Abstract
Escherichia coli is a genetically diverse species infecting hundreds of millions of people worldwide annually. We examined seven well-characterized E. coli pathogens causing urinary tract infections, gastroenteritis, pyelonephritis and haemorrhagic colitis. Their transport proteins were identified and compared with each other and a non-pathogenic E. coli K12 strain to identify transport proteins related to pathogenesis. Each pathogen possesses a unique set of protein secretion systems for export to the cell surface or for injecting effector proteins into host cells. Pathogens have increased numbers of iron siderophore receptors and ABC iron uptake transporters, but the numbers and types of low-affinity secondary iron carriers were uniform in all strains. The presence of outer membrane iron complex receptors and high-affinity ABC iron uptake systems correlated, suggesting co-evolution. Each pathovar encodes a different set of pore-forming toxins and virulence-related outer membrane proteins lacking in K12. Intracellular pathogens proved to have a characteristically distinctive set of nutrient uptake porters, different from those of extracellular pathogens. The results presented in this report provide information about transport systems relevant to various types of E. coli pathogenesis that can be exploited in future basic and applied studies.
Keywords: Escherichia coli, Pathogenesis, Transporters, Toxins, Iron acquisition, Intra vs. extracellular pathogens
1. Introduction
Escherichia coli is a major source of infant mortality, adult diarrhea and urinary tract infections worldwide [1,2]. Uropathogenic strains are responsible for up to 80% of urinary tract infections (UTIs) and a majority of pyelonephritis cases [3]. Enteroaggregative and enterotoxic pathovars have been the leading causes of travellers’ diarrhea, which has significantly contributed to infant mortality [4]. Enterohaemorrhagic serotypes are currently the leading cause of haemorrhagic colitis, which can lead to fatalities [5].
Some probiotic strains of E. coli including the E. coli Nissle and O83 strains contribute beneficially to human physiology [7,8]. Current research on probiotic E. coli in humans has proven to be clinically useful in the treatment of colitis and inflammatory bowel disease (IBD) syndromes and in combating pathogenic E. coli strains [9,10]. E. coli can have tremendously varied effects on humans and other animals, a consequence of its genetic diversity.
Prominent types of E. coli pathogens have been identified based on varying mechanisms of infection. There are five types of diarrheagenic strains that pertain specifically to the alimentary system: enteropathogenic (EPEC), enterohaemorrhagic (EHEC), enterotoxic (ETEC), enteroinvasive (EIEC), and enteroaggregative (EAEC). Diffusely adherent (DAEC) strains can infect both the urinary tract and the gastrointestinal tract due in part to their diverse mechanisms of host adhesion [6]. In our study, seven representative E. coli pathogens as well as E. coli K12 were examined. We use the first three letters/numbers of these strain designations as abbreviations to refer to these strains. Table 1 presents these abbreviations as well as basic information about the eight strains of E. coli examined.
Table 1.
Seven E. coli pathovars, the non-virulent E. coli K12 strain, and their basic traits.
| Strain | Abbr. | Genome size (Mbp) | # of proteins | Host location | Pathological condition | Pathovar type |
|---|---|---|---|---|---|---|
| 55989 | 559 | 5.15 | 4759 | Extracellular | Diarrhea | EAEC |
| ABU 83972 | ABU | 5.13 | 4796 | Intracellular | Bacteremia | UPEC |
| APEC O1 | APE | 5.50 | 4853 | Intracellular | Respiratory infection (avians) | AIEC |
| CFT073 | CFT | 5.23 | 5369 | Intracellular | Urinary tract infection & pyelonephritis | UPEC |
| E24377A | E24 | 5.25 | 4991 | Extracellular | Enterotoxicity | ETEC |
| UMN026 | UMN | 5.36 | 5014 | Extra/intracellular | Urinary tract infection | UPEC |
| O157:H7 | O15 | 5.70 | 5477 | Extracellular | Haemorrhagic colitis | EHEC |
| K12 MG1655 | K12 | 4.64 | 4290 | Extracellular | None | Non-virulent |
The general paradigm of the pathogenic mechanism associated with E. coli virulence involves (1) adhesion, (2) protein injection into host cells, (3) subversion of signaling mechanisms and (4) colonization leading to impaired immune responses, membrane potential disruption and cytoskeletal manipulation [11]. During the injection stage, E. coli typically secretes effectors into host cells using one or more protein secretion systems, either to escape the immune response or to alter signaling pathways. Host membrane potential disruption and apoptosis induction are employed by several pathovars.
Virulence genes are often clustered in “pathogenicity islands” that reflect horizontal transfer [12]. Horizontal gene transfer has allowed E. coli to survive under a variety of in vivo conditions, increasing the virulence of existing pathogens and creating new ones [13,14]. Because the pan genome of E. coli far exceeds the core genome, perhaps by as much as 20-fold [15], this increase in genetic plasticity allows for fluctuations in gene content, further contributing to divergence of E. coli pathogenic mechanisms [16].
2. Methods
2.1. G-BLAST search of E. coli proteomes
The proteomes of eight E. coli strains were screened for homologs of all proteins contained in the Transporter Classification Database (TCDB; www.tcdb.org) as of September, 2012 using a program designed for this purpose, G-BLAST [17]. The program retrieves information for both the genome query and the TC top hit sequences, TC#, protein size in number of amino acyl residues, number of predicted TMSs using the HMMTop 2.0 Program, e-value for the query and hit proteins, regions of sequence similarity and regions of TMS overlap. FASTA-formatted protein sequences from the completed genome were used. Each putative open-reading frame was used as a query in the BLASTP software to search for homologous proteins in TCDB. The low complexity filter was not used as it is normally of value only for larger datasets including proteins with multiple repeat elements. In addition, each open reading frame [18] was scanned with the HMMTOP 2.0 program [19] to predict the number of putative transmembrane segments (TMSs). The Web-based Hydropathy, Amphipathicity and Topology (WHAT) program [20] was used with a window size of 19 residues and an angle of 100° to display the hydropathy plot for individual proteins in order to resolve the differences in the numbers of TMSs between the proteins retrieved and their TCDB homologs. The plot generated by WHAT allows the user to judge if a program such as HMMTOP has missed a TMS or has predicted a TMS inappropriately. A cut-off value of 0.0001 was used with the G-BLAST program so proteins retrieved with larger values (greater sequence divergence) were not recorded. Proteins with no predicted TMSs were eliminated so that only integral membrane proteins, primarily multi-spanning membrane proteins, were retrieved. Proteins with only an N-terminal signal sequence are numerous because these proteins include almost all periplasmic, outer membrane and secreted proteins that are exported via the general secretory pathway (Sec) or twin arginine translocase (TAT). The topological prediction programs often miss these TMSs, recording them to have zero TMSs. Consequently, the numbers retrieved were not reliable and were therefore not always recorded. For example, single TMS proteins such as extracytoplasmic solute binding receptors of ABC transport systems were often predicted to lack a TMS, and therefore these proteins were not included in our study of the integral membrane transport proteins.
2.2. Identification of distant transport protein homologs
Proteins retrieved between the values of 0.0001 and 0.1 were examined manually to determine the likelihood that these proteins were members of recognized transport protein families, or if they might comprise representatives of novel families of putative transport proteins. A total of 82 non-orthologous homologous proteins were retrieved using the 0.0001–0.1 cutoff, but only 10 proved to be recognizable transport proteins. These were incorporated into TCDB. The 10 proteins were manually examined by conducting searches as follows. (1) TC-BLAST searches provided preliminary evidence for family assignment. (2) NCBI BLAST searches provided confirmation or refutation of family assignment based on the conserved domain database (CDD) and hits obtained with values to the query sequence of less than 1 × 10−7. (3) Topological analyses revealed similarities and differences between the query sequence and members of the assigned family. (4) Proteins proving to represent new potential families were included in TC subclass 9.B.
Candidate proteins were subsequently examined in greater detail to estimate their substrate specificities. On the basis of the numbers and locations of TMSs as well as degrees of sequence similarity with entries of known function in TCDB, transport proteins were classified into families and subfamilies of homologous transporters according to the classification system presented in TCDB. Regions of sequence similarity were examined using the WHAT program which shows hydropathy plots to ensure that homology was in a transmembrane region of 3 or more TMSs and not only in hydrophilic domains. Proteins encoded within single multicistronic operons were often identified in order to gain evidence for multicomponent systems and to help deduce functions. Operon analyses (genome context, a.k.a., synteny analyses) were performed for candidate proteins with assigned or unassigned transport functions as described in Castillo and Saier (2010) and Reddy et al. (2012) [21,22].
2.3. Overview of programs used
Transport proteins thus obtained were systematically analyzed for unusual properties using published [17] and unpublished in-house software. Among the programs described by Reddy and Saier [17], used in this report, were the GSAT, Protocol1, Protocol2, TSSearch, SSearch and GBlast programs. Unpublished software was used to tabulate information according to TC# or other criteria such as substrate type. Unusual characteristics of the query E. coli sequences were identified based in part on topologies that differed from corresponding family members in TCDB as well as e-values obtained with G-BLAST. Unusual properties can result from events such as genetic deletion and fusion, sometimes resulting in the gain or loss of extra domains or the generation of multifunctional proteins. Such results can be reflective of the actual protein sequence, but they can also be artifactual, due to sequencing errors or incorrect initiation codon assignment. In the latter cases, but not the former, the protein sequences were either corrected when possible or eliminated from our study.
Orthologous relationships among homologs were estimated by measuring the percent identities. We initially used an arbitrary cutoff of 95% identity, a value appropriate in view of the similarities of the strains considered. Homologs with lower percent identities were examined manually, revealing occasional sets of orthologs that exhibited less than 95% identity (See Table S1 for orthologous assignments).
3. Results
3.1. Overview of transporter types
According to the transporter classification system, there are five defined classes of transport systems (classes 1–5) as well as a class of transporter auxiliary proteins (class 8) and a class of incompletely defined transporters (class 9). These will be considered in sequence.
3.1.1. Pore-forming proteins
Eight E. coli genomes were analyzed for the occurrence of transport proteins using the Transporter Classification Database (TCDB; www.tcdb.org [23]) and the G-Blast program [17]. The results are summarized according to TC subclass in Table 2, Fig. 1, and Table S1. Examining the total number of transport proteins present in these 8 genomes, we see that E. coli K12 has the fewest at 1128. The most found in any genome is 1287, 159 more than in K12. However, the seven pathogenic species combined contained 838 unique transport proteins lacking in K12.
Table 2.
Numbers of proteins (even numbered columns) and percent distribution of subclasses by TC number (odd numbered columns) for all eight strains of E. coli as well as numbers of transport proteins per strain (bottom) and all strains combined, with and without K12 (last four columns).
