Abstract
The multiple-turnover ability of a series of locked nucleic acid (LNA)-based antisense oligonucleotides (AONs) in the RNase H-mediated scission reaction was estimated using a newly developed cell-free reaction system. We determined the initial reaction rates of AONs under multiple-turnover conditions and found that among 24 AONs tested, AONs with melting temperatures (Tm) of 40°C–60°C efficiently elicit multiple rounds of RNA scission. On the other hand, by measuring Tm with two 10-mer RNAs partially complementary to AONs as models of cleaved 5′ and 3′ fragments of mRNA, we found that AONs require adequate binding affinity for efficient turnover activities. We further demonstrated that the efficacy of a set of 13-mer AONs in mice correlated with their turnover efficiency, indicating that the intracellular situation where AONs function is similar to multiple-turnover conditions. Our methodology and findings may provide an opportunity to shed light on a previously unknown antisense mechanism, leading to further improvement of the activity and safety profiles of AONs.
Introduction
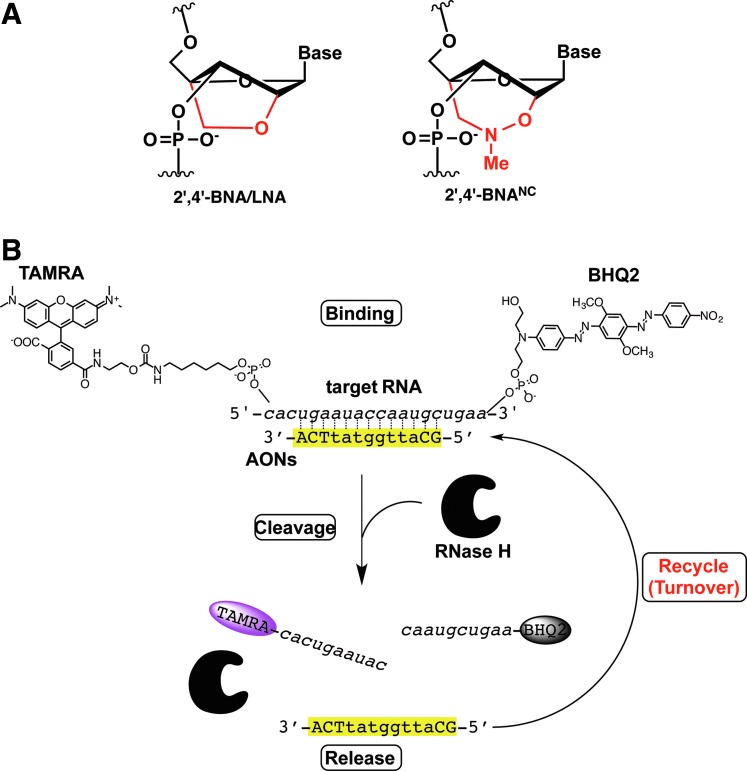
Antisense oligonucleotides (AONs) having specific configurations compatible with RNase H-inducible capacity have been developed over decades and have been shown to be very powerful and robust gene silencing materials in cultured cells and animals, as well as in humans (Crooke, 2007; Yamamoto et al., 2011). In particular, the “gapmer” configuration, which is a chimeric AON consisting of a central RNase H-recruitable DNA stretch pinched by affinity-enhancing modified nucleic acids with fully phosphorothioated (PS) internucleotide linkages, has shown great promise. Affinity-enhancing modified nucleic acids, such as MOE (2′-O-methoxyethyl RNA), 2′,4′-BNA/LNA (2′-O,4′-C-methylene bridged nucleic acid/locked nucleic acid) (Fig. 1A) (Obika et al., 1997; Obika et al., 1998; Singh et al., 1998), and other bridged nucleic acids (BNAs) (Hari et al., 2006; Miyashita et al., 2007; Seth et al., 2009; Prakash et al., 2010; Yahara et al., 2012) mostly interfere with RNase H activity, but when used in a chimeric gapmer, they assist in enhancing target binding and nuclease stability, greatly improving its potency without affecting RNase H capacity. Despite these innovations, very few products have been released on the market and some candidates in clinical trials have been dropped due to efficacy and safety issues.
FIG. 1.
