Summary
The NES1/MAD1 gene acts antagonistically with the EIN2-associated ORE1 signalling pathway to modulate the nitric oxide-induced Arabidopsis cotyledon senescence
Key words: Arabidopsis, cotyledon, induced senescence, NES1/MAD1, nitric oxide, ORE1/AtNAC2.
Abstract
After germination, cotyledons undertake the major role in supplying nutrients to the pre-photoautorophy angiosperm seedlings until they senesce. Like other senescence processes, cotyledon senescence is a programmed degenerative process. Nitric oxide can induce premature cotyledon senescence in Arabidopsis thaliana, yet the underlying mechanism remains elusive. A screen for genetic mutants identified the nes1 mutant, in which cotyledon senescence was accelerated by nitric oxide. Map-based cloning revealed that NES1 is allelic to a previously reported mitotic checkpoint family gene, MAD1. The nes1/mad1 mutants were restored to the wild type, in response to nitric oxide, by transforming them with pNES1::NES1. Ectopic expression of NES1 in the wild type delayed nitric oxide-mediated cotyledon senescence, confirming the repressive role of NES1. Moreover, two positive regulators of leaf senescence, the ethylene signalling component EIN2 and the transcription factor ORE1/AtNAC2/ANAC092, were found to function during nitric oxide-induced senescence in cotyledons. The block of ORE1 function delayed senescence and ectopic expression induced the process, revealing the positive role of ORE1. EIN2 was required to induce ORE1. Furthermore, the genetic interaction analysis between NES1 and ORE1 showed that the ore1 loss-of-function mutants were epistatic to nes1, suggesting the dominant role of ORE1 and the antagonistic role of NES1 during nitric oxide-induced cotyledon senescence in Arabidopsis.
Introduction
Cotyledons are formed during embryogenesis. In most plants, the function of the cotyledon is to provide nutrients for seedling establishment. During seedling development, the cotyledon is initially heterotrophic, then becomes photosynthetic, and eventually senesces. As an integral part of development, the senescence of cotyledons is a process that leads to nutrient recycling and ends in cell death, and is accompanied by colour changes, the dismantling of chloroplasts, and the degradation of DNA, RNA, and protein (Krul, 1974; Peterman and Siedow, 1985). Phytohormones such as cytokinin and ethylene affect cotyledon senescence, with cytokinin preventing chlorophyll breakdown and ethylene initiating the onset of the senescence process (Ananieva et al., 2008a ; Jing et al., 2008). Although cotyledon senescence has been studied for decades (McKersie et al., 1987; Rukes and Mulkey, 1993), its underlying regulatory network is unclear.
In contrast, leaf senescence is better understood (Lim et al., 2007). Based on transcriptome analysis, during natural leaf senescence, ~6–12% of Arabidopsis genes change expression (Buchanan-Wollaston et al., 2005; Breeze et al., 2011), which include >800 SAGs (senescence-associated genes). A number of SAGs have been well studied and established as markers. The favoured marker for monitoring age-dependent senescence is SAG12, whereas SAG13 and SAG14 are preferred for monitoring stress-induced senescence (Schippers et al., 2007). Nevertheless, deletion or overexpression of many individual SAGs affect senescence to a limited extent, although there are a few exceptions (Seo et al., 2011; Zhang and Gan, 2012), indicating the robust nature of the regulatory network. Certain transcription factors have been identified as positive regulators of age-dependent senescence in Arabidopsis by means of the loss-of-function mutant experiencing delayed leaf senescence, whereas others have been identified as negative regulators, in this case based on accelerated senescence in the loss-of-function mutant.
The better known positive regulators of leaf senescence are from the NAC (NAM, ATAF, and CUC) family. So far, a few have been well characterized, including AtNAP (Arabidopsis NAC domain containing protein 29) and ORE1/AtNAC2/ANAC092 (ORESARA1). Not only does a block of function delay senescence, but ectopic expression induces early senescence (Guo and Gan, 2006; Rauf et al., 2013). The control of the ORE1 transcript involves miR164 (microRNA164), which interacts with ORE1 mRNA to trigger its degradation. EIN2 (ethylene insensitive 2) and its downstream component EIN3 of the ethylene signalling pathway negatively block miRNA164 expression in an age-dependent manner, through the direct binding of EIN3 to the promoter of miRNA164, which allows ORE1 mRNA to accumulate (Kim et al., 2009; Li et al., 2013).
In addition to developmental senescence, various environmental stresses can induce or accelerate senescence. These environmental stresses may be biotic. Many of the stresses, including pathogen infection, drought, salinity, and extreme temperature (Bouchard and Yamasaki, 2008; Ma et al., 2008; Neill et al., 2008; Corpas et al., 2009; Zhao et al., 2009; Xuan et al., 2010), are known to increase the production of nitric oxide in plants. A bioactive gas, nitric oxide has been suggested to be a signalling component that mediates stress responses (Arasimowicz and Floryszak-Wieczorek, 2007). Under certain conditions, nitric oxide is able to interact with ethylene and cytokinin. In tobacco, ethylene accumulation in response to ozone treatment depends on nitric oxide (Ederli et al., 2006). In Arabidopsis, exposure to a high concentration of nitric oxide (48 ppm) results in ethylene accumulation (Magalhaes et al., 2000). Nitric oxide may directly interact with cytokinin in vivo (Liu et al., 2013), suggesting that nitric oxide represses endogenous cytokinin to some extent. In addition, nitric oxide represses the phosphorylation of the cytokinin signalling components AHP1 (histidine phosphotransfer protein 1) and ARR1 (response regulator 1) through the S-nitrosylation of AHP1, revealing the inhibitory effect of nitric oxide on cytokinin signalling (Feng et al., 2013). Inducing the burst of nitric oxide during environmental stress-triggered senescence processes may play a critical role in modulating the levels of ethylene and cytokinin and the related pathways, thereby inducing senescence.
The sixth rosette leaf is a favoured plant part in leaf senescence studies on Arabidopsis. This leaf naturally starts to turn yellow ~21 d after its initiation, and completes senescence by about day 30. A similar phenotype has been observed in cotyledons, and a majority of Arabidopsis cotyledons finish senescing within 28 d (Smith, 2001). For stress-induced senescence studies in cotyledons, seedlings 5 d after germination are chosen for treatment (Weaver and Amasina, 2001). To study nitric oxide-regulated cotyledon senescence, the nitric oxide donor SNP (sodium nitroprusside) was mixed into agar and this was added to the cover of the Petri plates used for the 5 d seedling treatment. As nitroprusside breaks down, the seedlings are exposed to nitric oxide. Due to its volatility, the nitric oxide is able to diffuse though the air to reach the seedlings and prevents them from being exposed to the breakdown products aquapentacyanoferrate [Fe(CN)5·H2O]3– and cyanide CN– (Frank et al., 1976; Arnold et al., 1984), which are confined to the liquid phase in the Petri plate cover. After 3 d of treatment, senescence in cotyledons started to become visible and a screen was carried out for mutants that were more sensitive to nitric oxide, with respect to early cotyledon senescence. Here, a molecular genetic analysis of NES1 (nitric oxide-induced early cotyledon senescence), which has been shown to be a negative regulator of nitric oxide-induced cotyledon senescence, is reported.
Materials and methods
Plant materials and treatment
All of the Arabidopsis mutagenic seeds, T-DNA insertion mutants, and transgenic plants used in this study were in a Col-0 (Columbia) background. For map-based cloning analysis, Ler (Landsberg erecta) was used as the pollen acceptor and the nes1-6 mutant was used as the pollen donor to create the F1 and then the F2 mapping populations. Five T-DNA insertional NES1-deficient mutants, nes1-1 (SALK_080570), nes1-2 (SALK_023425), nes1-3 (SALK_073889), nes1-4 (SALK_039008), and nes1-5 (SALK_130471), and the ORE1 deficient/overexpression lines ore1 (SALK_090154) and 35S::ORE1 (CS23887) were obtained and isolated from the ABRC (Arabidopsis Biological Resource Center). Before double mutant construction, nes1-2 was backcrossed three times to the Col-0 background. All plants were grown in a controlled growth chamber at 21–22 °C under cool-white fluorescent light (80–100 μmol m–2 s–1) in a long-day photoperiod (16h light/8h dark).
