Abstract
Despite their functional and structural diversity, G protein-coupled receptors (GPCRs) share a common mechanism of signal transduction via conformational changes in the seven-transmembrane (7TM) helical domain. New major insights into this mechanism come from the recent crystallographic discoveries of a partially hydrated sodium ion that is specifically bound in the middle of the 7TM bundle of multiple class A GPCRs. This review discusses the remarkable structural conservation and distinct features of the Na+ pocket in this most populous GPCR class, as well as the conformational collapse of the pocket on receptor activation. New insights help to explain allosteric effects of sodium on GPCR agonist binding and activation, and sodium’s role as a potential co-factor in class A GPCR function.
Keywords: GPCR activation, allosteric modulation, biased signaling, conserved pocket, sodium ion, water binding
Early phenomena attributed to a specific sodium-dependent modulation of GPCR function
G protein-coupled receptors (GPCRs) are the largest superfamily of membrane proteins in the human genome and have key roles in human physiology and in the action of more than 30% of therapeutic drugs [1]. The binding of a ligand stabilizes conformational changes in the receptor, which trigger the activation of intracellular (IC) effectors such as G proteins and arrestins [2], leading to a cascade of cellular responses. Although all GPCRs share a common seven-transmembrane (7TM) architecture, they can be divided into four major classes in humans, A, B, C, and Frizzled (F) (see Glossary), that have very little sequence homology between them [3,4]. Out of 826 human GPCRs, more than 700 belong to Class A [3,4] and have several highly conserved functional motifs in their 7TM domains, including D(E)RY in helix II, FxxCWxP in helix VI, NPxxY in helix VII, and the ‘hydrogen bond network’ between helices I, II, III, VI, and VII [5]. Although the importance of these motifs in GPCR signaling was known for years, their precise roles in signal transduction are only now being revealed, principally due to technological breakthroughs leading to a flurry of GPCR structures being captured in different activation states and complexes [6,7]. One of the most exciting major findings concerns the crystallographic discovery in class A GPCRs of a conserved allosteric binding site for a sodium ion [8], an essential ion that is implicated in many physiological functions.
The first hints of a specific allosteric effect of Na+ on class A GPCR function can be traced to a study performed 40 years ago [9]. This seminal work found that Na+ negatively modulates agonist binding to the opioid receptors, without significantly affecting the binding affinity of antagonists. In the absence of other biochemical assays, this ‘sodium effect’ was employed to differentiate opioid agonist from opioid antagonist candidate drugs [10]. Later, similar biochemical phenomena were observed for more than 20 diverse GPCRs, including adrenergic, dopaminergic, serotonergic, neurotensin, and other receptors (Table 1). These allosteric effects were usually described at physiologically relevant Na+ concentrations (~140 mM), supporting its biological role. Follow-up mutagenesis studies implicated a conserved acidic D2.50 (Ballesteros–Weinstein numbering [11]) residue in helix II as being critical for the sodium-dependent effects, suggesting that Na+ acts via binding at a specific site within the helical bundle. Moreover, substitution of D2.50 with uncharged residues (Table 1) dramatically reduced agonist-dependent signaling of some GPCRs, while maintaining ligand binding and often basal signaling, thereby indicating a specific role for this putative ‘sodium site’ in agonist-mediated GPCR signal transduction. Only now, however, has high-resolution crystallography of GPCRs provided the critical structural insights that are essential for unraveling the molecular underpinnings of these biochemical phenomena, as well as for illuminating the functional and physiological consequences of sodium binding to GPCRs.
Table 1.
Published evidence for allosteric effects of sodium and amilorides, and their dependence on D2.50 mutations
| GPCR families | Na+ allosteric effect on ligand binding | D2.50 controls allosteric effects of Na+ | D2.50 mutation effects on G protein coupling and activation | Allosteric effect of amiloride and analogs |
|---|---|---|---|---|
| Adenosinea [89]b | A1 [79,90], A2Aa [8,30,79], A3 [79,81] | A1 [90], A2Aa [80], A3 [81] | – | A1 [79], A2Aa [8,30,79,80], A3 [79,81] |
| Adrenergica | α[91], α2A [92– 94] | α2A [92–94] | α2A [93,94] | α2 [82], α1 [83] |
| Dopamine [64,95]* | D2 [84,96] | D2 [84,96] | D2 [84,96] | [84], D1, D2, D3, D4 [85] |
| Muscarinic | – | M3 [45], M1 [97], M1, M2, M3, M4 [98] | [86] | |
| 5HT | – | 2A [46], 1B [87] | 1B [87] | |
| Opioida | [9,10,31], μ-OR [34], NOP [99,100], δ-OR [15] | δ-ORa [15,61] | All subtypes [48] | – |
| Somatostatin | SSTR2 [101] | SSTR2 [101] | – | – |
| Neurotensin | NTSR1 [19,20] | NTSR1 [19,20] | NTSR1 [19,20] | – |
| Gonadotropin- releasing hormone | GnRHR [24] | – | GnRHR [49] | GnRHR [24] |
| Urotensin | UTR [102] | UTR [102] | UTR [102] | – |
| Cannabinoid | CB1 [41] | CB1 [41] | CB1,CB2 [41,53] | – |
| Angiotensin | – | AT1R [36] | – | |
| Endothelin | – | ETAR, ETBR [39] | – | |
| Cholecystokinin | – | CCKB [103] | – | |
| Bradykinin | B2 [50] | B2 [50] | B2 [50] | – |
G protein-coupled receptor (GPCR) subfamilies and subtypes for which crystallographic evidence of Na+ binding in the conserved pocket have been obtained.
Review and/or modeling studies.
Identification of Na+ in the crystal structures of class A GPCRs
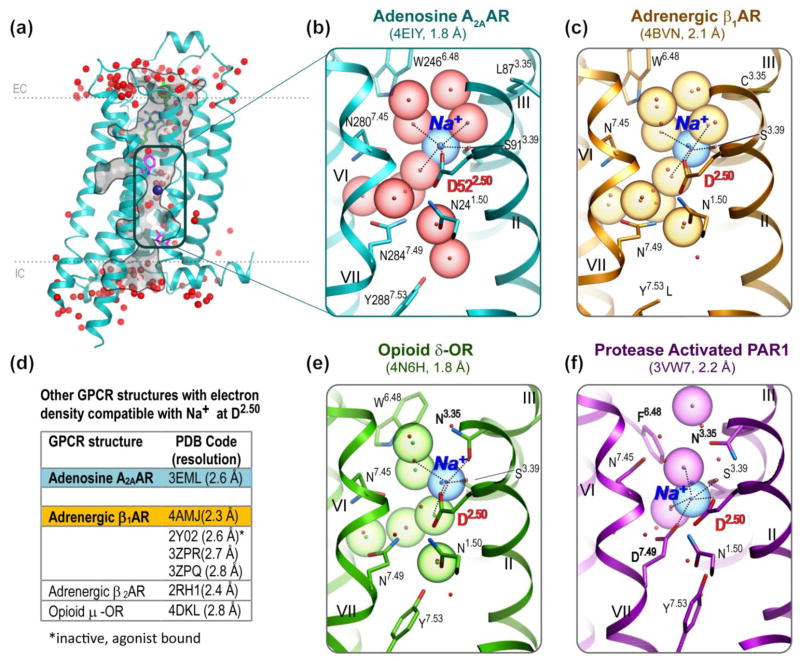
The recently solved high-resolution (1.8 Å) structure of the A2A adenosine receptor (A2AAR) [8] was the first to reveal a Na+/water cluster in the middle of the 7TM helical bundle, thereby providing a detailed description of a GPCR allosteric site (Figure 1A,B). The Na+ in the A2AAR is coordinated by two highly conserved residues, D2.50 and S3.39, and three water molecules. These water molecules belong to a nearly continuous water-filled passage connecting the A2AAR extracellular (EC) and IC sides. Importantly, the reliable detection of Na+ in this allosteric site was enabled by high-resolution crystal structures and the unambiguous location of at least five oxygen atoms, a hallmark of a Na+ coordination shell. Sodium ions can be identified then by their characteristic coordination geometry and short distances (2.2–2.6 Å) to the oxygen atoms [12]. Although a retrospective analysis of the earlier medium resolution structures (2.4–2.9 Å) suggests that some spherical electron densities in close proximity to D2.50 (previously modeled as water) are compatible with Na+ (Figure 1D), these structures could not provide unequivocal evidence for sodium at this site.
