Abstract
Strong promoters were isolated from Flavobacterium johnsoniae in a promoter-trap vector incorporating a gfp reporter system, and were used to express fluorescent protein markers (including GFP, YFP, mOrange and mStrawberry) and insecticidal protein genes in Flavobacterium strains. Sequence analysis of trapped DNA fragments showed conserved Bacteroidetes promoter motifs (TTG-N19-TAnnTTTG) located upstream of putative open reading frames. Plasmids harboring these genomic DNA fragments from F. johnsoniae promoted strong production of fluorescent proteins in Flavobacterium hibernum but not in E. coli. The most potent promoter (PompA) identified in this work was cloned upstream of genes encoding fluorescent proteins, and these were co-expressed in Flavobacterium strains. The p42 and p51 genes (binary toxins from Bacillus sphaericus) when translationally fused to the 3’-end of gfp showed strong expression. Flavobacteria expressing these genes exhibited toxicity against larvae of the mosquitoes Culex quinquefasciatus, Anopheles gambiae, and Ochlerotatus triseriatus. However, transformants with the transcriptional fusion construct between cry11A with p20 from Bacillus thuringiensis did not express Cry11A protein indicating that constitutive expression of cry11A may be problematic in Flavobacterium.
Keywords: expression system, Flavobacterium, larvicidal protein
INTRODUCTION
With the completion of several genome sequences (Duchaud et al., 2007; Xie et al., 2007; Xu et al., 2003), Bacteroidetes genetics is rapidly advancing. Molecular genetic studies of Flavobacterium have been a major part of this advance (Chen et al., 2007a; Nelson & McBride, 2006; Vingadassalom et al., 2005). The entire Flavobacterium psychrophilum genome sequence has been recently published (Duchaud et al., 2007), the F. johnsoniae genome is publicly available (Accession No. CP000685), and the sequencing of the genome of several other Flavobacterium strains is nearly complete. F. johnsoniae has several properties that make it a convenient model for genetic studying in Bacteroidetes: ubiquitous distribution in natural environments, versatile metabolism (Bernardet et al., 1996; Peterson et al., 2006), rapid growth in simple media, the largest genome size among Bacteroidetes so far (Accession No. CP000685), and amenability to genetic manipulation (McBride & Kempf, 1996; McBride et al., 2003; McBride & Braun, 2004). However, genetic tools to study Flavobacterium and related organisms are still underdeveloped. It is now well documented that genes in Bacteroidetes (e.g., antibiotic resistance genes, genes required for plasmid replication, and others) are usually not expressed when transferred into proteobacteria, and that proteobacterial genetic elements do not function well in Bacteroidetes (Alvarez et al., 2004; Chen et al., 2007b; Chen et al., 2007c; McBride & Kempf, 1996). Differences in mechanisms for control of gene expression at either transcription or translation levels to those used by proteobacteria are now evident (Vingadassalom et al., 2005). Bacteroidetes have unique promoter elements with −33/-7 consensus sequences (TTG/TAnnTTTG) separated by spacer of variable length (generally 17–23 nucleotides) (Chen et al., 2007b; Chen et al., 2007c; Vingadassalom et al., 2005). These elements are not homologous to those in the well-studied E. coli σ70 promoters (Eskin E, 2003; Harley & Reynolds, 1987). Most of the progress in understanding Bacteroidetes gene transcription initiation has been achieved by using purified RNA synthesis components in vitro. However, in vivo confirmation of these results has been limited by the lack of versatile genetic systems (Vingadassalom et al., 2005).
Flavobacteria are prominent members of the bacterial community in natural mosquito habitats (Xu et al., 2008). They impact the development of mosquito larvae in tree hole ecosystems and similar container habitats. As such, flavobacteria and related groups might be used as expression vehicles for larvicidal toxin genes. They present a favorable target for toxin expression because they are found in the feeding zones of surface-browsing and filter feeding mosquito larvae, and because they are likely involved in fundamental transformations of allocthonous organic matter that drives these small ecosystems (Kaufman et al., 2001; Xu et al., 2008). Many important disease-vectoring mosquitoes breed in such small discrete aquatic habitats that are close to human populations (Laird, 1988). The development of novel biological insecticide constructs are necessary to improve efficacy of existing toxins, reduce the potential for the development of resistance in insects, and to broaden the range of susceptible mosquito species (Federici et al., 2003; Park et al., 2005). The use of recombinants to express toxins of mosquitocidal bacilli (e.g. Baccilus. thuringiensis and B. sphaericus) is proving valuable in all of these aspects (Federici et al., 2003; Shao et al., 2001). Although ubiquitous in nature and found in soil and aquatic environments (Kirchman, 2002), B. thuringiensis or B. sphaericus, unlike Flavobacteria, do not remain in the feeding zones in the water column or propagate vegetatively in biofilms (Kaufman et al., 2001; Kaufman et al., 2002). Their persistence as biopesticides is also relatively short due to UV inactivation. We believe for these reasons that flavobacteria could be exploited as sustainable toxin expression vehicles for many types of larval mosquito habitats.
In this study, we sought to develop a strong expression system for insecticidal proteins in Flavobacterium. We have isolated several strong promoters from a well characterized strain of F. johnsoniae and demonstrated the potential for differential toxin gene expression using these promoters coupled with autofluorescent protein (AFP) production. These constructs give the simultaneous expression of the toxins and AFPs, thus allowing gene and strain tracking. Moreover, tagging Flavobacterium strains with specific fluorescent markers will allow future studies of how these strains interact with each other, with other bacterial species, and with mosquito larvae in container habitats.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
Plasmids used in this study are listed in Table 1. F. hibernum strain W22 was isolated from a water-filled tree hole in an American beech tree located near the Michigan State University campus. F. johnsoniae UW101 (ATCC17061) was obtained from Dr. Mark McBride of the University of Wisconsin-Milwaukee. Strains of E. coli were routinely grown in Luria Bertani broth (LB) or on LB agar plates at 37°C, and Flavobacterium strains at 26 °C in casitone yeast extract (CYE) medium as previously described (McBride & Kempf, 1996). Liquid cultures were incubated with shaking at 200 rpm. Solid CYE medium contained 20 g of agar per liter. Ampicillin was added (100 µg/ml) for plasmid selection in E. coli and erythromycin (Em) (100 µg/ml) for plasmid selection in Flavobacterium.
TABLE 1.