| Subclass | 559 | % | CFT | % | ABU | % | APE | % | E24 | % | UMN | % | O15 | % | K12 | % | All | % | All-K12 | % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.A | 42 | 3.4 | 37 | 2.9 | 41 | 3.2 | 41 | 3.3 | 42 | 3.5 | 39 | 3.1 | 42 | 3.47 | 39 | 3.5 | 323 | 3.3 | 284 | 3.3 |
| 1.B | 94 | 7.6 | 103 | 8.0 | 108 | 8.4 | 91 | 7.3 | 89 | 7.4 | 95 | 7.5 | 96 | 7.93 | 67 | 5.9 | 743 | 7.5 | 676 | 7.7 |
| 1.C | 15 | 1.2 | 18 | 1.4 | 20 | 1.6 | 15 | 1.2 | 11 | 0.9 | 15 | 1.2 | 21 | 1.74 | 9 | 0.8 | 124 | 1.3 | 115 | 1.3 |
| 1.E | 5 | 0.4 | 6 | 0.5 | 4 | 0.3 | 6 | 0.5 | 3 | 0.2 | 7 | 0.6 | 14 | 1.16 | 4 | 0.4 | 49 | 0.5 | 45 | 0.5 |
| 2.A | 318 | 25.6 | 325 | 25.4 | 319 | 24.8 | 315 | 25.3 | 309 | 25.7 | 319 | 25.2 | 299 | 24.71 | 311 | 27.6 | 2515 | 25.5 | 2204 | 25.2 |
| 2.C | 5 | 0.4 | 5 | 0.4 | 4 | 0.3 | 4 | 0.3 | 3 | 0.2 | 4 | 0.3 | 3 | 0.25 | 4 | 0.4 | 32 | 0.3 | 28 | 0.3 |
| 3.A | 358 | 28.8 | 378 | 29.5 | 386 | 30.0 | 369 | 29.7 | 353 | 29.4 | 390 | 30.8 | 369 | 30.50 | 340 | 30.1 | 2943 | 29.8 | 2603 | 29.8 |
| 3.B | 4 | 0.3 | 3 | 0.2 | 5 | 0.4 | 5 | 0.4 | 5 | 0.4 | 4 | 0.3 | 4 | 0.33 | 4 | 0.4 | 34 | 0.3 | 30 | 0.3 |
| 3.D | 69 | 5.5 | 66 | 5.2 | 64 | 5.0 | 59 | 4.7 | 69 | 5.7 | 72 | 5.7 | 68 | 5.62 | 69 | 6.1 | 536 | 5.4 | 467 | 5.3 |
| 4.A | 56 | 4.5 | 62 | 4.8 | 62 | 4.8 | 61 | 4.9 | 55 | 4.6 | 61 | 4.8 | 51 | 4.21 | 49 | 4.3 | 457 | 4.6 | 408 | 4.7 |
| 4.B | 2 | 0.2 | 4 | 0.3 | 4 | 0.3 | 4 | 0.3 | 2 | 0.2 | 2 | 0.2 | 2 | 0.17 | 2 | 0.2 | 22 | 0.2 | 20 | 0.2 |
| 4.C | 10 | 0.8 | 17 | 1.3 | 16 | 1.2 | 11 | 0.9 | 9 | 0.7 | 11 | 0.9 | 7 | 0.58 | 8 | 0.7 | 89 | 0.9 | 81 | 0.9 |
| 4.D | 9 | 0.7 | 10 | 0.8 | 12 | 0.9 | 9 | 0.7 | 8 | 0.7 | 7 | 0.6 | 9 | 0.74 | 8 | 0.7 | 72 | 0.7 | 64 | 0.7 |
| 5.A | 43 | 3.5 | 44 | 3.4 | 44 | 3.4 | 45 | 3.6 | 42 | 3.5 | 43 | 3.4 | 43 | 3.55 | 45 | 4.0 | 349 | 3.5 | 304 | 3.5 |
| 5.B | 3 | 0.2 | 2 | 0.2 | 2 | 0.2 | 2 | 0.2 | 2 | 0.2 | 4 | 0.3 | 3 | 0.25 | 3 | 0.3 | 21 | 0.2 | 18 | 0.2 |
| 8.A | 37 | 3.0 | 45 | 3.5 | 43 | 3.3 | 40 | 3.2 | 39 | 3.2 | 42 | 3.3 | 37 | 3.06 | 33 | 2.9 | 316 | 3.2 | 283 | 3.2 |
| 8.B | 1 | 0.1 | 1 | 0.1 | 1 | 0.1 | 1 | 0.1 | 1 | 0.1 | 1 | 0.1 | 1 | 0.08 | 1 | 0.1 | 8 | 0.1 | 7 | 0.1 |
| 9.A | 87 | 7.0 | 64 | 5.0 | 63 | 4.9 | 77 | 6.2 | 71 | 5.9 | 66 | 5.2 | 60 | 4.96 | 42 | 3.7 | 530 | 5.4 | 488 | 5.6 |
| 9.B | 86 | 6.9 | 91 | 7.1 | 89 | 6.9 | 88 | 7.1 | 89 | 7.4 | 85 | 6.7 | 81 | 6.69 | 90 | 8.0 | 699 | 7.1 | 609 | 7.0 |
| total | 1244 | 100 | 1281 | 100 | 1287 | 100 | 1243 | 100 | 1202 | 100 | 1267 | 100 | 1210 | 100 | 1128 | 100 | 9862 | 100 | 8734 | 100 |
Fig. 1.
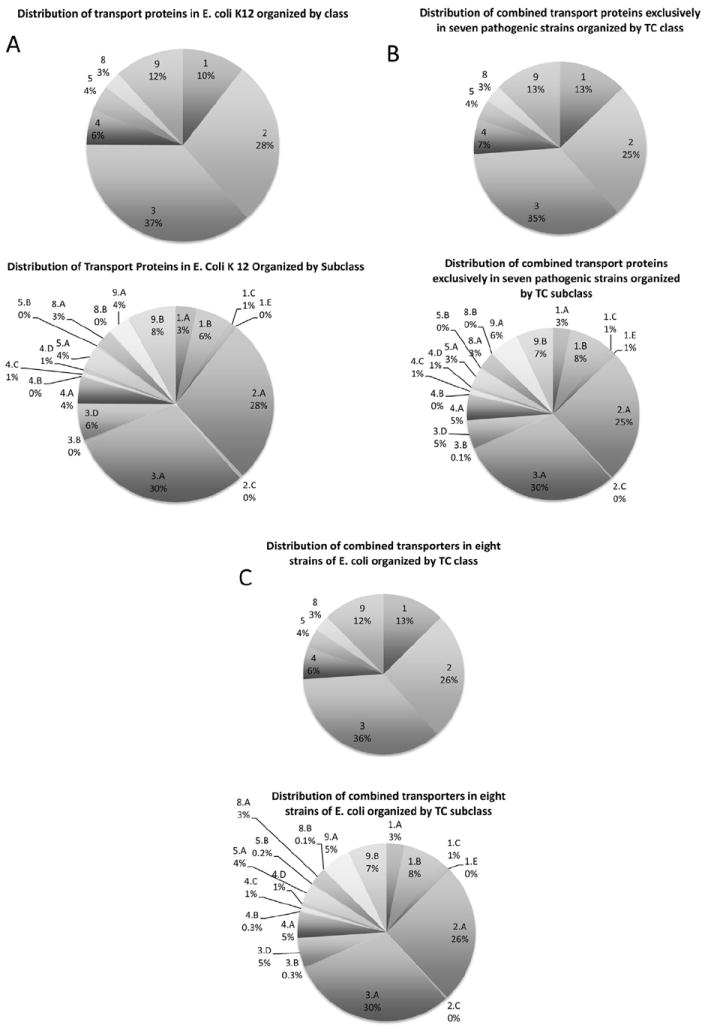
Distribution of transport proteins according to TC class (top) & TC subclass (bottom) for E. coli K12 str. MG1655 (top) and the seven pathogenic strains combined (bottom). Similar distributions for the eight E. coli strains individually and combined are presented in Supplementary Figs. S1A–S1I.
TC subclass 1.A in TCDB includes α-type channels. The eight strains have 37–42 such channel proteins. Surprisingly, however, the pathogenic organisms possess 24 non-orthologous channel proteins lacking in K12 (Table 2). It appears unlikely that α-type channels are major contributors to E. coli pathogenesis (Fig. 1).
TC subclass 1.B includes outer membrane β-type porins. In this subclass, we find striking differences between the organisms, where K12 has far fewer than any of the other strains. The largest number of these porins is 108 observed in ABU, and the seven pathogens possess 151 porins proteins non-orthologous to those in K12 (Table 2). This observation suggests that porins contribute to pathogenesis as confirmed and explained below.
TC subclass 1.C includes pore-forming toxins. K12 has only nine such proteins, and E24 has just two more, but all the others have between 15 and 20 such toxins, and 51 toxins, non-orthologous to those in K12, were identified (Table 2). As expected, an array of toxins characterizes each of the pathogens. It should be noted that toxins that do not form transmembrane pores were not included in our study.
Small α-type channel-forming holins (TC subclass 1.E.) function in release of auto-lysins which allow phage particle release when phage-encoded, or self-destruction by a suicide-type process when chromosomally encoded [17]. The E. coli strains have 3–7 holins each. However, 19 of these proteins are non-orthologous to those found in K12 (Table 2). It is possible therefore that chromosomally encoded holins play important roles in pathogenicity, possibly in programmed cell death as an aid to biofilm formation and communal life [24-26].
3.1.2. Secondary carriers
The largest number of transport systems, but not the largest number of transport proteins, includes secondary carriers belonging to TC subclass 2.A. The eight strains have anywhere from 309 to 325 such systems. K12 has 311, and 96 proteins non-orthologous to those in K12 were identified, many of which are likely to contribute to specific types of pathogenicity.
3.1.3. Primary active transporters
TC subclass 3.A consists of pyrophosphate-hydrolysis driven primary active transporters, usually multi-component systems. K12 has the fewest such systems (340), while UMN has the most (390), and 155 proteins found in pathogens appear to have no orthologs in K12. Many of these systems play essential roles in virulence. The strains examined have 3–5 decarboxylation driven sodium pumps, which probably play a housekeeping function in the maintenance of ionic homeostasis. Subclass 3.D, ion (H+ or Na+)-pumping electron carriers, also exhibit little variation with only seven proving to be non-orthologous to those of K12.
3.1.4. Group translocators
TC class 4 includes group translocators that modify their substrates during transport. Subclass 4.A includes the proteins of the multi-component sugar transporting phosphotransferase system (PTS) [27]. While K12 has the fewest such systems (49), both CFT and ABU have 13 more (62). In fact, 43 PTS proteins proved to be non-orthologous to those found in K12. These systems, likely to be important virulence factors, will be analyzed below.
Each of the eight different strains has between 7 and 12 TC subclass 4.D group translocating glycosyl transferases (4.D [28,29]). Interestingly, the pathogens possess eight sets of orthologs not found in K12, suggesting that unique glycosyl transferases may serve different functions in the different pathogens. Some of them could function in the production of capsules, known to be virulence factors in many pathogenic bacteria [30].
3.1.5. Transmembrane electron carriers
Electron-carriers (TC class 5) that transfer electrons from one side of the membrane to the other, thereby influencing cellular energetics [31], do not vary in numbers appreciably between the eight strains.
3.1.6. Poorly characterized transporters
Tremendous variation is observed between the numbers of poorly characterized transporters found in subclass 9.A. K12 has the fewest, with only 42 such proteins while 559 has over twice this number with 87. Most of the pathogens have between 60 and 70 such proteins, and 94 of these proteins are lacking in K12. This shows that the pathogens possess large variable numbers of poorly characterized transporters that are likely to serve highly specific functions within the cell. It is probable that they remain poorly characterized because they are not found in E. coli K12 and other commonly studied model bacteria. Elucidation of their functions and mechanisms of action is likely to prove particularly interesting and important for specific aspects of E. coli pathogenesis. In contrast to the situation observed for subclass 9.A, the strains examined have relatively little variation in the numbers of subclass 9.B members. The smallest number is found in O15 (81), and the largest number is found in CFT (91) with K12 having 90. Nevertheless, the pathogens collectively have 26 such proteins lacking in K12.
3.2. Pathogenicity-related transport protein analyses
Transport proteins likely to be important in pathogenicity include (1) toxins, (2) trans-envelope protein secretion systems, (3) outer membrane protein secretion systems, and (4) outer membrane iron-siderophore receptors that function with cytoplasmic membrane ABC-type iron uptake porters. The transport proteins that fall into these four classes are summarized in Tables 3-6, respectively.
Table 3.
Toxins identified in E. coli pathovars and K12.
| Family | TC ID | Function | 559 | ABU | APE | CFT | E24 | UMN | O15 | K12 |
|---|---|---|---|---|---|---|---|---|---|---|
| HlyE | 1.C.10.1.1 | Pore-forming hemolysin, HlyE | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 |
| RTX toxin | 1.C.11.1.3 | Pore-forming hemolysin, HlyA | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| IIITCP | 1.C.36.1.1 | T3SS: pore-forming complex, EspB/D | 0 | 0 | 0 | 0 | 0 | 2 | 0 | |
| IIITCP | 1.C.36.3.1 and 3.2 | T3SS: pore-forming complex, SipBD | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| IIITCP | 1.C.36.5.1 | T3SS: pore-forming complex, SseBCD | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Shiga toxin B | 1.C.54.1.1 | Shiga toxin B, St-B | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Clostridial cytotoxin | 1.C.57.3.2 and 3.3 | Pore formation; necrosis in host, Cnf | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| S-PFT | 1.C.75.1.1 | Pore-forming hemolysin, ShlA | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| Cytotoxic major fimbrial subunit (Mrx-A) | 1.C.80.1.1 | Adhesive fimbriae (pore formation) | 2 | 4 | 3 | 5 | 2 | 2 | 4 | 2 |
| Cytolethal distending toxin | 1.C.98.1.1 | Cell cycle arrest/apoptosis, CdtABC | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
Table 6.