Figure 1.
Na+ and water cluster detected in G protein-coupled receptor (GPCR) structures. (A) The high-resolution A2A adenosine receptor (A2AAR) structure [8] shown with waters (red spheres) and Na+ (blue sphere) in the narrow passage connecting the extracellular (EC) and intracellular (IC) sides of the receptor. (B) Close-up of the A2AAR allosteric pocket comprising Na+ and a cluster of 10 water molecules. Acidic residue D2.50 and all residue positions of the pocket involved in polar interaction with the cluster are shown as sticks. Roman numerals show numbering of the transmembrane helices. (C) Close-up of the β1AR allosteric pocket [13], colored orange. Ten water molecules and the Na+ position from A2AAR are shown for comparison as red dots and a blue dot, respectively. Note the Y3437.53L mutation. (D) A list of medium-resolution structures (2.3–2.8 Å) [18,75,104–107] with electron densities in the proximity of D2.50 that are potentially compatible with sodium binding. (E) Close-up of the δ-opioid receptor (δ-OR) allosteric pocket [15], colored green. (F) Close-up of the protease-activated receptor (PAR1) allosteric pocket [14], colored magenta.
Within 18 months of the discovery of the Na+ in the A2AAR structure, direct crystallographic evidence for sodium in an identical position was found in inactive-state GPCR structures of an adrenergic receptor (β1AR) [13], a protease-activated receptor (PAR1) [14], and an opioid receptor (δ-OR) [15] (Figure 1C–F). These crystal structures together represent three of the four major branches (α, δ, γ) in the class A GPCR tree [4]. In each of these high-resolution structures, the Na+ is coordinated by a salt bridge to D2.50 together with four additional polar interactions with receptor side chains and water molecules. The most striking similarities can be found between the A2AAR (PDB identifier: 4EIY) and β1AR (PDB identifier: 4BVN) [13] structures. Not only are all residues of the pocket conserved chemically and conformationally with RMSDALL_ATOM = 0.8 Å (except Y7.53L, which was introduced as a thermostabilizing mutation in β1AR [13]), but also the positions of Na+ and eight water molecules of the cluster are preserved to within 0.3 Å RMSD. Considering that A2AAR and β1AR share only ~32% sequence identity in their 7TM domains and have different ligands, this high level of structural conservation of the Na+ and water cluster is truly remarkable and suggests its key functional role. Interestingly, pharmacological analysis in the same study [13] shows that high Na+ concentrations do not impact agonist binding in b1AR, suggesting that Na+ specific binding can be easily overlooked by classical allosteric effect assessment.
Distinct types of sodium pockets in inactive GPCRs
The common binding of the Na+ in 7TM bundles, however, does not require an identical pocket structure to that found in the A2AAR and β1AR. Interestingly, opioid receptors (in the γ-branch) present a structural variation on the Na+/water coordination motif, whereas conformations of the 15 conserved residues of the pocket remain similar to A2AAR and β1AR. Indeed, the 1.8 Å resolution structure of the δ-OR [15] (PDB identifier: 4N6H) reveals the pivotal importance of another position, 3.35, for sodium binding in opioid receptors (Figures 1E and 2C). Although in many GPCR structures the 3.35 position has a hydrophobic residue that points towards the lipidic membrane interface, the δ-OR structure has an N3.35 side chain pointed to the interior of the sodium pocket. The oxygen of the N3.35 side chain in the δ-OR structure thereby directly coordinates the Na+ (at 2.45 Å distance) by occupying the same spatial positions as a water molecule found in the A2AAR structure. At the same time, the nitrogen of the N3.35 side chain replaces another water molecule seen in the A2AAR structure, and participates in a water-mediated hydrogen bonding network. Notably, N3.35 has the same conformation in the other inactive-state opioid receptor structures [16–18], and is conserved in 78 human GPCRs (mostly of the γ- and δ-branches), where it can also be involved in Na+ coordination.
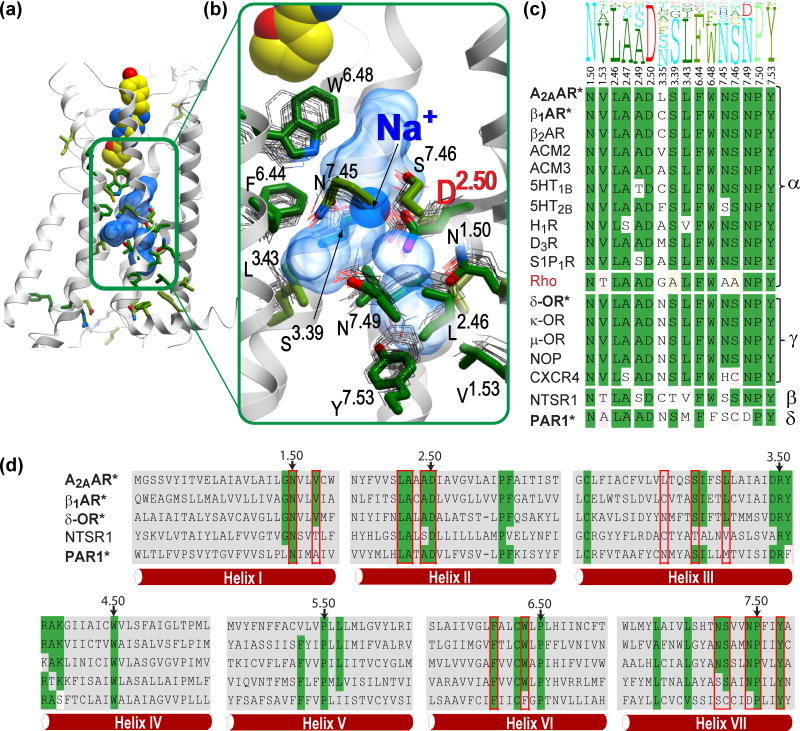
Figure 2.
Structural and sequence conservation of the Na+ and water pocket in G protein-coupled receptors (GPCRs). (A) Overview of the A2A adenosine receptor (A2AAR) crystal structure, showing residues with higher than 50% conservation in all non-olfactory class A GPCRs as sticks with green carbons. (B) A close-up of the central allosteric pocket (transparent blue surface), showing the side chains located within 5 Å from the 10 waters of the sodium ion/water cluster (green sticks: A2AAR; gray thin lines: the corresponding side chains of the overlaid GPCR crystal structures in inactive state). The helix VII backbone in the foreground has been removed for clarity. (C) Sequence conservation of the 16 pocket residues. The top part shows the residue conservation profile in all non-olfactory class A GPCRs, where the height of the residue letter represents the share of the residue in this position. The bottom part shows individual residues in all available class A crystal structures, with conserved residues highlighted in green. Greek letters on the right denote the four major branches of class A GPCRs. Receptors with sodium binding determined by a high-resolution crystal structure are in bold and marked with ‘*’. Rhodopsin, which lacks a Na+ binding pocket, is shown in red. (D) Sequence alignment in seven-transmembrane helices of representative class A GPCRs from all four major branches. Residues of the Na+ pocket are highlighted by red boxes, and conserved residues are highlighted in green. The most conserved residues in each helix (X.50) are marked by arrows.
The crystal structure of inactive PAR1 (a member of the δ-branch of GPCRs) [14] represents a dramatic deviation in the pocket configuration (Figure 1F), with seven residues of the pocket (1.53, 3.43, 3.35, 6.48, 7.45, 7.46, and 7.49) being different from those in the A2AAR. Moreover, in the PAR1 structure the Na+ is shifted by about 1.5 Å towards the IC side and is coordinated by acidic side chains two residues, D2.50 and D7.49. Although the 7.49 position of the NPxxY motif is conserved as N7.49 in 86% of all class A GPCRs, one can notice that other 7% of class A (PAR1 and other 51 receptors) have Asp side chains in both the 2.50 and 7.49 positions. Interestingly, many other δ-branch GPCRs and most of the 380 human olfactory receptors have a second acidic residue in the pocket located in a different position, 3.39. Those GPCRs with two negatively charged acidic side chains within the ion coordination shell are likely to have distinctive ion-binding properties and might potentially accommodate not only monovalent but also divalent cations.