Strains and plasmids used in this study.
| Strains or plasmids | Relevant characteristics and/or plasmid construction a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 (Nalr) relA1Δ (lacIZYA-argF)U169 deoR(Ф8dlacZΔM15) | Clontech |
| S17-1 | hsdR17 (rK mK) recA RP4-2 (Tcr::Mu-Kmr::Tn7 Strr) | (Simon, 1983) |
| JM109 | F' [traD36 proAB+ lacIqlacZΔM15]/recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi-1 mcrA Δ(lac-proAB) | Promega |
| F. johsoniae UW101 | ATCC 17061 | (McBride & Baker, 1996) |
| F. hibernum W22 | Wild type | This lab |
| Plasmids | ||
| pGEM-T easy vector | Cloning vector, Apr | Promega |
| pKEN2 | Source of gfpmut3; Apr | (Cormack et al., 1996) |
| pDsRed2 | DsRed2 gene; Apr | Clontech |
| pYFP | yfp gene template; Apr | (Shaner et al., 2005) |
| pmOrange | mOrange gene template; Apr | (Shaner et al., 2005) |
| pmStrawberry | mStrawberry gene template; Apr | (Shaner et al., 2005) |
| pSCH143 | E. coli-Flavobacterium shuttle plasmid; Apr (Emr) | (Chen et al., 2007a) |
| pSCH144 | Promoter-trap plasmid, derived from pSCH143; Apr (Emr) | (Chen et al., 2007a) |
| pFj29 | Expression vector carrying with ompA promoter; Apr (Emr) | (Chen et al., 2007a) |
| pMMB736 | Source of cry11A and p20 genes; Apr | (Xu et al., 2001b) |
| p45S1 | Source of p42 and p51 genes; Emr, Apr | (Park et al., 2003) |
| pSCH210 | cry11A expression plasmid; Apr (Emr) | This study |
| pSCH342 | yfp gene expression plasmid; Apr (Emr) | This study |
| pSCH343 | DsRed2 gene expression plasmid; Apr (Emr) | This study |
| pSCH443 | mStrawberry gene expression plasmid; Apr (Emr) | This study |
| pSCH445 | mOrange gene expression plasmid; Apr (Emr) | This study |
| pSCH175 | p20 gene expression plasmid; Apr (Emr) | This study |
| pSCH196 | cry11A+p20 gene expression plasmid; Apr (Emr) | This study |
| pSCH506 | gfp: p42 on T-easy vector; Apr | This study |
| pSCH512 | gfp p42 expression plasmid; Apr (Emr) | This study |
| pSCH529 | gfp p51 on T-easy vector; Apr | This study |
| pSCH532 | gfp p51 expression plasmid; Apr (Emr) | This study |
Apr, ampicillin; Emr, erythromycin. Unless indicated otherwise, antibiotic resistance phenotypes are those expressed in E. coli. Antibiotic resistance phenotypes listed in parentheses are those expressed in Flavobacterium strains but not in E. coli.
Recombinant DNA methods
Genomic DNA extractions were performed with an extraction kit (Promega, Madison, WI) and plasmid DNA was purified with the QIAprep spin miniprep kit (Qiagen, Germantown, MD). DNA ligations, restriction endonuclease digestions, and agarose gel electrophoresis were performed according to standard techniques (Sambrook, 1989). DNA transformation experiments with E. coli were carried out by the calcium chloride method and with Flavobacterium strains by electroporation as described previously (Chen et al., 2007c). PCR amplifications were performed with the Failsafe PCR system (Epicenter technology, Madison, WI). PCR products were separated on 0.7 to 1.0% (wt/vol) agarose gels, and the bands were purified with the QiaQuick gel extraction system (Qiagen). Ligation mixtures were transformed into E. coli JM109 (Promega), and transformants were plated on LB agar plates with ampicillin for selection. Resistant colonies were isolated and screened for the presence of plasmids. The plasmids were then introduced into Flavobacterium strains. To isolate the strong promoters, we used a promoter-trap strategy as previously described (Chen et al., 2007c). Briefly, a F. johnsoniae genomic library was constructed by ligating Sau3AI-digested chromosomal DNA fragments (with limited size between 500–1,500 bps) into the unique BamHI site of the plasmid pSCH144 (Table 1). In order to avoid restriction barriers, pSCH144 was extracted from F. johnsoniae and the above ligation mixture was directly electroporated into F. johnsoniae.
The various AFP reporters were amplified by PCR using the above plasmids as templates according to the standard procedures. The primers are listed in Table 2. These PCR products were first inserted into a T-easy vector individually. Fluorescent Flavobacterium colonies were screened under epifluorescence microscopy. The reporter genes were excised from T-easy vectors (Promega) using BamHI and SphI and inserted into a downstream region under promoter ompA (PompA) in pFj29 (Table 1), replacing gfp. PompA was chosen because it has most potent ability to drive expression of heterologous genes and was further characterized in one of our related studies (Chen et al., 2007a). The resultant plasmids expressing yfp, Dsred2, mStrawberry and mOrange under promoter ompA were designated as pSCH342, pSCH343, pSCH443 and pSCH445, respectively.
TABLE 2.
Primers used in this study
| Primers | Sequence (5’-3’) a |
|---|---|
| DsRed2F | CGCGGATCCTTTAAGAAGGAGATATACATATGGCGTCCGAGAACGTC |
| DsRed2R | GCATGCCGGCCGCTACAGGAACAGGTGGTG |
| mYFPF | CGCGGATCCTTTAAGAAGGAGATATACATATGGCCTCCTCCGAGGACGT |
| mYFPR | GCATGCTTAGGCGCCGGTGGAGTGGCGGCCCTCG |
| mOrangeF | CGCGGATCCTTTAAGAAGGAGATATACATATGGTGAGCAAGGGCGAG |
| mOrangeR | GCATGCCGGCCGCTTTACTTGTACAGCTCGTCCATG |
| mStrawberry | CGCGGATCCTTTAAGAAGGAGATATACATATGGTGAGCAAGGGCGAG |
| mStrawberry | GCATGCCGGCCGCTTTACTTGTACAGCTCGTCCATG |
| P20F | CGCGGATCCAATTAATAATTAAAATTTATGACAGAAAATGGAG |
| P20R | ACATGCATGCGCTTACAGACAAGCTGTGAC |
| Cry11aF | CGCGGATCCTTTAAGAAGGAGATATACATATGGAAGATAGTTCTTTAGATAC |
| Cry11Adetect | CCTCAATAATCCCCCTATATTC |
| Cry11aRBamH | CGCGGATCCCTACTTTAGTAACGGATTAATTTGCG |
| Cry11aRSphI | ACATGCATGCCTTTAGTAACGGATTAATTTGCGTCG |
| Cry11aRsma | TCCCCCGGGCTTTAGTAACGGATTAATTTGCG |
| GFPfusionF | CGCGGATCCTTTAAGAAGGAGATATACATATGAGTAAAGGAGAAG |
| GFPfusionR | TCCCCCGGGTTTGTATAGTTCATCCATGCC |
| P42F | TCCCCCGGGATGAGAAATTTGGATTTTATTGATTC |
| P42R | ACATGCATGCTTAGTGATGGTGATGGTGATGGTTTTGATCATCTGTAATAATCTTTG |
| P50F | TCCCCCGGGATGTGCGATTCAAAAGACAATTCTGGC |
| P50R | ACATGCATGCCTTTATCACTGGTTAATTTTAGGTATTAATTC |
Restriction sites on the primers are underlined.