Recycling of antisense oligonucleotides in antisense reaction. (A) Structures of bridged nucleic acids (BNAs). (B) Schematic illustration of cell-free Förster resonance energy transfer (FRET)-based turnover monitoring system used in this study. Color images available online at www.liebertpub.com/nat
More recently, Straarup et al. successfully improved efficacy of an earlier LNA-based gapmer targeting apolipoprotein B-100 (apoB) by trimming its conventional long-strand [16∼20 nucleotides (nt)] and utilizing the resulting shorter LNA gapmers (∼13 nt) (Straarup et al., 2010). Our group also independently reproduced and extended this observation by using newly developed 2′,4′-BNANC chemistry and supported the unusual notion that a drug with weaker binding has stronger silencing activity (Fig. 1A) (Yamamoto et al., 2012b). One possible explanation for this finding is that shorter AONs accelerate the reaction to a greater degree than conventional AONs via turnover mechanisms. Stanton et al. recently observed that melting temperatures (Tm) of greater than 80°C showed reduced silencing activity and explained this finding as a result of an inability to recycle AONs in cells (Stanton et al., 2012). However, to the best of our knowledge, there is no experimental evidence of AON turnover, despite its anticipated importance as in RNAi mechanisms (Hutvagner and Zamore, 2002). If turnover of AONs was demonstrated in antisense mechanisms, a more favorable configuration or chemistry that accelerates AON turnover may be discovered, and this discovery could lead to additional insights into strategies for further improving the activity and safety of AONs. Most previous works related to antisense reaction kinetics have been conducted to determine whether duplexes of interest have an ability to elicit RNase H, and are thus performed under excess amounts of AON/RNA duplex over RNase H in cell-free systems (single-turnover conditions for AON) (Crooke et al., 1995; Lima and Crooke, 1997; Vester et al., 2008; Stanton et al., 2012). In the present study, to investigate whether AONs are recyclable in antisense reactions, we devised a cell-free reaction system, in which synthetic 20-mer target RNA conjugated with a pair of FRET (Förster resonance energy transfer) dyes and Escherichia coli–derived RNase H are both in excess over AONs (multiple-turnover conditions). In this system, an increase in fluorescence from the FRET donor is observed after binding, cleavage, and release proceeds sequentially (Fig. 1B). The main problem facing previous works is the potential difficulty in separating affinity issues from length issues, because affinity usually varies as a function of strand length. In this study, we utilized LNA, which enables us to freely modify AON affinity without changing length; thus, we prepared a series of LNA-based apoB-targeting AONs with a central focus on 13-mer AONs (Table 1), one of which (ApoB-13a) had been previously characterized as highly potent in vitro and in vivo (Straarup et al., 2010; Yamamoto et al., 2012b).
Table 1.
Oligonucleotides Used in This Study
| No. | Sequence ID | Sequence | Tm (°C) |
|---|---|---|---|
| 1 | ApoB-20a | 5′-TTCAGcattggtattCAGTG-3′ | 76±0.4 |
| 2 | ApoB-20b | 5′-ToToCoAoGcattggtattCoAoGoToG-3′ | 79±0.6 |
| 3 | ApoB-20c | 5′-totocoaogocoaototogogotoaototocoaogotog-3′ | 59±0.6 |
| 4 | ApoB-16a | 5′-CAGcattggtatTCAG-3′ | 66±0.5 |
| 5 | ApoB-14a | 5′-AGCattggtatTCA-3′ | 62±0.6 |
| 6 | ApoB-14b | 5′-AgCattggtatTcA-3′ | 58±0.4 |
| 7 | ApoB-13a | 5′-GCattggtatTCA-3′ | 59±0.5 |
| 8 | ApoB-13b | 5′-GoCattggtatToCoA-3′ | 62±0.1 |
| 9 | ApoB-13c | 5′-gCattGgtatTCA-3′ | 63±0.5 |
| 10 | ApoB-13d | 5′-GcattggtaTTCA-3′ | 58±0.8 |
| 11 | ApoB-13e | 5′-GCattggtattCA-3′ | 55±0.5 |
| 12 | ApoB-13f | 5′-GoCattggtattCoA-3′ | 57±0.1 |
| 13 | ApoB-13g | 5′-GCattggtatTcA-3′ | 58±0.6 |
| 14 | ApoB-13h | 5′-gcattggtatTCA-3′ | 48±0.7 |
| 15 | ApoB-13i | 5′-GCAttggtattca-3′ | 50±0.5 |
| 16 | ApoB-12a | 5′-GCattggtatTC-3′ | 52±0.6 |
| 17 | ApoB-12b | 5′-GCattggtaTtC-3′ | 53±0.5 |
| 18 | ApoB-12c | 5′-GoCattggtatToC-3′ | 54±0.5 |
| 19 | ApoB-11a | 5′-CAttggtatTC-3′ | 39±0.5 |
| 20 | ApoB-11b | 5′-CoAttggtatToC-3′ | 41±0.4 |
| 21 | ApoB-10a | 5′-CattggtatT-3′ | 28±0.4 |
| 22 | ApoB-10b | 5′-CAttggtaTT-3′ | 33±0.6 |
| 23 | ApoB-10c | 5′-CoAttggtaToT-3′ | 36±0.5 |
| 24 | ApoB-10d | 5′-cattggtATT-3′ | 29±0.5 |
| 25 | DL-MRNA | 5′-R-cacugaauaccaaugcugaa-Q-3′ | |
| 26 | NL-MRNA | 5′-cacugaauaccaaugcugaa-3′ | |
| 27 | MRNA-1 | 5′-cacugaauac-3′ | |
| 28 | MRNA-2 | 5′-caaugcugaa-3′ |
Upper case, lower case, lower italic, and superscript circle indicate locked nucleic acid (LNA), DNA, RNA, and phosphodiester linkage, respectively. All internucleotide linkages are phosphorothioated unless otherwise noted. All RNAs numbered 25, 26, 27, and 28 have phosphodiester internucleotide linkages. Melting temperatures (Tm) are shown as mean±SD.