The β-branch of class A GPCRs is currently represented by only one crystal structure of the NT1 neurotensin receptor (NTSR1), which was solved in an ‘active-like’ state. As discussed below, the sodium site is collapsed in the known active-like state GPCR structures, and therefore sodium binding in this NTSR1 structure is not expected [19]. At the same time, biochemical experiments [20] provide convincing evidence for D2.50-dependent Na+ binding and allosteric effects at NTSR1 [19,20] and other β-branch receptors, such as the gonadotropin releasing hormone (GnRHR) and urotensin receptors (Table 1). The key polar residues coordinating the Na+ and water cluster (positions 1.50, 2.50, 3.39, 7.46, 7.49, and 7.53) are also conserved in NTSR1 (Figure 2C), supporting a putative Na+ binding site. Variations in some non-polar residues corresponding to the pocket, however, suggest distinct features of the Na+ pocket of β-branch receptors that remain to be crystallographically characterized.
It is likely that other structural variations can be found in the sodium pocket across all class A GPCR families, which could significantly affect both the sodium-binding profile and the sodium-dependent allosteric effects. For example, access to the sodium pocket can vary dramatically, from a rather open passage in the opioid [15,18] and muscarinic [21,22] receptor structures to a more restricted passage in A2AAR and β1AR. The presented examples, however, suggest that despite differences in detail, a specific sodium-binding site may be a common feature that is preserved in many receptors from all four major branches of class A GPCRs, in which D2.50 and the key polar side chains of the pocket are conserved.
Conservation of Na+ binding across class A GPCR families and branches
Sequence analysis of all class A GPCRs confirms an exceptionally high conservation of the pocket (Figure 2 and Figure S1 in the supplementary material online). In fact, the pocket (as defined in A2AAR) combines 15 of the 34 residue positions that are conserved in the majority of the non-olfactory class A GPCRs (Figure 2C). The pocket encompasses the three previously identified conserved motifs: (i) F6.44 and W6.48 of the FxxCWxP motif in helix VI; (ii) N7.49 and Y7.53 of the NPxxY motif in helix VII; and (iii) a 3D cluster of conserved polar residues in helices I, II, III, VI, and VII that is sometimes referred to as a hydrogen bonding network [5]. The only conserved cluster that is not a part of the sodium pocket is the ‘DRY’ motif in helix III located closer to the IC side of GPCRs. Remarkably, all 15 residues of the A2AAR sodium pocket are exactly conserved in 45 GPCRs, mostly in the aminergic and adenosine subfamilies, but also in sphingosine-1 phosphate and some orphan receptors, as well as in the γ-branch opioid and somatostatin receptors (Figure S1 in the supplementary material online). A simple estimation shows that the concomitant conservation of this group of residues is about ~2,500 fold higher than expected if the residues of the pocket mutated independently (Table S2 in the supplementary material online), suggesting strong evolutionary pressure for conservation of the pocket configuration as a whole. Most other receptors in α– and γ-branches have only minor variations in the pocket, with at least 12 residues of the pocket preserved as in A2AAR (Figure S1 in the supplementary material online).
Across all four branches of the non-olfactory class A GPCRs, the residues of the sodium pocket vary more significantly, although seven key acidic and polar side chains remain conserved. These highly conserved positions include D2.50 (90% conserved as Asp), N1.50 (97% Asn), S3.39 (75% Ser), N7.45 (70% Asn, or 90% as any polar side chain), S7.46 (66% Ser and 75% as any polar side chain), N7.49 (75% Asn, and 20% as Asp), and finally Y7.53 (89% Tyr). The variations between known Na+ binding GPCRs are mainly observed in non-polar residues of the pocket, suggesting that a GPCR’s compatibility with sodium binding is largely defined by these key polar positions.
The olfactory receptors also apparently have highly conserved pockets that are compatible with Na+, although binding and allosteric effects of Na+ in this large family of GPCRs remains to be experimentally established. All olfactory receptors have an acidic residue 2.50 (83% as Asp and 17% as Glu) and a conserved pattern of polar residues. This pattern is, however, quite distinct from non-olfactory receptors: for example, the olfactory GPCRs have an acidic residue 3.39 (80% as Glu and 14% as Asp), and a histidine at 6.44 (98% as His).
It is also important to note, that in the other human GPCR classes (B, C, and F), none of the residues corresponding to the sodium pocket is conserved. Apparently, these functionally and evolutionary distinct GPCR classes do not have a common Na+ binding site in their 7TM domains, although this does not exclude a possibility that some of these GPCRs are modulated by Na+ binding in the other parts of the receptor.
Class A GPCRs lacking a putative Na+ pocket have distinctive properties
Our analysis also suggests that those 36 of the class A receptors (~5%) that lack acidic residues D(E)2.50 (Table S1 in the supplementary material online) may have functional properties that are distinct from other class A GPCRs and, in most cases, do not possess ligand-modulated signal transduction. Thus, of these 36 proteins, 26 are described as ‘pseudogene’, ‘non-signaling’, ‘decoy’, ‘constitutively active orphan’, or ‘putative/probable’ GPCRs, as annotated by the IUPHAR or UniProt databases. Among these 26 proteins, there are also three orphan LGR receptors (LGR4–6) that act via binding to Frizzled receptors [23], as well as the NT2 neurotensin receptor (NTSR2). Unlike its NTSR1 homolog, which is fully functional, NTSR2 lacks the D2.50 side chain, and although it still binds neurotensin, NTSR2 is not modulated by allosteric sodium and does not signal in response to neurotensin [19,20].
The next group of nine receptors in Table S1 that lack D(E)2.50 may still have a normal signal transduction capability in response to diffusible ligands due to an alternative acidic residue supporting sodium binding. This group comprises six olfactory receptors that have another acidic residue in the pocket, E3.39, as well as three receptors with either D3.39, D7.49 or D7.50 residues, all in the sodium pocket. Among these receptors is GnRHR, which has a neutral N2.50 side chain, but an acidic D7.49 side chain. Indeed a recent study shows that sodium has a modest (but significant) allosteric effect on GnRHR [24], suggesting that an alternative acidic residue in the pocket can still support sodium binding.
This remarkable correlation between the absence of the allosteric Na+ binding site on the one hand and the lack of an established ability of the receptor to signal in response to small molecule ligands on the other hand further corroborates importance of the allosteric Na+ for class A GPCRs. Another interesting exception that only confirms this rule is discussed in the next section.
Lack of Na+ binding in visual opsins
Rhodopsin (OPSD) and other visual opsins (OPSB, OPSG, and OPSR) represent an interesting exception that deserves a closer look. The last line in Table S1 shows the absence of D2.50 or any other acidic side chain in the allosteric pocket of blue opsin (OPSB). Moreover, all four opsins lack polar side chains in the other two crucial sodium-coordinating positions, 3.39 and 7.45 (Figure S1 in the supplementary material online), which is likely to abolish specific Na+ binding. Analysis of the high-resolution (2.2 Å) crystal structure of bovine rhodopsin [25,26] also suggests an absence of sodium binding in the 7TM pocket. Indeed, among four water molecules found in proximity of the D2.50 side chain in this structure, none has distances and coordination that are compatible with a sodium ion. Moreover, access from the aqueous EC space into the 7TM bundle core in the rhodopsin structure [25] (and probably in homologous opsins) is completely blocked by the EC loops and N terminus, which makes Na+ binding in the allosteric pocket even less likely.
The visual opsins therefore present a rare case of signaling class A GPCRs that apparently lack specific sodium binding. However, the function of these receptors also presents a major exception among all human class A GPCRs in that opsins are activated by photochemical 11-cis to all-trans isomerization of a covalently bound retinal, rather than by a diffusible ligand. Such photochemical isomerization provides abundant energy for the receptor activation (ΔG ~ 35 kcal/mol [27]), which is several fold higher than the energy contributed by small diffusible ligands. This observation suggests that the presence of the allosteric sodium may not be required for receptors activated by large chemical energy such as opsins, while being critical for effective signaling by small diffusible ligands in most class A GPCRs.