Outer membrane iron-siderophore receptors.
| Family | TC ID | Receptor specificity | 559 | ABU | APE | CFT | E24 | UMN | O15 | K12 |
|---|---|---|---|---|---|---|---|---|---|---|
| OMR | 1.B.14.1.1 | Ferric coprogen, FhuE | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1.B.14.1.2 | Ferrichrome, FhuA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 1.B.14.1.3 | Ferric enterobactin, IroN | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 1.B.14.1.4 | Catecholate, CirA | 1 | 2 | 2 | 3 | 1 | 1 | 2 | 1 | |
| 1.B.14.1.9 | Catecholate, Flu | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 1.B.14.1.10 | Ferroxamine, FoxA | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | |
| 1.B.14.1.12 | Ferric citrate, FecA | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | |
| 1.B.14.1.13 | Ferric receptor, CfrA | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |
| 1.B.14.1.15 | Ferrichrome, FcuA | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 1.B.14.2.2 | Heme, HemA | 0 | 2 | 1 | 2 | 0 | 1 | 1 | 0 | |
| 1.B.14.3.1 | Cobalamin, BtuB | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 1.B.14.4.1 | Cupric ion, OprC | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | |
| 1.B.14.7.2 | Ferric-yersiniabactin, FyuA | 2 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | |
| 1.B.14.9.3 | Ferrichrome/aerobactin, IutA | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | |
| 2.C.1.1.1 | TonB | 1 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | |
| 2.C.1.2.1 | TolA | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 1 |
3.2.1. Toxins identified in E. coli strains
Table 3 summarizes the toxins identified in the eight E. coli strains included in this study. In addition to the pore-forming subunits that function in conjunction with type III secretion systems (IIITCP), seven families of toxins are represented. These will be discussed sequentially according to their TC numbers.
HlyE is a well-characterized toxin found in E. coli, Salmonella and Shigella species. Its high resolution X-ray structure is available, and models of its membrane-inserted homooligomeric pore complex have been proposed [32,33]. Five of the eight E. coli strains examined possess this toxin.
Another hemolysin is the E. coli Hemolysin A (HlyA), part of the RTX superfamily of pore-forming proteins [34]. The general mechanism proposed for HlyA is as follows: the toxin (1) recognizes and binds to β 2-integrin receptors on host cell membranes, (2) inserts into the host membrane forming nonselective pores, and (3) transports a toxin that inhibits host protein kinase B, leading to apoptosis of the host cell [35]. This toxin was found only in two intracellular strains, both of which have HlyE as well.
Type 3 Secretion System (T3SS) pore-forming proteins (IIITCP) are typically involved in attaching/effacing mechanisms in enteropathogenic strains [15]. The EspB/D complex and other homologous pore-forming protein pairs are secreted into the host cell and oligomerize to form translocation pores at the site of contact. EspB with EspD, for example, provide translocation functions for E. coli effector proteins and inhibit host phagocytosis by altering host cell cytoskeletal functions [36]. IIITCP family proteins were found in two pathovars.
The prophage-encoded shiga toxin (Stx) is a heteromeric protein with a glycolipid-recognizing, pore-forming, pentameric B subunit and a single A chain with N-glycosidase activity that results in cleavage of a single adenine in the 28S rRNA, resulting in blockage of protein synthesis [37]. Two StxB pore-forming toxins appear to be encoded in the O15 genome, although no such toxin was identified in the other strains.
Clostridial cytotoxins (CCT) are tripartite proteins with N-terminal catalytic domains, C-terminal receptor regions and central channel-forming domains [38]. Upon endocytosis into the host cell, the channel-forming domain is activated by acidic pH, which induces conformational changes promoting insertion into the endosomal membrane, concomitant with channel formation. The catalytic domain of CCT is translocated across the membrane, resulting in inactivation of Rho family GTPases and reorganization of the host actin cytoskeleton [39]. Two homologs of the clostridial cytotoxin were identified (see Table 3).
Serratia pore-forming toxins (S-PFT) (1.C.75) are exported through a two partner secretion system (TPS; TC# 1.B.20) [40]. Three organisms possess this type of pore-forming hemolysin, ABU, CFT and O15. The former two have huge toxins of 3216 amino acids that are nearly identical. By contrast, the O15 homolog is 1270 amino acids in length and corresponds in sequence to the N-terminal portion of the larger toxins.
Toxins of the Cytolethal Distending Toxin (CDT) family are tripartite protein complexes, CdtABC, that induce DNA cleavage, cell cycle arrest, and apoptosis in host epithelial and immune cells [41]. The catalytic subunit of the complex possesses phosphodiesterase activity that cleaves the host DNA, causing G2/M cell cycle arrest [42]. CdtA and C are dimeric subunits that deliver CdtB to the host cell cytoplasm [43]. The complete CdtA/B/C trimeric toxin system (1.C.98.1.1) was found in only one of the eight E. coli strains, APE.
3.2.2. Trans-envelope protein secretion systems in pathogenicity
Table 4 summarizes the various protein secretion systems present in the E. coli strains examined. These include (1) type I secretion systems (T1SS; ABC; TC#3.A.1), (2) type II secretion systems (T2SS; SEC-SRP complexes; TC#3.A.5) for secretion across the inner membrane, acting together with the Main Terminal Branch (MTB; TC#3.A.15) for secretion across the outer membrane, (3) type III secretion systems (T3SS; Fla; Path; TC#3.A.6), (4) type IV secretion systems (T4SS; Conj; Vir; TC#3.A.7), and (5) type VI secretion systems (T6SS; TC#3.A.23) [44].
Table 4.
Pathogenicity-related transenvelope protein secretion systems.
| Family | TC ID | Function | 559 | ABU | APE | CFT | E24 | UMN | O15 | K12 |
|---|---|---|---|---|---|---|---|---|---|---|
| T1SS | 3.A.1.105.3 and 105.4 | Drug exporter 1 | 8 | 8 | 8 | 8 | 7 | 8 | 8 | 8 |
| 3.A.1.109.4 | Protein exporter 1 | 0 | 4 | 0 | 4 | 0 | 2 | 2 | 0 | |
| 3.A.1.110.1 | Protein exporter 2 | 0 | 3 | 0 | 4 | 0 | 0 | 0 | 0 | |
| 3.A.1.113.3 | Peptide 3 exporter | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 3.A.1.134.7 | Peptide 7 exporter | 15 | 14 | 13 | 14 | 13 | 13 | 14 | 13 | |
| T2SS | 3.A.5.1.1 | SECeSRP complex | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| GSP | ||||||||||
| MTB | 3.A.15.1.1 | Pullulanase secretion system | 8 | 13 | 14 | 8 | 8 | 7 | 0 | 8 |
| 3.A.15.2.1 | Pilin secretion/fimbrial assembly system | 5 | 4 | 4 | 3 | 4 | 5 | 4 | 4 | |
| 3.A.15.3.1 | LSP/fimbrial assembly system | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |
| T3SS | 3.A.6.1.1 | Type III secretion system complex | 4 | 1 | 1 | 1 | 6 | 8 | 13 | 1 |
| 3.A.6.2.1 | Flagellar protein export complex | 11 | 11 | 11 | 11 | 11 | 17 | 11 | 10 | |
| T4SS | 3.A.7.3.1 | Pertussis toxin exporter | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| 3.A.7.7.1 | Trs DNA transfer protein complex | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| 3.A.7.11.1 | Type IV beta-proteobacterial DNA secretion system | 2 | 1 | 4 | 1 | 2 | 2 | 3 | 1 | |
| 3.A.7.13.2 | Plasmid pLS20 conjugation system | 1 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | |
| 3.A.7.14.2 | Conjugation system plasmid DNA IntP/TcpA-J | 5 | 4 | 3 | 5 | 5 | 4 | 5 | 5 | |
| T6SS | 3.A.23.1.1 | T6SS VasA-L | 19 | 8 | 18 | 7 | 17 | 13 | 13 | 0 |
| 3.A.23.2.1 | T6SS EvpA-P | 19 | 5 | 6 | 5 | 6 | 2 | 2 | 1 |
Type I secretion systems (T1SSs) identified are members of the ABC Protein Exporter-1 (TC#3.A.1.109) and Protein Exporter-2 (TC#3.A.1.110) families within the ABC functional superfamily [45]. ABC exporters consist of two transmembrane domains and two cytosolic ATP-hydrolyzing domains. T1SS efflux involves ATP hydrolysis-coupled translocation of the substrate across the two bacterial membranes in a single energy-coupled step, utilizing an outer membrane factor (OMF; TC#1.B.17) and a membrane fusion protein (MFP; TC#8.A.1) to couple cytoplasmic membrane transport with that across the periplasm and outer membrane ([46,47]). Protein exporter families of this type were found in abundance in the pathovars.
Type III secretion systems are found in numerous Gram-negative bacterial strains and are used for secretion of flagellar and pathogenicity effector proteins. Effector proteins can be exported across both bacterial membranes as well as the host cell membrane in a single step. The type III protein secretion system includes a needle complex, the injectisome, which attaches to the host cell membrane [48-51]. Several of these systems were present in E. coli pathogens (Table 4).
As expected, the complete general secretory pathway (TC#2.A.5.1.1) and the complete flagellar protein export complex (TC#3.A.6.2.1) were found in all strains. The UMN pathovar appears to possess six additional key components of the flagellar system (Table 4).
T4SSs vary tremendously with respect to sequence divergence and numbers of constituents required for function. An examination of potential T4SS components in the eight E. coli strains revealed the presence of potential subunits, suggesting that some of these organisms utilize this mechanism to promote pathogenesis.
Type VI secretion systems are found in multiple Gram-negative bacteria. Key constituents include an ATPase protein, ClpV, a phage tail-like protein that spans the outer membrane, and a “tail-spike” protein, VgrG, which penetrates the host membrane and dissociates from the complex to allow contact-dependent translocation of proteins into the host cell cytoplasm [52]. In the E. coli strains studied here, two sets of complete T6SS complex homologs were identified (VasA-L of 3.A.23.1.1 and EvpA-P of 3.A.23.2.1) in several strains.
3.2.3. Outer membrane protein secretion systems
Table 5 summarizes the outer membrane protein secretion systems present in the eight E. coli strains studied. These include members of the following families: Autotransporter-1 (TC#1.B.12), Autotransporter-2 (TC#1.B.40), outer membrane factors (OMF; TC#1.B.17), fimbrial usher proteins (FUP; TC#1.B.11), two partner secretion systems (TPS; TC#1.B.20), secretins (TC#1.B.22), outer membrane protein insertion porins (OmpIP; TC#1.B.33), curli fiber subunits (CsgA; TC#1.B.48), and putative Autotransporter-3 (Invasins; TC#1.B.54).
Table 5.
Outer membrane protein secretion systems.
| Family | TC ID | Function | 559 | ABU | APE | CFT | E24 | UMN | O15 | K12 |
|---|---|---|---|---|---|---|---|---|---|---|
| FUP | 1.B.11.2.1 and 2.2 | Type π fimbrial usher protein, PapC and YbgQ | 3 | 3 | 3 | 4 | 3 | 3 | 2 | 1 |
| 1.B.11.3.2, 3.3, 3.5. 3.6 and 3.9 | Type γ fimbrial usher protein, MrkC, EcpC, YcbS, YraJ, and FimD | 8 | 8 | 5 | 6 | 6 | 8 | 7 | 6 | |
| 1.B.11.4.1 | Type α fimbrial usher protein, CfaC | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | |
| AT-1 | 1.B.12.1.1 | Adhesin, AidA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| 1.B.12.1.2 | Virulence factor, IcsA (VirG) | 2 | 2 | 0 | 2 | 2 | 0 | 1 | 0 | |
| 1.B.12.1.3 | Fibronectin binding protein, MisL | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 0 | |
| 1.B.12.2.1 | Pertactin, Ptt | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | |
| 1.B.12.2.2 | Tracheal colonization factor, TcfA | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | |
| 1.B.12.2.3 | Resistance to killing proteins, BrkA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| 1.B.12.2.4 | Putative adhesin/protease/pertactin | 2 | 3 | 1 | 3 | 0 | 1 | 0 | 0 | |
| 1.B.12.4.2 and 4.3 | Serine proteases, EspP and Tsh | 3 | 3 | 1 | 3 | 0 | 1 | 1 | 0 | |
| 1.B.12.5.5 | Virulence factor, BigA | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 1.B.12.8.2 | Auto-aggregation adhesin, Flu | 1 | 3 | 1 | 3 | 2 | 1 | 1 | 1 | |
| 1.B.12.8.3 | Adhesin, TibA | 2 | 2 | 2 | 1 | 2 | 3 | 3 | 1 | |
| OMF | 1.B.17.1.1 and 1.4 | Hemolysin, drug and siderophore export protein, TolC | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 |
| 1.B.17.3.9 | Multidrug resistance outer membrane export protein, MdtP | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| TPS | 1.B.20.1.3 and 1.4 | Two-partner secretion exporter, CdiB and OptB | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 |
| Secretin | 1.B.22.1.1 | PulD secretin | 1 | 2 | 2 | 1 | 1 | 1 | 0 | 1 |
| 1.B.22.3.3 | YscC secretin | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | |
| 1.B.22.4.2 | HofQ secretin | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 1.B.22.5.1 | Phage gene IV protein secretin | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| OMIP | 1.B.33.1.3 | Outer membrane insertion complex | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| AT-2 | 1.B.40.1.2 | Hemagglutinin | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| 1.B.40.1.3 | Adhesin, YadB | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| 1.B.40.1.5 | Adhesin, Cha | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 1.B.40.2.3 | Adhesin, UpaG | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | |
| CsgA | 1.B.48.1.1 | Curli fiber subunit, CsgA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| AT-3 | 1.B.54.1.1 | Intimin/Invasin (putative Autotransporter-3), Eae | 2 | 5 | 4 | 4 | 4 | 5 | 4 | 2 |
Autotransporters are virulence factors that insert into the outer bacterial membrane to form transmembrane β-barrels that export their extracellular protein domains. An Autotransporter-1 protein consists of an N-terminal cleavable secretory signal, an exported passenger domain of variable lengths, and a C-terminal 250–300 amino acyl residue domain that inserts into the outer membrane, giving rise to a 12 TMS β-barrel structure [53].