apoB, apolipoprotein B-100; dl-mrna, dual-labeled complementary RNA probe; nl-mrna, non-labeled complementary 20-mer RNA.
Materials and Methods
Oligonucleotides
All oligonucleotides listed in Table 1 were purchased from Gene Design Inc.
Thermal denaturation experiments
Thermal denaturation experiments were carried out on SHIMADZU UV-1650 and UV-1800 spectrometers equipped with a Tm analysis accessory. For duplex formation, equimolecular amounts of target RNA and each AON were dissolved in 10 mM sodium phosphate buffer (pH=7.2) containing 100 mM (1.0 M for MRNA-1, MRNA-2) NaCl to give a final strand concentration of 2.0 μM. Duplex samples were then annealed by heating at 90°C, followed by slow cooling to room temperature. Melting profiles were recorded at 260 nm from 0°C to 95°C at a scan rate of 0.5°C/minute. Melting temperatures were obtained as maxima of the first derivative of the melting curves.
Turnover experiments
Dual-labeled complementary RNA probe (DL-MRNA) and non-labeled complementary 20-mer RNA (NL-MRNA) were combined in a 1:3 molar ratio. The intended amounts of the resulting mixture and AON were added to RNase H reaction buffer (New England Biolabs). The reaction was initiated by addition of 1 μL of the intended concentrations of E. coli RNase H (Takara) to 199 μL of reaction mixture. Fluorescence intensity was recorded once every 15 seconds for 15 minutes at 555 nm (ex) and 590 nm (em) using a fluorescence microplate reader (Molecular Devices). The initial turnover rates (v0) were calculated by fitting a linear regression line to the data for the first 0–60 seconds and then converted the resulting slopes expressed as RFU/second into v0 (nM/second) by using a conversion factor, 6.15 (RFU/nM), determined by experiments shown in Supplementary Fig. S1 (Supplementary Data are available online at www.liebertpub.com/nat).
In vivo pharmacological experiments
All animal procedures were performed in accordance with the guidelines of the Animal Care Ethics Committee of the National Cerebral and Cardiovascular Center Research Institute. All animal studies were approved by an institutional review board. All C57BL/6J mice (CLEA Japan) were male, and studies were initiated when animals were 8 weeks of age. Mice were maintained on a 12-hour light/12-hour dark cycle and fed ad libitum. Mice received a single treatment of AONs administered subcutaneously at a dose of 0.75 mg/kg. At the time of sacrifice, mice were anesthetized and livers were harvested and snap frozen until subsequent analysis. Whole blood was collected and subjected to serum separation for subsequent analysis.
mRNA quantification
Total RNA was isolated from mouse liver tissues using TRIzol Reagent (Life Technologies Japan) in accordance with the manufacturer's instructions. Gene expression was evaluated using a two-step quantitative reverse transcription-polymerase chain reaction (RT-PCR) method. Reverse transcription of RNA samples was performed using a High-Capacity cDNA Reverse-Transcription Kit (Life Technologies), and quantitative PCR was performed using TaqMan Gene Expression Assays (Life Technologies Japan). Messenger RNA levels of apoB were normalized against GAPDH mRNA levels. For murine apoB and GAPDH, TaqMan gene expression assays were used (assay IDs: Mm01545156_m1 and Mm99999915_g1, respectively).
AON quantification in liver
Assay was performed as described previously (Yamamoto et al., 2012a). Template DNA: 5′-gaatagcgatgaataccaatgc-3′ with biotin at the 3′ end; ligation probe DNA: 5′-tcgctattc-3′ with phosphate at the 5′ end and digoxigenin at the 3′ end.
Serum chemistry
Assay kits (#439-17501; WAKO) were used to measure serum levels of total cholesterol.
Statistics
Pharmacological studies were performed with more than three mice per treatment group. All data are expressed as means±standard deviation (SD). P<0.05 was considered to be statistically significant in all cases. Statistical comparisons were performed by Dunnett's or Bonferroni's multiple comparison tests.