Activation involves structural rearrangements in the Na+ pocket
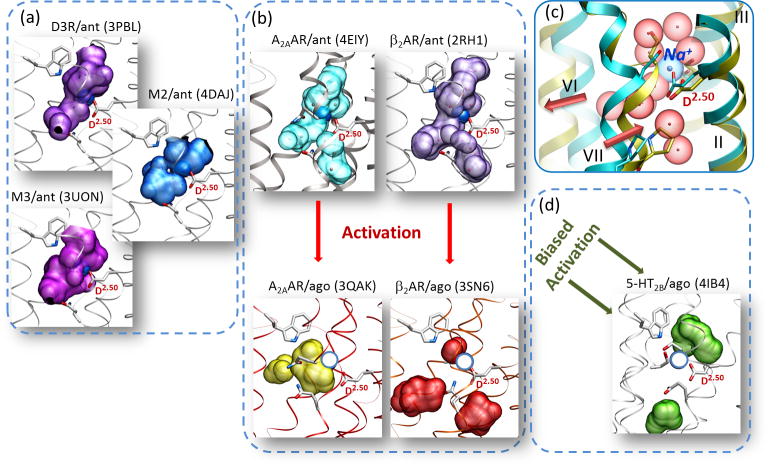
Further evidence for a key functional role of the Na+ cluster in the modulation of conformational transitions comes from an analysis of active state structures of class A GPCRs (Figure 3). Our comparison of inactive- and active-state crystal structures of A2AAR and β2AR [28,29] reveals that the Na+ and water pocket collapses in size from ~200 to <70 Å3 due to the activation-related movements of the TM helices. In particular, an inward movement of helix VII at the NPxxY motif and an outward movement of helix VI (both associated with receptor activation [28,29]) are the key rearrangements that are responsible for the pocket collapse. Conformational analysis and molecular dynamics studies for A2AAR suggest that such a collapsed pocket in the activated states of GPCRs is incompatible with Na+ binding [30]. Similar observations regarding the collapse of the Na+ pocket can be made by examining the other recently solved crystal structures of either fully or partially activated GPCRs (Figure 3). In all of these structures, the sodium pocket is reduced in size and often split into smaller cavities, which do not provide adequate coordination for a Na+ and its hydration shell waters. The shapes and locations of the residual pockets vary between these activated structures, probably reflecting differences between receptor types and activation stages. Note, however, that the most pronounced helical shift, as well as the most dramatic split and relocation of the pocket, has been observed in the fully activated complex of β2AR with G protein [28,29], suggesting that Na+ relocation is a critical part of the GPCR activation process.
Figure 3.
Activation involves collapse of the sodium pocket in class A G protein-coupled receptors (GPCRs). (A) Activation-related conformational changes, in particular inward movement of helix VII and outward movement of helix VI, cause collapse and relocation of the allosteric pocket in A2A adenosine receptor (A2AAR), which makes it incompatible with binding of the Na+ and water cluster. (B) Collapse of the pocket between inactive and activated states found in A2AAR structures (PDB identifiers 4EIY and 3QAK, respectively) is even more pronounced in β2-adrenergic receptors ((PDB identifiers 2RH1 and 3SN6). (C) Most other inactive class A GPCR structures have pockets that are compatible with sodium binding. (D) The pocket is also collapsed in the arrestin-biased activated structure of 5HT2B (right bottom panel). Allosteric pockets are shown by semitransparent surfaces, and the D2.50 side chain is labeled in all panels. The position of Na+ in the A2AAR-antagonist complex (PDB identifier 4EIY) superimposed on the other inactive structures is shown as dark blue spheres, whereas Na+ superimposed on the active-state structures is indicated as empty circles in light blue.
Functional studies and challenges
Although biochemical evidence for the allosteric effects of sodium on agonist binding exists for a number of diverse GPCRs (Table 1), a detailed understanding of the functional sodium effect on GPCR signaling in living cells is complicated by several factors. Some evidence for sodium’s impact on GPCR function was obtained by direct measurements of G protein binding to isolated cell membranes [31–33] or G protein-mediated signaling in whole cells [34,35] as a function of Na+ concentration. However, the presence of sodium-conducting ion channels and transporters, and an overall dependence of cell signaling on physiological concentrations of Na+, can profoundly affect a reliable delineation of the direct functional effects of Na+ on GPCRs.
Mutagenesis of the conserved sodium-coordinating residues provides an important alternative for evaluation of the functional effects of sodium. The majority of such mutagenesis studies have been focused on the 2.50 position, showing that in various class A GPCRs, D2.50 replacement by an uncharged residue can abolish or drastically reduce agonist-induced G protein binding and activation [36–43] (see also [44]). Some studies have also demonstrated that D2.50 substitutions can abrogate the allosteric effects of G protein on agonist binding [37,45]. In many of these cases, it was shown that D2.50 mutants can retain surface expression, and maintain (or even improve) agonist binding properties [45], suggesting that D2.50A and D2.50N mutants can maintain a correct fold for binding while affecting specific aspects of signal transduction. Mutagenesis studies have also been performed on the other sodium coordinating residues, 7.49, 7.45, and 3.39, by changing them to non-polar side chain residues. Similar to D2.50 substitutions, these substitutions resulted in the disruption of normal ligand dependent signaling [43,46,47], suggesting that Na+ itself is a major component of the signaling mechanism.
The importance of allosteric Na+ binding is further supported by gain-of-function experiments, in which the introduction of acidic residues in the sodium pocket partially restores signaling function in D2.50N mutants. Such studies were performed for the 5-HT2A serotonin receptor, showing that whereas the D2.50N mutant abrogated coupling to G protein, a D2.50N/N7.49D double mutant capable of binding Na+ regained most of the functional activity [46]. Similarly, a D2.50N/N7.49D double mutant partially restored function for the μ-opioid receptor [48], whereas in sodium-dependent GnRHR, these residues are already reversed as N2.50 and D7.49 in the wild type protein [49].
The constitutive activity of GPCRs can be dramatically affected by Na+ concentration as well as by mutations in D2.50 and other sodium-coordinating residues. Most of the published results are consistent with the notion that Na+ stabilizes the inactive state and thereby reduces basal G protein activity [35,50–52], although the effects may vary somewhat between GPCRs [13,53] and between functional readouts of signaling [20]. An interesting example is provided by viral GPCRs (vGPCRs), which are close homologs to human receptors but lack characteristic residues of the sodium pocket [54]. Thus, a vGPCR from Kaposi’s sarcoma-associated herpes virus (KSHV) is closely related to chemokine receptors but has mutations in the key conserved residues of the sodium pocket (2.50, 3.39, 7.45, and 7.49) that render it constitutively active.
Allosteric sodium site residues modulate signaling bias
Apart from G protein mediated signaling, GPCR function involves other independent pathways, including those mediated by β-arrestin [55]. Selective up- or down-regulation of these pathways by ligands or mutations often leads to so-called biased signaling or functional selectivity, which is of key importance for GPCR biology and pharmacology [56–59]. Several studies suggested that changes in the allosteric sodium pocket can also lead to pronounced signaling bias: for example, in the angiotensin 1 (AT1R) receptor [60]. More recently, dramatic signaling bias effects of the sodium site were observed for δ-OR signaling [15]. Site-directed mutagenesis and functional assays in this study show that D2.50A substitution, as well as some other substitution of the sodium-coordinating residues, such as N7.45A and N7.49A, can still retain G protein-mediated signaling for at least some agonists, as observed previously [61]. This study found, however, that these mutations transform classical δ-opioid antagonists, such as naltrindole, into potent β-arrestin-biased agonists [15]. A special role was also described for the N3.35 side chain, which participates in Na+ coordination in δ-OR (Figure 1E). Substitution of this residue either reduces (N3.35V) or abrogates (N3.35A) the allosteric effects of sodium on ligand binding, while at the same time dramatically augmenting constitutive arrestin-mediated signaling.
These data support the notion that the allosteric sodium in the δ-OR has a more profound impact on regulating β-arrestin efficacy, whereas its effects on canonical G-protein signaling may depend on specific ligands and specific residue mutations. Therefore, sodium-coordinating residues and sodium itself can act as selective ‘efficacy switches’ for distinct GPCR functional pathways.