Five-day-old seedlings, grown on agar plates with half-strength Murashige and Skoog (1/2 MS) medium supplemented with 0.6% (w/v) sucrose and 0.7% (w/v) agar, were used for the treatments with SNP, a chemical donor of nitric oxide when exposed to light, which was mixed in 10ml of 1% (w/v) agar medium and added only on the inside cover of Petri dishes.
Measurements of chlorophyll content and fluorescence
Samples were taken and weighed at the indicated times, then placed into 5ml of 90% (v/v) acetone for extraction. The chlorophyll content of each sample was assayed by measuring the absorbance at 652, 665, and 750nm using a spectrophotometer. For fluorescence measurement, samples were taken every 30min after 20 μM SNP application, with 30min dark incubation before measurement at room temperature. The F v/F m ratio was measured by MAXI Version (IMAGING-PAM M-Series, Germany) as described previously (Wingler et al., 2004).
Electrolyte leakage
For electrolyte leakage, 5-day-old seedlings were first treated with 20 μM nitroprusside for 24h. The excised cotyledons were then incubated in Milli-Q water and the conductivity of the solution was assayed with a conductivity cell (DJS-1C, GongGongPin Co., China). Finally, the samples were boiled for 5min and a second reading was taken. Electrolyte leakage is expressed as a percentage of the maximum.
Gene expression analysis by RT-PCR
The mRNA levels were measured by quantitative real-time PCR (qRT-PCR). Total RNA was extracted from the seedlings using Trizol Reagent (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer’s instructions. DNase was used to purify the RNA samples. The overall quality of the total RNA was monitored by formaldehyde–RNA denaturing electrophoresis and the A 260/280 ratio (~2.0) using a NanoDrop Instrument spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The concentration of total RNA was recorded. First-strand cDNA was synthesized with 2 μg of total RNA using M-MLV reverse transcriptase and an oligo(dT)15 primer (Promega) in a 20 μl mixture. The reaction was then diluted 2-fold with water and the cDNA was used as a template for both semi-quantitative RT-PCR and qRT-PCR. For the semi-quantitative PCR analysis, 1 μl of each reverse transcription reaction was used per PCR in a final volume of 20 μl. Semi-quantitative RT-PCR was performed by gene-specific primers under controlled conditions (Supplementary Table S2 available at JXB online). The reactions were repeated three times. Ubiquitin primers (AtUBQ) were used as the standard (Li et al., 2007). Real-time PCR was performed using the SYBR green PCR master mix and the Bio-Rad iQ5 real-time PCR detection system. ACT2 was used as the standard (Czechowski et al., 2005). The primer sequences are given in Supplementary Table S3. All reactions were repeated at least three times. Statistical analysis of the results of real-time PCR was performed using iQ5 software.
GUS staining
Seedlings were incubated with 10mM 5-bromo-4-chloro-indolyl-β-d-glucuronide (X-Glu) in 100mM phosphate buffer (pH 7.0) at 37 °C overnight. Chlorophyll was removed from leaves by clarification with 70% ethanol before observation. The whole β-glucuronidase (GUS) staining process was carried out according to the protocol of Jefferson (1987).
Plasmid constructions and plant transformation
Specific primers, designed upstream of the open reading frame, were pNES1-EcoRI-F (5′-CCGGAATTCCGTCTTGCCAAAAA GCCAT-3′) and pNES1-BamHI-R (5′-GCGGGATCCGTCTGCG TCGAGAAATTAGGG-3′). The length of the cloned promoter was ~1100bp and included EcoRI and BamHI restriction sites, as shown by the underlining. To obtain the NES1 gene, primers were designed at both ends of the open reading frame. The primers were NES1-BamHI-F (5′-CGCGGATCCATGATTTTGAGAACTCCG-3′) and NES1- NcoI-R (5′-GTACCCATGGGATATAGCGTCCGACGGTTG-3′). The cloned CDS (coding sequence) (~2200bp) was amplified from cDNA, including BamHI and NcoI enzyme sites. The promoter fragment was separately recombined into pCAMBIA1301 and pCAMBIA1302 to obtain NES1::GUS and pNES1::GFP (green fluorescent protein). The CDS of the NES1 gene was then recombined into pNES1::GFP to obtain pNES1::NES1::GFP. To obtain pNES1::NES1 and 35S::NES1, the primers for the CDS were modified through replacement of the restriction sites, which were NES1-NcoI-F (5′-GTACCCATGGATGATTTTGAGAACTCCG-3′) and NES1-NheI-R (5′-GATGCTAGCTCAATATAGCGTCCGAC-3′).
Transgenic lines were generated using the Agrobacterium tumefaciens LBA4404 vacuum infiltration method. Seeds of the first-generation transgenic line T1 from infiltrated plants were germinated on 1/2 MS medium containing 25mg l–1 hygromycin B. Several lines were obtained for each transformation and at least three generations of resistance screening were performed for preparation of material.
Western blot
Samples of cotyledons were collected and ground in liquid nitrogen, and then incubated with an extraction buffer [0.1M TRIS-HCl, pH 8.3, 5mM dithiothreitol (DTT), 5mM EDTA, and protease inhibitor]. The Bradford protein assay was used for quantification and normalization. Proteins were resolved under reducing conditions by using 10% SDS–polyacrylamide gels. The proteins were transferred onto ployvinylidene difluoride (PVDF) membranes (Immobilon-P from Millipore), which were incubated separately with a primary GFP antibody (Roche, diluted 1:5000) and then a secondary peroxidase-conjugated anti-mouse antibody (Santa Cruz, diluted 1:10 000) for 2h at room temperature in TBS (20mM TRIS-HCl, pH 7.8, 180mM NaCl) supplemented with 4% (w/v) skimmed milk powder. After incubation, the membranes were washed twice (10min each) with TBS containing 0.05% Tween-20. After the final wash, membrane-associated peroxidase activity was visualized by using the ECL kit (Amersham Pharmacia).
Results
Map-based cloning
Chemically mutagenized, light-grown seedlings were screened for enhanced cotyledon senescence in response to treatment with nitric oxide, and a line, named nes1-6, was isolated. With exposure to 20 μM SNP, cotyledon senescence in the mutant was markedly enhanced compared with the wild type (Fig. 1A, B). Ferricyanide was used as a control to exclude the side effects of the SNP breakdown products. In this condition, the two genotypes showed little difference from the non-treated seedlings (Supplementary Fig. S2 at JXB online), confirming the accelerated cotyledon senescence was caused by gaseous nitric oxide.