The p20 gene of B. thuringiensis was amplified from plasmid pMMB736 (Xu et al., 2001a) using primer P20F and P20R with a BamHI site at the 5’-end containing an engineered E. coli ribosome binding site (RBS) (Table 2), and a SphI site at the 3’-end. The PCR fragment was cloned into T-easy vector. The p20 gene fragment was next excised from T-easy vectors using BamHI and SphI and ligated to the expression vector pFj29, digested by the same restriction enzymes. The ligation mixture was electroporated into F. johnsoniae. Transformants with Em resistance were selected and the genetic organization confirmed by sequencing, resulting in the plasmid pSCH175. The F. johnsoniae strain carrying this plasmid was designated SCH175. The gene cry11A of B. thuringiensis was also amplified by PCR from plasmid pMMB736 DNA using primer Cry11aF and Cry11aRBamH (Table 2). The amplification generated a cry11A amplicon with an E. coli RBS. It also added BamHI sites on both ends. This PCR fragment was cloned into the T-easy vector and sequenced. The cry11A gene was next excised from T-easy vectors using BamHI and inserted into the same site in the pSCH175. The transformants with expected insertion were screened using the primers P20F and Cry11detec (Table 2), leading to plasmid pSCH196.
The translational fusion constructs were built as follows. The gfp reporter was amplified using forward primer GFPfusionF, containing a BamHI restriction enzyme site and an engineered E. coli RBS, and the reverse primer GFPfusionR contained a SmaI site and no stop codon. To construct a translational fusion between the reporter gfp and the p42 gene from plasmid p45S1 (Park et al., 2003), the latter was amplified using forward primer P42F with SmaI site and reverse primer P42R with SphI. The two fragments were digested by SmaI and then ligated. The ligation mixture was diluted and used as template to create an in-frame fusion by PCR using primers GFPfusionF and GFPfusionR. The gfp::p42 gene fragment was first cloned into the T-easy vector, leading to plasmid pSCH506. The translational fusion was released from vector pSCH506 by restriction enzymes BamHI and SphI and then inserted at the same sites on plasmid pFj29, resulting in plasmid pSCH512. Vector pSCH512 was introduced into F. johnsoniae by conjugation and a transconjugant with green fluorescence was selected and named strain SCH512. The gfp::p51 translational fusion was constructed using the same strategy as for gfp::p42 and cloned into T-easy vector, leading to plasmid pSCH529. The insert was released from pSCH529 by BamHI and SphI, and inserted into the same sites on plasmid pFj29. The ligation mixture was eletroporated into F. johnsoniae directly. A transformant with green fluorescence was selected and designated as strain SCH532.
Epifluorescence microscopy
Cells tagged with AFPs were visualized with an Olympus Provis AX70 microscope, with appropriate filters, mercury lamp, and DP-50 digital camera linked to an external PC. Cell suspensions were prepared as follows. Cells were grown overnight in CYE broth, pelleted by centrifugation and re-suspended in 3% ρ-formaldehyde (PFA) for 20 min. Cells in PFA were then centrifuged again and the pellets resuspended in sterile distilled water. These suspensions (10–20 µl) were placed onto glass slides, covered with coverslips (22 × 22 mm), blotted to remove excess liquid and the edges of the coverslip were sealed with clear nail polish.
Determination of AFP reporter activity
Cells harboring sequence-confirmed constructs were cultured in CYE media and quantitative analysis of AFP production was performed using a SpectraMax M5 spectrophotometer (Molecular Devices, Sunnyvale, CA). Aliquots (200 µl) of cultures were centrifuged, washed with 0.1 M phosphate buffered saline (PBS, pH 7.4), diluted in the same buffer to an OD600 of ~0.4 and subjected to fluorescence determination in a 96-well plate (Costar, Corning, NY). GFP fluorescence was determined at excitation wavelength 490 nm, emission wavelength 530 nm, cutoff 515 nm; YFP fluorescence at excitation wavelength 495 nm, emission wavelength 530 nm, cutoff 515 nm; mStrawberry fluorescence at excitation wavelength 560 nm, emission wavelength 600 nm, cutoff 590 nm; and mOrange fluorescence at excitation wavelength 540 nm, emission wavelength 580 nm, cutoff 570 nm. Cultures of untransformed strains were used as the blanks for calculation of the relative units of fluorescence. The growth of each constructed strain was monitored in triplicate over a 48-hr period by determination of optical density at 600 nm and by fluorescence.
Protein extraction and Western blotting
Cells in the log-phase of growth were harvested by centrifugation. Tris buffer (50 mM Tris-Hcl, 10 mM dithiothreitol, pH 6.8) was added, and the suspension was sonicated (three bursts of 15 s each on a Branson sonifier at 60% output). The cell contents were dissolved using Tris buffer, 8 M urea, and 1% SDS with 1X sample loading buffer, boiled for 10 min and subjected to a Western blot analysis as described before (Xu et al., 2001a). Briefly, proteins were separated on 12% SDS-PAGE, transferred onto PVDF-membrane and reacted with the anti-Cry11A or anti-P20 antiserum generated by immunization of rabbits (Xu et al., 2001a) or with anti-GFP antiserum (BD Biosciences, San Jose, CA). Peroxidase-conjugated goat anti-rabbit immunoglobulin G was used as secondary antibody, and the reaction was developed with Super Signal (Pierce, Rockford, Ill.).
DNA sequence analysis
Each construct was sequenced by the dideoxy termination method using an automated sequencing system (Applied Biosystems, Foster City, CA). GenBank database searches were carried out using the National Center for Biotechnology Information BLAST web server (http://www.ncbi.nlm.nih.gov/BLAST). Multiple sequence alignments were carried out with the ClustalW program (http://www.ebi.ac.uk/clustalw/) and later adjusted manually. Genome-scale DNA pattern searches were performed using PatScan (http://www-unix.mcs.anl.gov/compbio/PatScan/) (Dsouza, 1997) and regulatory sequence analysis tools (RSAT) (http://rsat.scmbb.ulb.ac.be/rsat/) (van Helden, 2003).
Bioassays
Bioassays were conducted on late instar larvae of three mosquito species, Ochlerotatus triseriatus (MSU lab strain), Anopheles gambiae (Kisumu strain) and Culex quinquefasciatus (obtained from Benzon Research, Pennsylvania, benzonresearch.com). Larvae were held in culture tubes in groups of 4 with 2 ml of distilled water for at least 2 h before addition of cells. Flavobacterium strains SCH512 (gfp::p42) and SCH532 (gfp::p51) were cultured to the log-phase and the cell densities were estimated by measuring OD600 and viable cell count (plating on CYE agar plates). The cells were then mixed at the ratio 1:1. Aliquots of the mixed cells were washed and serially diluted in phosphate buffer (pH 7.2) to give a range of cells added to the larval bioassay of 4 × 102 to 4 × 107 per assay tube. An identical series was prepared using the strain carrying pFj29 and was added to larval bioassay tubes as controls. Each mosquito species times cell density combination was replicated 6 times. Bioassay tubes were incubated at 26 deg. C in the dark. Mortality was assessed after 24 hours.