Possible mechanisms of sodium as a co-factor in GPCR signaling
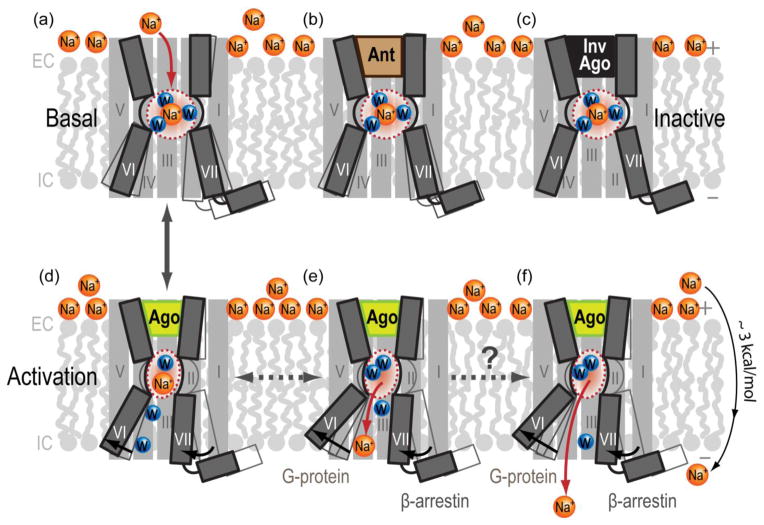
An essential role for the Na+ and water cluster in GPCR function is supported by: (i) the structural features of Na+ binding in the center of the 7TM bundle in the inactive state receptors; (ii) an exceptionally high conservation of the pocket in ligand activated class A GPCRs; (iii) dramatic activation-related changes in the Na+ pocket; and (iv) strong allosteric effects of the Na+ on constitutive and ligand-dependent GPCR signaling. One key aspect of the Na+ interactions with GPCRs is that, unlike most allosteric modulators, Na+ is present at high physiological concentrations (~140 mM) in most EC environments, which ensures near-saturation of the specific Na+ binding sites in GPCRs with EC50(Na+) < 50 mM [30]. This omnipresence in class A GPCRs and involvement in major aspects of signaling suggest a role of sodium as a co-factor that is essential for proper receptor function. Although the exact nature of Na+ involvement in signal transduction has only just begun to be understood, we suggest several hypothetical mechanisms (Figure 4) that can be further tested experimentally and computationally:
Figure 4.
Hypothetical mechanisms of Na+ involvement in GPCR activation. (A) In the inactive apostate receptors, sodium gains access from the extracellular side into the conserved allosteric pocket, where it forms a network of ionic and polar interactions as a part of Na+ and water cluster. (B) Antagonist (Ant) or (C) inverse agonist (InvAgo) binding in the orthosteric pocket is compatible with allosteric Na+ binding and can further stabilize the inactive state. (D) GPCR activation by agonists (Ago), as seen in crystal structures of activated receptors, involves an inward movement of helix VII and an outward movement of helix VI, leading to a partial collapse/reshaping of the allosteric pocket. The resulting collapse of an optimal Na+/water cluster potentiates displacement of Na+ along the internal passage towards the intracellular side of membrane, where it can either (E) engage in transient interactions with other conserved acidic residues [e.g., D(E)3.49 of the D(E)RY motif] and G proteins or (F) exit the protein altogether, traversing the membrane along the Na+ electrochemical gradient, estimated at about 3 kcal/mol.
Na+ bound at D2.50 along with the water cluster stabilize the inactive state [8,30] (Figure 4a,b,c), creating a potential barrier that diminishes basal activity [52] and reduces agonist affinity (see refs in Table 1). In some GPCRs, e.g. b1AR, Na+ appears to stabilize ligand-free receptor without affecting the equilibrium between inactive and active states [13].
Agonist binding disrupts the Na+/water cluster (Figure 4d) and relocates Na+ towards the intracellular side (Figure 4e), thus reducing the energy barrier for receptor activation, and, thus, facilitating signaling. Disruption of the cluster also allows formation of a new pattern of hydrogen bonds in the pocket, which may stabilize active states of the receptor.
The Na+ and water cluster facilitates coupling between the inward movement of helix VII (which compresses the pocket) and the outward movement of helix VI (which allows the Na+ to escape towards the cytoplasm). Note that helix VI has been associated with G protein signaling, whereas helix VII has been associated with β-arrestin signaling in previous studies of GPCRs [63]. Disruption of the sodium cluster (Figure 4E,F) may facilitate the inward movement of helix VII and uncouple it from the outward movement of helix VI, thereby differentially affecting activation of G protein and arrestin pathways, as observed in δ-OR mutants [15].
The Na+ enters the binding pocket from the EC solvent [64], thereby following a strong concentration gradient and electrostatic potential (Figure 4A). By contrast, entrance of the Na+ from the cytoplasm is very unlikely because it goes against the electrochemical potential and the Na+ would have to overcome repulsion of the highly positively charged protein interface on the IC side of GPCRs. On receptor activation and collapse of the sodium pocket, the ion is dislocated towards the IC side (Figure 4E), where it can interact with D3.49 of the D(E)RY motif in helix III, freeing R3.50 for interactions with G protein [65].
On receptor activation and pocket relocation, the Na+ can be further released into cytoplasm (Figure 4F). The entrance for the new ion from the EC solvent would be blocked by the bound agonists, thereby locking the activated receptor into a sodium-free state until agonist dissociation. Such a ‘locking’ mechanism can help to explain the surprising stability of specific ligand-induced activated states, which have been described crystallographically in several GPCRs [19,28,66,67]. It may also be implicated in persistent signaling of some receptors, including the emerging evidence for signaling of internalized GPCRs in early endosomes [68–70].
The transmembrane transfer of Na+ is promoted by both membrane electrostatic potentials and a Na+ concentration gradient (with a combined ΔG ~ 3 kcal/mol [71]). Because it is coupled to conformational changes in the receptor’s 7TM bundle, Na+ transfer can provide an energy source, assisting GPCR signaling by small molecules (Figure 4F).
The Na+ translocation may also help to explain an emerging evidence for the voltage sensing and the gating currents in GPCRs coupled to activation of the receptors [72–74]. Mechanisms of such voltage-dependence in GPCRs are likely to be different from those observed in ion channels, and remain largely unknown.
The above working hypotheses are not mutually exclusive and may work concomitantly. Thus, for example, the sodium cluster can serve as a soft barrier up to a certain stage in activation, its disruption and potentially dislocation of Na+ into the cytoplasm would leave the receptor in a quasi-stable activated state. Though the mechanistic illustrations in Figure 4 are oversimplified, theoretical predictions of potential measurable effects of Na+ may aid the planning of experiments to solve outstanding questions on sodium’s role in GPCR function (Box 1).
Box 1. Outstanding questions.
Which of the remaining untested, human, class A G protein-coupled receptors (GPCRs) specifically bind Na+ at physiological concentrations, and how do the variations in the pocket affect sodium affinity and allosteric effects?
What exactly is the functional role (or roles) of the Na+ in GPCR signal transduction, basal signaling, and coupling/decoupling between G protein and β-arrestin signaling pathways?
When the sodium site collapses on receptor activation, where does the Na+ go?
If the Na+ is transported across the membrane following the membrane electrochemical potential, how could such energy coupling benefit small molecule signaling via GPCRs?
Can other monovalent ions (e.g., K+) bind to the D2.50 pocket in a physiologically/therapeutically relevant way considering the lower concentrations of these ions in the extracellular solvent?
How does the presence of two acidic residues in the pocket (e.g., in protease-activated receptor (PAR1), olfactory receptors, and other receptors) change ion affinity and selectivity, and could it support the binding of divalent ions (e.g., Ca2+) at physiological concentrations?
How does changing concentrations of Na+ (e.g., in neuron synapses or endosomes that carry internalized GPCRs [68,70]) affect the spatio-temporal profile of GPCR signaling?
Could the Na+ site confer a key evolutionary advantage to class A GPCRs, the youngest but most sprawling of all human GPCR classes [88]?
Practical implications for GPCR studies
The knowledge of sodium’s impact on GPCR function may have immediate practical implications for GPCR structural and functional biology, beyond their early use as a screen for ligand agonism activity [9]. Thus, the allosteric effects of sodium can inform the choice of optimal salt conditions for structural studies. For example, antagonist-bound inactive state receptors have been crystallized in high NaCl concentrations [8,75], whereas crystallization of agonist-bound receptors in activated states favors low Na+ conditions [19]. Rationally designed mutations in the sodium pocket can also modulate the receptor’s ability to adopt an inactive, active, or biased signaling state [15], enhance surface expression in cells [76], or stabilize a specific state for structure-based drug discovery and biochemical screening assays. It is also clear that structural knowledge of Na+ binding and its importance in GPCR conformational changes should be taken into account in the efforts to build predictive atomistic models of GPCR activation mechanisms [77], and the first steps in this direction are already yielding interesting insights [30,64,78].