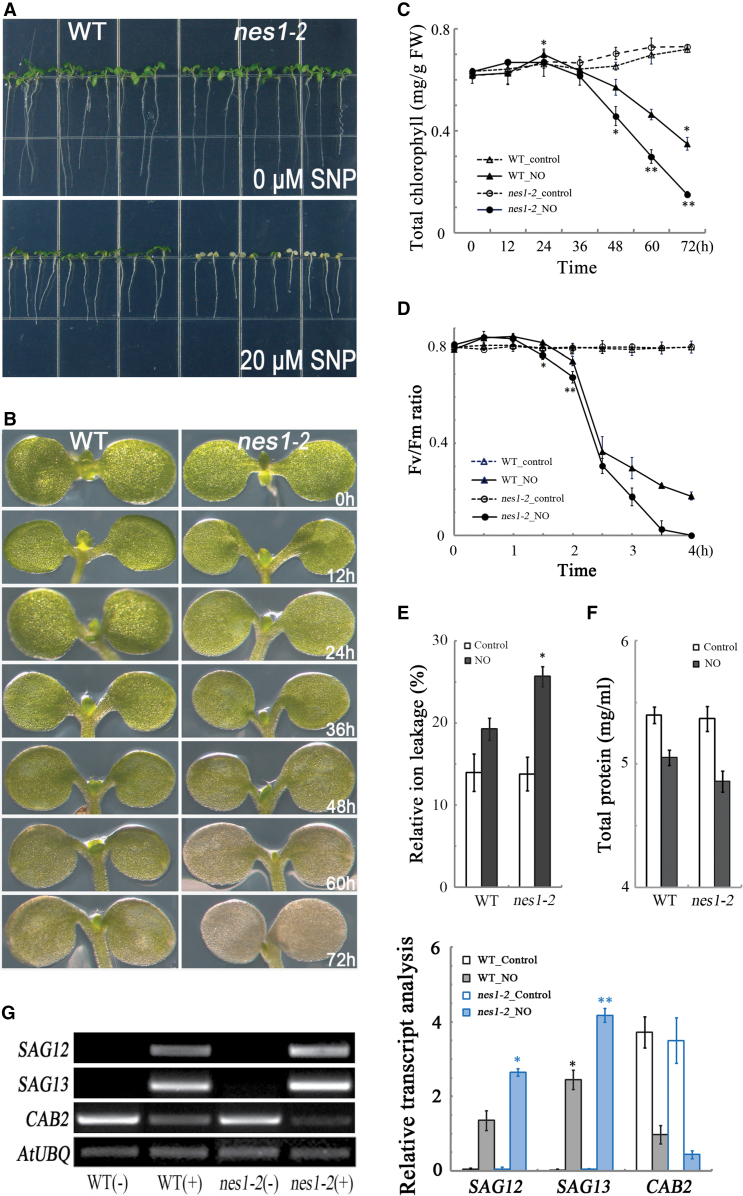
Fig. 1.
Nitric oxide-induced cotyledon senescence is accelerated in nes1-2. (A) Phenotypic comparison of 5-day-old wild type (WT) and nes1-2 treated as indicated for 72h. (B) Higher magnification images of plants treated as in (A) with 20 μM nitroprusside. (C) Chlorophyll content as a function of time for plants treated as in (A). Data are the means of 20 seedlings ±SE with four replicates for each time point. (D) Chlorophyll fluorescence parameter (F v/F m) for plants treated with nitric oxide. Data are means of five seedlings ±SE, with four replicates for each time point. (E) Protein content of plans treated as in (A) with 20 μM nitroprusside for 24h. (F) Electrolyte leakage of plants treated as in (E). Data are means of five seedlings ±SE, with three replicates for each time point. (G) Semi-quantitative RT-PCR analysis of the indicated genes for plants treated as in (E). The intensity of bands was statistically analysed. Data are mean values ±SE, with three replicates for each sample. Asterisks indicate significant differences (*P < 0.05; **P ≤ 0.01).
Based on a mapping population from a cross between the mutant nes1-6 (Col-0) and the wild type (Ler), the location of NES1 was narrowed down to an interval (~20kb) between markers CER435084 and CER435911 (Supplementary Fig. S3A at JXB online), which contained three genes. T-DNA insertion mutants were all obtained, but only At5g49880 had a similar phenotype. For this locus, five T-DNA insertion alleles were obtained (Supplementary Fig. S3B) in which transcripts of At5g49880 were all detectable, except that there was sharply reduced abundance of nes1-2 and nes1-3 (Supplementary Fig. S3D). Although these five alleles showed enhanced cotyledon senescence under nitric oxide, nes1-2 and nes1-3 were the two strongest alleles. The insertion site of nes1-2 was 125bp upstream of the start codon, in the same orientation as the gene. The site for nes1-3 was in the eighth exon, 2511bp downstream of the start codon, in the opposite direction.
Genetic analysis of the originally chemically mutagenized allele, nes1-6, revealed that this allele was recessive (Supplementary Table S1 at JXB online). Analyses of F1 progeny from crosses between nes1-6 and either nes1-2 or nes1-3 revealed similar accelerated cotyledon senescence to nes1-6 (Supplementary Fig. S3C), confirming that nes1-6 and the other five T-DNA insertion mutants were nes1 lines. Finally, the nitric oxide-enhanced cotyledon senescence phenotype in nes1-6 and nes1-2 was rescued by transforming either line with the full-length At5g49880 coding sequence driven by its native promoter (Fig. 3D). Taken together, it is concluded that the nitric oxide-induced early cotyledon senescence in the nes1 lines is caused by mutations in At5g49880.
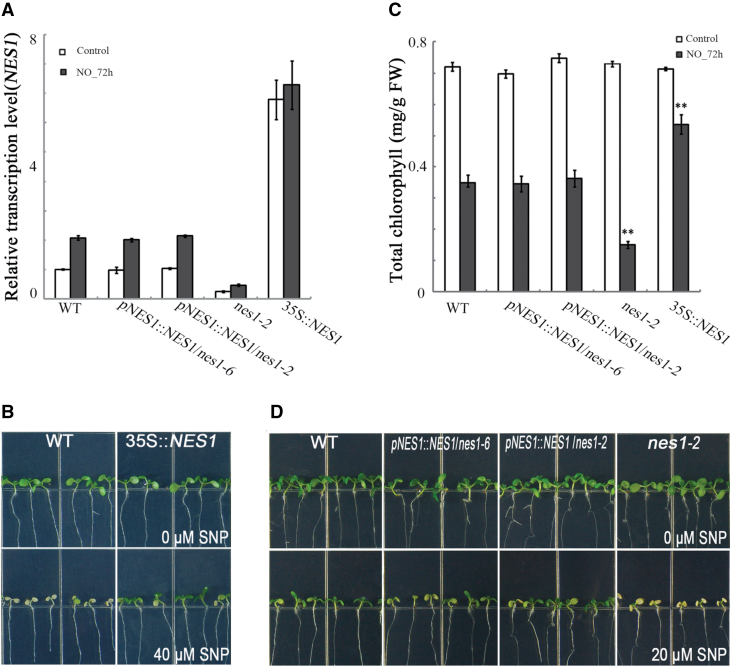
Fig. 3.
NES1 as a repressor in nitric oxide-induced cotyledon senescence. (A) Relative transcriptional quantification of NES1 in transgenic plants. Seedlings treated as in Fig. 1E. (B) The NES1-overexpressing seedlings (35S::NES1) treated with 40 μM nitroprusside. (C) Chlorophyll content at 72h of plants treated as in Fig. 1A. Data are mean values of 20 seedlings ±SE, with three replicates for each sample. (D) Complementation of the transgenic plants treated as in Fig. 1A. Asterisks indicate significant differences (*P < 0.05; **P ≤ 0.01).
At5g49880 has been previously annotated as a spindle assembly checkpoint protein MAD1 (mitotic arrest deficient 1). Interestingly, a role in cell cycle control was established for the closely related protein MAD2, but loss-of-function mitotic arrest phenotypes for mad1 were not reported (Ding et al., 2012). It appears that this gene has acquired a function in the pathway regulating senescence.
Mutant phenotype
Because of its strong phenotype, the T-DNA insertion mutant nes1-2/mad1 was used for further analysis. To establish a suitable timing for the nitric oxide treatment, cotyledon growth was first characterized (Supplementary Fig. S1A at JXB online). Cotyledon expansion was sustained and roughly isotropic for the first 10 d, and full size was attained a few days later (Supplementary Fig. S1B). The nes1-2 line behaved similarly. To minimize interference from true leaves, which emerged on day 5, day 5 after germination was chosen as the time for the nitric oxide treatment.
A typical characteristic of senescence, whether developmental or stress induced, is the loss of chlorophyll. Treating 5-day-old seedlings with 20 μM nitroprusside for 72h caused the cotyledons to become slightly bleached in the wild type but substantially more so in the mutant (Fig. 1A). A time course showed that the cotyledons in the mutant were visibly less green by 60h of treatment, and to an even greater extent after 72h (Fig. 1B). This visual impression was confirmed by measuring the chlorophyll content (Fig. 1C), which illustrated that the mutant had started to lose chlorophyll by 48h of treatment and showed that chlorophyll loss appeared to be accelerated in the mutant by 60h compared with the wild type. To examine the photosynthetic apparatus with greater temporal precision, chlorophyll fluorescence was used to report the F v/F m parameter. At time zero, this parameter was indistinguishable in the two genotypes and, interestingly, increased by ~10% over the first hour of treatment (Fig. 1B). After that, the F v/F m ratio decreased steeply and after 2.5h had decreased significantly more in the mutant than in the wild type, indicating that the senescence programme needed only a few hours to be induced and that it happened more rapidly in the mutant.