RESULTS
Screening of strong promoters by promoter-trap
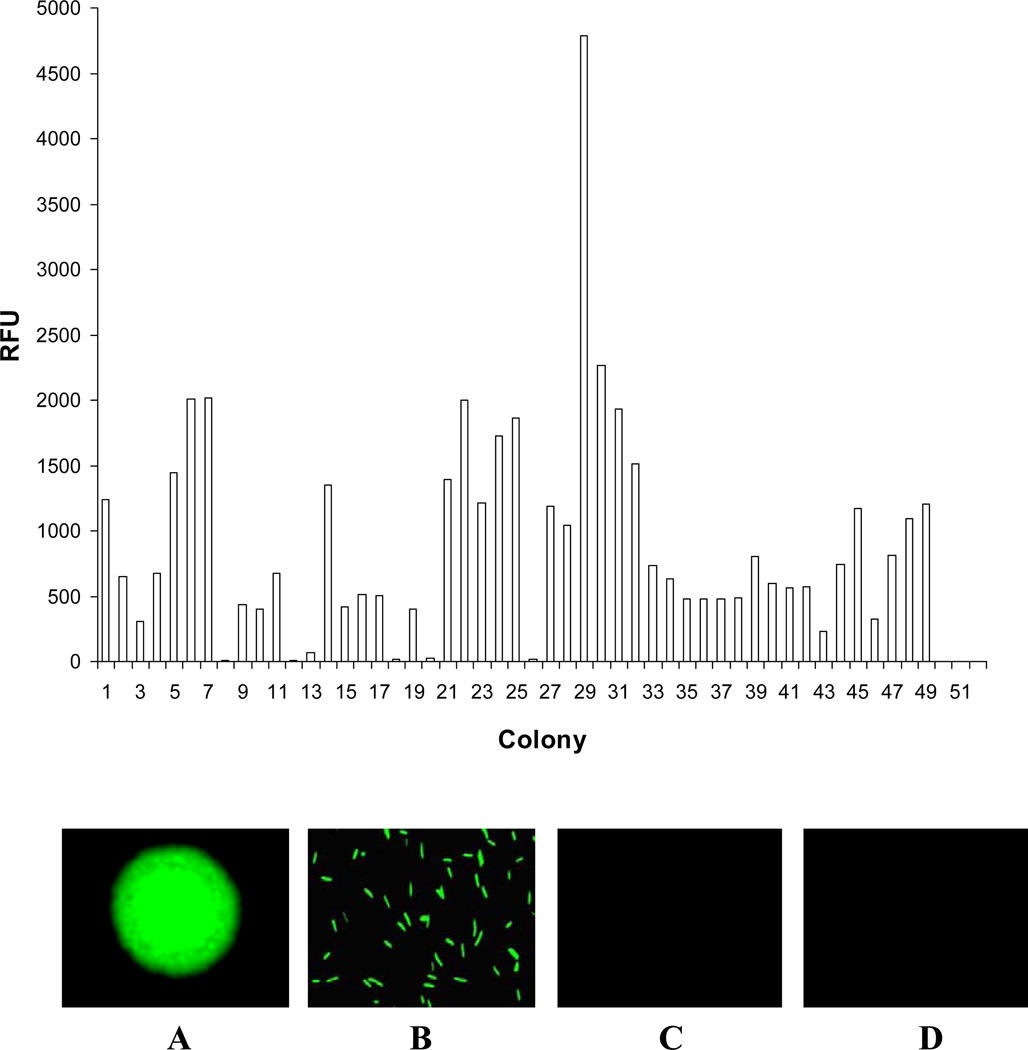
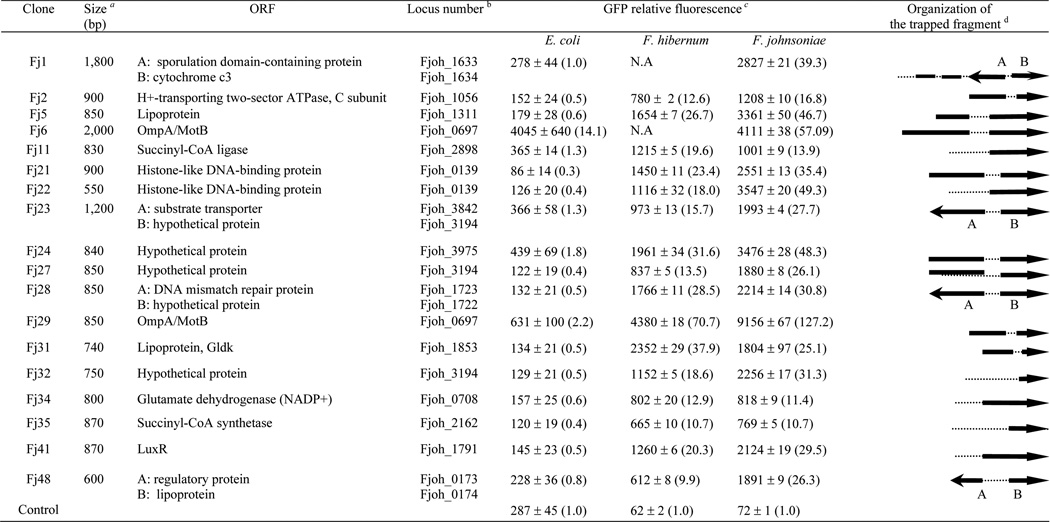
Our initial promoter trapping efforts yielded 49 colonies with strong GFP fluorescence out of at least 9,000 colonies (Fig. 1) and 18 isolates with uniform colony morphology and with the highest levels of gfp expression were chosen for further study (Fig. 2). Nucleotide sequence determination and restriction enzyme analysis indicated that the chromosomal inserts ranged from 550 to 2,200 bp (Fig. 2). The nucleotide and predicted amino acid sequences of these inserts were used to search the GenBank databases, and putative identification of the inserts was made based on homology data (Fig. 2). Most of the inserts had annotated ORFs and showed a high degree of similarity (ranging from 32% to 89%) with coding sequences from various bacterial genes. Their functions are generally well established in bacteria, and several are either very important or essential to cellular viability (Fig. 2).
FIG. 1. Comparison of GFP fluorescence emitted by the selected F. johnsoniae transformants.
Upper panel: Cultures were incubated overnight and quantified with fluorescence spectrometer as described in Materials and Methods. Lower panel: demonstration of GFP production in F. johnsoniae. A: Fj29 colony; B: Fj29 cells; C: SCH144 colony; D: SCH144 cells.