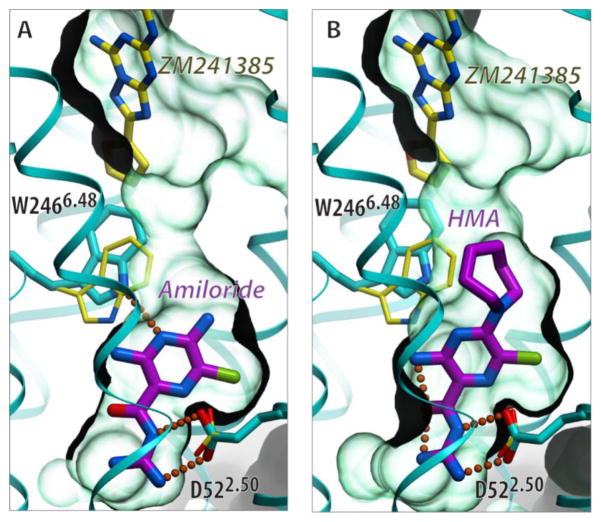
The sodium pocket may also serve as a target for allosteric and bitopic ligands with unique functional features. The size and properties of the sodium-binding pocket in A2AAR/β1AR and other GPCR structures suggest that it can accommodate small (MW~200–300 Da) molecules carrying a positively charged group. Indeed, this might be the case for the diuretic drug amiloride and its derivatives, for which several studies (Table 1) demonstrated binding to several GPCRs from class A α- and β-branches. The binding of amilorides to GPCRs was shown to (i) compete with Na+, (ii) disappear on D2.50 mutations, and (iii) allosterically reduce agonist binding [24,79–87]. Flexible docking of amilorides into the A2AAR sodium pocket [8,30] suggests their snug fit into the pocket, with the positively charged guanidine group forming a salt bridge to D2.50 (Figure 5). The predicted position of the amiloride scaffold is further compatible with the binding of the derivatives with bulky N5 substitutions, which protrude towards the orthosteric ligand binding pocket and may impact not only agonist but also some antagonist binding [24,30].
Figure 5.
Predicted binding of amilorides in the conserved allosteric Na+ pocket. The flexible docking poses of amiloride (A) and 5-(N,N-Hexamethylene)amiloride (HMA) (B) in the sodium pocket of inactive A2AAR suggest that both ligands optimally fit the sodium cavity with only slight conformational changes from the crystal structure (PDB identifier: 4EIY). Neither amiloride ligand (shown with carbon atoms colored magenta) makes direct contact with the orthosteric antagonist ZM241385 (with yellow carbons, positioned as in 4EIY). Their impact on ZM241385 binding can be mediated by the shift in the W2466.48 side chain, which is predicted to be especially pronounced for HMA, in agreement with the stronger negative modulation of antagonist binding by HMA. The receptor is shown by a cyan cartoon with the carbon atoms of the side chains of D522.50 and W2466.48 colored cyan when shown in the crystal structure conformation and colored in yellow for the flexible model conformation. The binding cavity is shown as a semitransparent surface in light green.
Although the affinities of known amilorides to GPCRs are in the micromolar range, more effective allosteric or bitopic molecules targeting the allosteric sodium pocket could have novel functional properties desirable for tool compounds or potential therapeutic applications.
Concluding remarks
The ‘sodium effect’ on GPCR agonist binding has captivated researchers for more than 40 years. Only now, however, have high-resolution crystallographic studies revealed a common structural basis for Na+-specific binding in the center of the 7TM helical bundle, which explains this and other sodium effects on GPCRs. The observed activation-related collapse of the sodium pocket implicates a specific role for the Na+ in the signal transduction mechanism, where the ion translocates towards or into the cytoplasm. The exceptionally high evolutionary conservation of the sodium binding site in most class A GPCRs (and its absence in light-activated visual opsins, non-signaling GPCRs, or GPCRs of other classes) also points to a critical role of the Na+ in the function of those class A GPCRs that are activated by small diffusible ligand binding in the 7TM domain. These crystallographic and evolutionary insights, combined with biochemical evidence, support the physiological omnipresence of sodium as a key co-factor in GPCR functional mechanisms. The robust 3D structural platform in combination with biochemical, biophysical, and computational approaches opens a path towards deciphering further specific details and variations in the Na+-dependent mechanisms in about 680 receptors of class A GPCRs. Some of the remaining key questions regarding the functional role of sodium are challenging, but the answers could help to clarify GPCR signaling mechanisms and enable the discovery of new allosteric and bitopic ligands with distinct functional properties.
Supplementary Material
Highlights.
A Na binding site is found in high-resolution structures of several diverse GPCRs
This allosteric site in the middle of 7TM domain is highly conserved in class A GPCRs
Specific binding of Na+ explains its biochemical and functional effects on GPCRs
Collapse of the pocket and Na+ relocation are involved in GPCR signaling
Acknowledgments
This work was supported by the NIGMS PSI:Biology grants U54 GM094618 (V.K., V.C., and R.C.S.), U19MH82441 (B.L.R.), and R01DA017204 (B.L.R.). We thank K. Kadyshevskaya for assistance with figure preparation, A. Walker for assistance with manuscript preparation, C. Tate, K. Jacobson, A. IJzerman, and M. Audet for helpful discussions, and C. Tate for providing unreleased coordinates of the 4BVN structure.
Glossary
- Allosteric modulation
modification of the orthosteric ligand binding and/or receptor signaling by another ligand or ion that binds to a distinct (allosteric) site
- Ballesteros–Weinstein residue numbering
uses the X.YY format to denote the transmembrane helix number (X) and residue position (YY) relative to the most conserved residue in this helix (X.50). The numbering is used to refer to structurally equivalent residue positions in different G protein-coupled receptors (GPCRs)
- Biased signaling (or functional selectivity)
preferential signaling via certain signaling pathways by specific ligands (or in a mutant receptor), as compared to endogenous ligands (or wild-type receptors). In many cases, signaling bias was observed between G protein and β-arrestin-dependent pathways
- Bitopic ligand
ligand binding to both allosteric and orthosteric sites
- Co-factor
a non-protein chemical compound that is required for the protein’s biological activity
- Constitutive (or basal) activity
receptor signaling in the absence of a bound ligand
- GPCR classes and branches
about 826 human GPCRs are classified into four classes (A, B, C, and F) on the basis of sequence similarity. Class A comprises about 715 GPCRs, subdivided into α, β, γ and δ branches (or groups), as well as a separate group of ~380 olfactory receptors
- Orthosteric ligand
ligand that binds to the same site of the receptor as the endogenous agonist
Footnotes
Supplementary data associated with this article can be found at http://dx.doi.org/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rask-Andersen M, et al. The druggable genome: evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]
- 2.Audet M, Bouvier M. Restructuring g-protein- coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 4.Fredriksson R, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 5.Nygaard R, et al. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Katritch V, et al. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katritch V, et al. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pert CB, et al. Opiate agonists and antagonists discriminated by receptor binding in brain. Science. 1973;182:1359–1361. doi: 10.1126/science.182.4119.1359. [DOI] [PubMed] [Google Scholar]
- 10.Snyder SH, Pasternak GW. Historical review: opioid receptors. Trends Pharmacol Sci. 2003;24:198–205. doi: 10.1016/S0165-6147(03)00066-X. [DOI] [PubMed] [Google Scholar]
- 11.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 12.Kuppuraj G, et al. Factors governing metal-ligand distances and coordination geometries of metal complexes. J Phys Chem B. 2009;113:2952–2960. doi: 10.1021/jp807972e. [DOI] [PubMed] [Google Scholar]
- 13.Miller-Gallacher JL, et al. The 2.1 Å resolution structure of a beta1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS One. doi: 10.1371/journal.pone.0092727. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenalti G, et al. Molecular control of delta-opioid receptor signalling. Nature. 2014;506:191–196. doi: 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson AA, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JF, et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin S, et al. Pivotal role of an aspartate residue in sodium sensitivity and coupling to G proteins of neurotensin receptors. Mol Pharmacol. 1999;55:210–215. doi: 10.1124/mol.55.2.210. [DOI] [PubMed] [Google Scholar]
