Abstract
Endogenous mitochondrial genes encode critical oxidative phosphorylation components and their mutation results in a set of disorders known collectively as mitochondrial encephalomyopathies. There is intensive interest in modulating mitochondrial function as organelle dysfunction has been associated with numerous disease states. Proteins encoded by the mitochondrial genome cannot be genetically manipulated by current techniques. Here we report the development of a mitochondrial-targeted RNA expression system (mtTRES) utilizing distinct non-coding leader sequences (NCLs) and enabling in vivo expression of small mitochondrial-targeted RNAs. mtTRES expressing small chimeric antisense RNAs were used as translational inhibitors (TLIs) to target endogenous mitochondrial protein expression in vivo. By utilizing chimeric antisense RNA we successfully modulate expression of two mitochondrially-encoded proteins, ATP6 and COXII, and demonstrate the utility of this system in vivo and in human cells. This technique has important and obvious research and clinical implications.
Keywords: Mitochondrial disorders, translation inhibitors, non-coding small chimeric RNA, mitochondrial encoded proteins
Introduction
Mutations in the mitochondrial genome cause a set of devastating disease conditions categorized as primary respiratory chain diseases, also known as mitochondrial encephalomyopathies (MEs) (DiMauro and Schon, 2003). Mitochondrial gene therapy has been proposed as a treatment for ME, however, this approach remains controversial as there are limited preclinical data demonstrating efficacy and evidence suggesting this approach may have significant limitations (Manfredi et al., 2002, Bokori-Brown and Holt, 2006, Perales-Clemente et al., 2011).
Endogenously encoded mitochondrial proteins function within large well-characterized respiratory complexes that perform oxidative phosphorylation (OXPHOS). The mitochondrial genome is known to harbor hundreds of pathogenic mutations, including ones affecting all of the tRNA genes and over 260 distinct coding mutations. The vast majority of protein-coding gene mutations associated with human mitochondrial disease are missense mutations, accounting for ~225 of the pathogenic mitochondrial mutations (www.mitomap.org), implying that mutant protein is usually capable of being expressed in the disease state. We previously discovered and characterized a Drosophila model of ME with an endogenous missense mutation in the ATP6 gene affecting the F1Fo-ATPsynthase (complex V) (Celotto et al., 2006a, Palladino, 2010). Twenty-one distinct human missense mutations exist within the ATP6 gene, fourteen of which have been shown to cause human MEs including Familial Bilateral Striatal Necrosis (FBSN), Neuropathy, Ataxia, and Retinitis Pigmentosa (NARP), or Maternally Inherited Leigh’s Syndrome (MILS) (Noji et al., 1997, Stock et al., 1999, D'Aurelio et al., 2010). ATP61 mutant flies contain a missense mutation with high mutant heteroplasmy and exhibit phenotypes analogous to human symptoms including locomotor and progressive neural dysfunction, seizures, myodegeneration, and reduced longevity (Palladino, 2010).
Competition with mutant protein for incorporation into mature respiratory complexes is likely a major obstacle to a viable mitochondrial gene therapy: a fact that has largely been ignored. This competition may explain the controversial allotopic expression results and remains a formidable obstacle to the treatment of MEs resulting from any endogenous mitochondrial missense mutation. A method to specifically reduce expression of mitochondrial-encoded genes is not known.
Several RNAs are naturally imported into the mitochondria from the cytoplasm and detailed studies have provided critical insight into the import process and import substrates (Schneider and Marechal-Drouard, 2000, Tarassov et al., 2007, Lithgow and Schneider, 2010). Although the exact mechanism of RNA import into mitochondria is unknown, several pathways have been suggested to mediate mitochondrial RNA import (Mahapatra and Adhya, 1996, Wang et al., 2010, Schneider, 2011). We have identified a nuclear encoded mitochondrial 5S rRNA isoform and engineered a novel vector to express small RNAs in vivo. We developed a mitochondrial-targeted translational inhibition (TLI) approach using small chimeric RNAs to regulate endogenous mitochondrial protein expression. Here we demonstrate the efficacy of mitochondrial-targeted TLIs by targeting two distinct loci encoding essential proteins of two different OXPHOS complexes, one in vivo and the other in vitro. The ability to selectively modulate mitochondrial protein expression in animals represents an important technological advance with obvious research and clinical applications.
Material and methods
Engineering mtTRES and mtTRES-TLI constructs
The mtTRES vector was created using the available pUAST-attB vector as a backbone (Bischof et al., 2007). A StuI site was added by site directed mutagenesis 5’ to the attP integration site using Quick Change Lightning (Invitrogen, USA). The 5S rRNA RNAPIII promoter (AE013599.4) and termination (AE013599.4) sequences were PCR amplified from wild type Drosophila genomic DNA and directionally inserted using standard cloning methods and the HindIII-EcoRI and StuIKpnI cloning sites, respectively. For the mammalian mtTRES vector the human U6 promoter (NT_010194.17) was PCR amplified from pSilencer 2.1 (Invitrogen, USA), purified and inserted in place of the fly RNAP III promoter. The EcoRI-EagI cloning sites were used to insert NCLs. The 5S rRNAmt variant was identified as the most abundant mitochondrial isoform by clonal analyses (88%) from three independent cloning events and sequence analysis of 135 clones. The 5S rRNAmt was the major mitochondrial isoform in all three independent clonal populations (Supplementary material: Figure S1 and GenBank: CR33451). The 5S rRNAmt sequence was synthesized with flanking EcoRI-EagI cloning sites (GeneWiz, South Plainfield NJ, USA). The MRP and RNAseP (RNP) oligonucleotides were annealed and directionally cloned into EcoRIEagI cloning sites using published sequences (Wang et al., 2012b). TLI complementary sequences were synthesized as oligonucleotides, annealed and directionally cloned into EagIKpnI cloning sites. TLI-5Smt::ATP6(a) is 25 bases long, whereas TLI-5Smt::ATP6(b) is 26 nucleotides in length and the complementary region is shifted 3 nucleotides 5’. All oligonucleotides were commercially synthesized by IDT (Coralville IA, USA). The final constructs were sequence verified (GeneWiz, South Plainfield NJ, USA).
Drosophila transgenesis, longevity and locomotor assays
mtTRES vectors allow site-directed PhiC31-mediated attP/B transgenesis. We used the VK00027 attP insertion site and flies bearing the VK27 attP chromosome are the control for all transgenic experiments. DNA injections were performed by Genetic Services (Cambridge MA, USA) and successful transgenesis events were identified using whitemc+. Homozygous transgenic strains were tested. Previously established methods were used to test longevity (Palladino et al., 2003) and locomotor assays (Fergestad et al., 2006, Fergestad et al., 2008).
