35–37 Å away from its expected CP26 cross-linked site Glu-96 (Fig. 5, A and E). The 15-amino acid extension of PsbP, currently unresolved in the literature (see above), might eventually allow for some shifting of the PsbP position toward CP26, but still some distance would remain. A possibility remains whereby a secondary binding site exists, to which PsbP may transiently interact, during the PSII assembly process. Indeed, previous studies have suggested that two PsbP per PSII are required for the full activation of the oxygen-evolving activity (43), and truncation of nine N-terminal residues of PsbP increase the amount of PsbP bound to PSII in the reconstitution experiments using salt-washed PSII membranes (44).
In conclusion, we have shown that PsbP and PsbQ are capable of multiple interactions with PSII intrinsic subunits as well as other light-harvesting proteins present within the larger, stable PSII supercomplex form. This indicates that PsbP and PsbQ not only replace the function of PsbU and PsbV structurally but have evolved the ability to stabilize the overall LHCII-PSII supercomplex in green plants. It is plausible that an evolutionary change of the PSII extrinsic proteins, to increase the efficiency of maintaining the water-splitting microenvironment in higher plants, might be related to the modification in the outlying PSII light-harvesting antenna system, including the subunit CP26. Because the association of the peripheral antennae to PSII is a dynamic process, these lumenally bound, extrinsic PsbP and PsbQ proteins are likely to also have a direct role in adjusting the photosynthetic light reactions to rapidly changing environmental conditions. It is clear that high resolution (>10 Å resolution) direct structural analysis, by crystallography or three-dimensional cryo-TEM, is required to reveal the detailed extrinsic lumenal relationships between PsbP, PsbQ, CP26, PsbR, and PsbO, the extrinsic loops of CP43 and CP47, and the whole LHCII-PSII supercomplex.
Supplementary Material
This work was supported in part by a grant from Japan Science and Technology Agency PRESTO (to Ke. I.) and by Grant-in-aid for Young Scientists (B) from 18770032JSPS (to Ke. I.).

This article contains supplemental Tables S1–S8.
- PSII
- photosystem II
- Chl
- chlorophyll
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- sulfo-NHS
- N-hydroxysulfosuccinimide
- LHC
- light-harvesting complex
- PDB
- Protein Data Bank
- TEM
- transmission electron microscopy.
REFERENCES
- 1. Renger G., Renger T. (2008) Photosystem II: The machinery of photosynthetic water splitting. Photosynth. Res. 98, 53–80 [DOI] [PubMed] [Google Scholar]
- 2. Vinyard D. J., Ananyev G. M., Dismukes G. C. (2013) Photosystem II: the reaction center of oxygenic photosynthesis. Annu. Rev. Biochem. 82, 577–606 [DOI] [PubMed] [Google Scholar]
- 3. Bricker T. M., Roose J. L., Fagerlund R. D., Frankel L. K., Eaton-Rye J. J. (2012) The extrinsic proteins of photosystem II. Biochim. Biophys. Acta 1817, 121–142 [DOI] [PubMed] [Google Scholar]
- 4. Umena Y., Kawakami K., Shen J.-R., Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 5. Shen J.-R., Inoue Y. (1993) Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32, 1825–1832 [DOI] [PubMed] [Google Scholar]
- 6. Shen J.-R., Qian M., Inoue Y., Burnap R. L. (1998) Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37, 1551–1558 [DOI] [PubMed] [Google Scholar]
- 7. Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 8. Thornton L. E., Ohkawa H., Roose J. L., Kashino Y., Keren N., Pakrasi H. B. (2004) Homologs of Plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in cyanobacterium Synechosistis 6803. Plant Cell 16, 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ohta H., Suzuki T., Ueno M., Okumura A., Yoshihara S., Shen J.-R., Enami I. (2003) Extrinsic proteins of photosystem II: an intermediate member of PsbQ protein family in red algal PS II. Eur. J. Biochem. 270, 4156–4163 [DOI] [PubMed] [Google Scholar]