- 21.Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 24.Heitman LH, et al. Amiloride derivatives and a nonpeptidic antagonist bind at two distinct allosteric sites in the human gonadotropin-releasing hormone receptor. Mol Pharmacol. 2008;73:1808–1815. doi: 10.1124/mol.107.043521. [DOI] [PubMed] [Google Scholar]
- 25.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 26.Angel TE, et al. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci U S A. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada T, et al. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 28.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez-de-Teran H, et al. the role of a sodium ion binding site in the allosteric modulation of the A2A adenosine G protein-coupled receptor. Structure. 2013;21:2175–2185. doi: 10.1016/j.str.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa T, et al. Spontaneous association between opioid receptors and GTP-binding regulatory proteins in native membranes: specific regulation by antagonists and sodium ions. Mol Pharmacol. 1990;37:383–394. [PubMed] [Google Scholar]
- 32.Williams AJ, et al. Somatostatin5 receptor-mediated [35S]guanosine-5′-O-(3-thio)triphosphate binding: agonist potencies and the influence of sodium chloride on intrinsic activity. Mol Pharmacol. 1997;51:1060–1069. doi: 10.1124/mol.51.6.1060. [DOI] [PubMed] [Google Scholar]
- 33.Lin H, et al. Assays for enhanced activity of low efficacy partial agonists at the D(2) dopamine receptor. Br J Pharmacol. 2006;149:291–299. doi: 10.1038/sj.bjp.0706866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yabaluri N, Medzihradsky F. Regulation of mu-opioid receptor in neural cells by extracellular sodium. J Neurochem. 1997;68:1053–1061. doi: 10.1046/j.1471-4159.1997.68031053.x. [DOI] [PubMed] [Google Scholar]
- 35.Selley DE, et al. Effects of sodium on agonist efficacy for G-protein activation in mu-opioid receptor-transfected CHO cells and rat thalamus. Br J Pharmacol. 2000;130:987–996. doi: 10.1038/sj.bjp.0703382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bihoreau C, et al. Mutation of Asp74 of the rat angiotensin II receptor confers changes in antagonist affinities and abolishes G-protein coupling. Proc Natl Acad Sci U S A. 1993;90:5133–5137. doi: 10.1073/pnas.90.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perlman JH, et al. Interactions between conserved residues in transmembrane helices 1, 2, and 7 of the thyrotropin-releasing hormone receptor. J Biol Chem. 1997;272:11937–11942. doi: 10.1074/jbc.272.18.11937. [DOI] [PubMed] [Google Scholar]
- 38.Wang CD, et al. Site-directed mutagenesis of the serotonin 5-hydroxytrypamine2 receptor: identification of amino acids necessary for ligand binding and receptor activation. Mol Pharmacol. 1993;43:931–940. [PubMed] [Google Scholar]
- 39.Rose PM, et al. Aspartate mutation distinguishes ETA but not ETB receptor subtype-selective ligand binding while abolishing phospholipase C activation in both receptors. FEBS Lett. 1995;361:243–249. doi: 10.1016/0014-5793(95)00164-5. [DOI] [PubMed] [Google Scholar]
- 40.Prossnitz ER, et al. Binding of low affinity N-formyl peptide receptors to G protein. Characterization of a novel inactive receptor intermediate. J Biol Chem. 1995;270:10686–10694. doi: 10.1074/jbc.270.18.10686. [DOI] [PubMed] [Google Scholar]
- 41.Tao Q, Abood ME. Mutation of a highly conserved aspartate residue in the second transmembrane domain of the cannabinoid receptors, CB1 and CB2, disrupts G-protein coupling. The Journal of pharmacology and experimental therapeutics. 1998;285:651–658. [PubMed] [Google Scholar]
- 42.Fanelli F, et al. Activation mechanism of human oxytocin receptor: a combined study of experimental and computer-simulated mutagenesis. Mol Pharmacol. 1999;56:214–225. doi: 10.1124/mol.56.1.214. [DOI] [PubMed] [Google Scholar]
- 43.Ringholm A, et al. Pharmacological characterization of loss of function mutations of the human melanocortin 1 receptor that are associated with red hair. The Journal of investigative dermatology. 2004;123:917–923. doi: 10.1111/j.0022-202X.2004.23444.x. [DOI] [PubMed] [Google Scholar]
- 44.Strange PG. Signaling mechanisms of GPCR ligands. Curr Opin Drug Discov Devel. 2008;11:196–202. [PubMed] [Google Scholar]
- 45.Li B, et al. Random mutagenesis of the M3 muscarinic acetylcholine receptor expressed in yeast: identification of second-site mutations that restore function to a coupling-deficient mutant M3 receptor. J Biol Chem. 2005;280:5664–5675. doi: 10.1074/jbc.M411623200. [DOI] [PubMed] [Google Scholar]
- 46.Sealfon SC, et al. Related contribution of specific helix 2 and 7 residues to conformational activation of the serotonin 5-HT2A receptor. J Biol Chem. 1995;270:16683–16688. doi: 10.1074/jbc.270.28.16683. [DOI] [PubMed] [Google Scholar]
- 47.Barak LS, et al. The conserved seven-transmembrane sequence NP(X)2,3Y of the G-protein-coupled receptor superfamily regulates multiple properties of the beta 2-adrenergic receptor. Biochemistry. 1995;34:15407–15414. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 48.Xu W, et al. Functional role of the spatial proximity of Asp114(2.50) in TMH 2 and Asn332(7.49) in TMH 7 of the mu opioid receptor. FEBS Lett. 1999;447:318–324. doi: 10.1016/s0014-5793(99)00316-6. [DOI] [PubMed] [Google Scholar]
- 49.Flanagan CA, et al. The functional microdomain in transmembrane helices 2 and 7 regulates expression, activation, and coupling pathways of the gonadotropin-releasing hormone receptor. J Biol Chem. 1999;274:28880–28886. doi: 10.1074/jbc.274.41.28880. [DOI] [PubMed] [Google Scholar]
- 50.Quitterer U, et al. Na+ ions binding to the bradykinin B2 receptor suppress agonist-independent receptor activation. Biochemistry. 1996;35:13368–13377. doi: 10.1021/bi961163w. [DOI] [PubMed] [Google Scholar]
- 51.Seifert R, Wenzel-Seifert K. Unmasking different constitutive activity of four chemoattractant receptors using Na+ as universal stabilizer of the inactive (R) state. Receptors Channels. 2001;7:357–369. [PubMed] [Google Scholar]
- 52.Seifert R, Wenzel-Seifert K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn-Schmiedeberg’s archives of pharmacology. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 53.Nie J, Lewis DL. Structural domains of the CB1 cannabinoid receptor that contribute to constitutive activity and G-protein sequestration. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:8758–8764. doi: 10.1523/JNEUROSCI.21-22-08758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montaner S, et al. Molecular mechanisms deployed by virally encoded G protein-coupled receptors in human diseases. Annu Rev Pharmacol Toxicol. 2013;53:331–354. doi: 10.1146/annurev-pharmtox-010510-100608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 56.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 57.Reiter E, et al. Molecular Mechanism of beta-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urban JD, et al. Functional selectivity and classical concepts of quantitative pharmacology. The Journal of pharmacology and experimental therapeutics. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 59.Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011;51:117–144. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- 60.Bonde MM, et al. Biased signaling of the angiotensin II type 1 receptor can be mediated through distinct mechanisms. PLoS One. 2010;5:e14135. doi: 10.1371/journal.pone.0014135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bot G, et al. Mutagenesis of the mouse delta opioid receptor converts (-)-buprenorphine from a partial agonist to an antagonist. The Journal of pharmacology and experimental therapeutics. 1998;284:283–290. [PubMed] [Google Scholar]
- 62.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu JJ, et al. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selent J, et al. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comput Biol. 2010;6:e1000884. doi: 10.1371/journal.pcbi.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhorov BS, Ananthanarayanan VS. Signal transduction within G-protein coupled receptors via an ion tunnel: a hypothesis. J Biomol Struct Dyn. 1998;15:631–637. doi: 10.1080/07391102.1998.10508980. [DOI] [PubMed] [Google Scholar]