Western blotting and antisera production
Standard methods were used for western blot analyses (Celotto et al., 2012). Briefly, flies were carbon dioxide anesthetized and snap frozen in liquid nitrogen. Thoraces from 8 flies were dissected and homogenized in sample buffer (125 µl), heated at 95 °C for 5 min, loaded into the wells of an SDS-PAGE gel. Antisera was generated to fly ATP6 protein using purified HKEFKTLLGPSGHNGS peptide (hc17), immunized New Zealand rabbits and antigen affinity purification (NeoBioSci, Cambridge MA, USA). Anti-ATP6 antibody recognition specificity of hc17 peptide was confirmed by Southern Blot and ELISA (by NeoBioSci, Cambridge MA, USA). Western blotting identifies a single ~25kDa protein that enriches with mitochondria. Competitive ELISA (kit by Cell Biolabs Inc., USA) using fly lysates and increasing concentrations of hc17 peptide was used to further validate the specificity of the anti-ATP6 antibody (Supplementary material, Figure S2). ATP6 antisera is used 1:2000. Anti-COXII antibodies (Proteintech, Chicago IL, USA) and anti-SOD2 antibodies (LSBio, Seattle WA, USA) were used at 1:2500 and 1:2000, respectively. Anti-ATP-alpha (a5-c antibody, Developmental Studies Hybridoma Bank, University of Iowa, USA) was used as a loading control. ATPalpha is a nuclear encoded plasma membrane protein (the catalytic subunit of the Na+/K+ ATPase). For HeLa cells, 1× 106 cells were electroporated and harvested after ~48–72 hr for western blot. GAPDH (1:3000) (Abcam, USA) was used as a loading control. Secondary detection was performed using anti-rabbit (1:4000) (Biorad, USA) and anti-mouse (1:10000) (Biorad, USA) HRP conjugated antibodies. For all Western blots sub-saturation images have been quantified. In some cases, darker exposures of the quantified images are used in the figures.
RNA isolation and Quantitative RT–PCR
RNA was extracted from 12 whole flies, using 250 µl Trizol (Invitrogen, San Diego, USA) and the RNeasy mini kit (Qiagen, Valencia, USA). RNA was eluted in 100 µl dH2O and quantified. 5 µg RNA was used to perform a reverse transcription reaction (Superscript RT, Invitrogen). Quantitative Real-Time PCR [Mx3000P QPCR System, Stratagene] was performed using standard techniques with normalization to RP49 expression (Celotto et al., 2006b). Only DNA-free cDNA samples were used. In a total reaction of 25 µl, 12.5 µl 2X-SYBR Green Supermix (Qiagen, Valencia, USA), 2 µl of cDNA and 400 nM each of forward and reverse primers (ATP6, COXII) were used. Fold change (FC) was determined using the equation, FC=2−Δ(ΔCt). All QPCR experiments were performed with four biological replicates and the data were normalized to mRNA expression levels of RP49.
Isolation of mitochondria from HeLa cells
Mitochondria were isolated using standard differential centrifugation procedure. In short, 24 million cells were trypsinized and homogenized by Dounce homogenizer. Nuclear fraction was pelleted at 1000g for 15 min. The supernatant was then centrifuged at 10000g for 15min. The pellet contained the enriched mitochondria.
In vitro transcription and radiolabeling
Primers were designed to amplify 5Smt and TLI-5Smt::COXII sequences from previously engineered mtTRES-5Smt and mtTRES-5Smt-TLI::COXII plasmids. T7+5s_For (TAATACGACTCACTATAGGGGCCAACGACCATACCACGCTGAATAC) and 5s_Rev (AGGCCAACAACACGCGGT GTTC) primers were used for 5S DNA amplification. For TLI- 5Smt::COXII, T7+5s_For and COX2_Rev (TCCAAAAAATCTTAATGGCACATGCAGC) primers were used.
Using Thermo Scientific TranscriptAid T7 High Yield Transcription Kit and [α-32P] adenosine 5’-triphosphate (MP Biomedicals) in vitro transcription was performed, as per the manufacturer’s instructions. Unincorporated [α-32P] ATP was removed using NucAway™ Spin Columns (Ambion Inc. Austin, Texas). Specific activities of radiolabeled RNA products were quantified by LS6500 Multi-Purpose Scintillation Counter (Beckman Coulter) and equal amounts used in the mitochondrial import assay.
Mitochondrial RNA import assay
Mitochondrial RNA import assay was modified from (Magalhaes et al., 1998, Bhattacharyya et al., 2002, Wang et al., 2012a). In short, mitochondrial pellets were suspended with RNA probes in the import buffer (200 µl final volume) containing 0.25M sucrose, 2 mM KH2PO4, 50 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 5 mM L-methionine, 1 mg/ml BSA, 5 mM ATP, 2 mM DTT, 20 mM succinate, 50 mM HEPES, [pH 7.1]. The mixture was incubated for 20–30 minutes at room temperature. Mitochondria were spun at 11000g for 5 min and washed once with wash buffer (0.6 M sorbitol, 20 mM Tris, [pH 8.0]). To remove RNA that was not imported in the mitochondria, the pellet was spun again and resuspended in 200 µl nuclease buffer containing 25 µg/ml of micrococcal nuclease (New England Biolabs Inc.) and incubated for 30 min at 27°C. Mitochondria were collected, solubilized in SDS buffer at 65°C for 5 min, RNA was purified using TRIzol® reagent (Life Technologies) and resolved by denaturing polyacrylamide urea gel (National Diagnostics). Autoradiography was performed using phosphor imager and gel was scanned using Image Quant software.