Results and Discussion
Our first goal was to prepare bioactive AONs with a variety of binding affinities for the target RNA. We designed and synthesized 24 AONs, as shown in Table 1. Most of the AONs were 10- to 20-mer LNA/DNA chimeras with full or partial PS backbones. DNA stretches on these AONs were kept in the 6- to 10-nt range, which is expected to be sufficient for eliciting RNase H of both E. coli and mammalian origins (Monia et al., 1993; Kurreck et al., 2002). ApoB-13c containing LNA in the center of the gap was prepared as a non-cleavable negative control. We next determined Tm values of all AONs with the NL-MRNA (Table 1). As expected, Tm values of these AONs were uniformly and broadly distributed from approximately 30° to 80°C under the indicated buffer conditions.
In order to investigate turnover activities of AONs, we developed a cell-free fluorescent turn-on system. DL-MRNA labeled with reporter dye (TAMRA) and quencher (BHQ2) on the 5′ and 3′ termini, respectively, was designed and prepared to detect RNA scission. Using this probe, we first evaluated the turnover activity of previously validated ApoB-13a. In the presence of an 80- to 240-fold molar excess of complementary RNA, 10 nM ApoB-13a was pre-incubated at 37°C in a 96-well microplate before addition of RNase H. It should be noted that the target complementary RNA used here consists of one-quarter dual-labeled DL-MRNA and three-quarters non-labeled NL-MRNA to avoid undesirable quenching or other interactions that may affect fluorescence (Supplementary Table S1). After addition of 60 units per well RNase H to the reactions, fluorescence intensities of TAMRA were measured (excitation=555 nm and emission=590 nm) every 15 seconds for 15 minutes (Supplementary Fig. S1A). Fluorescence of the reporter dye increased over time. The initial reaction rates also increased as a function of RNA concentration, where the initial reaction rates were determined from the slope of the initial linear portions (0–60 seconds) of the plots of fluorescence intensity versus time. In contrast, time-dependent fluorescence changes in TAMRA were not seen in an ApoB-13a-lacking control. We also observed very low background fluorescence levels in the control group, which indicates efficient quenching of TAMRA fluorescence by BHQ2, despite these dyes being 20 nt distant from one another. The fluorescence increases reached a plateau at 5 minutes. We confirmed that this indicates the completion of degradation of all target RNAs and then attempted to estimate the conversion coefficient between fluorescence intensity and concentration of RNA (Supplementary Figs. S1B, S2). By plotting fluorescence intensities at 15 minutes as a function of RNA concentration, we found high linear correlations between fluorescence intensity and concentration and determined 6.15 (RFU/nM) as a conversion factor. To determine the required amount of RNase H in this system, we performed further tests with various amounts of RNase H (2–180 units/well). A near maximum reaction rate could be obtained when at least 60 units/well of RNase H were added to the reaction, and confirmed that the rate-determining step of this reaction was not the scission step, but was the recycling step under these conditions (Supplementary Fig. S3). Taken together, these results are consistent with the recycling of ApoB-13a during this cell-free antisense reaction (see also Supplementary Fig. S4; Supplementary Table S2), and this system is useful for rapid screening of multiple-turnover activities of a series AONs.
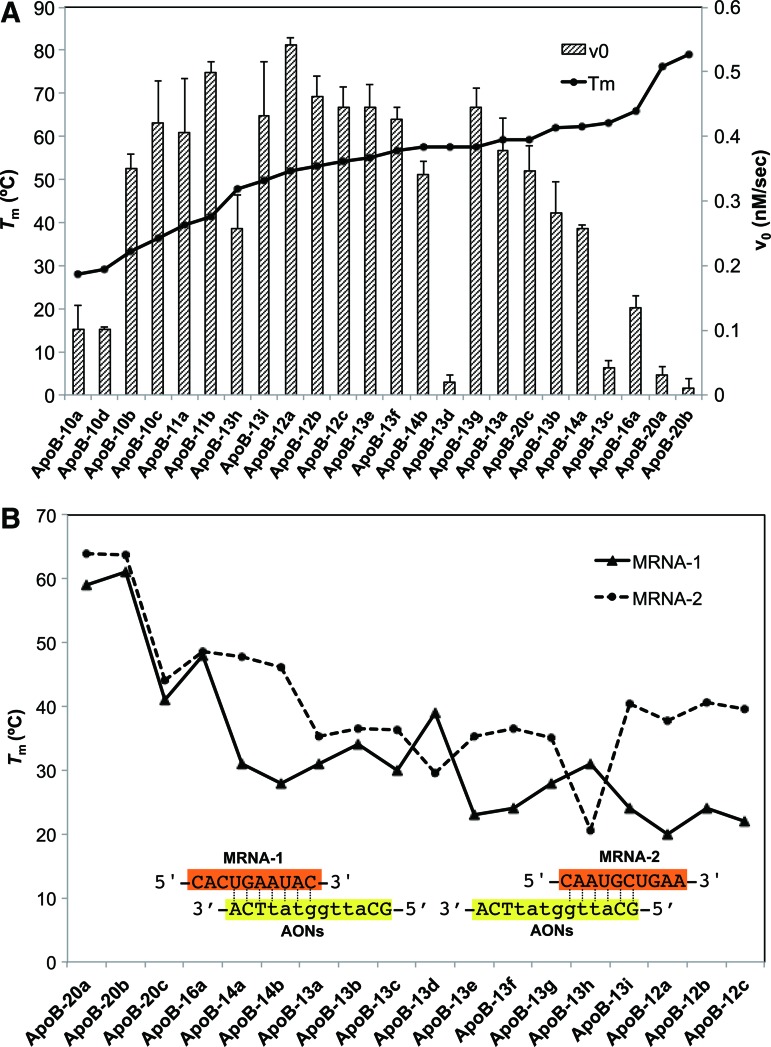