Autotransporter adhesins (e.g., AidA; TC#1.B.12.1.1) were present in all eight E. coli strains examined, but virulence factor-associated autotransporters were present in only certain pathogens. For example, fibronectin binding proteins and a tracheal colonization factor autotransporter were identified only in pathovars (Table 5). Overall, the most Autotransporter-1 proteins were found in ABU with sixteen homologs; fourteen were found in 559, twelve in CFT and UMN, ten in APE and E24, eight in O15, and only five in K12.
Autotransporter-2 family proteins have trimeric structures organized into three domains: an N-terminal head that adheres to the host cell membrane, a stalk, and a C-terminal anchor, rich in glycine, which forms a β-strand domain that oligomerizes to form a pore for auto-transport [54]. Haemagglutinins, a dissimilar adhesin, YadB [55], and other Autotransporter-2 family members were identified in specific pathovars (Table 5). Interestingly, AT-2 family proteins are far less prevalent than AT-1 family proteins in all strains and are lacking in K12.
Invasins or intimins are also called Autotransporters-3, but a function in autotransport is not well established [56]. The N-terminal domain serves as an anchor and inserts a pore-like β-barrel in the outer membrane. The C-terminus contains folds that bind to Tir (translocated intimin receptor) and β-1 integrins on host cells, leading to pathogenesis in enterohaemorrhagic and enteropathogenic strains [57,58]. In our study, intimin/invasin proteins were identified across all serotypes and were found in abundance in six of the seven pathogens. The UPEC strains have more intimins/invasins than the enteropathogenic strains.
Outer membrane factors (OMFs; 1.B.17) and membrane fusion proteins (MFPs; TC#8.A.1) function with Major Facilitator Superfamily (MFS) type porters, Resistance-Nodulation Cell-Division [59] family proteins (2.A.6), ATP-Binding Cassette (ABC) porters and Aromatic Acid Exporters (AAE, TC#2.A.85) to form complexes that export various drugs, oligosaccharides, proteins, aromatic acids and siderophores across both the inner and outer membranes in a single energy-coupled step ([60-64]). OMF proteins form homotrimeric, 12-stranded β-barrel complexes that form holes in the outer membrane for translocation. The OMF component, TolC (TC#1.B.17.1.1), was identified in all eight strains. TolC-dependent exporters mediate translocation of hemolysins, drugs and various siderophores [65,66]. Additional TolC-like proteins were found in the pathogens but not in K12.
The Fimbrial Usher Proteins of the FUP family function in conjunction with periplasmic chaperone proteins in fimbrial assembly [67]. The usher proteins contain central domains that span the outer membrane 24 times to form β-barrel pores. The C-terminal domains act as “plugs” that lie within the lumen of the β-barrels [68]. These plug domains, along with helical folds, regulate channel opening and export of proteins that contribute to fimbrial assembly [69]. FUPs were identified in all strains of E. coli but were present in greater numbers in the pathovars (Table 5).
Homologs of a single two-partner secretion (TPS) system (ShlB; TC#1.B.20.1.1) were identified. The oligomeric β-barrel channel of ShlB serves in secretion of the effector protein across the outer membrane [71]. These TPS systems were found exclusively in O15, where two paralogues were present, and in ABU, where a single protein was identified.
A secretin (TC#1.B.22) consists of an N-terminal periplasmic domain and a C-terminal domain responsible for forming large channels via homomultimeric ring structures [72]. Secretins participate in various transport processes such as fimbrial protein export, T2SS, T3SS, and phage export [73,74], and as expected, a number of these were identified in the pathogens (Table 5). Also as expected, outer membrane protein (OMP) insertion porins (OmpIP or BAM, TC#1.B.33), required for OMP insertion, were ubiquitously present, and all six required subunits were identified in all eight strains.
3.2.4. Iron scavenging outer membrane receptors
Table 6 lists outer membrane receptors of TC family 1.B.14, most of which are involved in the uptake of iron-siderophore complexes. This table also includes a cobalamin receptor and a copper receptor and indicates the occurrence of the TonB protein, which together with other proteins, is involved in OMR energization. TolA homologs involved in outer membrane stabilization and colicin uptake are also tabulated. For some of the OMRs, a single ortholog was found in all eight organisms (e.g., receptors for ferric copragen, ferrichrome, catecholate, and cobalamin). Some organisms possess more than one gene encoding a specific type of iron complex uptake receptor. These include: 1.B.14.1.4 (catecholate) where four of the organisms have a single gene encoding this system, but three organisms have two, and one (CFT) has three (Table 6). A similar situation was observed for 1.B.14.7.2, an iron pesticin uptake receptor. Six additional types of outer membrane receptors specific for iron complexes were found in some of these E. coli strains but not others. Similarly, only four of the pathovars possess the copper receptor (TC#1.B.14.4.1). All in all, each of these organisms possesses nine to fifteen iron uptake receptors except for K12 and E24, which have only six. Interestingly, the uropathogenic strains all possess more (ten to fifteen) iron complex receptors than the other serovars.
Table 6 also includes the energizing modules of family 2.C.1, TonB, which energizes OMR-mediated uptake, and TolA, which provides outer membrane stability and mediates colicin uptake [Table S1[75]]. It can be seen that while E. coli K12 has just one of each of these two proteins, four of the pathogens have two TonB’s, five of them have two TolA’s, and one (559) has three TolA’s. TonB, but not TolA, associates with ExbB–ExbD or TolQ–TolR to energize uptake of iron-siderophore complexes and other large molecules across the outer membranes of Gram-negative bacteria via outer membrane receptors (OMR; TC#1.B.14). The dissimilar Tol complex includes six proteins, TolA, TolB, YbgF, Pal and the pmf-dependent energizer, TolQR. This complex is involved in the maintenance of outer membrane stability and facilitates late stages of cell division.
3.2.5. Inner membrane iron uptake systems
Table 7 summarizes inner membrane iron uptake systems identified in the eight E. coli strains. These systems function to transport iron from the periplasm into the cytoplasm of the bacterium, cooperating with the outer membrane iron-scavenging OMR complexes, which import iron from the surrounding extracellular environment into the periplasm. Various mechanisms are employed for inner membrane transport of iron/iron-chelate complexes. These include the CorA heavy metal ion uptake channels (1.A.35), the zinc–iron permeases (ZIP (2.A.5)), metal ion transporters (Nramp (2.A.55)), GTPase-coupled ferrous iron uptake systems (FeoB (9.A.8)), and ATP-binding Cassette (ABC (3.A.1)) transporters.
Table 7.
Inner membrane iron uptake systems.
| Family | TC ID | Function | 559 | ABU | APE | CFT | E24 | UMN | O15 | K12 |
|---|---|---|---|---|---|---|---|---|---|---|
| CorA | 1.A.35.2.1 | Divalent cation transport | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| ZIP | 2.A.5.5.1 | Pmf-mediated M2+ uptake | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Nramp | 2.A.55.3.1 | Divalent cation:H+ symport | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| FeoB | 9.A.8.1.1 and 1.9 | GTPase-coupled ferrous iron uptake | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| FeT | 3.A.1.10.3 | Ferric iron uptake | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| FeCT | 3.A.1.14.1 | Ferric iron-dicitrate uptake | 4 | 4 | 0 | 0 | 4 | 4 | 0 | 4 |
| 3.A.1.14.2 | Ferric iron-enterobactin uptake | 4 | 8 | 4 | 8 | 4 | 4 | 4 | 4 | |
| 3.A.1.14.3 | Ferric iron-hydroxamate uptake | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |
| 3.A.1.14.8 | Ferric-vibrioferrin uptake | 0 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | |
| 3.A.1.14.18 | Heme uptake | 0 | 3 | 3 | 3 | 0 | 3 | 3 | 0 | |
| 3.A.1.14.21 | Heme uptake | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | |
| MZT | 3.A.1.15.7 | Mn2+ and Fe2+ uptake | 0 | 5 | 4 | 5 | 0 | 4 | 0 | 0 |
| BIT | 3.A.1.20.1 | Iron uptake complex | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| Total | 16 | 32 | 21 | 30 | 16 | 24 | 19 | 16 |
Zinc–Iron Porters (ZIP) have about eight TMSs with histidinerich repeats near the N- and C-termini [76]; a cation symport mechanism may be operative [77]. A single such transport protein was found in each of the eight strains examined (Table 7). Metal ion transporter (Nramp) homologs generally function by a symport mechanism in a 1:1 ratio of ferrous ion and H+ although stoichiometrics have been reported to be variable [78]. A single Nramp protein was found in each of the eight strains of E. coli examined (Table 7). Another iron influx system, FeoB (TC#9.A.8), has N-terminal catalytic domains with sequences homologous to GTP-binding domains [79]. Their C-termini catalyze transport, possibly coupled to K+-activated GTPase activity. GTP hydrolysis has been reported to be required for transport, and if there is direct energy coupling, the mechanism may resemble that of ABC transporters ([80-83]). Two FeoB homologs were identified in each of the eight strains of E. coli.
Four types of inner membrane ABC iron-uptake systems were identified: ferric porters (TC#3.A.1.10), iron chelate porters (TC#3.A.1.14), manganese/zinc/iron chelate porters (TC#3.A.1.15), and homologs of the Brachyspira iron transporters (TC#3.A.1.20). A complete four-subunit ferric-citrate uptake system (TC#3.A.1.14.1) was identified in five of the eight strains. Complete ferric-enterobactin and ferric-hydroxamate uptake systems were identified in all eight strains of E. coli examined. However, a second complete ferric-enterobactin system (TC#3.A.1.14.2) was identified in two strains.
Homologs of the Shigella heme uptake system (ShuTUV (3.A.1.14.18)) were present in five pathogenic strains as complete systems. Complete systems corresponding to the Salmonella Mn2+ and Fe2+ uptake system (3.A.1.15.7) were identified in four pathovars (see Table 7).
Fig. 2 reveals the correlation between numbers of OMRs and the number of iron ABC uptake systems. The juxtaposition of the two datasets demonstrated that the UPEC strains, ABU and CFT, contain the greatest numbers of both OMRs and inner membrane iron-siderophore transporters. E24 and K12 contain the smallest numbers of both types of systems. A clear correlation between the numbers of iron-siderophore OMRs and inner membrane transporters is evident for the eight strains of E. coli (RSQ value = 0.90). The slope of the trendline (SLOPE = 1.85) confirms the positive correlation between the two sets of values (see Fig. 3).
Fig. 2.
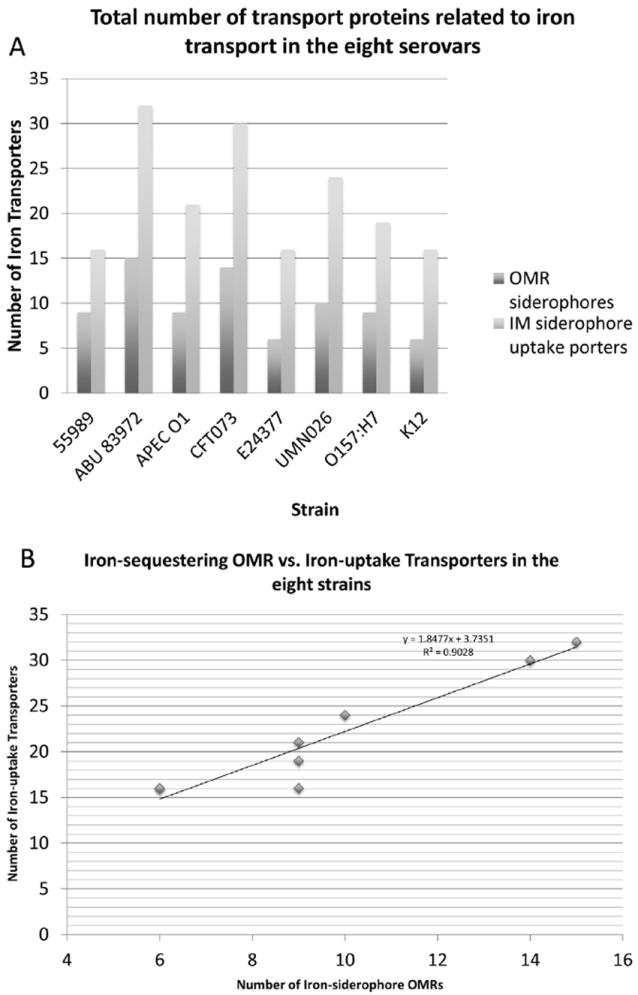
Total number of proteins associated with iron transport in the eight E. coli strains in which (A) shows the relationship between the serovars (x-axis) and number of iron-siderophore transporters (y-axis). The OMR-type transporters are represented in dark gray while the ABC iron-siderophore uptake porters are presented in light gray. (B) Correlation between iron-sequestering OMRs (x-axis) and ABC iron-uptake transporters (y-axis) for the eight strains of E. coli examined. The best-fit line has a slope of 1.85 and a R2 value is 0.90.