Results
Generation of mtTRES for TLI in vivo
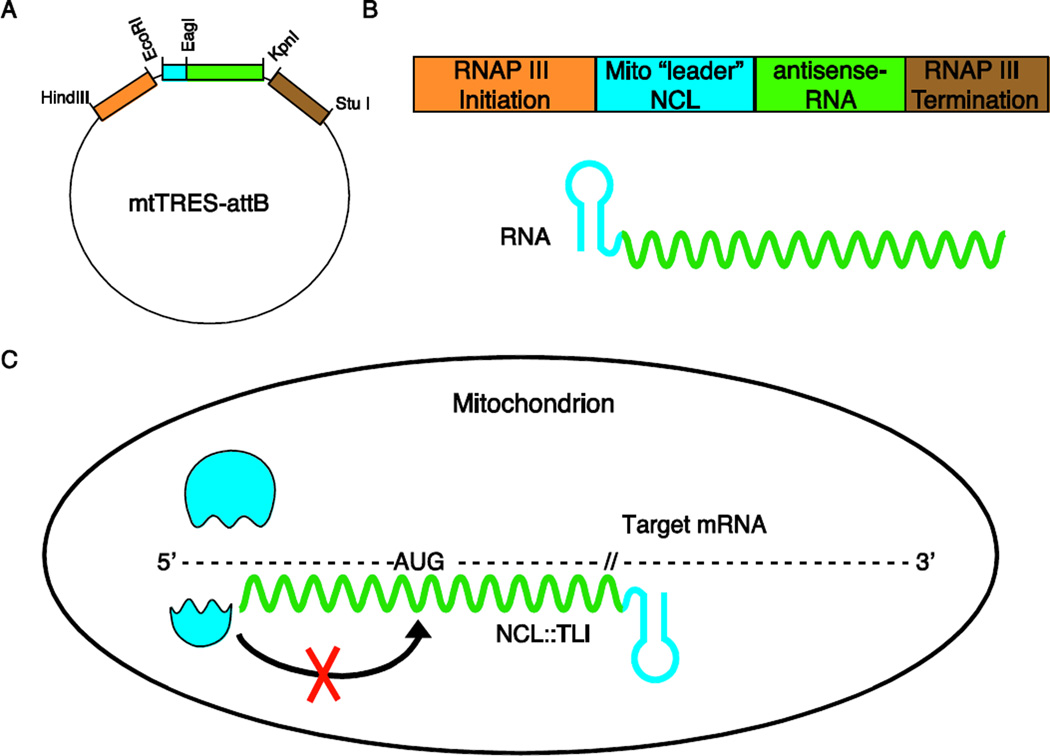
The metazoan mitochondrial genome contains 37 genes that are critical for electron transport chain function. These include 13 protein-coding genes and RNAs to facilitate their expression: 22 tRNAs and 2 rRNAs (12S and 16S) (Anderson et al., 1981, Bibb et al., 1981). All other proteins and RNAs functioning within the mitochondrion are imported from the cytoplasm. It has previously been described that 5S rRNAs are expressed from large nuclear gene arrays (~100–200 genes) and that 5S rRNAs can readily be found within mitochondria from flies to humans (Benhamou and Jordan, 1976, Artavanis-Tsakonas et al., 1977, Yoshionari et al., 1994, Magalhaes et al., 1998, Entelis et al., 2001, Smirnov et al., 2008, Smirnov et al., 2010). Although these gene arrays encode many 5S rRNA isoforms, we identified a single common mitochondrial 5S rRNA variant (5S rRNAmt) representing the majority of 5S rRNAs within Drosophila mitochondria. To enable studies in vivo, we developed the mtTRES (mitochondrial Targeted RNA Expression System) vector using 5S rRNAmt as a non-coding leader sequence (NCL) and employing RNAP III promoter and termination elements (Figure 1). The RNAPIII promoter was selected due to its ability to direct transcription of rRNAs, tRNAs and other small non-coding RNAs (Dieci et al., 2007).
Figure 1. Design of the mtTRES attB transgenesis vector.
(A) Restriction enzyme map of the mtTRES-attB vector. (B) Cartoon describing the linear 5’ to 3’ order of required components for allotopic RNA expression: RNAP III specific initiation and termination (orange and brown, respectively), non-coding leader sequence (NCL) RNA (blue) and the antisense RNA (TLI) (green). The subsequent RNA transcribed will be a chimeric NCL-TLI RNA. (C) Cartoon demonstrating the proposed mechanism of translational inhibition. The complementary sequence competes with the small subunit of the ribosome for binding thus inhibiting docking to the target RNA at the start codon (AUG).
We asked whether we might utilize this mitochondrial RNA targeting system to modulate expression of endogenous mitochondrial genes in vivo. To test this we generated transgenic mtTRES animals capable of expressing chimeric RNAs consisting of an NCL and a sequence complementary to a mitochondrial mRNA, specifically targeting the known translational start site (Figure 1B, C). Translational inhibition/repression has been demonstrated to be functional within the cytosol by antagonizing small ribosomal subunit docking and lowering translational efficiency (Doyle et al., 2001) but has never been demonstrated in mitochondria.
mtTRES ATP6 TLIs phenocopy ATP61 longevity and locomotor dysfunction
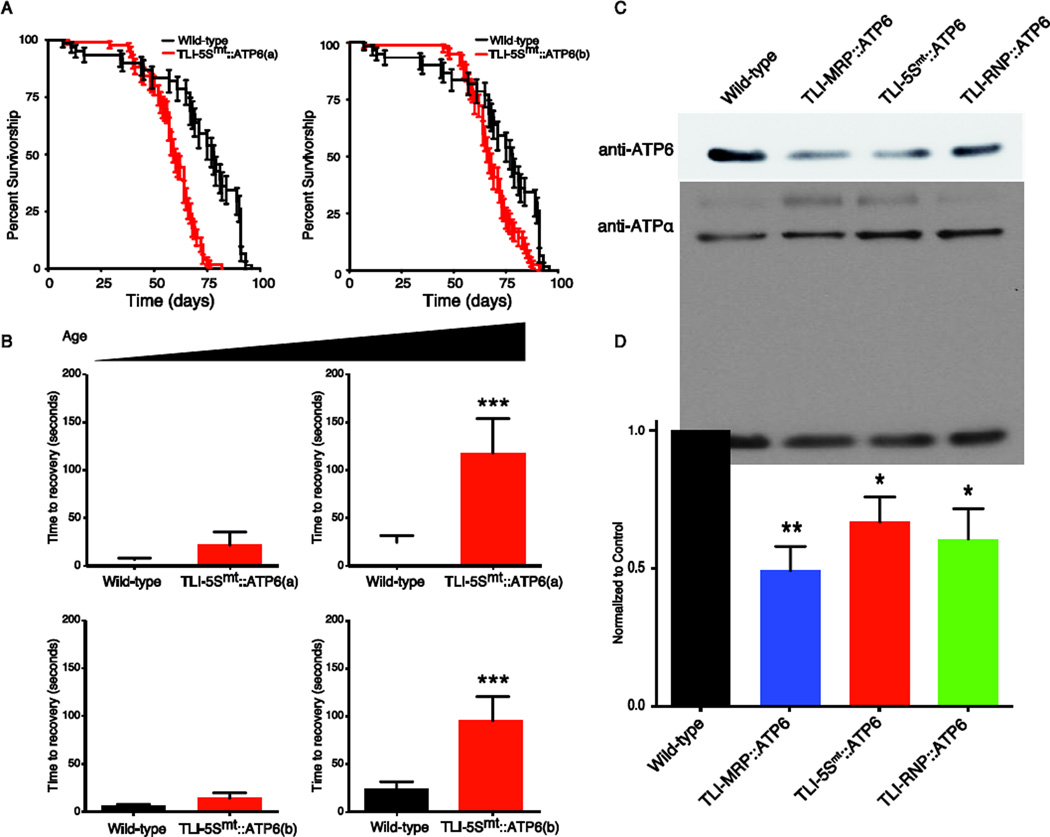
Numerous ATP6 missense mutations are associated with human disease and our detailed understanding of ATP61 mutant phenotypes prompted us to initially ask whether we could functionally knock down ATP6 expression in vivo. We utilized mtTRES to generate two independent transgenic ATP6 TLIs designated as TLI-5Smt::ATP6(a) and TLI-5Smt::ATP6(b). Lifespan assays were performed to test whether TLI-5Smt::ATP6 TLIs affect the longevity of flies. We observed a significant decrease in the longevity of flies expressing TLI-5Smt::ATP6(a) or TLI-5Smt::ATP6(b) compared to wild type control flies (Figure 2A). These data demonstrate that mtTRES TLIs targeting ATP6 reduce longevity consistent with a loss of ATP6 function in vivo.