In addition to losing chlorophyll, progression of senescence is determined by measuring other senescence parameters, such as loss of total protein content and increase of ion leakage, associated with cell death. Like leaves, cotyledons treated with nitric oxide to induce senescence also lost protein by 24h and the decrease was larger in the mutant than in the wild type (Fig. 1F). Likewise, ion leakage was induced and the extent of the leakage was significantly greater in the mutant (Fig. 1E).
To assay senescence on a molecular level, the expression of two SAGs, SAG12 encoding a cysteine protease and SAG13 encoding an alcohol dehydrogenase, both of which were up-regulated during senescence, was examined together with the expression of a photosynthetic gene, CAB2 (chlorophyll a/b-binding protein 2), which was down-regulated. In the absence of treatment, both SAG transcripts were hardly detectable but CAB2 was expressed strongly. The two genotypes appeared to be indistinguishable. However, with nitric oxide for 24h, the up-regulation of SAG12 and SAG13 was significantly stronger in nes1-2. The down-regulation of CAB2 was apparent in the wild type and even more so in the mutant (Fig. 1G). Thus, taken together, nes1-2 accelerates the progression of nitric oxide-induced senescence in cotyledons.
Expression pattern of NES1
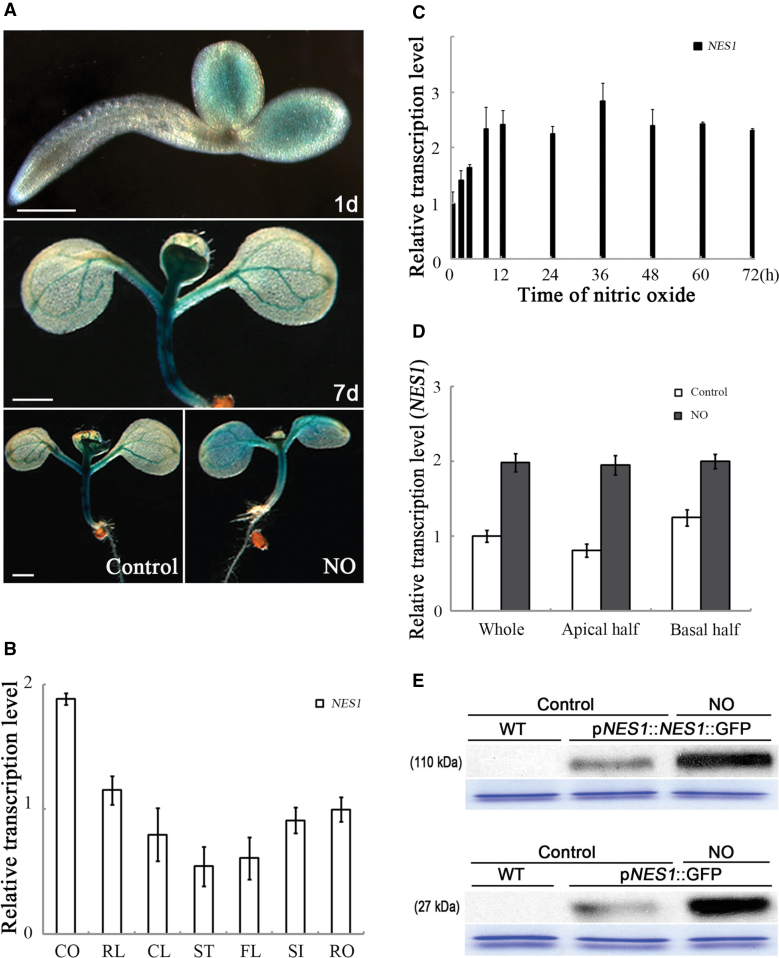
To examine the expression of NES1, transgenic plants carrying a native NES1 promoter–GUS gene fusion were first prepared. On the first day after germination, GUS activity was mainly detected in cotyledons, and, on the seventh day, GUS was more widely expressed throughout the seedling and concentrated in the vasculature (Fig. 2A). The GUS expression was strongly increased when 5-day-old seedlings were treated with 20 μM nitroprusside for 2 d. To examine the expression of NES1 further, quantitative real-time RT-PCR was used, which allowed the detection of the native message. NES1 was expressed in all organs, although the relative transcript levels were slightly higher in the cotyledons (1.88), compared with slightly lower levels in the stems (0.54) and the flowers (0.61), than the standard in the roots of 1.00 (Fig. 2B). When 5-day-old seedlings were treated with 20 μM nitroprusside, the message level of NES1 in the cotyledons rose linearly within 12h and then remained steady up to at least 72h (Fig. 2C). When 5-day-old cotyledons were bisected into apical and basal halves, the latter had a slightly higher level of NES1 basic expression. However, after a 24h treatment with 20 μM nitroprusside, the expression was raised to essentially the same level in both regions (Fig. 2D). Finally, to extend the analysis of expression to the protein level, a translational fusion was made between NES1 and GFP, driven by the NES1 promoter (pNES1::NES1::GFP). As a control, GFP alone driven by the NES1 promoter (pNES1::GFP) was used. Based on probing a western blot with anti-GFP antibody, the constructs were expressed in untreated seedlings, and both were strongly up-regulated by treatment with nitroprusside, with no detectable signal in the wild-type control (Fig. 2E). Taking these results together, NES1 appears to be widely and constitutively expressed and strongly up-regulated by nitric oxide.
Fig. 2.
Expression of NES1. (A) GUS staining of transgenic seedlings carrying pNES1::GUS at the indicated times. The two lower panels show 5-day-old seedlings treated or not with 20 μM nitroprusside for 48h. Bars=100 μm (top) and 1mm (middle and bottom). (B) Organ-specific expression of NES1, with quantitative real-time PCR. CO, cotyledons; RL, rosette leaves; CL, cauline leaves; ST, stems; FL, flowers; SI, siliques; RO, roots. (C) The NES1 transcript abundance as a function of treatment time. Seedlings treated as in Fig. 1B. Transcript quantified as in (B). (D) The NES1 expression in apical and basal halves of the cotyledon. Seedlings treated as in Fig. 1E. (E) Western blot. Blots were probed with an anti-GFP primary antibody. Seedlings treated as for Fig. 1E. The loading control shows a band at ~50kDa stained with Coomassie blue.
NES1 as a negative regulator of nitric oxide-induced cotyledon senescence
To assay further the function of NES1 in nitric oxide-induced cotyledon senescence, NES1-overexpressing transgenic lines were generated by transformation of 35S::NES1. Three transgenic lines were obtained and tested, and only the strongest line was used in the experiment. In this line, the NES1 message level was almost 10 times higher than in the wild type, but was not further induced by nitroprusside (Fig. 3A). When the line was exposed to as much as 40 μM nitroprusside, the cotyledons remained green whereas those of the wild type appeared strongly bleached (Fig. 3B). The visual impression was confirmed by measuring the cotyledon chlorophyll content 72h after 20 μM nitroprusside treatment (Fig. 3C). The overexpression lines, driven by the 35S promoter, expressed the coding sequence of NES1. To determine whether this led to artefacts, the coding sequence from the native promoter (pNES1::NES1) was expressed and introduced into either the nes1-6 or the nes1-2 background. These lines were indistinguishable from the wild type in terms of mRNA level (Fig. 3A), chlorophyll content (Fig. 3C), and overall appearance (Fig. 3D), thereby validating the use of the coding sequence in the overexpression lines. These results are consistent with NES1 being a repressor of nitric oxide-induced cotyledon senescence.