- 10. Nagao R., Moriguchi A., Tomo T., Niikura A., Nakajima S., Suzuki T., Okumura A., Iwai M., Shen J.-R., Ikeuchi M., Enami I. (2010) Binding and functional properties of five extrinsic proteins in oxygen-evolving photosystem II from a marine centric diatom, Chaetoceros gracilis. J. Biol. Chem. 285, 29191–29199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Las Rivas J., Roman A. (2005) Structure and evolution of the extrinsic proteins that stabilize the oxygen-evolving engine. Photochem. Photobiol. Sci. 4, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 12. Calderone V., Trabucco M., Vujicić A., Battistutta R., Giacometti G. M., Andreucci F., Barbato R., Zanotti G. (2003) Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep. 4, 900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ifuku K., Nakatsu T., Kato H., Sato F. (2004) Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep. 5, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balsera M., Arellano J. B., Revuelta J. L., de las Rivas J., Hermoso J. A. (2005) The 1.49 Å resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J. Mol. Biol. 350, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 15. Kopecky V., Jr., Kohoutova J., Lapkouski M., Hofbauerova K., Sovova Z., Ettrichova O., González-Pérez S., Dulebo A., Kaftan D., Smatanova I. K., Revuelta J. L., Arellano J. B., Carey J., Ettrich R. (2012) Raman spectroscopy adds complementary detail to the high-resolution x-ray crystal structure of photosynthetic PsbP from Spinacia oleracea. PLoS One 7, e46694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nield J., Barber J. (2006) Refinement of the structural model for the photosystem II supercomplex of higher plants. Biochim. Biophys. Acta 1757, 353–361 [DOI] [PubMed] [Google Scholar]
- 17. Ifuku K., Ido K., Sato F. (2011) Molecular functions of PsbP and PsbQ proteins in the photosystem II supercomplex. J. Photochem. Photobiol. B 104, 158–164 [DOI] [PubMed] [Google Scholar]
- 18. Tomita M., Ifuku K., Sato F., Noguchi T. (2009) FTIR evidence that the PsbP extrinsic protein induces protein conformational changes around the oxygen-evolving Mn cluster in photosystem II. Biochemistry 48, 6318–6325 [DOI] [PubMed] [Google Scholar]
- 19. Kakiuchi S., Uno C., Ido K., Nishimura T., Noguchi T., Ifuku K., Sato F. (2012) The PsbQ protein stabilizes the functional binding of the PsbP protein to photosystem II in higher plants. Biochim. Biophys. Acta 1817, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 20. Ido K., Ifuku K., Yamamoto Y., Ishihara S., Murakami A., Takabe K., Miyake C., Sato F. (2009) Knockdown of the PsbP protein does not prevent assembly of the dimeric PSII core complex but impairs accumulation of photosystem II supercomplexes in tobacco. Biochim. Biophys. Acta 1787, 873–881 [DOI] [PubMed] [Google Scholar]
- 21. Allahverdiyeva Y., Suorsa M., Rossi F., Pavesi A., Kater M. M., Antonacci A., Tadini L., Pribil M., Schneider A., Wanner G., Leister D., Aro E. M., Barbato R., Pesaresi P. (2013) Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 75, 671–684 [DOI] [PubMed] [Google Scholar]
- 22. Ido K., Kakiuchi S., Uno C., Nishimura T., Fukao Y., Noguchi T., Sato F., Ifuku K. (2012) The conserved His-144 in the PsbP protein is important for the interaction between the PsbP N-terminus and the Cyt b559 subunit of photosystem II. J. Biol. Chem. 287, 26377–26387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ifuku K., Sato F. (2001) Importance of the N-terminal sequence of the extrinsic 23 kDa polypeptide in photosystem II in ion-retention in oxygen-evolution. Biochim. Biophys. Acta 1546, 196–204 [DOI] [PubMed] [Google Scholar]