Figure 2. Translational inhibitors exhibit reduced longevity, mechanical stress sensitivity and lower steady state protein expression in vivo.
(A) TLI-5Smt::ATP6(a) (red) has a 24% reduction in survival as compared to the wild-type animals (black) (n=95, p<0.0001). TLI- 5Smt::ATP6(b) (red) displays a 15% reduction in survival as compared to wild-type animals (in black) (n=83, p<0.0001). (B) TLI-5Smt::ATP6(a) animals exhibit a progressive increase in mechanical stress sensitivity (day 5 and day 50 shown). (C) TLI-NCL::ATP6(a) fly extracts were probed with anti-ATP6 antibody to examine steady state protein levels. The expression levels were normalized to ATPalpha, the plasma membrane Na/K ATPase catalytic subunit (upper panel). (D) Quantitation of western blots show 50% decrease in TLI-MRP::ATP6(a), 34% decrease in TLI-5Smt::ATP6(a) and 40% decrease in TLI-RNP::ATP6(a). Unpaired t-test was used as statistical test; *p<0.01, **p<0.001, ***=p< 0.0001; mean ± SEM, n=3–4.
TLI-5Smt::ATP6(a) and TLI-5Smt::ATP6(b) flies were tested for conditional locomotor function in response to sensory hyperstimulation (bang sensitivity), a progressive seizure-related phenotype resulting from loss of ATP6 function in vivo (Celotto et al., 2006a). Young TLI-5Smt::ATP6(a) and TLI-5Smt::ATP6(b) animals (day 5) were aphenotypic; however, aged animals (day 50) exhibited conditional locomotor impairment compared to wild type control animals (Figure 2B). Strikingly, both TLI-5Smt::ATP6 transgenic strains phenocopy the conditional locomotor dysfunction observed in ATP61, including the progressive nature of this mitochondrial seizure-related phenotype. Importantly, ATP61 is of extremely high mutant heteroplasmy (98%) and results in severe locomotor and longevity phenotypes, whereas, TLI-5Smt::ATP6 results in an ~50% knockdown and the observed phenotypes are qualitatively similar but less severe, as would be expected for the hypomorphic condition.
Mitochondrial TLIs modulate protein levels
To more directly test the ability of mtTRES TLI’s to modulate protein expression we performed western blotting with TLI-5Smt::ATP6(a) transgenic fly lysates. Western blotting demonstrated a 34% reduction in ATP6 levels compared to lysates from wild type control animal (Figure 2C, D). These data demonstrate the mtTRES TLI approach is capable of endogenous mitochondrial protein modulation in vivo.
mtTRES TLI using distinct NCLs in vivo
Previously two small RNAs, MRP and RNP, were shown to be actively imported into mammalian mitochondria in vitro (Wang et al., 2010), suggesting their utility as NCLs. We generated two additional transgenesis vectors for in vivo animal studies, mtTRESMRP and mtTRESRNP. As an additional test of the functionality of the mtTRES system we generated TLI-MRP:: ATP6 and TLI-RNP::ATP6 that express chimeric RNAs targeting ATP6 mRNAs for TLI using mtTRESMRP and mtTRESRNP, respectively. ATP6 protein levels were examined in TLI-MRP::ATP6 and TLI-RNP::ATP6 animal extracts and were shown to be significantly reduced similar to TLI-5Smt::ATP6 (Figure 2C, D). Together these data demonstrate the ability to reduce steady state ATP6 protein levels using several independent constructs in vivo. Importantly these experiments utilize three distinct NCL sequences, including two discovered in mammals (Chang and Clayton, 1987, 1989, Puranam and Attardi, 2001).
mtTRES TLIs modulate expression independent of RNA stability
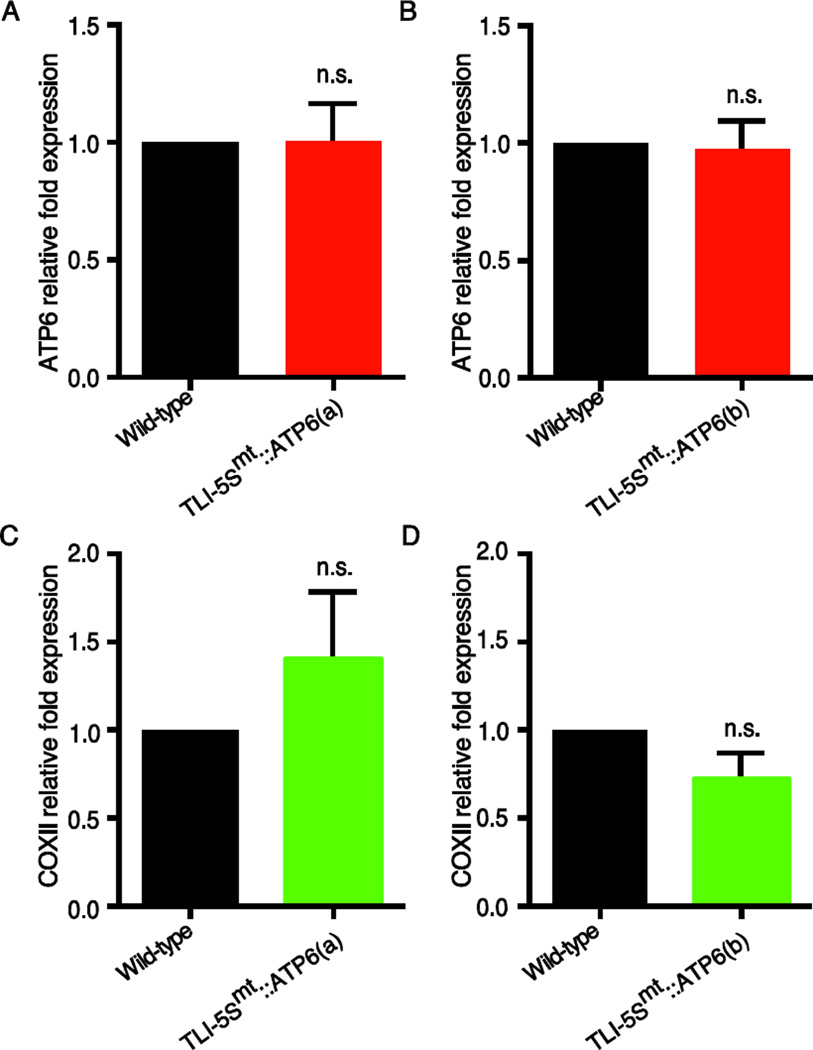
mtTRES TLIs are designed to function by antagonizing translation and reducing steady state protein levels by an RNA-stability independent mechanism. To test whether these chimeric mitochondrial targeted RNAs are modulating protein levels by regulating RNA stability, we performed qRT-PCR analyses on total RNA from TLI-5Smt::ATP6(a), TLI-5Smt::ATP6(b), and wild type control animals to determine whether RNA levels of the targeted gene were altered. No changes in ATP6 RNA levels were observed (Figure 3A, B). We also examined whether TLI-5Smt::ATP6(a) or TLI-5Smt::ATP6(b) altered the RNA levels of another mitochondrial expressed gene, COXII, and found no significant changes in COXII transcript levels with either of the TLIs (Figure 3C, D). Together these data are consistent with a translational inhibition/repression mechanism of action that is independent of alterations in RNA stability.