ORE1 positively regulates nitric oxide-induced cotyledon senescence
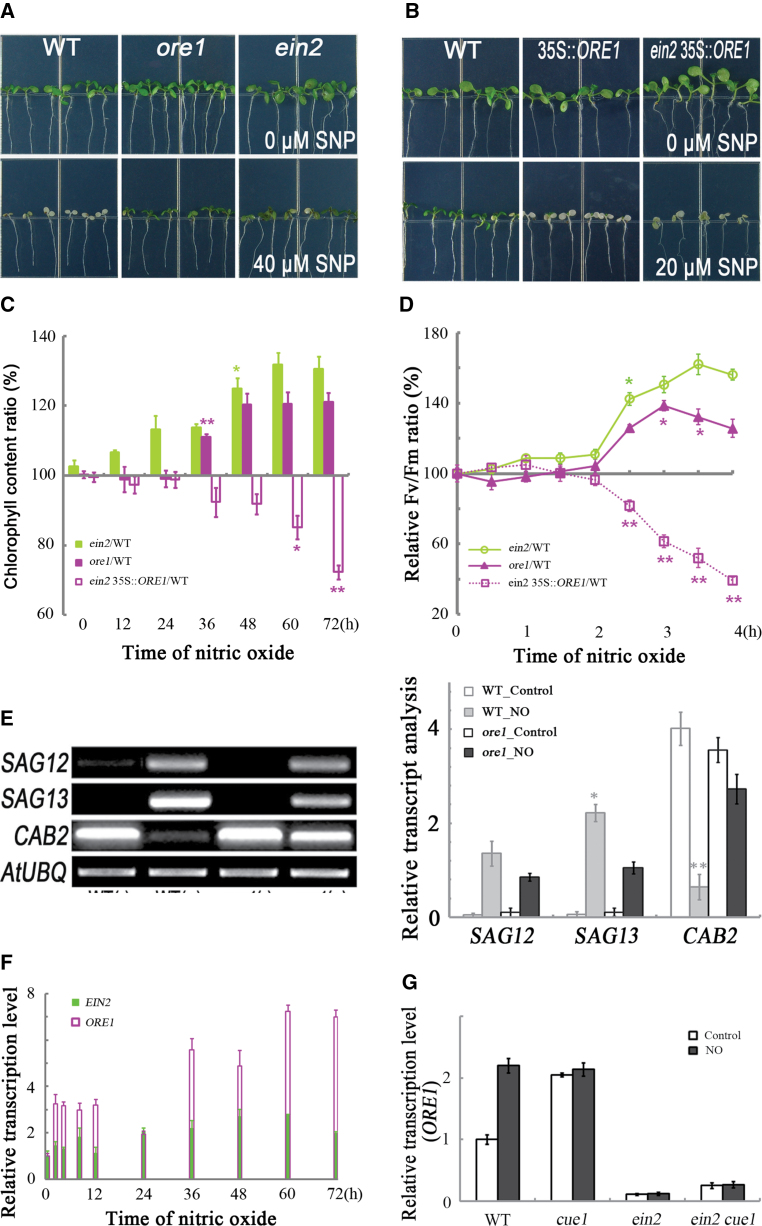
During developmental senescence in Arabidopsis leaves, the increasing expression of the transcription factor ORE1 acts as a key positive regulator in inducing senescence-associated downstream genes (Rauf et al., 2013). To explore the role of ORE1 in nitric oxide-regulated cotyledon senescence, the response to nitric oxide in ore1 was checked and it was found that nitric oxide-induced cotyledon senescence was delayed in the mutant (Fig. 4A). Conversely, transgenic plants overexpressing ORE1 in the wild-type background (35S::ORE1) had accelerated cotyledon senescence under nitric oxide treatment (Fig. 4B). These phenotypes were confirmed by measuring the cotyledon chlorophyll content and F v/F m ratio (Fig. 4C, D). Finally, in ore1, nitric oxide treatment changed the levels of SAG12, SAG13, and CAB2 to a lesser extent than in the wild type (Fig. 4E).
Fig. 4.
EIN2 is required for nitric oxide to induce ORE1 during cotyledon senescence. (A) The ein2 and ore1 seedlings were treated as in Fig. 3B. (B) ORE1-overexpressing seedlings (35S::ORE1 and ein2 35S::ORE1) treated as in Fig. 1A. (C) Chlorophyll content as a function of time for plants treated as in (B). Data are the means of 20 seedlings ±SE, with four replicates for each time point. (D) Chlorophyll fluorescence parameter (F v/F m) for plants treated with nitric oxide. Data are means of five seedlings ±SE, with four replicates for each time point. (E) Semi-quantitative RT-PCR analysis of the indicated genes for plants treated as in Fig. 1E. The intensity of bands was statistically analysed. Data are mean values ±SE, with three replicates for each sample. (F) The EIN2 and ORE1 transcript abundance as a function of treatment time. Seedlings treated as in Fig. 1B. (G) The ORE1 expression in cue1 and ein2. Seedlings treated as in Fig. 1E. Asterisks indicate significant differences (*P < 0.05; **P ≤ 0.01).
ORE1 induction requires EIN2 during developmental leaf senescence (Kim et al., 2009). Therefore, experiments were conducted to determine whether this is also true for nitric oxide-induced senescence using the cotyledon system. First, the phenotype of ein2 was checked to determine that it was nitric oxide-mediated late cotyledon senescence (Fig. 4A). Transgenic plants overexpressing ORE1 in the ein2 background were then found to present early cotyledon senescence under nitric oxide treatment (Fig. 4B). When chlorophyll (Fig. 4C) and the F v/F m ratio were quantified (Fig. 4D), ein2 was affected more strongly than ore1. After nitric oxide treatment, the chlorophyll content decreased in the wild type (Fig. 1C). In ein2 and ore1 mutants, the percentage of chlorophyll and the F v/F m ratio rose linearly with respect to the wild type after nitric oxide treatment. This reflected that the ein2 and ore1 mutants prevented the loss of chlorophyll and of electron transport capacity with similar kinetics. However, the overexpression of ORE1 in the ein2 background reversed the phenotype of the ein2 mutants, and caused a more rapid decline of the chlorophyll content and F v/F m ratio, compared with the wild type, under nitric oxide treatment. This was also supported by the molecular evidence (Fig. 4F). Indeed, ein2 essentially had a limited background level of the ORE1 message in cotyledons. However this was not increased by nitric oxide treatment (Fig. 4G), meaning that the ORE1 induction relied on EIN2. As an alternative to nitroprusside treatment, the cue1 (CAB underexpressed) mutant was also examined, which has elevated levels of endogenous nitric oxide (He et al., 2004). Consistently, in cue1, the ORE1 transcript level was increased even in the absence of nitroprusside treatment, while in cue1 ein2, the level of ORE1 was low and not increased by treatment. Evidently EIN2 is required for nitric oxide to induce ORE1, at least in cotyledons. These results confirm the positive EIN2-associated ORE1 pathway during nitric oxide-induced cotyledon senescence.