FIG. 2. Description of clones containing putative promoter regions and comparisons of the relative promoter activities in F. johnsoniae F. hibernum and E. coli.
a The insert size was determined by restriction digestion analysis with KpnI and BamHI.
b ORFs were identified with Gene Finder. The locus number for ORFs was given based on F. johnsoniae genome sequence (Accession No. CP000685).
c Strains carrying pSCH144 (promoterless gfp) were used as the negative control. The relative fluorescence values were determined as described in the Methods and Materials. The promoter activity was normalized to that of the promoter clones carrying pSCH144 (defined as 1.0), as shown in parentheses. Triplicate samples were used, and the standard deviations are shown. NA, not applicable.
d The solid arrows represent the direction of putative ORFs with the putative promoter(s), the single lines represent coding regions without promoter(s), and the dashed lines represent noncoding fragments. DNA inserts are not drawn to scale relative to each other.
Comparisons of promoter activity in F. johnsoniae, F. hibernum and E. coli
Application of the conserved promoter motifs to predict the location of other related promoter sequences has been shown to be successful in the F. johnsoniae genome (Chen et al., 2007a). For further examination of trapped F. johnsoniae genomic fragments, a string pattern search was conducted using the same strategy. 13 hits (among the 18 sequenced trapped fragment) were identified to carry a putative conserved −7 promoter sequences (TAnnTTTG), including two well characterized ones (Fj29 and Fj31) (Fig. 2 and Fig. 3). The putative −33 region (TTG) were also found upstream of the −7 region with a spacer ranging from 18 to 22 bp. The average GC content in these putative promoter sequences is ~25%. They are located upstream within 300 bp of the ORFs. Since these fragments have the ability to drive high expression of reporter genes, most of them likely function as authentic promoters. Plasmids harboring strong promoters of F. johnsoniae were extracted and transformed into E. coli and F. hibernum W22 in order to test the relative strength of each promoter in these strains. SCH144 (promoterless gfp) was used as negative controls. Strong expression, ranging from 9.9- to 70.7-fold and 10.7- to 127.2-fold levels of the control, was observed in F. hibernum strain W22 and F. johnsoniae respectively. In general, promoters of F. johnsoniae exhibited lower expression in F. hibernum to some extent (Fig. 2). In contrast, most of the above isolated promoters (except PompA on pFj6 and pFj29) functioned poorly in E. coli as shown in Fig. 2 although copy numbers of the plasmid in E. coli are more than 10-fold higher than in Flavobacterium strains (Chen et al., 2007c). Transformants could not be obtained in F. hibernum when pFj1 or pFj6 were introduced by electroporation or conjugational transfer, indicating that these gene fragments had toxic effects when they were over-expressed in F. hibernum.
FIG. 3. Alignment of putative promoters functional in Flavobacterium.
A string scan was performed in the trapped gene fragments using the conserved −7 promoter motif (TAnnTTTG). The consensus sequences derived from this alignment is given at the bottom. It was defined as nucleotides that are present at any given position in more than 50% of the sequences. The −7 and −33 consensus regions are capitalized and boldface.
Using various AFP reporters to tag Flavobacterium strains
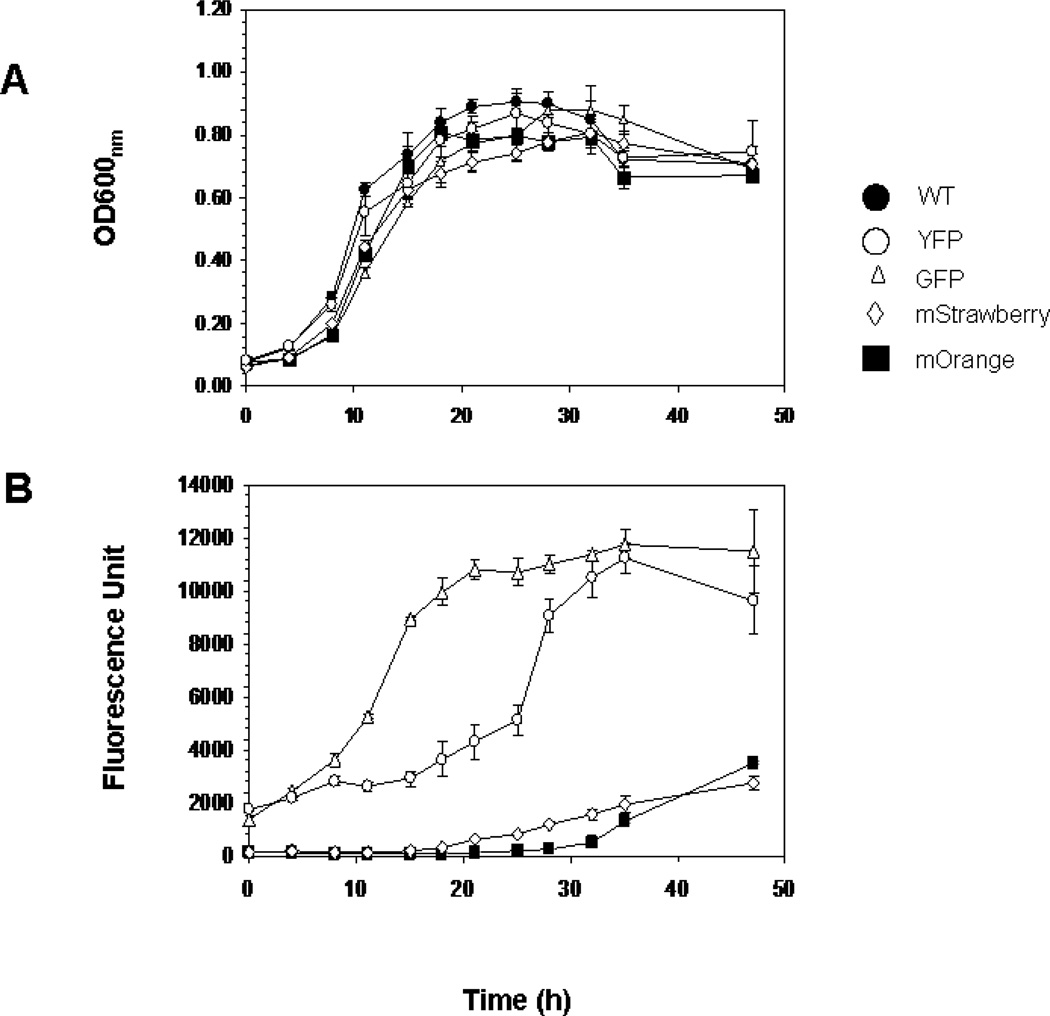
Different fluorescent protein genes encoding DsRed2, mStrawberry, mOrange and YFP were inserted downstream of the most potent promoter, PompA, individually and introduced them into Flavobacterium strains. Stable fluorescent transformants were successfully obtained with all of above constructs except DsRed2. No difference in the growth rate between the cells expressing fluorescent proteins and those of the wild type F. johnsoniae were detected (Fig. 4). GFP fluorescence increased with the cell growth and reached peak level when cells started to enter the stationary phase (~20 h). However, the maximum fluorescence for YFP, mOrange and mStrawberry was recorded at 35, 48 and 48 hr under our testing conditions, respectively (Fig. 4). The highest GFP production was observed at 26 °C and pH 6.0, and plasmids containing fluorescent protein genes were stable in F. johnsoniae for at least 60 generations without the antibiotic selection (data not shown). To demonstrate feasibility of fluorescent reporters for studying bacteria-mosquito larvae interactions, we fed mosquito larvae with the F. johnsoniae tagged with different reporters and observed them with epifluorescence microscopy. The cells were ingested rapidly (ca 5 min.) and quickly filled the whole larvae including the midgut where the bacteria were usually digested as food source (Fig. 5).