Figure 3. TLIs function by an RNA stability independent mechanism.
Fold change in transcript levels determined by qRT-PCR. Fold change mRNA expression of ATP6 (red) and COXII (green) is shown relative to wild type controls (black). (A and C) TLI-5Smt::ATP6(a). (B and D) TLI-5Smt::ATP6(b). All transcript levels were normalized to RP49 expression. One-way ANOVA was performed to test significance; n.s. is p > 0.05; mean ± SEM, n=9 (3 biological and 3 technical repeats of each sample).
Mitochondrial TLIs specifically knockdown target proteins
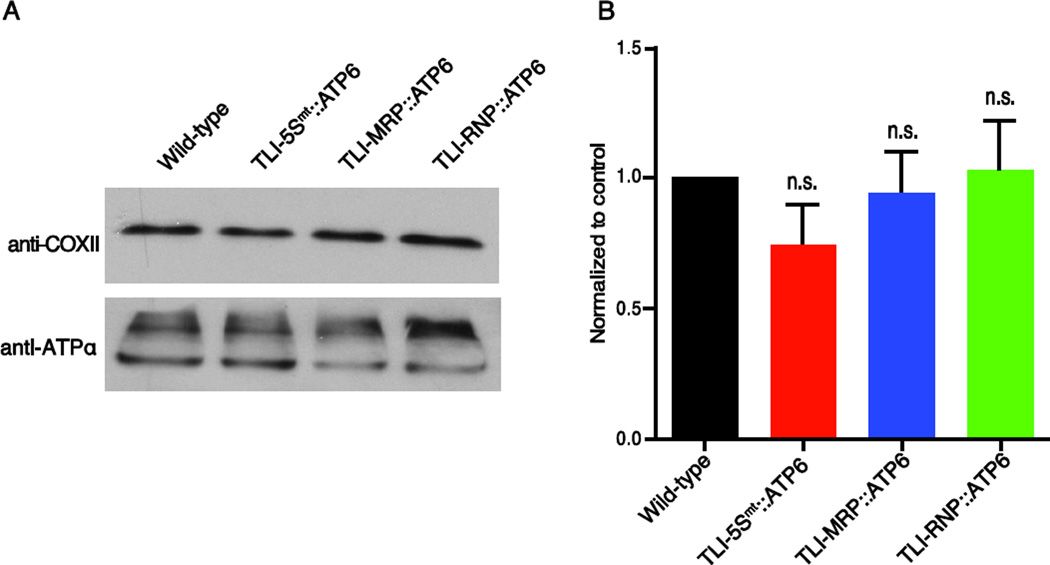
The chimeric RNAs in the present study are predicted to knockdown mitochondrial protein expression levels by specifically inhibiting the docking of the small subunit of mitoribosomes on target mRNA akin to the cytosolic mechanism of action (Doyle et al., 2001). To test the specificity of TLI-NCL::ATP6 chimeric RNAs, we examined COXII protein levels by western blot (Figure 4). TLI-NCL::ATP6 chimeric RNAs were able to modulate ATP6 protein levels, however, COXII protein was not altered (Figure 4A, B). These data suggest that TLINCL:: ATP6 chimeric RNAs do not globally alter translation and modulate target mitochondrial gene expression specifically.
Figure 4. Translational inhibitors knockdown target protein specifically.
(A) TLI-NCL:: ATP6(a) fly extracts were probed with anti-COXII antibody to examine steady state protein levels. The expression levels were normalized to ATPα (upper panel). (B) Quantitation of western blots showing no significant change in the COXII expression levels in NCL::ATP6(a). One-way ANOVA with multiple comparisons was used to test significance; n.s. is p > 0.05; mean ± SEM, n=3.
Mitochondrial TLIs in vitro, import assay and scrambled control
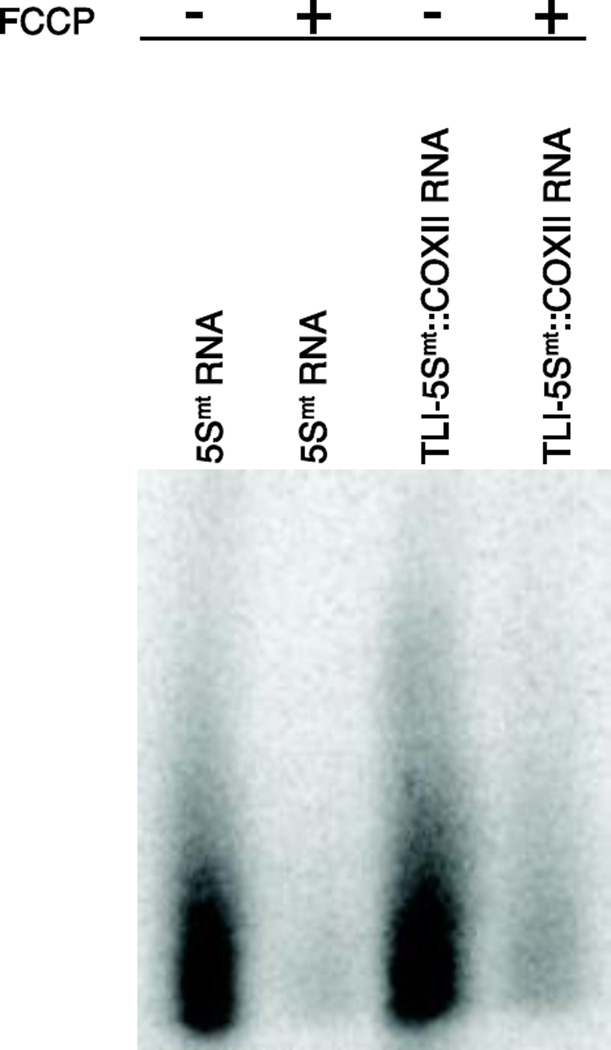
We developed a mammalian version of the mtTRES vector to examine efficacy of import and mtTRES-TLI constructs in human cells. We created a series of TLI-NCL::COXII constructs designed to target human COXII mRNAs. For chimeric TLI RNAs to regulate expression of endogenous mitochondrial proteins via translational repression they must be efficiently imported into mitochondria. We directly examined mitochondrial import of 5Smt rRNA and chimeric TLI-5Smt::COXII RNAs using an established import assay (Figure 5). RNA import was dependent on mitochondrial membrane potential, as evidenced by a lack of import when mitochondria were treated with FCCP (Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone) prior to adding the RNA probes. These data demonstrate robust import of the 5Smt rRNA used as an NCL and the chimeric TLI-5Smt::COXII RNAs and suggest our constructs might function well as TLIs in mammalian systems.