- 66.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calebiro D, et al. Signaling by internalized G-protein-coupled receptors. Trends Pharmacol Sci. 2010;31:221–228. doi: 10.1016/j.tips.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 69.Mullershausen F, et al. Persistent signaling induced by FTY720-phosphate is mediated by internalized S1P1 receptors. Nature chemical biology. 2009;5:428–434. doi: 10.1038/nchembio.173. [DOI] [PubMed] [Google Scholar]
- 70.Irannejad R, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lodish H, et al. Molecular Cell Biology. W. H. Freeman; 2000. [Google Scholar]
- 72.Ben-Chaim Y, et al. Movement of ‘gating charge’ is coupled to ligand binding in a G-protein-coupled receptor. Nature. 2006;444:106–109. doi: 10.1038/nature05259. [DOI] [PubMed] [Google Scholar]
- 73.Ben Chaim Y, et al. Voltage affects the dissociation rate constant of the m2 muscarinic receptor. PLoS One. 2013;8:e74354. doi: 10.1371/journal.pone.0074354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rinne A, et al. Voltage regulates adrenergic receptor function. Proc Natl Acad Sci U S A. 2013;110:1536–1541. doi: 10.1073/pnas.1212656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang K, et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. doi: 10.1038/nature13083. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dror RO, et al. Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan S, et al. The role of water and sodium ions in the activation of the mu-opioid receptor. Angewandte Chemie (Int Edn) 2013;52:10112–10115. doi: 10.1002/anie.201302244. [DOI] [PubMed] [Google Scholar]
- 79.Gao ZG, et al. Differential allosteric modulation by amiloride analogues of agonist and antagonist binding at A(1) and A(3) adenosine receptors. Biochem Pharmacol. 2003;65:525–534. doi: 10.1016/s0006-2952(02)01556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao ZG, Ijzerman AP. Allosteric modulation of A(2A) adenosine receptors by amiloride analogues and sodium ions. Biochem Pharmacol. 2000;60:669–676. doi: 10.1016/s0006-2952(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 81.Gao ZG, et al. Identification of essential residues involved in the allosteric modulation of the human A(3) adenosine receptor. Mol Pharmacol. 2003;63:1021–1031. doi: 10.1124/mol.63.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Howard MJ, et al. Interactions of amiloride with alpha- and beta-adrenergic receptors: amiloride reveals an allosteric site on alpha 2-adrenergic receptors. Mol Pharmacol. 1987;32:53–58. [PubMed] [Google Scholar]
- 83.Leppik RA, et al. Allosteric interactions between the antagonist prazosin and amiloride analogs at the human alpha(1A)-adrenergic receptor. Mol Pharmacol. 2000;57:436–445. doi: 10.1124/mol.57.3.436. [DOI] [PubMed] [Google Scholar]
- 84.Neve KA. Regulation of dopamine D2 receptors by sodium and pH. Mol Pharmacol. 1991;39:570–578. [PubMed] [Google Scholar]
- 85.Hoare SR, et al. Regulation of human D(1), d(2(long)), d(2(short)), D(3) and D(4) dopamine receptors by amiloride and amiloride analogues. Br J Pharmacol. 2000;130:1045–1059. doi: 10.1038/sj.bjp.0703370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dehaye JP, Verhasselt V. Interaction of amiloride with rat parotid muscarinic and alpha-adrenergic receptors. General pharmacology. 1995;26:155–159. doi: 10.1016/0306-3623(94)00153-e. [DOI] [PubMed] [Google Scholar]
- 87.Pauwels PJ. Competitive and silent antagonism of recombinant 5-HT1B receptors by amiloride. General pharmacology. 1997;29:749–751. doi: 10.1016/s0306-3623(97)00008-6. [DOI] [PubMed] [Google Scholar]
- 88.Krishnan A, et al. The origin of GPCRs: identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS One. 2012;7:e29817. doi: 10.1371/journal.pone.0029817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim SK, et al. Modeling the adenosine receptors: comparison of the binding domains of A2A agonists and antagonists. J Med Chem. 2003;46:4847–4859. doi: 10.1021/jm0300431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barbhaiya H, et al. Site-directed mutagenesis of the human A1 adenosine receptor: influences of acidic and hydroxy residues in the first four transmembrane domains on ligand binding. Mol Pharmacol. 1996;50:1635–1642. [PubMed] [Google Scholar]
- 91.Tsai BS, Lefkowitz RJ. Agonist-specific effects of monovalent and divalent cations on adenylate cyclase-coupled alpha adrenergic receptors in rabbit platelets. Mol Pharmacol. 1978;14:540–548. [PubMed] [Google Scholar]
- 92.Horstman DA, et al. An aspartate conserved among G-protein receptors confers allosteric regulation of alpha 2-adrenergic receptors by sodium. J Biol Chem. 1990;265:21590–21595. [PubMed] [Google Scholar]
- 93.Ceresa BP, Limbird LE. Mutation of an aspartate residue highly conserved among G-protein-coupled receptors results in nonreciprocal disruption of alpha 2-adrenergic receptor-G-protein interactions. A negative charge at amino acid residue 79 forecasts alpha 2A-adrenergic receptor sensitivity to allosteric modulation by monovalent cations and fully effective receptor/G-protein coupling. J Biol Chem. 1994;269:29557–29564. [PubMed] [Google Scholar]
- 94.Wilson MH, et al. The role of a conserved inter-transmembrane domain interface in regulating alpha(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol Pharmacol. 2001;59:929–938. doi: 10.1124/mol.59.4.929. [DOI] [PubMed] [Google Scholar]
- 95.Neve KA, et al. Modeling and mutational analysis of a putative sodium-binding pocket on the dopamine D2 receptor. Mol Pharmacol. 2001;60:373–381. doi: 10.1124/mol.60.2.373. [DOI] [PubMed] [Google Scholar]
- 96.Neve KA, et al. Pivotal role for aspartate-80 in the regulation of dopamine D2 receptor affinity for drugs and inhibition of adenylyl cyclase. Mol Pharmacol. 1991;39:733–739. [PubMed] [Google Scholar]
- 97.Lameh J, et al. Hm1 muscarinic cholinergic receptor internalization requires a domain in the third cytoplasmic loop. J Biol Chem. 1992;267:13406–13412. [PubMed] [Google Scholar]
- 98.Suga H, Ehlert FJ. Effects of asparagine mutagenesis of conserved aspartic acids in helix 2 (D2.50) and 3 (D3.32) of M1-M4 muscarinic receptors on the irreversible binding of nitrogen mustard analogs of acetylcholine and McN-A-343. Biochemistry. 2013;52:4914–4928. doi: 10.1021/bi4003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ardati A, et al. Interaction of [3H]orphanin FQ and 125I-Tyr14-orphanin FQ with the orphanin FQ receptor: kinetics and modulation by cations and guanine nucleotides. Mol Pharmacol. 1997;51:816–824. doi: 10.1124/mol.51.5.816. [DOI] [PubMed] [Google Scholar]
- 100.Mahmoud S, et al. Modulation of silent and constitutively active nociceptin/orphanin FQ receptors by potent receptor antagonists and Na+ ions in rat sympathetic neurons. Mol Pharmacol. 2010;77:804–817. doi: 10.1124/mol.109.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kong H, et al. Mutation of an aspartate at residue 89 in somatostatin receptor subtype 2 prevents Na+ regulation of agonist binding but does not alter receptor-G protein association. Mol Pharmacol. 1993;44:380–384. [PubMed] [Google Scholar]
- 102.Proulx CD, et al. Mutational analysis of the conserved Asp2.50 and ERY motif reveals signaling bias of the urotensin II receptor. Mol Pharmacol. 2008;74:552–561. doi: 10.1124/mol.108.045054. [DOI] [PubMed] [Google Scholar]
- 103.Jagerschmidt A, et al. Mutation of Asp100 in the second transmembrane domain of the cholecystokinin B receptor increases antagonist binding and reduces signal transduction. Mol Pharmacol. 1995;48:783–789. [PubMed] [Google Scholar]
- 104.Warne T, et al. Crystal structures of a stabilized beta1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure. 2012;20:841–849. doi: 10.1016/j.str.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moukhametzianov R, et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a {beta}1-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:8228–8232. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Christopher JA, et al. Biophysical fragment screening of the beta1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. J Med Chem. 2013;56:3446–3455. doi: 10.1021/jm400140q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.