NES1 and ORE1 in nitric oxide-induced cotyledon senescence
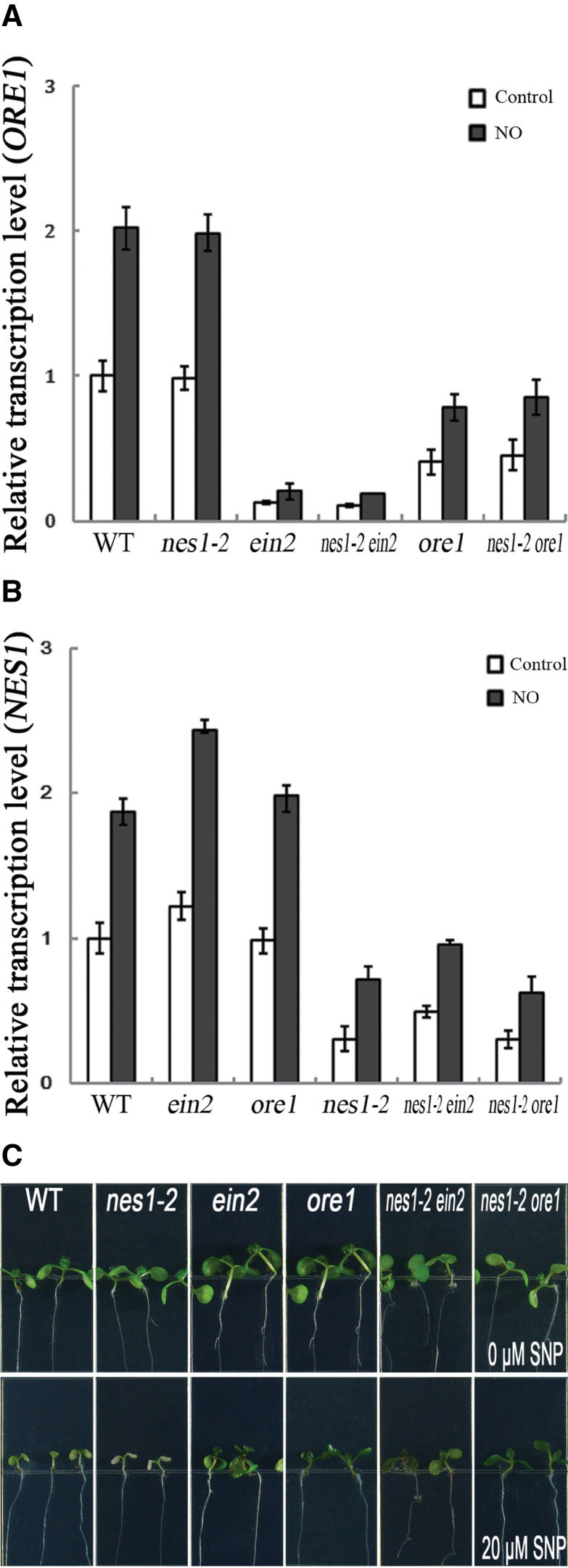
To explore the relationship between NES1 and ORE1 in the regulation of nitric oxide-induced cotyledon senescence, nes1-2 ein2 and nes1-2 ore1 double mutants were generated, and the NES1, EIN2, and ORE1 mutants were further analysed. Nitric oxide-regulated cotyledon senescence was accelerated in the nes1-2 mutant (Fig. 1A) and ORE1 expression was indistinguishable in both the nes1-2 mutant and the wild type (Fig. 5A). This implied that NES1, without interference from ORE1, was responsible for the accelerated senescence in the nes1-2 mutant. In addition, nitric oxide-delayed cotyledon senescence in the ore1 mutants (Fig. 4A), together with NES1 expression indistinguishable in the ore1 mutant and the wild type (Fig. 5B), suggested that ORE1 mainly contributed to the delayed cotyledon senescence in the ore1 mutant. The NES1 transcript level was slightly higher in the ein2 mutant than that in the ore1 mutant and the wild type. In the ein2 mutant, transcription of ORE1 declined significantly (Fig. 5A) and NES1 increased slightly (Fig. 5B) compared with that in the wild type. Alternations of these genes co-operatively contribute to the delay of nitric oxide-mediated cotyledon senescence in the ein2 mutant (Fig. 4A).
Fig. 5.
EIN2 and ORE1 are epistatic to NES1 during nitric oxide-induced cotyledon senescence. (A) Relative transcriptional quantification of ORE1 in the mutants. Seedlings treated as in Fig. 1E. Transcript quantified as in Fig. 2B. (B) The relative transcriptional quantification of NES1. Seedlings treated as in Fig 1E. Transcript quantified as in (A). (C) Phenotype analysis of the mutants. Seedlings treated as in Fig. 1A.
These results illustrated that NES1 and ORE1 did not affect each other’s expression in either induced or delayed senescence, indicating that there may be limited direct interaction between NES1 and the EIN2-associated ORE1 pathway during nitric oxide-induced cotyledon senescence. However, the test of responses to nitric oxide in the double mutant nes1-2 ein2 and nes1-2 ore1 showed that both ein2 and ore1 were epistatic to nes1-2, in terms of visual inspection of cotyledon colour (Fig. 5C) and the ORE1 transcripts (Fig. 5A). These results imply that the delayed cotyledon senescence in the nes1-2 ein2 and nes1-2 ore1 double mutants is mostly driven by the EIN2-associated ORE1 pathway.
Discussion
Early cotyledon senescence leads to various alterations in the physiological homeostasis of plants, which can result in plant seedlings failing to initiate and ultimately cause agricultural losses (Zhou and Brown, 2006; Chandler, 2008), but the molecular mechanism of cotyledon senescence is currently uncertain. Previous studies have focused on characterizing age-dependent cotyledon senescence in different plant species, such as soybean, cucumber, cotton, and pumpkin (Bick and Strehler, 1971; Kanazawa et al., 2000; Xie et al., 2008; Ananieva et al., 2008b ) and light irradiation accelerated cotyledon senescence (Watanabe et al., 1994; Wilhelmová et al., 2004). In this study, the phenotype of nitric oxide-induced cotyledon senescence was investigated and the nes1 mutant was characterized as having early cotyledon senescence which was induced by nitric oxide.
Nitric oxide is claimed to serve as a biological mediator (Besson-Bard et al., 2008), in a wide array of physiological processes, including senescence (Cam et al., 2012). Early studies have described age-dependent senescence as being associated with a significant decrease in intrinsic nitric oxide generation (Corpas et al., 2004), and in vitro nitric oxide fumigation was found to delay the senescence process (Mishina et al., 2007). Both of these results suggest that nitric oxide acts as a negative regulator in leaf senescence. In contrast, during salt-triggered senescence, the level of endogenous nitric oxide sharply increased (David et al., 2010). This suggests that the role of nitric oxide may differ in developmental and stress-induced senescence. One explanation is that the function of nitric oxide is strongly dependent on its concentration (Procházková and Wilhelmová, 2011). This explanation has been supported by a study of leaf senescence, which shows that small doses of nitric oxide delay senescence, while large doses accelerate the process (Selcukcan and Cevahir, 2008). These results emphasize that the dose-dependent action of nitric oxide is a double-edged sword (Colasanti and Suzuki, 2000).
To study the function of nitric oxide in cotyledon senescence, SNP was used to release nitric oxide. In these experimental conditions, the concentration of nitric oxide in sealed Petri plates initially undergoes complex transients and requires at least 2h to reach a stable level (Ferrero et al., 1999). Therefore, the dynamic behaviour observed for the F v/F m ratio probably reflects these dynamics. However, the sharp and significantly greater decrease in the mutant is clear (Fig. 1D).
The transcription factor ORE1 not only initiates developmental leaf senescence, but also plays a role in stress-induced leaf senescence, such as heat stress, oxidative stress, salt stress, and drought stress (Harding et al., 1990; Woo et al., 2004; Valente et al., 2009; Balazadeh et al., 2010; Wagstaff et al., 2010). All of these stresses are coupled with a massive accumulation of nitric oxide (Bouchard and Yamasaki, 2008; Lozano-Juste and León, 2010). As was shown in this study, during nitric oxide-accelerated cotyledon senescence, EIN2 and ORE1 play important positive roles. Therefore, during senescence triggered by an environmental stimulus, nitric oxide is probably involved in transmitting the stress signal to the EIN2-associated ORE1 pathway.
Previous studies have revealed that EIN2 functions as a positive regulator of ethylene- and dark-induced senescence (Kim et al., 2009; Chen et al, 2012). Recently, dark-accelerated senescence in the nitric oxide-deficient mutant nos1/noa1 was reverted by EIN2 mutation (Niu and Guo, 2012), which suggests that EIN2 is involved in nitric oxide-regulated leaf senescence. Although the mutation of NOS1/NOA1 is associated with a significant reduction in nitric oxide production, the role of NOS1/NOA1 in biosynthesis is still controversial (Guo et al., 2003; Zemojtel et al., 2006; Moreau et al., 2008). The present study not only confirms the role of EIN2 in nitric oxide-regulated senescence but also reveals the positive function of EIN2-associated ORE1 in nitric oxide-induced cotyledon senescence (Fig. 4).