Figure 5. In vitro import of radiolabeled RNA into mitochondria.
[α-32P] labeled 5Smt RNA and TLI-5Smt::COXII RNA were transcribed in vitro and incubated with equal amounts of mitochondria isolated from HeLa cells in presence or absence of FCCP (+ or −). The pellet was treated with nuclease to digest non-imported RNAs and SDS treatment was performed to produce mitoplasts. Extracted RNAs were resolved using urea polyacrylamide gels and analyzed using Image quant (Storm 860 Molecular Imager). The experiment was repeated 3 times with a representative image shown.
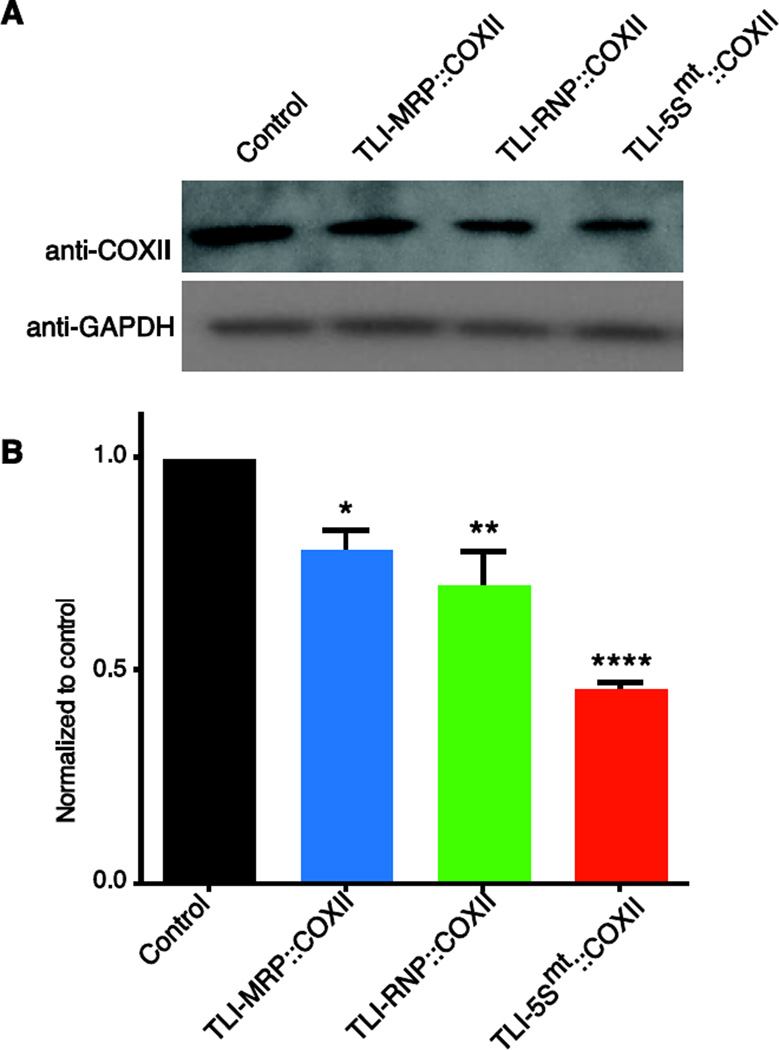
We functionally tested our mammalian TLI-NCL::COXII constructs using each of the three NCLs previously tested in vivo. All three TLI-NCL::COXII constructs significantly decreased COXII protein levels (Figure 6). These data demonstrate the ability of mtTRES TLI constructs to modulate protein levels in human cells using multiple NCL targeting signals, although the TLI-5Smt::COXII reliably gave the most significant knockdown.
Figure 6. Translational inhibitors decrease steady state protein levels in HeLa cells.
(A) HeLa cells were transfected with mtTRES plasmids expressing TLIs directed to human mitochondrial COXII RNAs. The cells were harvested at ~48–72 hrs and analyzed by western blot. (B) Quantification of steady-state COXII shown relative to the control plasmid (black) in cells transfected with mammalian mtTRES plasmids TLI-MRP::COXII, TLI-RNP::COXII and TLI-5Smt::COXII revealed reduced expression of 22%, 30% and 55%, respectively. GAPDH was the loading control. One-way ANOVA with multiple comparisons was performed to test significance; * is p < 0.03, ** is p < 0.0005; **** is p < 0.0001; mean ± SEM, n=3.
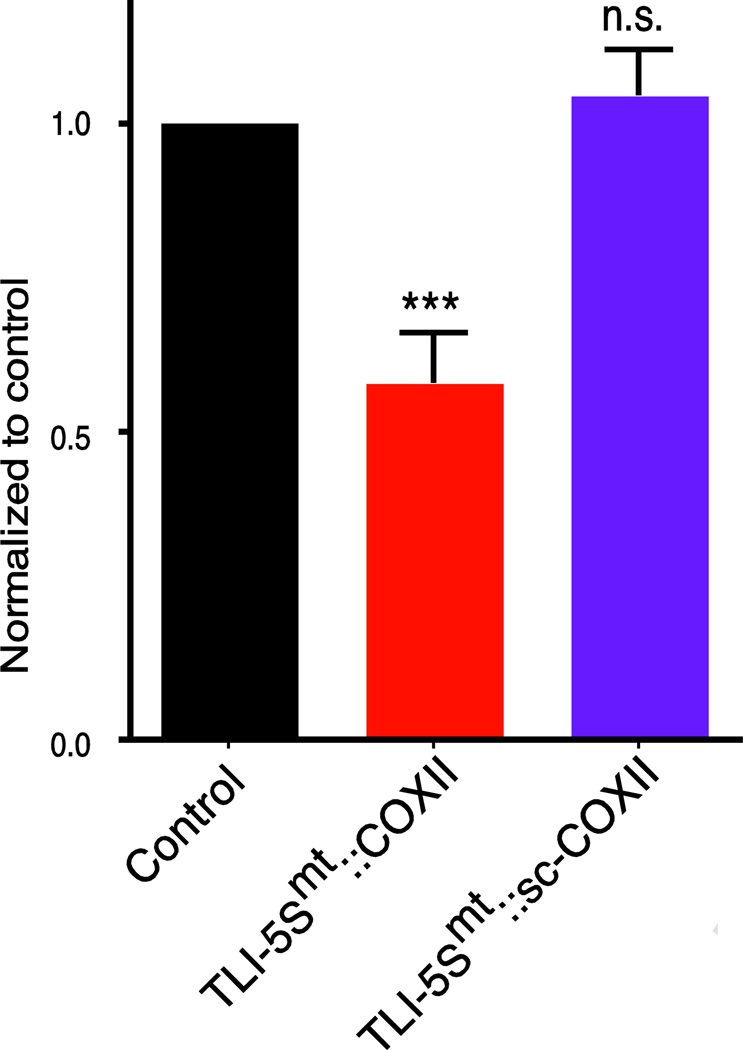
To further test the specificity of the mitochondrial TLI we generated a vector with a scrambled COXII complementary sequence (TLI-5Smt::scCOXII) and repeated our analysis with this additional control. Western blot data shows that TLI-5Smt::COXII reduced COXII protein, as it had previously, however, TLI-5Smt::sc-COXII (scrambled) does not alter the target protein (Figure 7). Together these data demonstrate the utility of mtTRES-TLI constructs in human cells.