We next aimed to measure a set of initial rates of AONs (Table 1). The initial velocities were determined as described above. At a concentration of 10 nM, each AON was incubated in the presence of 800 nM complementary RNA (DL-MRNA:NL-MRNA=1:3) and 60 units/well RNase H. Time-dependent fluorescence changes were measured and initial rates were subsequently determined. The observed initial rates were rearranged in ascending order of Tm values of corresponding AONs and are shown in Fig. 2A. As expected, we found an inverted U-shaped relationship between initial rate and Tm values on the whole: AONs with high (>60°C) and low (<30°C) Tm values have relatively small initial rates, while AONs having Tm of 40°C–60°C showed efficient turnover in this system. This implies that AONs having higher multiple-turnover activities are more potent than conventional long LNA gapmers with extraordinarily high affinity. On the other hand, ApoB-12a showed the highest turnover ability among AONs tested, but ApoB-12a was shown to be less potent than ApoB-13a in vivo (Straarup et al., 2010). Thus, interpreting these data, we must take into account the differences between experimental buffer conditions used here and physiological conditions. For instance, it is known that longer PS-DNAs are more likely to form stronger undesirable complexes with proteins and inactivate RNase H to reduce their efficacy (Gao et al., 1992; Watanabe et al., 2006). As in this system, there are limited accompanying components such as inorganics, proteins, and lipids, and effects including such length-dependent factors may not have been considered. Among the four 10-mer LNAs, ApoB-10a and ApoB-10d showed marked inefficient turnover activity when compared with ApoB-10b and ApoB-10c, indicating a lack of binding affinity. In contrast, the two 20-mer AONs showed inefficient turnover activities when compared with ApoB-20c, probably due to slow product release.
FIG. 2.
(A) Relationship between turnover rate and gross or local affinity. Initial rates (v0, bar graph) of turnover reaction rearranged in ascending order of melting temperature (Tm) values (black line) of corresponding antisense oligonucleotides (AONs). Data are means±standard deviation (SD). (B) Melting temperatures with MRNA-1 (solid) and MRNA-2 (dotted). All experiments here were repeated at least three times. Color images available online at www.liebertpub.com/nat
Surprisingly, turnover activities of ApoB-13d and ApoB-13h were exceptionally low, although their Tm values were in an active range (Fig. 2A). To better understand this observation, we further measured Tm values of AONs with two additional 10-mer RNAs (MRNA-1, MRNA-2) (Fig. 2B). MRNA-1 and MRNA-2 were prepared as models for the cleaved products of RNase H and correspond to the 5′ and 3′ halves of NL-MRNA, respectively. The results showed that melting temperatures for MRNA-1 were lower than those for MRNA-2, but this trend was reversed in ApoB-13d and ApoB-13h. This potential affinity bias may explain the difference in efficiency of turnover activity. The consensus sequence for the preferred RNase H cleavage sites is unknown; instead, a strong positional preference for cleavage has been observed in a family of enzymes. For example, human RNase H1 has been shown to preferentially cleave the RNA part of RNA/DNA hybrid several nucleotides away from the 5′-RNA/3′-DNA terminus, probably due to the binding directionality of the enzyme (Lima et al., 2007a; Lima et al., 2007b). It has been predicted that the hybrid binding domain of RNase H binds the 5′-RNA/3′-DNA flank of the hybrid relative to the catalytic domain, which is strongly supported by the crystal structure of human RNase H with RNA/DNA hybrids (Nowotny et al., 2007). Despite some reported differences between E. coli and human RNase H such as a minimal gap size required for activation of RNase H (Monia et al., 1993; Crooke et al., 1995), the high similarity of the structures of human RNase H1 and E. coli RNase H1 suggests that this positional directionality for cleavage encourages ApoB-13d to produce longer RNA segments than fragments produced by other AONs, thereby forming very stable duplexes with the 3′ half of ApoB-13d and eventually decelerating product release, as well as turnover. Collectively, the requirements for high turnover rates would be adequate binding affinity of the duplex formed by using several bases of AON (for the rapid release of cleaved mRNA), as well as moderate binding affinity of full-length AON/mRNA duplex (for efficient target capture). In contrast, the affinity of the 5′ half of ApoB-13h is thought to be too low to form stable duplexes for cleavage.