Fig. 3.
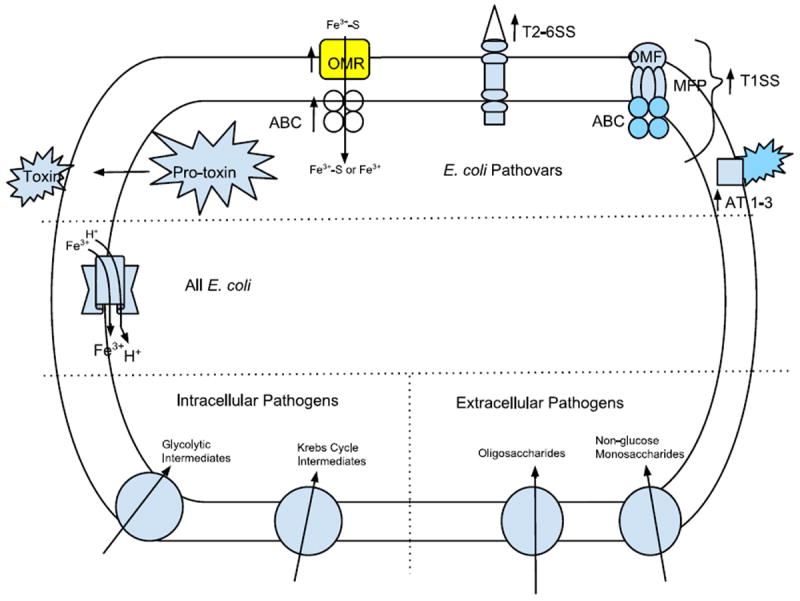
Schematic view of an E. coli cell showing transport proteins of particular importance for pathogenesis. All E. coli cells have the same secondary carriers for iron uptake which therefore do not play a specific role in pathogenesis (center left). Systems preferentially present in pathogens (top) often lacking in E. coli K12, include pro-toxins, ABC-type iron uptake systems which are proposed to function with their iron-specific outer membrane receptors (OMRs), transenvelope protein secretion systems types 1–6 and autotransporters of the types AT1-3 transporters specifically found in extracellular vs. intracellular pathogens are illustrated in the bottom right and left, respectively. See text for details.
3.2.6. Nutrient transporters: intracellular vs. extracellular pathogens
We identified numerous nutrient uptake porters that are uniquely found only in the three intracellular pathogens or only in the extracellular pathogens (Table 8). Transporters found exclusively in the intracellular pathovars include certain α-type channels (TC#1.A), β-barrel porins (1.B), holins (1.E), MFS porters (2.A.1), multidrug resistance (MDR), RND-type efflux pumps (2.A.6), ABC porters (3.A.1), PTS sugar uptake transporters (4.A), and various TC class 9.B proteins. Thus, the APE, CFT and ABU strains, all primarily intracellular pathogens, possess many transport systems that take up metabolic products produced via host intracellular metabolic processes. Examples include a phosphoglycerate:Pi antiporter (2.A.1.4.2) that takes up glycolytic intermediates, 2- and 3-phosphoglycerates as well as phosphoenolpyruvate, a pmf-dependent acetate uptake system (TC#1.A.14.2.2), an ATP-dependent ribose porter (3.A.1.2.1) and a phosphoenolpyruvate (PEP)-dependent d-tagatose/d-psicose/d-sorbose PTS uptake system (4.A.2.1.9). While ribose is a substrate of RNA biosynthesis and a product of RNA degradation, the latter 2-keto hexoses are intermediates of several intracellular metabolic pathways and of nutritional value ([84,85]). These intracellular pathogens also uniquely possess a citrate:H+ symporter (2.A.1.6.1), a fumarate–malate transporter (2.A.13.1.2), a citrate:succinate antiporter (2.A.47.3.2), a tripartite tricarboxylate (citrate, isocitrate and cisaconitate) transporter (2.A.80.1.1), and a putative C4-dicarboxylate transporter (9.B.50.1.2), all of which are specific for intermediates in the Krebs (TCA) cycle, and none of which are found in the extracellular pathovars. In addition to Krebs cycle metabolism, C4-dicarboxylates can be generated via multiple pathways including amino acid deamination and lipolysis.
Table 8.
Metabolite transporters unique to intracellular vs. extracellular pathogens.
| Family | TCID | Function | Exclusively in 2 of 3 intracellular pathovars | Exclusively in all 3 intracellular pathovars | Exclusively in extracellular pathovars |
|---|---|---|---|---|---|
| E-ClC | 1.A.13.2.3 | Chloride efflux | x | ||
| TEGT | 1.A.14.2.2 | Acetate transport | x | ||
| FNT | 1.A.16.1.2 | Formate transport | x | ||
| OMF porin | 1.B.6.1.2 | Solute diffusion | x | ||
| Tsx | 1.B.10.1.1 | Nucleoside-specific channel | x | ||
| FUP | 1.B.11.2.1 | Type π fimbrial export | x | ||
| 1.B.11.2.2 | Putative fimbrial export | x | |||
| 1.B.11.3.3 | Type γ fimbrial export | x | |||
| 1.B.11.3.5 | Type γ fimbrial export | x | |||
| AT-1 | 1.B.12.1.1 | Adhesin export | x | ||
| 1.B.12.2.1 | Pertactin (adhesin) export | x | |||
| 1.B.12.2.2 | Tracheal colonization factor export | x | |||
| 1.B.12.4.1 | Protease export | x | |||
| 1.B.12.8.2 | Adhesin export | x | |||
| 1.B.12.8.3 | Adhesin/invasin export | x | |||
| OMF | 1.B.17.3.4 | Metal cation (Ag+) export | x | ||
| OMA | 1.B.18.1.2 | Polysaccharide export | x | ||
| Gene IV secretin (OMS) | 1.B.22.5.1 | Phage export | x | ||
| OmpL porin | 1.B.35.2.2 | Small molecule transport | x | ||
| AT-2 | 1.B.40.1.2 | Hemagglutinin export | x | ||
| 1.B.40.1.3 | Adhesin (YadB) export | x | |||
| AT-3 (Int/Inv) | 1.B.54.1.1 | γ-Intimin export | x | ||
| 1.B.54.1.2 | Invasin export | x | |||
| IIITCP (EspB/D) | 1.C.36.1.1 | Host pore for T3SS | x | ||
| ST-B | 1.C.54.1.1 | STX pore-forming virulence factor | x | ||
| Cnt2 | 1.C.57.3.3 | Pore-forming virulence factor | x | ||
| Lambda holin | 1.E.2.1.4 | Holin; autolysis | x | ||
| OMR | 1.E.14.1.9 | Holin; autolysis | x | ||
| MFS, SP | 2.A.1.1.3 | Monosaccharide uptake | x | ||
| MFS; DHA1 | 2.A.1.2.7 | Drug efflux | x | ||
| MFS; DHA1 | 2.A.1.2.52 | Drug efflux | x | ||
| MFS; DHA2 | 2.A.1.3.18 | Efflux of signaling molecules | x | ||
| MFS; OPA | 2.A.1.4.2 | Glycolytic intermediate uptake | x | ||
| MFS; MHS | 2.A.1.6.1 | Tricarboxylate uptake | x | ||
| 2.A.1.6.9 | Metabolite uptake | x | |||
| MFS; ACS | 2.A.1.14.2 | Hexuronate uptake | x | ||
| MFS; AAHS | 2.A.1.15.2 | Aromatic acid uptake | x | ||
| MFS; CP | 2.A.1.17.1 | Cyanate uptake | x | ||
| GPH | 2.A.2.1.1 | Disaccharide uptake | x | ||
| 2.A.2.3.9 | Glycoside uptake | x | |||
| APC | 2.A.3.1.13 | Putrescine (polyamine) uptake | x | ||
| 2.A.3.2.7 | Arginine uptake; acid homeostasis | x | |||
| 2.A.3.7.2 | Glutamate uptake; acid homeostasis | x | |||
| 2.A.3.8.17 | Amino acid derivative uptake | x | |||
| RND | 2.A.6.2.19 | Drug export | x | ||
| 2.A.6.2.6 | Drug/autoinducer efflux | x | |||
| Betaine/carnitine/choline symporter | 2.A.15.2.3 | Organoamine uptake | x | ||
| CPA1 | 2.A.36.3.1 | Na+:H+ antiport | x | ||
| CNT | 2.A.41.2.10 | Nucleoside uptake | x | ||
| DASS | 2.A.47.3.2 | Citrate uptake; succinate export | x | ||
| 2.A.47.4.2 | Arsenite/antimonite export | x | |||
| SulP | 2.A.53.3.8 | Bicarbonate uptake | x | ||
| TRAP-T | 2.A.56.1.5 | Rhamnogalacturonide uptake | x | ||
| 2.A.56.1.6 | Sialic acid uptake | x | |||
| DcuC | 2.A.61.1.2 | Dicarboxylate antiport | x | ||
| PST | 2.A.66.2.10 | Lipopolysaccharide precursor export | x | ||
| AbgT | 2.A.68.1.1 | p-Aminobenzoyl-glutamate uptake | x | ||
| TTT | 2.A.80.1.1 | Tricarboxylate uptake | x | ||
| ABC; CUT2 | 3.A.1.2.1 | Ribose uptake | x | ||
| 3.A.1.2.8 | Sugar uptake | x | |||
| 3.A.1.2.11 | Sugar alcohol uptake | x | |||
| 3.A.1.2.13 | Autoinducer-2 uptake | x | |||
| ABC; PepT | 3.A.1.5.12 | Rhamnose oligosaccharide uptake | x | ||
| 3.A.1.5.20 | Oligopeptide uptake | x | |||
| ABC; LPT | 3.A.1.125.2 | Lipoprotein export | x | ||
| ArsAB | 3.A.4.1.1 | Arsenite export | x | ||
| NDH | 3.D.1.9.1 | H+/K+ exchange (hydrogenase) | x | ||
| NFO | 3.D.6.1.2 | H+ export; NADH:ferredoxin oxido-reductase | x | ||
| PTS; Glc | 4.A.1.1.3 | Maltose uptake | x | ||
| 4.A.1.1.9 | Glucose uptake | x | |||
| 4.A.1.1.10 | Glucose uptake | x | |||
| PTS; Fru | 4.A.2.1.3 | 2-O-α-Mannosyl-d glycerate uptake | x | ||
| 4.A.2.1.6 | Mannose uptake | x | |||
| 4.A.2.1.9 | Keto sugar uptake | x | |||
| PTS; Gut | 4.A.4.1.2 | Sugar alcohol (glucitol) uptake | x | ||
| PTS; Gat | 4.A.5.1.3 | Sugar alcohol (galactitol) uptake | x | ||
| PnuC | 4.B.1.1.4 | Coenzyme precursor (thiamin) uptake | x | ||
| DUF318 | 9.B.28.1.1 | Unknown | x | ||
| YiaW; DUF3302 | 9.B.32.1.2 | Unknown | x | ||
| UIT3 | 9.B.50.1.2 | Possible dicarboxylate uptake | x | ||
| DUF805 | 9.B.124.1.1 | Unknown | x | ||
| DUF805 protein | 9.B.124.1.4 | Unknown | x |
Two of the intracellular pathogens, APE and CFT, possess an arginine/agmatine antiporter (2.A.3.2.7), which allows these cells to survive under acidic conditions, for example, as exist inside lysosomes [86]. These intracellular pathogens also possess a probable antimonite/arsenite resistance exporter (2.A.47.4.2). Arsenite can be passively taken up by host cells via aquaglyceroporins (TC#1.A.8) [87]. Additionally, an ATP-dependent nucleoside group translocator (4.B.1.1.1) that synthesizes NMN, an intermediate in NAD synthesis, and a thiamin porter (4.B.1.1.4), involved in the synthesis of thiamin pyrophosphate (TPP), were found in these strains but not in the extracellular pathogens (Table 8).