FIG. 4. Growth and fluorescence production characteristics of the AFP reporter strains in CYE medium compared to that of the wild type.
(A) Strains Fj29 (GFP), SCH342 (YFP), SCH443 (mStrawberry), SCH445 (mOrange) and WT were adjusted to the same cell density and cultured for 48 hrs. (B) Fluorescence determination for AFP reporter strains and WT as described in the Methods and Material. Values in Panels A and B are means of data from triplicate cultures (with standard deviation).
FIG. 5. Demonstration of the Flavobacterium cells tagged with AFP reporters and their ingestion by mosquito larvae.
Strains carrying plasmid pSCH144, pFj29, pSCH342, pSCH443 were cultured to the log phase, washed with PBS and then fed to mosquito larvae. The larvae samples were observed under the epifluorescent microscopy as described in the Methods and Materials after feeding for 5 mins.
Heterologous expressin of larvicidal protein genes in F. johnsoniae
To express cry11A, p20 or the cluster cry11A+p20 in F. johnsoniae we have inserted these genes into the plasmid pFj29 and introduced recombinant plasmids (pSCH210, pSCH175 and pSCH196, Table 1) into the strain F. johnsoniae UW101. The single transformants containing cry11A or p20 were recovered. However, no stable transformants bearing the cluster cry11A+p20 were obtained in F. johnsoniae. Western-blot with polyclonal anti-P20 antibodies confirmed that the P20 helper protein was produced in abundance in F. johnsoniae (Fig. 6). The synthesis of Cry11A, however, was not detectable (Fig. 6).
FIG. 6. Western blot assay of the recombinant Flavobacterium strains expressing the larvicidal genes.
(A). lane 1, proteins were extracted from F. johnsoniae carrying pSCH175 (p20); lane 2, proteins were extracted from F. johnsoniae carrying pSCH143 (negative control); anti-P20 antiserum was added. (B). lane1, proteins were extracted from E. coli strain carrying pMMB736 (positive control); lane2, proteins were extracted from F. johnsoniae carrying pSCH210 (cry11A); anti-Cry11A was added. (C). lane 1, proteins were extracted from F. johnsoniae carrying plasmid pSCH144 (negative control); lane2, proteins were extracted from F. johnsoniae carrying plasmid pSCH512 (gfp::P42); lane 3, proteins were extracted from F. johnsoniae carrying plasmid pSCH532 (gfp::p51); lane 4. proteins were extracted from F. johnsoniae carrying pFj29; anti-GFP antiserum was used.
We next constructed translational fusions between gfp and genes p42 and p51 of B. sphaericus, encoding the binary mosquito larvicidal proteins (42 and 51 kDa, respectively). Introduction of pSCH512 (gfp::p42) or pSCH532 (gfp::p51) into F. johnsoniae resulted into Em-resistant fluorescent F. johnsoniae colonies. Western blot analysis using the GFP antisera (Fig. 6) showed a dominant band with a molecular size of 70kDa, which is close to the theoretical values for GFP::P42 produced in the SCH512. However, three products were observed in strain SCH532 with molecular sizes of 80, 60, and 32 kDa, respectively, indicating that GFP::P51 protein was possibly processed at multiple sites. A single band with a molecular size of 27 kDa was present in control Fj29 and no bands were detected in negative control (Fig. 6).
Toxicity of recombinant p42 and p51 for mosquito larvae
We mixed strains with pSCH512 (gfp::p42) and pSCH532 (gfp::p51) in 1:1 ratios to test potential utilization of recombinant Flavobacterium strains as insecticides because both of P42 and P51 are required to obtain larvicidal function (Broadwell et al., 1990). The combination of SCH512 and SCH532 was toxic in a dose-dependent manner against the three mosquito species using whole cell assay methods (Fig. 7). Cx. quinquefasciatus was most susceptible, followed by An. gambiae and then Oc. triseriatus. Control survival was effectively 100%.
FIG. 7. Bioassay results of recombinant Flavobacterium with Bs constructs against larvae of 3 mosquito species.
Strain Fj29 was used as control. The mixed Flavobacterium strains SCH512 (gfp::p42) and SCH532 (gfp::p51) were prepared as described in Methods and Materials and fed to Oc. triseriatus An. gambiae, and Culex quinquefasciatus Oc. triseriatus (MSU strain) and An. gambiae (Kisumu strain). After 24 h, the results were recorded. Values are mean + one SE, n = 6.
DISCUSSION
In a previous study, we isolated several promoters with unique Bacteroidetes genetic elements from F. hibernum using a promoter-trap strategy (Chen et al., 2007c). However, F. hibernum strain W22 is a strain recently isolated from the environment. It is not well characterized and, at present, can not be considered a good representative of its genus. In the present study, several strong promoters were isolated from F. johnsoniae using a new promoter-probe vector (pSCH144) based on a green fluorescent protein system. These promoters also functioned well in F. hibernum strain W22 (Fig. 2) and exhibited sequences resembling the unique promoter motifs (Fig. 3), −33 region with TTG and −7 region with TAnnTTG, separating by 18–22 bp, dominant in other members of Bacteroidetes. Our results also indicated that the −7 region is more conserved than the −33 region (Fig. 3). The strongest promoter isolated in this work, PompA, was extensively dissected by mutational analysis (Chen et al., 2007a). The results showed that the −7 motif (TAnnTTTG) is more important than the −33 motif (TTG) in maximizing promoter activity. The space length and GC content are also critical. Collectively, results obtained by our research group and by others confirmed that transcriptional start signals in Flavobacterium are distinct from those in proteobacteria (Chen et al., 2007c).
Our research goals are to understand microbe-insect and microbe-microbe interactions that affect the production of mosquitoes in natural environments and to exploit these interactions as mosquito control strategies. Flavobacteria and related species are prominent in the water column and leaf surfaces in tree holes, serving as food sources for mosquito larvae (Xu et al., 2008). From this perspective, a Flavobacterium strain genetically modified to express and deliver (via ingestion in the larval mosquito alimentary canal) B. thuringiensis or B. sphaericus insecticidal proteins would fulfill these criteria. Toward this end, the efficient expression system developed here using the strong promoter, PompA, may provide a valuable tool.