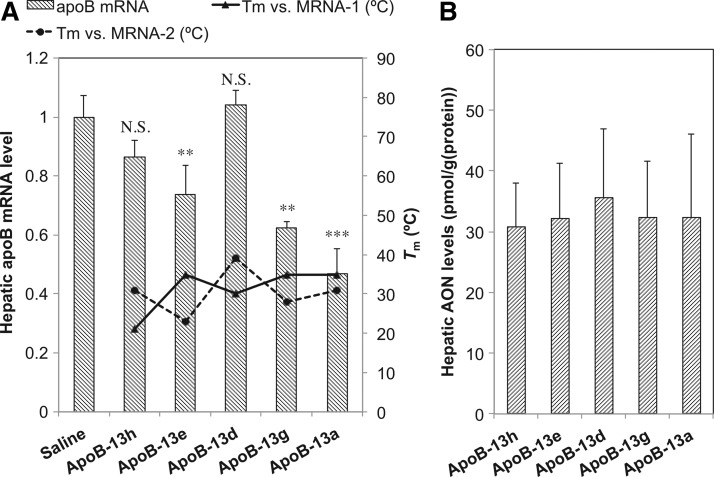
In order to explore the capacity of turnover under biological conditions, we attempted to evaluate the in vivo efficacy of AONs with identical length (ApoB-13a, -13d, -13e, -13g, and -13h), and to compare the Tm data relevant to turnover activity. We here adopted a low dosage of 0.75 mg kg–1, which had been confirmed as being sufficient to achieve knockdown of the target apoB mRNA for ApoB-13a, a positive control for in vivo screening, because even a single administration of a relatively low dose of 5 mg/kg ApoB-13a had been shown to reduce apoB mRNA by 97%, which made it difficult to discriminate differences in efficacy when AONs screened have similar high efficacy (Straarup et al., 2010). Mice (n=3/group) were dosed subcutaneously with 0.75 mg kg–1 ApoB-13a, -13d, -13e, -13g and -13h. After 48 hours post-injection, expression levels of apoB mRNA in the liver were analyzed. The apoB mRNA reduction is associated with the clinically relevant therapeutic phenotype characterized by reduced blood cholesterol concentration for the treatment of hypercholesterolemia. Expression levels were rearranged in ascending order of Tm values of corresponding AONs vs full-length NL-MRNA and are described in Fig. 3. The highest level of reduction in hepatic apoB mRNA was observed in ApoB-13a, while the lowest level of reduction was observed in ApoB-13d and -13h (Fig. 3A; Supplementary Table S3). Statistical significance was seen for ApoB-13a, -13e and -13g, but not for ApoB-13d and -13h. A similar-sized LNA phosphorothioate oligonulecotide without target sites on apoB mRNA was used as a control, showing no decrease in hepatic apoB mRNA and no potential toxicity (Supplementary Tables S3, S4).
FIG. 3.
Reduction of apoB mRNA in the livers (bar graph) of mice (n=3/group) receiving a single subcutaneous dose of 0.75 mg kg–1 of a series of 13-mer AONs (A) rearranged in ascending order of Tm values along with melting temperatures versus MRNA-1 (solid) and MRNA-2 (dashed). Dunnett's multiple comparison test, ***p<0.001; **p<0.01; N.S., not significant. (B) The mouse liver content of a series of 13-mer AONs. Bonferroni multiple comparison tests did not reveal any of arms to be significantly different across groups, p<0.05. Error bars represent group means+SD n=3.
The efficacy order of ApoB-13a>- 13g>- 13e>- 13h>- 13d appears to be unrelated to binding affinity to full-length NL-MRNA, but inefficiency of ApoB-13d and ApoB-13h in vivo were consistent with their slow turnover rates in the cell-free system, while other AONs are potent in vivo and show fast turnover rates in the cell-free system. Serum reduction levels in total cholesterol denoted the same tendency as mRNA reduction levels (Supplementary Fig. S5). As we selected AONs with an identical length of 13 nt, identical sequences and similar compositions for in vivo examination, the mouse liver content of these 13-mer AONs was measured and found to be almost identical (Fig. 3B). In addition to this, we speculate that AONs are not necessarily in vast excess of mRNA even in vivo. Sohlenius-Sternbeck has estimated a hepatocellularity number for humans, mice and other animal livers (SOHLENIUS-STERNBECK, 2006). The value for mice was estimated to be 135×106 cells per gram of liver, where livers of 8-week-old mice are 1.0 gram on average. On the other hand, we and others have calculated that approximately <10% of dosed oligonucleotides (<400 pmol for 0.75 mg/kg) reside in liver, even at 48–72 h post-dosing (Straarup et al., 2010; Yamamoto et al., 2012a).