Surprisingly, the intracellular pathogens possess a glucuronate/galacturonate porter (2.A.1.14.2), which is absent in the extracellular pathogens (Table 8). These metabolic products have been shown to be generated intracellularly when exogenous sugars such as lactose are metabolized by E. coli [88]. These compounds are present intracellularly in numerous organs of animals, and these E. coli strains utilize cytoplasmic UDP-glucuronyltransferases for mucopolysaccharide biosynthesis [89]. Additionally, a sialic acid uptake porter (2.A.56.1.6) is found exclusively in the intracellular pathogens. Although sialic acids are found in abundance in cell surface glycolipids and extracellular glycoproteins, N-acetylneuraminic acid, the main mammalian sialic acid, can be found intracellularly where it participates in glycolipid and glycoprotein biosynthesis [90]. Sialic acid-containing polysaccharide capsules are virulence factors in E. coli urinary tract infections and contribute to biofilm formation [91].
Intracellular E. coli strains lack various transporters present in extracellular pathogens that take up metabolites normally found extracellularly. For example, the latter, but not the former strains possess a rhamnose oligosaccharide porter (3.A.1.5.12), a melibiose permease (2.A.2.1.1) and a 2-O-α-mannosyl-d-glycerate porter (4.A.2.1.3). These first two oligosaccharides are constituents of glycolipid and glycoproteins found on the external surfaces of intestinal villus cells, while the last compound is a known intracellular osmolyte in some bacteria [92]. The extracellular strains, but not the intracellular strains, also have a xylose:H+ symporter (2.A.1.1.3), a mannose PTS porter (4.A.2.1.6), a glucitol PTS transporter (4.A.4.1.2), and a galactitol PTS permease (4.A.5.1.3). The former two sugars are found in abundance in cell surface polysaccharides [93,94] while the hexitols are common osmolytes in animals [95]. The α-glucoside-specific IICB permease of the PTS (4.A.1.1.10), which transports many glucosides is similarly found in the extracellular pathogens but lacking in intracellular strains (Table 8).
Other transporters lacking in intracellular but present in extracellular pathogens include an uptake system for the folate catabolite, p-aminobenzoyl-glutamate (2.A.68.1.1), and a cyanate/thiocyanate porter (2.A.1.17.1) ([96,97]). Thiocyanate and cyanate are found in extracellular environments, and although toxic, they can be used as a nitrogen source following destruction by cyanase, CynS [96]. E. coli cells lacking the cyn operon are more sensitive to the toxic effects of cyanate and thiocyanate than strains that possess it [98].
A putrescine uptake porter (2.A.3.1.13) was found to be present only in extracellular strains. Putrescine, like other positively charged polyamines, stabilizes the negatively charged DNA during replication, but it is also found in abundance in extracellular environments in mammals such as seminal fluid and urine, especially during episodes of protein breakdown [99,100]. Several MDR and iron-siderophore transporters are found exclusively in intracellular pathogens, while others are present in extracellular pathogens exclusively. These facts may reflect the natural cellular substrates of MDR pumps currently not recognized.
3.3. Proteins with e-values between 0.1 and 0.0001
Several proteins were identified that gave poor scores in the G-BLAST search, but which nevertheless proved to be members of TC families. Some of these are presented here in order of their TC numbers. (1) A putative invasin with similarity to proteins in the OmpA family (TC#1.B.6) was identified and given the TC number 1.B.6.2.13. (2) A homolog of pore-forming phage coat protein A was identified and assigned TC#1.B.53.1.4. (3) Several proteins proved to be homologous to the cytotoxic pore-forming oligomeric fimbrial subunit, MrxA. One such protein has been entered into TCDB under TC#1.C.80.1.2. (4) A holin of 108 aas was identified and entered into TCDB with TC#1.E.2.1.10. This protein was encoded by a gene within a prophage segment of the genome of UMN with two adjacent genes, a lysozyme inhibitor, and a lytic protein, probably a lysozyme. (5) An ABC porter, YnjBCD, was assigned TC#3.A.1.19.4. Although it is annotated as a sulfate/thiosulfate transporter, other members of this family transport vitamins. (6) Another protein designated YjiG proved to be a distant member of the FeoB family (TC#9.A.8). A nucleoside-recognition domain may be present in some of these homologs. YjiG was entered into TCDB with TC#9.B.156.1.1.
3.4. Unique characteristics of enterohaemorrhagic (EHEC) strain, O15
Strain O15 exhibited a number of characteristics that distinguished it from the other six pathogenic strains of E. coli examined. It has two-partner secretion (TPS) systems corresponding to TC#1.B.20.1.3 and 1.B.20.1.4. The former system plays a role in contact inhibition of growth [59] while the latter protein is a ShlB-type protein that contains a domain critical for substrate recognition and folding [101]. It probably secretes a hemolysin-like virulence factor [102] and has been entered into the database under 1.B.20.1.7.
This strain uniquely has an AT-2 type adhesin, most closely related to TC#1.B.40.1.3, as well as an intimin, most closely related to TC#1.B.54.1.1. These proteins were found in no other E. coli strain examined. Most surprisingly, we identified 14 hits of 1.B.6.2.1 and 1.B.6.2.2, all related to OmpX of the OmpA family. These proteins are in the Ail/Lom family, which in Yersinia species are multifunctional virulence factors that protect against complement [103]. Equally surprising was the unique discovery of 12 putative holins corresponding to TC#1.E.1.1.3. These holins may be related to phage-encoded shiga-toxins [104].
4. Discussion
4.1. Iron siderophore receptors and transporters contribute to E. coli pathogenesis in a host organism
E. coli is a facultative anaerobe that utilizes iron primarily for electron transport and cellular respiration. Succinate dehydrogenase and various cytochromes as well as NADH and ferric oxidase activities are diminished with an iron deficiency [105]. In humans, iron sequestration plays an important role in host defense against pathogens. Certain conserved eukaryotic genes encode siderophore/iron-uptake complexes that allow the host to upregulate iron sequestration in response to inflammatory signals accompanying infection [106]. Some of these genes play pivotal roles in host defense in Crohn’s disease, for their upregulation allows the host intestinal epithelial cells to sequester more iron [107]. In our study, the pathogenic E. coli strains examined proved to have greater numbers of iron-siderophore receptor-encoding genes than the nonpathogenic K12 strain.
Certain iron-siderophore receptors and ABC iron uptake systems are abundant only in pathovars (Tables 6 and 7). These receptors include a Fe3+ catecholate receptor (1.B.14.1.4) and the ferric yersiniabactin uptake receptor (1.B.14.7.2). The ferric yersiniabactin uptake receptor is known to be required for biofilm production in UPEC strains [108], possibly relevant to host inflammatory signaling. Ferri-enterobactin receptors (1.B.14.1.3) and ferrichrome receptors (1.B.14.1.15) were identified exclusively in UPEC strains (ABU and CFT). These receptors may serve specific functions in UPEC pathogenicity.
The increased numbers of iron-complex receptors in pathovars indicate targets for combating E. coli pathogenicity. Increases in siderophore receptor complexes may serve as compensatory mechanisms in response to host defense. Increased ferrichrome sequestration undoubtedly allows these pathovars to take up iron more readily, leading to increased survival in the host.
As shown in Fig. 2, increases in numbers of iron-sequestering outer membrane receptors correlate with increases in inner membrane iron ABC transport systems. The UPEC strains (ABU, CFT and UMN) have larger numbers of both iron-siderophore OMRs and inner membrane transporters. We speculate that because UPEC strains must survive in both the gastrointestinal and the urogenital tracts, they must possess a greater number of iron-siderophore transporters for cellular respiration and survival.
It has been shown that ABC porters (3.A.1) in general possess higher affinities for their substrates than secondary carriers (TC#2.A). Secondary carriers transporting metal cations typically exhibit Km values of ~20–800 μM [109] while primary active transporters may exhibit Km values ≤1 μM [110]. Interestingly, the majority of ABC iron transporters were found either exclusively in pathogenic strains or in increased numbers (Table 7), and the same strains possess the largest numbers of iron-siderophore OMRs. TC subclass 2.A iron uptake secondary carriers appear to be present in invariant numbers in all eight E. coli strains although the ABC iron-specific transporters differ greatly among the different serovars (Table 7). This suggests that the low affinity secondary iron carriers serve housekeeping functions outside of the body, where iron concentrations are relatively high, while the ABC transporters function in pathogenesis in the in vivo environment, where competition with the host is an important factor.
4.2. Pore-forming toxins
EHEC and EPEC pathovars typically operate by attaching and effacing (A/E) mechanisms. This type of invasion includes utilizing a Type III secretion system to inject effector proteins into host cells. One effector, EspF, plays an important role in tight junction disruption as well as inhibition of sodium and water uptake [111]. The EspB/D complex was identified in UMN (UPEC) and O15 (EHEC) and serves to initiate pore formation in the host cell plasma membrane as well as translocation of EspF into the cell. UPEC strains typically utilize Type I Secretion Systems for adhesion and entry through manipulating the host cell cytoskeleton using effectors that stimulate Rho-family GTPases and kinases [112]. Nevertheless, a complete T3SS was identified in UMN, suggesting that an A/E mechanism of invasion may occur in this pathovar.
The identification of Hemolysin A (HlyA) exclusively in the UPEC strains supports a previous finding that these strains can utilize haemolysins to inhibit protein kinase B signaling and lead to apoptosis of host cells [35]. Similarly, the exclusive presence of shiga toxin in O15 supports previous findings of EHEC pathogenicity [113]. The Stx pathway may provide an alternative pathway by which the EHEC strain can inhibit chemical signaling and immune responses in the host cell.
Homologs of the clostridial cytotoxin (CCT) form channels and inactivate Rho-type GTPases, leading to manipulation of the host cytoskeleton [39]. Predictably, such a toxin was identified in the UPEC strain, ABU, but more surprisingly, it was also found in O15. This may suggest an alternative mechanism by which EHEC strains manipulate the host’s cytoskeletal machinery since these pathovars typically utilize the mitochondrial-associated protein [69] to inhibit the host cell control protein 42 (CDC42), which provides regulatory functions in host cell actin dynamic control [114]. Since the homologs in ABU and O15 are 85% identical, the same mechanism is likely to be operative.
AIEC strains such as APE, have been cited in 36% of Crohn’s disease. These pathovars operate through necrosis factors that impair host immune responses, allowing the bacteria to colonize the ileal mucosa and lamina propria [115]. We identified the active subunit of Cytolethal Distending Toxin (CdtB) only in APE. The CdtB active subunit induces DNA double-strand breakage, leading to host cell cycle arrest in the G2/M phase [116]. This is preferable in the AIEC pathotype, presumably because pauses and irregularities in host cell cycle give the bacteria time to proliferate and colonize host cells. Rapid proliferation of these AIEC cells can then induce secretion of Tumor Necrosis Factor Alpha (TNF-α) that causes inflammation in host cells.
Serratia-type pore-forming toxins (S-PFT) were identified in ABU (UPEC), CFT (UPEC) and O15 (EHEC). These toxins exhibit properties vastly different from those of RTX pore-forming haemolysins, which are exported via two-partner secretion systems and are much larger in size [40]. The presence of two-partner secretion system(s) in both ABU and O15 confirmed the possibility of a novel Serratia-type Pore-forming Toxin (S-PFT) in both UPEC and EHEC strains (see Table 3).
4.3. Outer membrane protein secretion systems
Many Autotransporter-1 systems were identified in the seven pathovars examined. Examples include the fibronectin binding protein (1.B.12.1.3), a tracheal colonization factor (1.B.12.2.2), a firmicute homolog (1.B.12.2.4), and a Salmonella BigA homolog (1.B.12.5.5) (see Table 5). Fibronectin binding protein is known to assist in host cell adhesion by Salmonella typhimurium [117], providing evidence that a similar mechanism of adhesion may be utilized by UPEC, AIEC, ETEC and EAEC strains for host cell targeting. Interestingly, homologs of the Bordetella pertussis tracheal colonization factor were identified in all pathovars except for O15, providing clues as to how such strains target cilia-rich epithelia in the intestines.
Several Autotransporter-2 systems were found exclusively in the pathovars. Homologs of the membrane-anchored hemagglutinin, identified in UMN and O15, may play roles in bacterium-host cell adhesion [18]. Trimeric adhesins such as Cha (1.B.40.1.5) and UpaG (1.B.40.2.3) were also identified exclusively in pathovars, and they are known to function in adhesion to the host extracellular matrix [118]. YadB homologs (1.B.40.1.3) function by a similar mechanism, and one was identified in O15, suggesting that it may function specifically in EHEC strains.
