Summary
The EIN2-mediated senescence signalling pathway coordinates the expression of genes during leaf senescence via the gene regulatory network involving EIN3 and senescence-associated NAC TFs.
Key words: Arabidopsis, EIN2-mediated senescence signalling, EIN3, gene, NAC transcription factor.
Abstract
Leaf senescence is a finely tuned and genetically programmed degeneration process, which is critical to maximize plant fitness by remobilizing nutrients from senescing leaves to newly developing organs. Leaf senescence is a complex process that is driven by extensive reprogramming of global gene expression in a highly coordinated manner. Understanding how gene regulatory networks involved in controlling leaf senescence are organized and operated is essential to decipher the mechanisms of leaf senescence. It was previously reported that the trifurcate feed-forward pathway involving EIN2, ORE1, and miR164 in Arabidopsis regulates age-dependent leaf senescence and cell death. Here, new components of this pathway have been identified, which enhances knowledge of the gene regulatory networks governing leaf senescence. Comparative gene expression analysis revealed six senescence-associated NAC transcription factors (TFs) (ANAC019, AtNAP, ANAC047, ANAC055, ORS1, and ORE1) as candidate downstream components of ETHYLENE-INSENSITIVE2 (EIN2). EIN3, a downstream signalling molecule of EIN2, directly bound the ORE1 and AtNAP promoters and induced their transcription. This suggests that EIN3 positively regulates leaf senescence by activating ORE1 and AtNAP, previously reported as key regulators of leaf senescence. Genetic and gene expression analyses in the ore1 atnap double mutant revealed that ORE1 and AtNAP act in distinct and overlapping signalling pathways. Transient transactivation assays further demonstrated that ORE1 and AtNAP could activate common as well as differential NAC TF targets. Collectively, the data provide insight into an EIN2-mediated senescence signalling pathway that coordinates global gene expression during leaf senescence via a gene regulatory network involving EIN3 and senescence-associated NAC TFs.
Introduction
Leaf senescence is a well-orchestrated and genetically programmed cell death process that constitutes the final stage of leaf development (Lim et al., 2007). During leaf senescence, cells in a leaf undergo a dramatic transition in cellular metabolism and the degradation of cellular structures in an orderly manner, resulting in the recycling of nutrients to newly developing vegetative and reproductive organs (Nooden, 1988; Nam, 1997; Lim et al., 2007). Leaf senescence proceeds with the age of a leaf; however, it is also influenced by various endogenous factors, including phytohormones, and external environmental factors, such as salt stress, extreme temperatures, and pathogen attack (Thomas and Stoddart, 1980; Weaver et al., 1998; Dai et al., 1999; Quirino et al., 2000; Buchanan-Wollaston et al., 2005; Lim et al., 2007). Thus, leaf senescence is a very complicated process incorporating multiple developmental and environmental signals, which involves extensive reprogramming and modulation of gene expression.
Intensive genetic and genomic studies during the past decade have led to major advances in our understanding of leaf senescence at the molecular level. In particular, recent genome-wide transcriptome studies have uncovered a global picture of the leaf senescence process, which involves thousands of senescence-associated genes (SAGs) that are differentially expressed during leaf senescence (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Breeze et al., 2011). Given that the expression of >200 transcription factor (TF) genes is altered during leaf senescence (Buchanan-Wollaston et al., 2003; Balazadeh et al., 2008; Liu et al., 2011; Li et al., 2012; Guo, 2013) and TFs regulate the transcription of their target genes in a spatiotemporal-specific manner, gene regulatory networks composed of interactions between these TFs and their targets have been implicated in controlling leaf senescence. Indeed, by taking advantage of high-throughput and computational analyses, researchers have begun to identify gene regulatory networks involved in the leaf senescence process. For example, a gene regulatory network model has been reconstructed using selected SAGs from microarray-based temporal expression profiling during Arabidopsis leaf senescence (Breeze et al., 2011). The proposed network model predicts the effects of a plant-specific NAC (NAM/ATAF1,2/CUC2) TF, ORESARA1 (ORE1/NAC2/ANAC092), which is known to be one of the central positive regulators of leaf senescence, on the expression of multiple known downstream target genes and several stress-related TFs. Hickman et al. (2013) proposed a gene regulatory network model involving ANAC019, ANAC055, and ANAC072, based on high-throughput yeast one-hybrid (Y1H) assays and time-course gene expression data. Although initial attempts have been made to characterize gene regulatory networks important for leaf senescence, what the gene regulatory network involving TFs important for the control of leaf senescence is composed of and how it is operated have been largely unexplored.