Figure 7. TLI with scrambled complementary sequence do not alter COXII protein level.
TLI-5Smt::COXII reduced COXII protein expression in transfected HeLa cells, as previously. TLI-5Smt::scCOXII with an intact 5S NCL but a scrambled complementary sequence did not alter COXII protein levels. Quantitation of western blots showing no significant change in the expression levels of COXII in HeLa cells. One-way ANOVA with multiple comparisons was used to test significance; n.s. is p > 0.05, *** is p < 0.0004; mean ± SEM, n=3–6.
Conclusions/discussion
Mitochondrial dysfunction has been associated with the pathogenesis of numerous significant disease conditions (Wallace et al., 1988, Wallace, 2005, DiMauro and Schon, 2008, Wallace, 2010, 2012). The study of ME has been severely hampered by a limited number of animal models, especially those affecting endogenous mitochondrial genes (Palladino, 2010). Researchers have developed innovative methods to alter heteroplasmy using mitochondrial-targeted restriction enzymes (Xu et al., 2008) and reported manipulating mitochondrial DNA or general effects on mitochondrial translation via RNA import strategies (Kolesnikova et al., 2004, Mukherjee et al., 2008, Karicheva et al., 2011, Wang et al., 2012a, Bacman et al., 2013, Comte et al., 2013), however, the ability to directly manipulate the expression of mitochondrial-encoded proteins has remained elusive and has obvious basic and clinical applications. The identification of NCL sequences that direct RNAs to mitochondria enables a TLI approach using chimeric RNAs with a complementary element. The development of the mtTRES expression system enables expression of these chimeric RNAs in vivo. mtTRES utilizes RNAPIII promoter and termination elements such that NCLs and chimeric RNAs containing NCLs resemble natural substrates for RNA import. The demonstration that the mtTRES system is functional in vivo now enables the manipulation of mitochondrial genome expression, which opens up numerous avenues of investigation and is of immense value to the mitochondrial research and clinical communities.
These data demonstrate the general applicability of the approach by targeting two loci encoding proteins for which antibodies were available to verify functionality. TLI-NCL::ATP6 RNAs achieved ~ 40–50% reduction in steady state protein levels in vivo. In human cells TLI-NCL:: COXII achieved 20–50 % reduction in protein levels, dependent upon the NCL employed. Differences in reduction between the various NCLs and TLIs could be due to sequence selective effects or differences in the stability of chimeric RNA secondary structures affecting import or availability of the complementary RNA sequence for targeting.
Although these data demonstrate general applicability, there is notable constraint in the design of TLIs that has the potential to restrict its application. There are data suggesting antisense targeting for translational repression, at least in the cytosol, must be directed to the start codon to be effective, restricting construct design options. RNAP III is known to terminate within stretches of poly T, potentially restricting the use for some genes in some organisms where a poly A exists proximal to the start codon. Lastly, our data demonstrate differing levels of functionality within the NCL. Although we cannot fully explain these differences at this time, such sequence selectivity could reflect the fact that within the chimeric RNA the NCL must be recognized and the complementary sequence must still be accessible in its native structure. Certain NCL- complementary sequence combinations will form stable secondary structures that abrogate one of these functions in a manner that may not be fully predictable. Since the 5Smt NCL is larger and more highly structured with a lower delta G, it is predicted that this NCL will be more reliable but additional studies will be needed to fully test this prediction.
Mitochondrial-targeted TLIs were designed to function by antagonizing ribosome docking and lowering translational efficiency. Antisense RNAs are commonly employed to reduce expression of nuclear genes through a well-understood RNA interference mechanism that leads to target RNA degradation. We investigated this possibility by examining RNA levels of ATP6 and COXII and the data demonstrate normal RNA levels in vivo, arguing against an RNA-destabilizing mechanism of action. To demonstrate that the 5Smt RNA and TLI-5Smt::COXII chimeric RNA are being imported into the mitochondria with similar efficiency we performed a direct in vitro RNA import assay. Earlier studies have suggested that RNA import is dependent on mitochondrial membrane potential. We observed that the RNA import into the mitochondria was almost negligible in presence of FCCP, which uncouples and depolarizes mitochondria.
The data presented here demonstrate the utility and general applicability of the mtTRES system as well as the ability to engineer TLI chimeric RNAs to modulate endogenous mitochondrial gene expression. The capability to modulate mitochondrial gene expression will enable detailed studies of mitochondrial function and organelle dysfunction in vivo. Efforts to develop a mitochondrial gene therapy face a formidable challenge of competing with endogenous mutant protein expression. The mtTRES TLI system described here has the potential to accelerate the realization of an effective gene therapy for mitochondrial diseases.
Supplementary Material
Highlights.
We developed a novel series of mtTRES vectors that express small RNAs in vivo.
Chimeric mitochondrial-targeted RNAs function as translational inhibitors (TLIs).
We developed mtTRES TLI constructs that modulate expression of ATP6 in vivo.
Expression of human COXII is modulated by chimeric TLI RNAs in HELA cells.