Considering these conditions, each parenchymal cell may be exposed to AONs to a lesser extent than it is in a conventional in vitro transfection experiment. Furthermore, nonparenchymal cells such as Kupffer cells are thought to be more likely to ingest AONs than apoB-expressing parenchymal cells do and intracellular distribution of AONs via non-productive uptake further reduces the active form of AONs (Koller et al., 2011). In the light of this context, our in vivo multiple-turnover hypothesis is a compelling explanation for the in vivo activity of AONs. Of course, as there may exist differences such as intracellular distribution of AONs into productive versus less productive compartments, which can be influenced by small changes in chemistry and protein binding ability, it is necessary to continue gathering evidence. This hypothesis may also offer a new direction with regard to the remaining issues in antisense drug development; for example, inconsistency between in vitro gene silencing activity of AONs delivered in complex with transfection vehicles and in vivo activity of naked AONs (Stein et al., 2010; Zhang et al., 2011).
In the presence of transfection reagents, AONs are delivered quite efficiently to reaction sites, and consequently, might be placed under single-turnover conditions, while inefficient naked conditions may encourage multiple-turnover conditions. However, it should again be noted that it is difficult to monitor the intracellular turnover reaction in living cells and tissues due to their dynamic nature; thus, further experimental support is necessary to determine whether AONs are actually placed under multiple-turnover conditions at the intracellular antisense reaction site. Nevertheless, our study provides an important opportunity to shed light on the uncertain antisense mechanisms, and may lead to further improvement of the activity and safety profiles of AONs.
Supplementary Material
Acknowledgments
This work was supported by JSPS KAKENHI grant number 24890102 and the Advanced Research for Medical Products Mining Programme of the National Institute of Biomedical Innovation (NIBIO).
Author Disclosure Statement
No competing financial interests exist.
References
- CROOKE S.T., LEMONIDIS K.M., NEILSON L., GRIFFEY R., LESNIK E.A., and MONIA B.P. (1995). Kinetic characteristics of Escherichia coli RNase H1: cleavage of various antisense oligonucleotide-RNA duplexes. Biochem. J. 312,599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROOKE T.S. (2007). Antisense Drug Technologies: Principles, Strategies, and Applications. 2nd ed. (CRC Press Taylor & Francis Group, Boca Raton, FL: ) [Google Scholar]
- GAO W.Y., HAN F.S., STORM C., EGAN W., and CHENG Y.C. (1992). Phosphorothioate oligonucleotides are inhibitors of human DNA polymerases and RNase H: implications for antisense technology. Mol. Pharmacol. 41,223–229 [PubMed] [Google Scholar]
- HARI Y., OBIKA S., OHNISHI R., EGUCHI K., OSAKI T., OHISHI H., and IMANISHI T. (2006). Synthesis and properties of 2′-O,4-C-methyleneoxymethylene bridged nucleic acid. Bioorgan. Med. Chem. 14,1029–1038 [DOI] [PubMed] [Google Scholar]
- HUTVAGNER G., and ZAMORE P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297,2056–2060 [DOI] [PubMed] [Google Scholar]
- KOLLER E., VINCENT T. M., CHAPPELL A., DE S., MANOHARAN M., and BENNETT C. F. (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 39,4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURRECK J., WYSZKO E., GILLEN C., and ERDMANN V.A. (2002). Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30,1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIMA W.F., and CROOKE S.T. (1997). Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide-RNA hybrids. Biochemistry 36,390–398 [DOI] [PubMed] [Google Scholar]
- LIMA W.F., ROSE J.B., NICHOLS J.G., WU H.J., MIGAWA M.T., WYRZYKIEWICZ T.K., SIWKOWSKI A.M., and CROOKE S.T. (2007a). Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 71,83–91 [DOI] [PubMed] [Google Scholar]
- LIMA W.F., ROSE J.B., NICHOLS J.G., WU H.J., MIGAWA M.T., WYRZYKIEWICZ T.K., VASQUEZ G., SWAYZE E.E., and CROOKE S.T. (2007b). The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol. Pharmacol. 71,73–82 [DOI] [PubMed] [Google Scholar]
- MIYASHITA K., RAHMAN S.M.A., SEKI S., OBIKA S., and IMANISHI T. (2007). N-Methyl substituted 2 ′,4 ′-BNANC: a highly nuclease-resistant nucleic acid analogue with high-affinity RNA selective hybridization. Chem. Commun. 2007,3765–3767 [DOI] [PubMed] [Google Scholar]