EHEC and EPEC strains have been shown to use invasins/intimins to target translocated intimin receptors (Tir) and β-1 integrins on host extracellular matrices for adhesion purposes followed by stimulation of actin rearrangements via the N-Wasp pathway ([119,120]). UPEC strains, however, utilize type I pili to bind β-1 integrin on host cell extracellular matrices to initiate actin rearrangements [112]. The presence of invasins/intimins to target β-1 integrins reveals a potential novel mechanism of cell-adhesion and actin rearrangement utilized by these UPEC pathovars.
The MdtP outer membrane factor (OMF; TC#1.B.17.3.9) functions with the MdtO aromatic acid/drug exporter (2.A.85.6.1) and the auxiliary MFP protein, MdtN (8.A.1.1.3). This OMF was found in all pathovars but not in K12. This may indicate a multi-drug resistance mechanism found exclusively in the pathogenic strains that contributes to their resistance to antibiotics.
Fimbrial usher proteins (FUPs) serve important functions in the assembly of fimbriae [67]. As expected, these proteins were found ubiquitously in all strains including K12. However, greater numbers of the encoding genes were observed in all pathovars compared to K12, presumably contributing to pathogenesis by promoting bacterium-host cell adhesion [70].
4.4. Multiple pathovars contain T6SS constituents
Potentially complete VasA-L T6SSs were identified in multiple pathovars. Protein products of the icmF, hlc and clpV genes play essential roles in biofilm assembly and general motility [121]. In recent studies, the T6SS present in Pseudomonas aeruginosa and other bacterial cells were shown to engage in “T6SS dueling” with surrounding bacteria in competition for survival in the microbiome [122,123].
With the discovery of T6SSs in E. coli pathovars, we speculate that E. coli cells engage in T6SS-mediated competition in the human body. It is possible that pathovars of E. coli can utilize T6SSs both for offense and for defense in the host microbiome. Such a mechanism may prove valuable during invasion of the colon, where microbes are densely packed with other types of facultative/obligate anaerobes competing for limited metabolic resources.
4.5. Transporters in intracellular vs. extracellular pathogens
An interesting pattern revealed by our studies was the existence of different sets of nutrient uptake transporters, depending on the organismal site of proliferation. Intracellular vs. extracellular organisms were distinguishable on the basis of the nature of the substrates transported. In general, the intracellular pathovar-specific strains take up metabolites expected to be found inside host cells while the extracellular pathovars transport substrates that are normally found in abundance outside of host cells. Examples of this can be seen in transporters for glycolytic and Kreb cycle intermediates found only in intracellular pathogens as well as transporters for oligosaccharides found only in extracellular pathogens (Table 8). These facts indicate that these organisms have acquired transport systems through the frequent gain and loss of the encoding genes probably via horizontal transfer, to take advantage of nutrients available in their specific environments. Since intracellularity may be a relatively recent development, this suggests that the gain and loss of genes may have occurred with high frequency in response to immediate need. It seems probable that such characteristics (e.g., predilection for certain types of nutrient transporters) can be used for diagnostic purposes.
4.6. Unique characteristics of O15
In screening the data, we noticed that each strain examined has characteristics that distinguish it from all others. Particularly noteworthy was strain O15, which had numerous characteristics lacking in the other seven strains. One such example includes extra two-partner secretion systems as well as a unique AT-2 adhesin and an intimin. More striking was the occurrence of large numbers of OmpA-like Ail/Lom family proteins that probably serve multiple virulence functions, including protection against complement as has been documented in Yersinia species [103]. Additionally, this strain possessed numerous holins, all homologous to 1.E.1.1.3, a unique characteristic.
Supplementary Material
Acknowledgments
We thank Vamsee Reddy for writing a program for tabulating the results summarized in supplementary Table S1 as well as several of the programs used in this study. This work was supported by NIH grant GM077402. We also appreciate the support of the research associates of the Saier Laboratory Group. Finally, we thank the reviewers of this paper for their valuable suggestions for improvement.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.micpath.2014.03.008.
References
- 1.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc Natl Acad Sci U S A. 2010;107:17739–44. doi: 10.1073/pnas.1009491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams EJ, Embleton ND, Bythell M, Ward Platt MP, Berrington JE. The changing profile of infant mortality from bacterial, viral and fungal infection over two decades. Acta Paediatr. 2013;102:999–1004. doi: 10.1111/apa.12341. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Foxman B, Manning SD, Tallman P, Marrs CF. Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect Immun. 2000;68:2009–15. doi: 10.1128/iai.68.4.2009-2015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18:465–83. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen MB, Giannella RA. Hemorrhagic colitis associated with Escherichia coli O157:H7. Adv Intern Med. 1992;37:173–95. [PubMed] [Google Scholar]
- 6.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli, nature reviews. Microbiology. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 7.Kokesova A, Frolova L, Kverka M, Sokol D, Rossmann P, Bartova J, et al. Oral administration of probiotic bacteria (E. coli Nissle, E. coli O83, Lactobacillus casei) influences the severity of dextran sodium sulfate-induced colitis in BALB/c mice. Folia Microbiol. 2006;51:478–84. doi: 10.1007/BF02931595. [DOI] [PubMed] [Google Scholar]
- 8.Huebner C, Ding Y, Petermann I, Knapp C, Ferguson LR. The probiotic Escherichia coli Nissle 1917 reduces pathogen invasion and modulates cytokine expression in Caco-2 cells infected with Crohn’s disease-associated E. coli LF82. Appl Environ Microbiol. 2011;77:2541–4. doi: 10.1128/AEM.01601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonkers D, Penders J, Masclee A, Pierik M. Probiotics in the management of inflammatory bowel disease: a systematic review of intervention studies in adult patients. Drugs. 2012;72:803–23. doi: 10.2165/11632710-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Hancock V, Vejborg RM, Klemm P. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics. 2010;284:437–54. doi: 10.1007/s00438-010-0578-8. [DOI] [PubMed] [Google Scholar]
- 11.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–34. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 12.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–8. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokurenko EV, Gomulkiewicz R, Dykhuizen DE. Source-sink dynamics of virulence evolution. Nature reviews Microbiology. 2006;4:548–55. doi: 10.1038/nrmicro1446. [DOI] [PubMed] [Google Scholar]
- 14.Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 15.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nature reviews Microbiology. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 16.Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–3. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy VS, Saier MH., Jr BioV suite – a collection of programs for the study of transport protein evolution. FEBS J. 2012;279:2036–46. doi: 10.1111/j.1742-4658.2012.08590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mancini J, Weckselblatt B, Chung YK, Durante JC, Andelman S, Glaubman J, et al. The heat-resistant agglutinin family includes a novel adhesin from enteroaggregative Escherichia coli strain 60A. J Bacteriol. 2011;193:4813–20. doi: 10.1128/JB.05142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda M, Arai M, Lao DM, Shimizu T. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. Silico Biol. 2002;2:19–33. [PubMed] [Google Scholar]
- 20.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–2. [PubMed] [Google Scholar]
- 21.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–35. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castillo R, Saier MH. Functional promiscuity of homologues of the bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The transporter classification database: recent advances. Nucleic Acids Res. 2009;37:D274–8. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarzer C, Fu Z, Patanwala M, Hum L, Lopez-Guzman M, Illek B, et al. Pseudomonas aeruginosa biofilm-associated homoserine lactone C12 rapidly activates apoptosis in airway epithelia. Cell Microbiol. 2012;14:698–709. doi: 10.1111/j.1462-5822.2012.01753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong SH, Wang X, Wood TK. Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb Biotechnol. 2010;3:344–56. doi: 10.1111/j.1751-7915.2010.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durkee JW, Jr, Antich PP, Lee CE. Exact solutions to the multiregion time-dependent bioheat equation. I: solution development. Phys Med Biol. 1990;35:847–67. doi: 10.1088/0031-9155/35/7/004. [DOI] [PubMed] [Google Scholar]
- 27.Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–34. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis JK. Combining polysaccharide biosynthesis and transport in a single enzyme: dual-function cell wall glycan synthases. Front Plant Sci. 2012;3:138. doi: 10.3389/fpls.2012.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard C, McNamara JT, Azumaya C, Patel MS, Zimmer J. The hyaluronan synthase catalyzes the synthesis and membrane translocation of hyaluronan. J Mol Biol. 2012;418:21–31. doi: 10.1016/j.jmb.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 30.DeAngelis PL. Microbial glycosaminoglycan glycosyltransferases. Glycobiology. 2002;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- 31.Denoncin K, Collet JF. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid Redox Signal. 2013;19:63–71. doi: 10.1089/ars.2012.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzokov SB, Wyborn NR, Stillman TJ, Jamieson S, Czudnochowski N, Artymiuk PJ, et al. Structure of the hemolysin E (HlyE, ClyA, and SheA) channel in its membrane-bound form. J Biol Chem. 2006;281:23042–9. doi: 10.1074/jbc.M602421200. [DOI] [PubMed] [Google Scholar]
- 33.Hunt S, Moir AJ, Tzokov S, Bullough PA, Artymiuk PJ, Green J. The formation and structure of Escherichia coli K-12 haemolysin E pores. Microbiology. 2008;154:633–42. doi: 10.1099/mic.0.2007/011700-0. [DOI] [PubMed] [Google Scholar]
- 34.Davies RL, Whittam TS, Selander RK. Sequence diversity and molecular evolution of the leukotoxin (lktA) gene in bovine and ovine strains of Mannheimia (Pasteurella) haemolytica. J Bacteriol. 2001;183:1394–404. doi: 10.1128/JB.183.4.1394-1404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiles TJ, Dhakal BK, Eto DS, Mulvey MA. Inactivation of host Akt/protein kinase B signaling by bacterial pore-forming toxins. Mol Biol Cell. 2008;19:1427–38. doi: 10.1091/mbc.E07-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iizumi Y, Sagara H, Kabe Y, Azuma M, Kume K, Ogawa M, et al. The enteropathogenic E. coli effector EspB facilitates microvillus effacing and antiphagocytosis by inhibiting myosin function. Cell Host Microbe. 2007;2:383–92. doi: 10.1016/j.chom.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Nakao H, Takeda T. Escherichia coli Shiga toxin. J Nat Toxins. 2000;9:299–313. [PubMed] [Google Scholar]
- 38.Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc Natl Acad Sci U S A. 2010;107:13467–72. doi: 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oswald E, Sugai M, Labigne A, Wu HC, Fiorentini C, Boquet P, et al. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci U S A. 1994;91:3814–8. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hertle R. The family of Serratia type pore forming toxins. Curr Protein Pept Sci. 2005;6:313–25. doi: 10.2174/1389203054546370. [DOI] [PubMed] [Google Scholar]
- 41.Eshraghi A, Maldonado-Arocho FJ, Gargi A, Cardwell MM, Prouty MG, Blanke SR, et al. Cytolethal distending toxin family members are differentially affected by alterations in host glycans and membrane cholesterol. J Biol Chem. 2010;285:18199–207. doi: 10.1074/jbc.M110.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elwell CA, Dreyfus LA. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol Microbiol. 2000;37:952–63. doi: 10.1046/j.1365-2958.2000.02070.x. [DOI] [PubMed] [Google Scholar]
- 43.Smith JL, Bayles DO. The contribution of cytolethal distending toxin to bacterial pathogenesis. Crit Rev Microbiol. 2006;32:227–48. doi: 10.1080/10408410601023557. [DOI] [PubMed] [Google Scholar]
- 44.Saier MH, Ma CH, Rodgers L, Tamang DG, Yen MR. Protein secretion and membrane insertion systems in bacteria and eukaryotic organelles. Adv Appl Microbiol. 2008;65:141–97. doi: 10.1016/S0065-2164(08)00606-0. [DOI] [PubMed] [Google Scholar]