Under the control of the ompA promoter, the expression level of various fluorescent protein genes, including gfp, yfp, mOrange and mStrawberry, was high enough for easy visualization in bacteria cultures and in larval guts. However, the construct for DsRed2 failed in F. johnsoniae. This result suggests that high amounts of the DsRed2 protein were toxic to F. johnsoniae. The failure to express DsRed2 has been seen in constructs of other bacteria and one of the possible reasons can be improper folding of the expressed products (Shaner et al., 2004). The construction of multiple fluorescent derivatives for the sequenced strain of F. johnsoniae will allow a wide variety of studies involving in situ detection of bacterial cells within larvae or the larval habitat. Plasmids constructed in this study also allowed to us to label cells with different fluorescent proteins having distinct excitation wavelengths, e.g., GFP vs mOrange. Furthermore, the introduced plasmids in F. johnsoniae were found to be stable without antibiotic selection, indicating that their applications for further insecticide development are promising. This will be invaluable when studying bacteria-bacteria interactions (such as the relative competiveness and fitness of different flavobacteria strains) in larval mosquito habitats. We believe this AFP expression system is likely to be functional in many other Flavobacterium strains because regulatory regions of promoter ompA are conserved among Flavobacterium strains (Chen et al., 2007a). Because some Flavobacterium species are pathogenic for fish (Decostere et al., 1999; Madetoja et al., 2003; Nematollahi et al., 2003), one important use of the described system could be construction of strains that may be used in studying pathogenic mechanisms employed by Flavobacterium or related strains. For instance, heterologous production of F. psychrophilum proteins in E. coli proved difficult because of codon usage bias (Devendra H. S. et al., 2008). Since F. psychrophilum and F. johnsoniae have similar genetic backgrounds (Alvarez et al., 2004; Duchaud et al., 2007), using F. johnsoniae expression system can be an alternate way to remedy challenges associated with expression and production of F. psychrophilum recombinant proteins. A direct application of this expression system has been recently conducted for fish pathogen Flavobacterium columnare by Staroscik et al. (Staroscik et al., 2008)
Our preliminary efforts to express genes encoding larvicidal proteins from B. thuringiensis or B. sphaericus met with variable results. To our knowledge, this is first study to develop an efficient expression system for Flavobacterium as a host for recombinant insecticides; results from B. sphaericus binary toxins were very favorable and conformed with predictions regarding the range of responses in the three mosquito species tested here. By contrast, the protein Cry11A was not produced in detectable quantities in F. johnsoniae, but the helper protein P20 was readily produced (Fig. 6). Since it has previously been documented that Cry11A production is dependent on its helper protein P20 (Xu et al., 2001a), it is possible that our failure to obtain transformants containing plasmids with the cry11A + p20 gene cluster was the result of Cry11A toxicity to F. johnsoniae. Cry11A has some antibacterial properties, most notably against Micrococcus spp. but not against Flavobacterium spp. (Yudina et al., 2003). The lack of stable Cry11A expression was initially surprising given the variety of gram-negative bacteria capable of Bacillus thuringiensis toxin expression (Liu et al., 1996), but this result likely re-emphasizes fundamental differences between Bacteroidetes and Proteobacteria. We are continuing to investigate whether or not constitutive expression of endotoxins from Bacillus thuringiensis in Flavobacterium is a viable approach in developing alternative and more potent mosquito larvicides.
Genes encoding the B. sphaericus binary toxins, p42 and p51 were readily expressed in F. johnsoniae and this provided an effective way to deliver the toxins to mosquito larvae. The dose required for 100% larval mortality of the susceptible Culex (ca 5.6 log10 cells/ml) was comparable to previous B. sphaericus studies (Promdonkoy et al., 2003), indicating that our recombinant constructs were quite potent as the initial study. As expected, the toxin doses from our constructs needed for potential control of An. gambiae and Oc. triseriatus were much higher than those required for Culex, as these species are less susceptible to toxins from B. sphaericus (Brown et al., 2004; Silva-Filha et al., 1997).
ACKNOWLEDGEMENTS
The authors thank Dr. Mark McBride (University of Wisconsin-Milwaukee) for generously providing E. coli-Flavobacterium shuttle plasmids and several bacteria strains. We gratefully acknowledge Dr. Brian Federici (University of California-Riverside) for plasmid p45S1 and Dr. Roger Tsien (University of California-San Diego) for providing AFP reporters. We also thank William Morgan and Blair Bullard for their assistance. This project was funded by NIH grant AI21884.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alvarez B, Secades P, McBride MJ, Guijarro JA. Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum . Appl Environ Microbiol. 2004;70:581–587. doi: 10.1128/AEM.70.1.581-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardet JF, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Cutting a gordian knot: Emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom nov (basonym, Cytophaga aquatilis Strohl and Tait 1978) IntJ Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- Broadwell A, Baumann L, Baumann P. Larvicidal properties of the 42 and 51 kilodaltonBacillus sphaericus proteins expressed in different bacterial hosts: Evidence for a binary toxin. Current Microbiology. 1990;21:361–366. [Google Scholar]
- Brown MD, Watson TM, Carter J, Purdie DM, Kay BH. Toxicity of VectoLex (Bacillus sphaericus) products to selected Australian mosquito and nontarget species. J Econ Entomol. 2004;97:51–58. doi: 10.1093/jee/97.1.51. [DOI] [PubMed] [Google Scholar]
- Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae . J Bacteriol. 2007a;189:5108–5118. doi: 10.1128/JB.00401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bagdasarian M, Kaufman MG, Walker ED. Organization of a partial S10 operon and its transcriptional analysis in Flavobacterium hibernum strain W22. FEMS Microbiol Lett. 2007b;267:38–45. doi: 10.1111/j.1574-6968.2006.00543.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Bagdasarian M, Kaufman MG, Walker ED. Characterization of strong promoters from an environmental Flavobacterium hibernum etrain using a green fluorescent protein-based reporter system. Appl Environ Microbiol. 2007c;73:1089–1100. doi: 10.1128/AEM.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Decostere A, Haesebrouck F, Charlier G, Ducatelle R. The association of Flavobacterium columnare strains of high and low virulence with gill tissue of black mollies (Poecilia sphenops) Vet Microbiol. 1999;67:287–298. doi: 10.1016/s0378-1135(99)00050-4. [DOI] [PubMed] [Google Scholar]