The transcription factor ORE1/AtNAC2 affects leaf senescence under a number of conditions, including age-dependent and stress-induced senescence. ORE1/AtNAC2 functions in nitric oxide-induced cotyledon senescence (Fig. 4A) and ORE1 may extend the role in age-dependent cotyledon senescence. The ORE1 transcript level in cotyledons was massively up-regulated by day 21 (Supplementary Fig. S4A at JXB online), which was approximately the time of onset of developmental senescence (Supplementary Fig. S1A). In contrast, this pronounced up-regulation of ORE1 had not occurred in leaves by day 21 (Supplementary Fig. S4A), where leaf senescence did not start until several days later. Together, these findings suggest that ORE1 plays a positive role in developmental cotyledon senescence. And the results of studies on ORE1/AtNAC2 with different model systems are quite similar to those demonstrated in Arabidopsis leaves and cotyledons. For example, RhNAC2 was found to regulate flower senescence in Rosa hybrida (Dai et al., 2012), MaNAC2 was shown to be involved in banana fruit ripening (Shan et al., 2012), and AtNAC2 was demonstrated to function in Arabidopsis silique senescence (Kunieda et al., 2008). In conclusion, ORE1/AtNAC2 acts as a key element in positively regulating whole-plant senescence during age-dependent and induced processes.
Besides the positive role of the EIN2-associated ORE1 regulatory pathway during senescence, negative regulation by NES1 plays an important role in the nitric oxide-induced cotyledon senescence process. During the development process, NES1 has little impact on age-dependent cotyledon senescence, as the nes1-2 mutants and 35S::NES1 transgenic plants showed similar developmental stages to the wild type. At the same time, just as the level of ORE1 rises in senescing cotyledons, NES1 was able to be gradually induced during the natural cotyledon ageing process. However, the patterns were quite different in leaves. The level of ORE1 only increased just before the initiation of senescence in cotyledons (Supplementary Fig. S4A at JXB online) and true leaves (Kim et al., 2009), compared with NES1, which was generally induced in both cotyledons and true leaves. Once NES1 was induced in senescent cotyledons, the up-regulation pattern was consequently observed in young leaves (Supplementary Fig. S4B). The gradual increase of NES1 in cotyledon developmental ageing implies that NES1 is affected during developmental cotyledon senescence. The consequent induction of NES1 in young leaves suggests that NES1 might have another function in true leaf development. NES1 was also identified as nuclear-located MAD1, which interacted with MAD2 to suppress premature cell division at the Arabidopsis root meristem (Ding et al., 2012). However, the accelerated senescence in the nes1-2 mutant and the nitric oxide-induced NES1 expression pattern are obvious (Figs 1A, 2C). In conclusion, NES1 negatively regulates nitric oxide-accelerated cotyledon senescence, but not developmental senescence in cotyledons.
During nitric oxide-triggered cotyledon senescence, ORE1 and NES1 are both induced (Figs 2C, 4F), and function antagonistically against each other. The interaction between them reveals two distinct profiles during induced cotyledon senescence. The EIN2-associated ORE1 is a senescent signal transducer, and the induced NES1 functions as a negative regulator modulating the process, resulting from the evolution and maintenance in accordance with rigorous controlled plant acclimation (Heil and Baldwin, 2002). The nitric oxide-accelerated cotyledon senescence in nes1-2 and delayed cotyledon senescence in 35S::NES1 revealed its dominant role in repression of senescence. In 35S::NES1, although ectopically overexpressed, the delayed senescence phenotype (Fig. 3B) was due to the large increase of NES1 (Fig 3A), which indicates that NES1 could be epistatic to ORE1, in a dose-dependent manner. Together with the finding that ORE1 was epistatic to NES1 in nitric oxide-induced cotyledon senescence (Fig. 5C), this shows the balance of ORE1 and NES1 might be a critical factor in determining the fate of plant cotyledons. Once NES1 was dominant, plant senescence would be postponed, even without the initiation of senescence.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Age-dependent cotyledon senescence.
Figure S2. Nitric oxide induces cotyledon senescence.
Figure S3. The fine genetic and physical map-based cloning of NES1.
Figure S4. The transcription level of ORE1 and NES1 during cotyledon and leaf development.
Table S1. Genetic analysis of allelic NES1.
Table S2. Primers for semi-quantitative RT-PCR.
Table S3. Primers for quantitative real-time PCR.
Acknowledgements
This work was supported by the Ministry of Science and Technology of China [2013CB967300, 2007CB948201 to YH]. DK was supported by the US National Science Foundation [MCB-1244303 to JMK]. We wish to thank Xiaojun Pan and Meifan Guo for their help in the mutant screening, Yonghong Li for assisting in preparing some of the experiments, and Lihong Xiao for helpful discussions. We are grateful to Tobias I. Baskin (University of Massachusetts-Amherst, USA) for his critical discussion and editing of the manuscript.
References
- Ananieva K, Ananiev ED, Doncheva S, Georgieva K, Tzvetkova N, Kamínek M, Motyka V, Dobrev P, Gajdosová S, Malbeck J. 2008. a Senescence progression in a single darkened cotyledon depends on the light status of the other cotyledon in Cucurbita pepo (zucchini) seedlings: potential involvement of cytokinins and cytokinin oxidase/dehydrogenase activity. Physiologia Plantarum 134, 609–623 [DOI] [PubMed] [Google Scholar]
- Ananieva K, Ananieva ED, Mishev K, Georgieva K, Tzvetkova N, Van Staden J. 2008. b Changes in photosynthetic capacity and polypeptide patterns during natural senescence and rejuvenation of Cucurbita pepo L. (zucchini) cotyledons. Plant Growth Regulation 54, 23–29 [Google Scholar]
- Arasimowicz M, Floryszak-Wieczorek J. 2007. Nitric oxide as a bioactive signaling molecule in plant stress responses. Plant Science 172, 876–887 [Google Scholar]
- Arnold WP, Longnecker DE, Epstein RM. 1984. Photodegradation of sodium nitroprusside: biologic activity and cyanide release. Anesthesiology 61, 254–260 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Wu A, Mueller-Roeber B. 2010. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana . Plant Signaling and Behavior 5, 733–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. 2008. New insights into nitric oxide signaling in plants. Annual Review of Plant Biology 59, 21–39 [DOI] [PubMed] [Google Scholar]
- Bick MD, Strehler BL. 1971. Leucyl transfer RNA synthetase changes during soybean cotyledon senescence. Proceeding of the National Academy of Sciences, USA 68, 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard JN, Yamasaki H. 2008. Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: a possible linkage between nitric oxide and the coral bleaching phenomenon. Plant and Cell Physiology 49, 641–652 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, et al. 2011. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. The Plant Cell 23, 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis . The Plant Journal 42, 567–585 [DOI] [PubMed] [Google Scholar]
- Cam Y, Pierre O, Boncompagni E, Hérouart D, Meilhoc E, Bruand C. 2012. Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytologist 196, 548–560 [DOI] [PubMed] [Google Scholar]
- Chandler JW. 2008. Cotyledon organogenesis. Journal of Experimental Botany 59, 2917–2931 [DOI] [PubMed] [Google Scholar]
- Chen GH, Liu CP, Chen SC, Wang LC. 2012. Role of ARABIDOPSIS A-FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2 . Journal of Experimental Botany 63, 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti M, Suzuki H. 2000. The dual personality of NO. Trends in Pharmacological Sciences 21, 249–252 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, et al. 2004. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiology 136, 2722–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Hayashi M, Mano S, Nishimura M, Barroso JB. 2009. Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiology 151, 2083–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Zhang C, Jiang X, Kang M, Yin X, Lü P, Zhang X, Zheng Y, Gao J. 2012. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiology 160, 2064–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, Yadav S, Bhatla SC. 2010. Sodium chloride stress induces nitric oxide accumulation in root tips and oil body surface accompanying slower oleosin degradation in sunflower seedlings. Physiologia Plantarum 140, 342–354 [DOI] [PubMed] [Google Scholar]
- Ding D, Muthuswamy S, Meier I. 2012. Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Molecular Biology 79, 203–216 [DOI] [PubMed] [Google Scholar]