- MONIA B.P., LESNIK E.A., GONZALEZ C., LIMA W.F., MCGEE D., GUINOSSO C.J., KAWASAKI A.M., COOK P.D., and FREIER S.M. (1993). Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 268,14514–14522 [PubMed] [Google Scholar]
- NOWOTNY M., GAIDAMAKOV S.A., GHIRLANDO R., CERRITELLI S.M., CROUCH R.J., and YANG W. (2007). Structure of human RNase h1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell 28,264–276 [DOI] [PubMed] [Google Scholar]
- OBIKA S., NANBU D., HARI Y., ANDOH J., MORIO K., DOI T., and IMANISHI T. (1998). Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 39,5401–5404 [Google Scholar]
- OBIKA S., NANBU D., HARI Y., MORIO K., IN Y., ISHIDA T., and IMANISHI T. (1997). Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C-3,-endo sugar puckering. Tetrahedron Lett. 38,8735–8738 [Google Scholar]
- PRAKASH T.P., SIWKOWSKI A., ALLERSON C.R., MIGAWA M.T., LEE S., GAUS H.J., BLACK C., SETH P.P., SWAYZE E.E., and BHAT B. (2010). Antisense oligonucleotides containing conformationally constrained 2′,4′-(N-methoxy)aminomethylene and 2′,4′-aminooxymethylene and 2′-O,4′-C-aminomethylene bridged nucleoside analogues show improved potency in animal models. J. Med. Chem. 53,1636–1650 [DOI] [PubMed] [Google Scholar]
- SETH P.P., SIWKOWSKI A., ALLERSON C.R., VASQUEZ G., LEE S., PRAKASH T.P., WANCEWICZ E.V., WITCHELL D., and SWAYZE E.E. (2009). Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 52,10–13 [DOI] [PubMed] [Google Scholar]
- SINGH S.K., NIELSEN P., KOSHKIN A.A., and WENGEL J. (1998). LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998,455–456 [Google Scholar]
- SOHLENIUS-STERNBECK A.K. (2006) Determination of the hepatocellularity number for human, dog, rabbit, rat and mouse livers from protein concentration measurements. Toxicol. In Vitro 20,1582–1586 [DOI] [PubMed] [Google Scholar]
- STANTON R., SCIABOLA S., SALATTO C., WENG Y., MOSHINSKY D., LITTLE J., WALTERS E., KREEGER J., et al. (2012). Chemical modification study of antisense gapmers. Nucleic Acid Ther. 22,344–359 [DOI] [PubMed] [Google Scholar]
- STEIN C.A., HANSEN J.B., LAI J., WU S., VOSKRESENSKIY A., HOG A., WORM J., HEDTJARN M., SOULEIMANIAN N., MILLER P., et al. (2010). Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 38,e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRAARUP E.M., FISKER N., HEDTJARN M., LINDHOLM M.W., ROSENBOHM C., AARUP V., HANSEN H.F., ORUM H., HANSEN J.B., and KOCH T. (2010). Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 38,7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VESTER B., BOEL A.M., LOBEDANZ S., BABU B.R., RAUNKJAER M., LINDEGAARD D., RAUNAK HR.DLICKA P.J., HOJLAND T., et al. (2008). Chemically modified oligonucleotides with efficient RNase H response. Bioorg. Med. Chem. Lett. 18,2296–2300 [DOI] [PubMed] [Google Scholar]
- WATANABE T.A., GEARY R.S., and LEVIN A.A. (2006). Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302). Oligonucleotides 16,169–180 [DOI] [PubMed] [Google Scholar]
- YAHARA A., SHRESTHA A.R., YAMAMOTO T., HARI Y., OSAWA T., YAMAGUCHI M., NISHIDA M., KODAMA T., and OBIKA S. (2012). Amido-bridged nucleic acids (AmNAs): synthesis, duplex stability, nuclease resistance, and in vitro antisense potency. Chembiochem 13,2513–2516 [DOI] [PubMed] [Google Scholar]
- YAMAMOTO T., HARADA-SHIBA M., NAKATANI M., WADA S., YASUHARA H., NARUKAWA K., SASAKI K., SHIBATA M.A., TORIGOE H., et al. (2012a). Cholesterol-lowering action of BNA-based antisense oligonucleotides targeting PCSK9 in atherogenic diet-induced hypercholesterolemic mice. Mol. Ther. Nucleic Acids 1,e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO T., NAKATANI M., NARUKAWA K., and OBIKA S. (2011). Antisense drug discovery and development. Future Med. Chem. 3,339–365 [DOI] [PubMed] [Google Scholar]
- YAMAMOTO T., YASUHARA H., WADA F., HARADA-SHIBA M., IMANISHI T., and OBIKA S. (2012b). Superior silencing by 22,42-BNANC-Based short antisense oligonucleotides compared to 22,42-BNA/LNA-based apolipoprotein B antisense inhibitors. J. Nucleic Acids, Article ID 707323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y., QU Z., KIM S., SHI V., LIAO B., KRAFT P., BANDARU R., WU Y., GREENBERGER L.M., and HORAK I.D. (2011). Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection. Gene Ther, 18,326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.