Acknowledgments
This work was supported by National Institutes of Health [grant numbers AG027453, NS078758, and GM103369 to M.J.P.,]; ASPET Dalton-Zannoni training grant to M.J.P., and the Dept. of Pharmacology & Chemical Biology, University of Pittsburgh, School of Medicine. We thank E. Burton (Dept. of Neurology, University of Pittsburgh), M. Tarpey (Dept. of Anesthesiology, University of Pittsburgh) and S. Shiva (Dept. of Pharmacology & Chemical Biology, University of Pittsburgh) for providing feedback for this project; B. Roland for critical suggestions for anti-ATP6 validation; T. Novak, J. Chaves, C. Mangiaracina, K. Gaitonde and N. Kotchey for technical assistance; B. Goodpaster (Division of Endocrinology and Metabolism, Dept. of Medicine, University of Pittsburgh), N. Pichaud (School of Biotechnology and Biomolecular Sciences, University of New South Wales, Australia) for technical advise; E. Hoffman (Dept. of Neurology, University of Pittsburgh) for assistance with electroporation; and K. Basler for the pUAST-attB vector.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors maybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Schedl P, Tschudi C, Pirrotta V, Steward R, Gehring WJ. The 5S genes of Drosophila melanogaster. Cell. 1977;12:1057–1067. doi: 10.1016/0092-8674(77)90169-6. [DOI] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nature medicine. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou J, Jordan BR. Nucleotide sequence of Drosophila melanogaster 5S RNA: evidence for a general 5S RNA model. FEBS letters. 1976;62:146–149. doi: 10.1016/0014-5793(76)80039-7. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Chatterjee S, Adhya S. Mitochondrial RNA import in Leishmania tropica: aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Molecular and cellular biology. 2002;22:4372–4382. doi: 10.1128/MCB.22.12.4372-4382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokori-Brown M, Holt IJ. Expression of algal nuclear ATP synthase subunit 6 in human cells results in protein targeting to mitochondria but no assembly into ATP synthase. Rejuvenation Res. 2006;9:455–469. doi: 10.1089/rej.2006.9.455. [DOI] [PubMed] [Google Scholar]
- Celotto AM, Frank AC, McGrath SW, Fergestad T, Van Voorhies WA, Buttle KF, Mannella CA, Palladino MJ. Mitochondrial encephalomyopathy in Drosophila. J Neurosci. 2006a;26:810–820. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Frank AC, Seigle JL, Palladino MJ. Drosophila model of human inherited triosephosphate isomerase deficiency glycolytic enzymopathy. Genetics. 2006b;174:1237–1246. doi: 10.1534/genetics.106.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Liu Z, Vandemark AP, Palladino MJ. A novel Drosophila SOD2 mutant demonstrates a role for mitochondrial ROS in neurodevelopment and disease. Brain Behav. 2012;2:424–434. doi: 10.1002/brb3.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- Comte C, Tonin Y, Heckel-Mager AM, Boucheham A, Smirnov A, Aure K, Lombes A, Martin RP, Entelis N, Tarassov I. Mitochondrial targeting of recombinant RNAs modulates the level of a heteroplasmic mutation in human mitochondrial DNA associated with Kearns Sayre Syndrome. Nucleic acids research. 2013;41:418–433. doi: 10.1093/nar/gks965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aurelio M, Vives-Bauza C, Davidson MM, Manfredi G. Mitochondrial DNA background modifies the bioenergetics of NARP/MILS ATP6 mutant cells. Hum Mol Genet. 2010;19:374–386. doi: 10.1093/hmg/ddp503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends in genetics : TIG. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annual review of neuroscience. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- Doyle DF, Braasch DA, Simmons CG, Janowski BA, Corey DR. Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry. 2001;40:53–64. doi: 10.1021/bi0020630. [DOI] [PubMed] [Google Scholar]
- Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA. 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. The Journal of biological chemistry. 2001;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- Fergestad T, Ganetzky B, Palladino MJ. Neuropathology in Drosophila membrane excitability mutants. Genetics. 2006;172:1031–1042. doi: 10.1534/genetics.105.050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Olson L, Patel KP, Miller R, Palladino MJ, Ganetzky B. Neuropathology in Drosophila mutants with increased seizure susceptibility. Genetics. 2008;178:947–956. doi: 10.1534/genetics.107.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karicheva OZ, Kolesnikova OA, Schirtz T, Vysokikh MY, Mager-Heckel AM, Lombes A, Boucheham A, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic acids research. 2011;39:8173–8186. doi: 10.1093/nar/gkr546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:799–817. doi: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes PJ, Andreu AL, Schon EA. Evidence for the presence of 5S rRNA in mammalian mitochondria. Molecular biology of the cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Adhya S. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. The Journal of biological chemistry. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J, Schon EA. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Mahata B, Mahato B, Adhya S. Targeted mRNA degradation by complex-mediated delivery of antisense RNAs to intracellular human mitochondria. Hum Mol Genet. 2008;17:1292–1298. doi: 10.1093/hmg/ddn017. [DOI] [PubMed] [Google Scholar]
- Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- Palladino MJ. Modeling mitochondrial encephalomyopathy in Drosophila. Neurobiol Dis. 2010;40:40–45. doi: 10.1016/j.nbd.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Bower JE, Kreber R, Ganetzky B. Neural dysfunction and neurodegeneration in Drosophila Na+/K+ ATPase alpha subunit mutants. J Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales-Clemente E, Fernandez-Silva P, Acin-Perez R, Perez-Martos A, Enriquez JA. Allotopic expression of mitochondrial-encoded genes in mammals: achieved goal, undemonstrated mechanism or impossible task? Nucleic acids research. 2011;39:225–234. doi: 10.1093/nar/gkq769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Molecular and cellular biology. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annual review of biochemistry. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- Schneider A, Marechal-Drouard L. Mitochondrial tRNA import: are there distinct mechanisms? Trends in cell biology. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- Smirnov A, Comte C, Mager-Heckel AM, Addis V, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. The Journal of biological chemistry. 2010;285:30792–30803. doi: 10.1074/jbc.M110.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Tarassov I, Mager-Heckel AM, Letzelter M, Martin RP, Krasheninnikov IA, Entelis N. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. Rna. 2008;14:749–759. doi: 10.1261/rna.952208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- Tarassov I, Kamenski P, Kolesnikova O, Karicheva O, Martin RP, Krasheninnikov IA, Entelis N. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell cycle. 2007;6:2473–2477. doi: 10.4161/cc.6.20.4783. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environmental and molecular mutagenesis. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nature reviews Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, 2nd, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, 3rd, Koehler CM, Teitell MA. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA, Koehler CM. Correcting human mitochondrial mutations with targeted RNA import. Proceedings of the National Academy of Sciences of the United States of America. 2012a;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA, Koehler CM. Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Sci U S A. 2012b;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, DeLuca SZ, O'Farrell PH. Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science. 2008;321:575–577. doi: 10.1126/science.1160226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshionari S, Koike T, Yokogawa T, Nishikawa K, Ueda T, Miura K, Watanabe K. Existence of nuclear-encoded 5S-rRNA in bovine mitochondria. FEBS letters. 1994;338:137–142. doi: 10.1016/0014-5793(94)80351-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.