- 45.Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- 46.Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 47.Paulsen IT, Park JH, Choi PS, Saier MH., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 48.Saier MH., Jr Protein secretion and membrane insertion systems in gram-negative bacteria. J Membr Biol. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 49.Mueller CA, Broz P, Cornelis GR. The type III secretion system tip complex and translocon. Mol Microbiol. 2008;68:1085–95. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- 50.Woestyn S, Allaoui A, Wattiau P, Cornelis GR. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–9. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clements A, Young JC, Constantinou N, Frankel G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes. 2012;3:71–87. doi: 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–13. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loveless BJ, Saier MH., Jr A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–23. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 54.Roggenkamp A, Ackermann N, Jacobi CA, Truelzsch K, Hoffmann H, Heesemann J. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J Bacteriol. 2003;185:3735–44. doi: 10.1128/JB.185.13.3735-3744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forman S, Wulff CR, Myers-Morales T, Cowan C, Perry RD, Straley SC. yadBC of Yersinia pestis, a new virulence determinant for bubonic plague. Infect Immun. 2008;76:578–87. doi: 10.1128/IAI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leo JC, Grin I, Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Phil Trans R Soc London Series B Biol Sci. 2012;367:1088–101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gal-Mor O, Gibson DL, Baluta D, Vallance BA, Finlay BB. A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog. 2008;4:e1000036. doi: 10.1371/journal.ppat.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wentzel A, Christmann A, Adams T, Kolmar H. Display of passenger proteins on the surface of Escherichia coli K-12 by the enterohemorrhagic E. coli intimin EaeA. J Bacteriol. 2001;183:7273–84. doi: 10.1128/JB.183.24.7273-7284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–8. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 60.Harley KT, Saier MH., Jr A novel ubiquitous family of putative efflux transporters. J Mol Microbiol Biotechnol. 2000;2:195–8. [PubMed] [Google Scholar]
- 61.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–25. [PubMed] [Google Scholar]
- 62.Dinh T, Paulsen IT, Saier MH., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–31. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong KK, Brinkman FS, Benz RS, Hancock RE. Evaluation of a structural model of Pseudomonas aeruginosa outer membrane protein OprM, an efflux component involved in intrinsic antibiotic resistance. J Bacteriol. 2001;183:367–74. doi: 10.1128/JB.183.1.367-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letoffe S, Delepelaire P, Wandersman C. Protein secretion in gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 1996;15:5804–11. [PMC free article] [PubMed] [Google Scholar]
- 65.Bleuel C, Grosse C, Taudte N, Scherer J, Wesenberg D, Krauss GJ, et al. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J Bacteriol. 2005;187:6701–7. doi: 10.1128/JB.187.19.6701-6707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrobial Agents Chemother. 2001;45:1126–36. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clegg S, Wilson J, Johnson J. More than one way to control hair growth: regulatory mechanisms in enterobacteria that affect fimbriae assembled by the chaperone/usher pathway. J Bacteriol. 2011;193:2081–8. doi: 10.1128/JB.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Henderson NS, So SS, Martin C, Kulkarni R, Thanassi DG. Topology of the outer membrane usher PapC determined by site-directed fluorescence labeling. J Biol Chem. 2004;279:53747–54. doi: 10.1074/jbc.M409192200. [DOI] [PubMed] [Google Scholar]
- 69.Mapingire OS, Henderson NS, Duret G, Thanassi DG, Delcour AH. Modulating effects of the plug, helix, and N- and C-terminal domains on channel properties of the PapC usher. J Biol Chem. 2009;284:36324–33. doi: 10.1074/jbc.M109.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–75. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guedin S, Willery E, Tommassen J, Fort E, Drobecq H, Locht C, et al. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J Biol Chem. 2000;275:30202–10. doi: 10.1074/jbc.M005515200. [DOI] [PubMed] [Google Scholar]
- 72.Brok R, Van Gelder P, Winterhalter M, Ziese U, Koster AJ, de Cock H, et al. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J Mol Biol. 1999;294:1169–79. doi: 10.1006/jmbi.1999.3340. [DOI] [PubMed] [Google Scholar]
- 73.Korotkov KV, Gonen T, Hol WG. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci. 2011;36:433–43. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLaughlin LS, Haft RJ, Forest KT. Structural insights into the type II secretion nanomachine. Curr Opin Struct Biol. 2012;22:208–16. doi: 10.1016/j.sbi.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jakes KS, Cramer WA. Border crossings: colicins and transporters. Annu Rev Genet. 2012;46:209–31. doi: 10.1146/annurev-genet-110711-155427. [DOI] [PubMed] [Google Scholar]
- 76.Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. J Biol Chem. 2006;281:24085–9. doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- 77.Gaither LA, Eide DJ. The human ZIP1 transporter mediates zinc uptake in human K562 erythroleukemia cells. J Biol Chem. 2001;276:22258–64. doi: 10.1074/jbc.M101772200. [DOI] [PubMed] [Google Scholar]
- 78.Techau ME, Valdez-Taubas J, Popoff JF, Francis R, Seaman M, Blackwell JM. Evolution of differences in transport function in Slc11a family members. J Biol Chem. 2007;282:35646–56. doi: 10.1074/jbc.M707057200. [DOI] [PubMed] [Google Scholar]
- 79.Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci U S A. 2002;99:16243–8. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koster S, Wehner M, Herrmann C, Kuhlbrandt W, Yildiz O. Structure and function of the FeoB G-domain from Methanococcus jannaschii. J Mol Biol. 2009;392:405–19. doi: 10.1016/j.jmb.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 81.Ash MR, Guilfoyle A, Clarke RJ, Guss JM, Maher MJ, Jormakka M. Potassium-activated GTPase reaction in the G protein-coupled ferrous iron transporter B. J Biol Chem. 2010;285:14594–602. doi: 10.1074/jbc.M110.111914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deshpande CN, McGrath AP, Font J, Guilfoyle AP, Maher MJ, Jormakka M. Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:399–404. doi: 10.1107/S1744309113005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ash MR, Maher MJ, Guss JM, Jormakka M. The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure. PloS One. 2011;6:e23355. doi: 10.1371/journal.pone.0023355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mu W, Zhang W, Feng Y, Jiang B, Zhou L. Recent advances on applications and biotechnological production of D-psicose. Appl Microbiol Biotechnol. 2012;94:1461–7. doi: 10.1007/s00253-012-4093-1. [DOI] [PubMed] [Google Scholar]
- 85.Boudebbouze S, Maguin E, Rhimi M. Bacterial L-arabinose isomerases: industrial application for D-tagatose production. Recent Pat DNA Gene Seq. 2011;5:194–201. doi: 10.2174/187221511797636248. [DOI] [PubMed] [Google Scholar]
- 86.Iyer R, Williams C, Miller C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol. 2003;185:6556–61. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mathews S, Rathinasabapathi B, Ma LQ. Uptake and translocation of arsenite by Pteris vittata L.: effects of glycerol, antimonite and silver. Environ Pollut. 2011;159:3490–5. doi: 10.1016/j.envpol.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 88.Rothe M, Alpert C, Loh G, Blaut M. Novel insights into E. coli’s hexuronate metabolism: KduI facilitates the conversion of galacturonate and glucuronate under osmotic stress conditions. PloS One. 2013;8:e56906. doi: 10.1371/journal.pone.0056906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr Drug Metab. 2000;1:143–61. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- 90.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–22. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 91.Goller CC, Seed PC. Revisiting the Escherichia coli polysaccharide capsule as a virulence factor during urinary tract infection: contribution to intracellular biofilm development. Virulence. 2010;1:333–7. doi: 10.4161/viru.1.4.12388. [DOI] [PubMed] [Google Scholar]
- 92.Robicsek F, Citron DS, Taylor FH, Sanger PW. The treatment of malignant hypertension due to ischemia of the solitary functioning kidney. Angiology. 1963;14:377–80. doi: 10.1177/000331976301400707. [DOI] [PubMed] [Google Scholar]
- 93.Rudd PM, Wormald MR, Dwek RA. Sugar-mediated ligand-receptor interactions in the immune system. Trends Biotechnol. 2004;22:524–30. doi: 10.1016/j.tibtech.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 94.Peter-Katalinic J. Methods in enzymology: O-glycosylation of proteins. Meth Enzymol. 2005;405:139–71. doi: 10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- 95.Edmands SD, Hughs KS, Lee SY, Meyer SD, Saari E, Yancey PH. Time-dependent aspects of osmolyte changes in rat kidney, urine, blood and lens with sorbinil and galactose feeding. Kidney Int. 1995;48:344–53. doi: 10.1038/ki.1995.302. [DOI] [PubMed] [Google Scholar]
- 96.Anderson PM, Sung YC, Fuchs JA. The cyanase operon and cyanate metabolism. FEMS Microbiol Rev. 1990;7:247–52. doi: 10.1111/j.1574-6968.1990.tb04920.x. [DOI] [PubMed] [Google Scholar]
- 97.Sung YC, Fuchs JA. Identification and characterization of a cyanate permease in Escherichia coli K-12. J Bacteriol. 1989;171:4674–8. doi: 10.1128/jb.171.9.4674-4678.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pedemonte N, Caci E, Sondo E, Caputo A, Rhoden K, Pfeffer U, et al. Thiocyanate transport in resting and IL-4-stimulated human bronchial epithelial cells: role of pendrin and anion channels. J Immunol. 2007;178:5144–53. doi: 10.4049/jimmunol.178.8.5144. [DOI] [PubMed] [Google Scholar]
- 99.Abdulhussein AA, Wallace HM. Polyamines and membrane transporters. Amino Acids. 2014;46(3):655–60. doi: 10.1007/s00726-013-1553-6. http://dx.doi.org/10.1007/s00726-013-1553-6. Epub 2013 Jul 13. [DOI] [PubMed] [Google Scholar]
- 100.Andersson AC, Henningsson S, Rosengren E. Increased formation of diamines and polyamines in the pregnant rat. J Physiol. 1978;285:311–24. doi: 10.1113/jphysiol.1978.sp012573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–81. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi PS, Dawson AJ, Bernstein HD. Characterization of a novel two-partner secretion system in Escherichia coli O157:H7. J Bacteriol. 2007;189:3452–61. doi: 10.1128/JB.01751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kolodziejek AM, Hovde CJ, Minnich SA. Yersinia pestis Ail: multiple roles of a single protein. Front Cell Infect Microbiol. 2012;2:103. doi: 10.3389/fcimb.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Neely MN, Friedman DI. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998;28:1255–67. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 105.Rainnie DJ, Bragg PD. The effect of iron deficiency on respiration and energy-coupling in Escherichia coli. J Gen Microbiol. 1973;77:339–49. doi: 10.1099/00221287-77-2-339. [DOI] [PubMed] [Google Scholar]
- 106.Wang L, Cherayil BJ. Ironing out the wrinkles in host defense: interactions between iron homeostasis and innate immunity. J Innate Immun. 2009;1:455–64. doi: 10.1159/000210016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Semrin G, Fishman DS, Bousvaros A, Zholudev A, Saunders AC, Correia CE, et al. Impaired intestinal iron absorption in Crohn’s disease correlates with disease activity and markers of inflammation. Inflamm Bowel Dis. 2006;12:1101–6. doi: 10.1097/01.mib.0000235097.86360.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hancock V, Ferrieres L, Klemm P. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology. 2008;154:167–75. doi: 10.1099/mic.0.2007/011981-0. [DOI] [PubMed] [Google Scholar]
- 109.Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 2010;22:904–17. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anderson DS, Adhikari P, Nowalk AJ, Chen CY, Mietzner TA. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J Bacteriol. 2004;186:6220–9. doi: 10.1128/JB.186.18.6220-6229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hodges K, Alto NM, Ramaswamy K, Dudeja PK, Hecht G. The enteropathogenic Escherichia coli effector protein EspF decreases sodium hydrogen exchanger 3 activity. Cell Microbiol. 2008;10:1735–45. doi: 10.1111/j.1462-5822.2008.01163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 2007;3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gobert AP, Vareille M, Glasser AL, Hindre T, de Sablet T, Martin C. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-kappaB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J Immunol. 2007;178:8168–74. doi: 10.4049/jimmunol.178.12.8168. [DOI] [PubMed] [Google Scholar]
- 114.Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, et al. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol biology. 2009;16:853–60. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1277–83. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- 116.Thelestam M, Frisan T. Cytolethal distending toxins. Rev Physiol Biochem Pharmacol. 2004;152:111–33. doi: 10.1007/s10254-004-0030-8. [DOI] [PubMed] [Google Scholar]
- 117.Tukel C, Akcelik M, de Jong MF, Simsek O, Tsolis RM, Baumler AJ. MarT activates expression of the MisL autotransporter protein of Salmonella enterica serotype Typhimurium. J Bacteriol. 2007;189:3922–6. doi: 10.1128/JB.01746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koretke KK, Szczesny P, Gruber M, Lupas AN. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J Struct Biol. 2006;155:154–61. doi: 10.1016/j.jsb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 119.Swimm AI, Kalman D. Cytosolic extract induces Tir translocation and pedestals in EPEC-infected red blood cells. PLoS Pathog. 2008;4:e4. doi: 10.1371/journal.ppat.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schuller S, Chong Y, Lewin J, Kenny B, Frankel G, Phillips AD. Tir phos-phorylation and Nck/N-WASP recruitment by enteropathogenic and enter-ohaemorrhagic Escherichia coli during ex vivo colonization of human intestinal mucosa is different to cell culture models. Cell Microbiol. 2007;9:1352–64. doi: 10.1111/j.1462-5822.2006.00879.x. [DOI] [PubMed] [Google Scholar]
- 121.de Pace F, Boldrin de Paiva J, Nakazato G, Lancellotti M, Sircili MP, Guedes Stehling E, et al. Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology. 2011;157:2954–62. doi: 10.1099/mic.0.050005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–94. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–5. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