- Devendra HS, Cain KD, Wiens GD, Cal DR. Challenges associated with heterologous expression of Flavobacterium psychrophilum proteins in Escherichia coli . Mar Biotechnol. 2008;10:719–730. doi: 10.1007/s10126-008-9111-z. [DOI] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Duchaud E, Boussaha M, Loux V other authors. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum . Nat Biotechnol. 2007;25:763–769. doi: 10.1038/nbt1313. [DOI] [PubMed] [Google Scholar]
- Eskin EKU, Gelfand MS, Pevzner PA. Genome-wide analysis of bacterial promoter regions. Pac Symp Biocomput. 2003;8:29–40. [PubMed] [Google Scholar]
- Federici BA, Park HW, Bideshi DK, Wirth MC, Johnson JJ. Recombinant bacteria for mosquito control. J Exp Biol. 2003;206:3877–3885. doi: 10.1242/jeb.00643. [DOI] [PubMed] [Google Scholar]
- Harley CB, Reynolds RP. Analysis of E. coli Pormoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman MG, Bland SN, Worthen ME, Walker ED, Klug MJ. Bacterial and fungal biomass responses to feeding by larval Aedes triseriatus (Diptera : Culicidae) J Med Entomol. 2001;38:711–719. doi: 10.1603/0022-2585-38.5.711. [DOI] [PubMed] [Google Scholar]
- Kaufman MG, Goodfriend W, Kohler-Garrigan A, Walker ED, Klug MJ. Soluble nutrient effects on microbial communities and mosquito production in Ochlerotatus triseriatus habitats. Aquat Microb Ecol. 2002;29:73–88. [Google Scholar]
- Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- Laird M. The natural history of larval mosquito habitats. London: Academic Press; 1988. [Google Scholar]
- Liu J-W, Yap WH, Thanabalu T, Porter AG. Efficient synthesis of mosquitocidal toxins in Asticcacaulis excentricus demonstrates potential of Gram-negative bacteria in mosquito control. Nat Biotech. 1996;14:343–347. doi: 10.1038/nbt0396-343. [DOI] [PubMed] [Google Scholar]
- Madetoja J, Nystedt S, Wiklund T. Survival and virulence of Flavobacterium psychrophilum in water microcosms. FEMS Microbiol Ecol. 2003;43:217–223. doi: 10.1111/j.1574-6941.2003.tb01061.x. [DOI] [PubMed] [Google Scholar]
- McBride M, Baker S. Development of techniques to genetically manipulate members of the genera Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga. Appl Environ Microbiol. 1996;62:3017–3022. doi: 10.1128/aem.62.8.3017-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Kempf MJ. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae . J Bacteriol. 1996;178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Braun TF, Brust JL. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J Bacteriol. 2003;185:6648–6657. doi: 10.1128/JB.185.22.6648-6657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Braun TF. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J Bacteriol. 2004;186:2295–2302. doi: 10.1128/JB.186.8.2295-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SS, McBride MJ. Mutations in Flavobacterium johnsoniae secDF result in defects in gliding motility and chitin utilization. J Bacteriol. 2006;188:348–351. doi: 10.1128/JB.188.1.348-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nematollahi A, Decostere A, Pasmans F, Haesebrouck F. Flavobacterium psychrophilum infections in salmonid fish. J Fish Dise. 2003;26:563–574. doi: 10.1046/j.1365-2761.2003.00488.x. [DOI] [PubMed] [Google Scholar]
- Park H-W, Bideshi DK, Federici BA. Recombinant strain of Bacillus thuringiensis producing Cyt1A, Cry11B, and the Bacillus sphaericus binary toxin. Appl Environ Microbiol. 2003;69:1331–1334. doi: 10.1128/AEM.69.2.1331-1334.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Shin SC, Kim CS, Lee HJ, Choi WS, Ahn YJ. Larvicidal activity of lignans identified in Phryma leptostachya Var. asiatica roots against three mosquito species. J Agric Food Chem. 2005;53:969–972. doi: 10.1021/jf048208h. [DOI] [PubMed] [Google Scholar]
- Peterson SB, Dunn AK, Klimowicz AK, Handelsman J. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl Environ Microbiol. 2006;72:5421–5427. doi: 10.1128/AEM.02928-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promdonkoy B, Promdonkoy P, Audtho M, Tanapongpipat S, Chewawiwat N, Luxananil P, Panyim S. Efficient expression of the mosquito larvicidal binary toxin gene from Bacillus sphaericus in Escherichia coli . CurrMicrobiol. 2003;47:383–387. doi: 10.1007/s00284-003-4035-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotech. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Meth. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shao Z, Liu Z, Yu Z. Effects of the 20-Kilodalton helper protein on Cry1Ac production and spore formation in Bacillus thuringiensis . Appl Environ Microbiol. 2001;67:5362–5369. doi: 10.1128/AEM.67.12.5362-5369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Filha MH, Nielsen-Leroux C, Charles J-F ois. Binding kinetics of Bacillus sphaericus binary toxin to midgut brush-border membranes of Anopheles and Culex sp. mosquito larvae. Eur J Biochem. 1997;247:754–761. doi: 10.1111/j.1432-1033.1997.00754.x. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Staroscik A, Hunnicutt D, Archibald K, Nelson D. Development of methods for the genetic manipulation of Flavobacterium columnare . BMC Microbiol. 2008;8:115. doi: 10.1186/1471-2180-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. An unusual primary sigma factor in the Bacteroidetes phylum. Mol Microbiol. 2005;56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- Xie G, Bruce DC, Challacombe JF& other authors. Genome sequence of the cellulolytic gliding bacterium. Cytophaga hutchinsonii Appl Environ Microbiol. 2007;73:3536–3546. doi: 10.1128/AEM.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Xu Y, Nagai M, Bagdasarian M, Smith TW, Walker ED. Expression of the p20 Gene from Bacillus thuringiensis H-14 Increases Cry11A Toxin Production and Enhances Mosquito-Larvicidal Activity in Recombinant Gram-Negative Bacteria. Appl Environ Microbiol. 2001a;67:3010–3015. doi: 10.1128/AEM.67.7.3010-3015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Nagai M, Bagdasarian M, Smith TW, Walker ED. Expression of the p20 gene from Bacillus thuringiensis H-14 increases Cry11A Toxin production and enhances mosquito larvicidal activity in recombinant Gram-negative Bacteria. Appl Environ Microbiol. 2001b;67:3010–3015. doi: 10.1128/AEM.67.7.3010-3015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chen S, Kaufman MG, Maknojia S, Bagdasarian M, Walker ED. Bacterial Community Structure in Tree Hole Habitats of Ochlerotatus triseriatus: Influences of Larval Feeding. Journal of the American Mosquito Control Association. 2008;24:219–227. doi: 10.2987/5666.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudina TG, Konukhova AV, Revina LP, Kostina LI, Zalunin IA, Chestukhina GG. Antibacterial activity of Cry- and Cyt-proteins from Bacillus thuringiensis ssp. israelensis . Can J Microbiol. 2003;49:37–44. doi: 10.1139/w03-007. [DOI] [PubMed] [Google Scholar]