- 24. Xu H., Freitas M. A. (2009) Mass Matrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu H., Hsu P. H., Zhang L., Tsai M. D., Freitas M. A. (2010) Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry, J. Proteome Res. 9, 3384–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 27. Pagliano C., Nield J., Marsano F., Pape T., Barera S., Saracco G., Barber J. (2013) Proteomic characterization and three-dimensional electron microscopy study of PSII-LHCII supercomplexes from higher plants. Biochim. Biophys. Acta 10.1016/j.bbabio.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 28. Suorsa M., Sirpiö S., Allahverdiyeva Y., Paakkarinen V., Mamedov F., Styring S., Aro E. M. (2006) PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J. Biol. Chem. 281, 145–150 [DOI] [PubMed] [Google Scholar]
- 29. Liu H., Frankel L. K., Bricker T. M. (2009) Characterization and complementation of a psbR mutant in Arabidopsis thaliana. Arch. Biochem. Biophys. 489, 34–40 [DOI] [PubMed] [Google Scholar]
- 30. Caffarri S., Kouril R., Kereïche S., Boekema E. J., Croce R. (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tohri A., Dohmae N., Suzuki T., Ohta H., Inoue Y., Enami I. (2004) Identification of domains on the extrinsic 23-kDa protein possibly involved in electrostatic interaction with the extrinsic 33-kDa protein in spinach photosystem II. Eur. J. Biochem. 271, 962–971 [DOI] [PubMed] [Google Scholar]
- 32. Nishimura T., Uno C., Ido K., Nagao R., Noguchi T., Sato F., Ifuku K. (2014) Identification of the basic amino acid residues on the PsbP protein involved in the electrostatic interaction with photosystem II. Biochim. Biophys. Acta 10.1016/j.bbabio.2013.12.012 [DOI] [PubMed] [Google Scholar]
- 33. Miyao M., Murata N. (1989) The mode of binding of three extrinsic proteins of 33 kDa, 23 kDa, and 18 kDa in the photosystem II complex of spinach. Biochim. Biophys. Acta 977, 315–321 [DOI] [PubMed] [Google Scholar]
- 34. Bricker T. M., Roose J. L., Zhang P., Frankel L. K. (2013) The PsbP family of proteins. Photosynth. Res. 116, 235–250 [DOI] [PubMed] [Google Scholar]
- 35. Meades G. D., Jr., McLachlan A., Sallans L., Limbach P. A., Frankel L. K., Bricker T. M. (2005) Association of the 17-kDa extrinsic protein with photosystem II in higher plants. Biochemistry 44, 15216–15221 [DOI] [PubMed] [Google Scholar]
- 36. Liu H., Zhang H., Weisz D. A., Vidavsky I., Gross M. L., Pakrasi H. B. (2014) MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamamoto Y. (1988) Organization of the oxygen-evolution enzyme complex studied by butanol/water phase partitioning of spinach photosystem II particles. J. Biol. Chem. 263, 497–500 [PubMed] [Google Scholar]
- 38. Bernier M., Carpentier R. (1995) The action of mercury on the binding of the extrinsic polypeptides associated with the water oxidizing complex of photosystem II. FEBS Lett. 360, 251–254 [DOI] [PubMed] [Google Scholar]
- 39. Yu H., Xu X., Britt R. D. (2006) The 33 kDa protein can be removed without affecting the association of the 23 and 17 kDa proteins with the luminal side of PS II of spinach. Biochemistry 45, 3404–3411 [DOI] [PubMed] [Google Scholar]
- 40. Nagao R., Suzuki T., Okumura A., Niikura A., Iwai M., Dohmae N., Tomo T., Shen J.-R., Ikeuchi M., Enami I. (2010) Topological analysis of the extrinsic PsbO, PsbP and PsbQ proteins in a green algal PSII complex by cross-linking with a water-soluble carbodiimide. Plant Cell Physiol. 51, 718–727 [DOI] [PubMed] [Google Scholar]
- 41. Yi X., Hargett S. R., Liu H., Frankel L. K., Bricker T. M. (2007) The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J. Biol. Chem. 282, 24833–24841 [DOI] [PubMed] [Google Scholar]
- 42. Yi X., Hargett S. R., Frankel L. K., Bricker T. M. (2006) The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J. Biol. Chem. 281, 26260–26267 [DOI] [PubMed] [Google Scholar]
- 43. Seidler A. (1994) Expression of the 23 kDa protein from the oxygen-evolving complex of higher plants in Escherichia coli. Biochim. Biophys. Acta 1187, 73–79 [DOI] [PubMed] [Google Scholar]
- 44. Miyao M., Fujimura Y., Murata N. (1988) Partial degradation of the extrinsic 23-kDa protein of the Photosystem II complex of spinach. Biochim. Biophys. Acta 936, 465–474 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.