- Ederli L, Morettini R, Borgogni A, Wasternack C, Miersch O, Reale L, Ferranti F, Tosti N, Pasqualini S. 2006. Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiology 142, 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wang C, Chen Q, Chen H, Ren B, Li X, Zuo J. 2013. S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nature Communications 4: 1529. [DOI] [PubMed] [Google Scholar]
- Ferrero R, Rodríguez-Pascual F, Miras-Portugal MT, Torres M. 1999. Comparative effects of several nitric oxide donors on intracellular cyclic GMP levels in bovine chromaffin cells: correlation with nitric oxide production. British Journal of Pharmacology 127, 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Johnson JB, Rubin SH. 1976. Spectrophotometric determination of sodium nitroprusside and its photodegradation products. Journal of Pharmaceutical Sciences 65, 44–48 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM. 2003. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103 [DOI] [PubMed] [Google Scholar]
- Guo YF, Gan SS. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612 [DOI] [PubMed] [Google Scholar]
- Harding SA, Guikema JA, Paulsen GM. 1990. Photosynthetic decline from high temperature stress during maturation of wheat: I. Interaction with senescence processes. Plant Physiology 92, 648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, et al. 2004. Nitric oxide represses the Arabidopsis floral transition. Science 305, 1967–1971 [DOI] [PubMed] [Google Scholar]
- Heil M, Baldwin IT. 2002. Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends in Plant Science 7, 61–67 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter 5, 387–405 [Google Scholar]
- Jing HC, Schippers JH, Hille J, Dijkwel PP. 2008. Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis . Journal of Experimental Botany 56, 2915–2923 [DOI] [PubMed] [Google Scholar]
- Kanazawa S, Sano S, Koshiba T, Ushimaru T. 2000. Changes in antioxidative enzymes in cucumber cotyledons during natural senescence: comparison with those during dark-induced senescence. Physiologia Plantarum 109, 211–216 [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057 [DOI] [PubMed] [Google Scholar]
- Krul WR. 1974. Nucleic acid and protein metabolism of senescing and regenerating soybean cotyledons. Plant Physiology 54, 36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunieda T, Mitsuda N, Ohme-Takagi M, Takeda S, Aida M, Tasaka M, Kondo M, Nishimura M, Hara-Nishimura I. 2008. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis . The Plant Cell 20, 2631–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KX, Wang YN, Han CY, Zhang WS, Jia HZ, Li JX. 2007. GA signaling and CO/FT regulatory module mediate salt-induced late flowering in Arabidopsis thaliana . Plant Growth Regulation 53, 195–206 [Google Scholar]
- Li Z, Peng J, Wen X, Guo H. 2013. ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis . The Plant Cell 25, 3311–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136 [DOI] [PubMed] [Google Scholar]
- Liu WZ, Kong DD, Gu XX, et al. 2013. Cytokinins can act as suppressors of nitric oxide in Arabidopsis . Proceeding of the National Academy of Sciences, USA 110, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J. 2010. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiology 152, 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA. 2008. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiology 148, 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JR, Monte DC, Durzan D. 2000. Nitric oxide and ethylene emission in Arabidopsis thaliana . Physiology and Molecular Biology of Plants 6, 117–127 [Google Scholar]
- McKersie BD, Lepock JR, Kruuv J, Thompson JE. 1987. The effects of cotyledon senescence on the composition and physical properties of membrane lipid. Biochimica et Biophysica Acta 508, 197–212 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J. 2007. Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis . Plant, Cell and Environment 30, 39–52 [DOI] [PubMed] [Google Scholar]
- Moreau M, Lee GI, Wang Y, Crane BR, Klessing DF. 2008. AtNOS1/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. Journal of Biological Chemistry 283, 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I. 2008. Nitric oxide, stomatal closure, and abiotic stress. Journal of Experimental Botany 59, 165–176 [DOI] [PubMed] [Google Scholar]
- Niu YH, Guo FQ. 2012. Nitric oxide regulates dark-induced leaf senescence through EIN2 in Arabidopsis . Journal of Integrative Plant Biology 54, 516–525 [DOI] [PubMed] [Google Scholar]
- Peterman TK, Siedow JN. 1985. Behavior of lipoxygenase during establishment, senescence, and rejuvenation of soybean cotyledons. Plant Physiology 78, 690–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procházková D, Wilhelmová N. 2011. Nitric oxide, reactive nitrogen species and associated enzymes during plant senescence. Nitric Oxide 24, 61–65 [DOI] [PubMed] [Google Scholar]
- Rauf M, Arif M, Dortay H, Matallana-Ramírez LP, Waters MT, Gil Nam H, Lim PO, Mueller-Roeber B, Balazadeh S. 2013. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Reports 14, 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukes KL, Mulkey TJ. 1993. Hormonal regulation of cotyledon senescence. Bioscience 19, 10–15 [Google Scholar]
- Schippers JH, Jing HC, Hille J, Dijkwel PP. 2007. Developmental and hormonal control of leaf senescence. In: Gan SS, ed. Senescence processes in plants. Oxford: Blackwell Publishing Ltd, 145–170 [Google Scholar]
- Selcukcan EC, Cevahir O. 2008. Investigation on the relationship between senescence and nitric oxide in sunflower (Helianthus annuus L.) seedlings. Pakistan Journal of Botany 40, 1993–2004 [Google Scholar]
- Seo PJ, Park JM, Kang SK, Kim SG, Park CM. 2011. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233, 189–200 [DOI] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Chen L, et al. 2012. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. Journal of Experimental Botany 63, 5171–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. 2001. Characterization of cotyledon development in Arabidopsis thaliana . http://www.reu.iastate.edu/2001/papers/JennySmith.pdf
- Valente MA, Faria JA, Soares-Ramos JR, et al. 2009. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. Journal of Experimental Botany 60, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff C, Bramke I, Breeze E, Thornber S, Harrison E, Thomas B, Buchanan-Wollaston V, Stead T, Rogers H. 2010. A specific group of genes respond to cold dehydration stress in cut Alstroemeria flowers whereas ambient dehydration stress accelerates developmental senescence expression patterns. Journal of Experimental Botany 61, 2905–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Hamada K, Yokoi H, Watanabe A. 1994. Biphasic and differential expression of cytosolic glutamine synthetase genes of radish during seed germination and senescence of cotyledons. Plant Molecular Biology 26, 1807–1817 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM. 2001. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiology 127, 876–886 [PMC free article] [PubMed] [Google Scholar]
- Wilhelmová N, Procházková D, Macháčková I, Vágner M, Srbová M, Wilhelm J. 2004. The role of cytokinins and ethylene in bean cotyledon senescence. The effect of free radicals. Biologia Plantarum 48, 523–529 [Google Scholar]
- Wingler A, Marès M, Pourtau N. 2004. Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytologist 161, 781–789 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Nam HG, Lim PO. 2004. The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant and Cell Physiology 45, 923–932 [DOI] [PubMed] [Google Scholar]
- Xie QE, Liu ID, Yu SX, Wang RF, Fan ZX, Wang YG, Shen FF. 2008. Detection of DNA ladder during cotyledon senescence in cotton. Biologia Plantarum 52, 654–659 [Google Scholar]
- Xuan Y, Zhou S, Wang L, Cheng Y, Zhao L. 2010. Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiology 153, 1895–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Fröhlich A, Palmieri MC, et al. 2006. Plant nitric oxide synthase: a never-ending story? Trends in Plant Science 11, 524–525 [DOI] [PubMed] [Google Scholar]
- Zhang K, Gan SS. 2012. An abscisic acid–AtNAP transcription factor–SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiology 158, 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Chen L, Zhang LL, Zhang WH. 2009. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiology 151, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Brown DC. 2006. High efficiency plant production of North American ginseng via somatic embryogenesis from cotyledon explants. Plant Cell Reports 25, 166–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.