It was previously reported that the trifurcate feed-forward pathway, which involves ETHYLENE-INSENSITIVE2 (EIN2/ORE2/ORE3), miRNA164 (miR164), and ORE1, regulates age-dependent leaf senescence and cell death (Kim et al., 2009). EIN2, a central signalling component required for ethylene responses (Alonso et al., 1999), induces ORE1 in an age-dependent manner. ORE1 is negatively regulated by miR164 in young leaves, which is relieved in old leaves due to the age-dependent down-regulation of miR164 by EIN2. In young Arabidopsis leaves, miR164 suppresses ORE1, which positively regulates leaf senescence. However, in old leaves, EIN2 suppresses miR164 and thereby induces ORE1 expression, which leads to leaf senescence. Based on the results of mathematical modelling and genetic analyses with the ein2 and ore1 mutants, it was further suggested that EIN2 utilizes another pathway that does not include ORE1. Recently, EIN3, a well-known key TF in the EIN2-mediated ethylene signalling cascade (Chao et al., 1997), has been shown to be involved in the trifurcate feed-forward pathway of age-dependent senescence and cell death (Li et al., 2013). EIN3 induces the accumulation of ORE1 transcript in an age-dependent manner by directly repressing miR164 transcription. However, how EIN2-mediated senescence signalling is transduced to ORE1 and how a gene regulatory network involving ORE1 is organized to regulate leaf senescence have not been investigated.
In this study, novel components in the trifurcate feed-forward pathway were identified and characterized to augment understanding of the composition, organization, and function of the gene regulatory networks that govern leaf senescence. As a first step toward expanding the trifurcate feed-forward pathway for leaf senescence, six senescence-associated NAC TFs were identified, including ORE1 and AtNAP/ANAC029, as candidate downstream targets of EIN2, and whether these NAC TF genes were acting downstream of EIN3 was further examined. Y1H and chromatin immunoprecipitation (ChIP) assays demonstrated that EIN3 directly bound to the promoters of the ORE1 and AtNAP genes. Transiently overexpressed EIN3 in Arabidopsis protoplasts was sufficient to activate the expression of ORE1 and AtNAP. Genetic and gene expression analyses in ore1 atnap double mutants revealed that ORE1 and AtNAP have partially additive functions in age-dependent and artificially induced leaf senescence. Using transient transactivation assays, it was further found that ORE1 and AtNAP regulate common as well as distinct NAC TF targets. Based on these data, a plausible model for an EIN2–EIN3–NAC TF gene regulatory cascade that has an important role in the control of leaf senescence is proposed. Collectively, the data provide insight into how the EIN2-mediated senescence signalling pathway coordinates global gene expression during leaf senescence via a gene regulatory network involving EIN3 and NAC TFs.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col) is the parent strain for all mutants used in this study. The ore1-2, ore1-3, ein2-34/ore3-1, ore9-1, and ore12-1 mutants were described previously (Woo et al., 2001; Kim et al., 2006; Kim et al., 2009). The ein3-1eil1-1 (ein3-like 1-1) (Alonso et al., 2003) mutant and the ein3 eil1 ebf1 (ein3-binding f-box1) ebf2 mutant containing estradiol-inducible EIN3-3XFLAG (iE/qm) (An et al., 2010) were kindly provided by H. Guo (Peking University, China). The 35S promoter (35Sp):EIN3-FLAG mutant was kindly provided by S.D. Yoo (Korea University, South Korea). The atnap T-DNA insertion line (SALK_005010C) was obtained from the Salk T-DNA insertion collection (Alonso et al., 2003). The genotype of each line was confirmed by PCR-based genotyping (Supplementary Table S1 available at JXB online). The 35Sp:EIN3-FLAG ore1-2 and 35Sp:EIN3-FLAG atnap were generated by genetic cross, and double homozygous lines were identified through PCR-based genotyping (Supplementary Table S1). Plants for physiological experiments were grown in an environmentally controlled growth room at 22 °C with 16h of light from a fluorescent light at 100 μmol m–2 s–1.
Assays of leaf senescence
Age-dependent leaf senescence was assayed as described by Woo et al. (2001). The photochemical efficiency of photosystem II (PSII) was deduced from the chlorophyll fluorescence (Oh et al., 1996) using an Imaging-PAM chlorophyll fluorometer (Heinz Walz GmbH, Germany). The ratio of the maximum variable fluorescence to the maximum yield of fluorescence, which corresponds to the potential quantum yield of the photochemical reactions of PSII, was used as a measure of the photochemical efficiency of PSII (John et al., 1995; Raggi, 1995; Oh et al., 1997). Chlorophyll was extracted from individual leaves by heating in 95% ethanol at 80 °C. The chlorophyll concentration per fresh weight of leaf tissue was calculated as described by Lichtenthaler (1987). For dark-induced leaf senescence experiments, the third or fourth rosette leaves of wild-type or mutant plants at 12 d of leaf age were carefully detached and incubated in 3mM MES buffer (pH 5.7) at 22 °C in the dark for the designated time. For hormone treatment, leaves were floated on 3mM MES buffer (pH 5.7) in the presence or absence of 50 μM 1-aminocyclopropane-1-carboxylic acid (ACC; Sigma-Aldrich, USA) or 50 μM methyl jasmonate (MeJA; Sigma-Aldrich, USA) for 5 d. All hormonal treatments were performed at 22 °C under continuous light.
RNA isolation and quantitative reverse transcription–PCR (qRT–PCR)
Total RNA was isolated from the third and fourth rosette leaves using WelPrep total RNA isolation reagent (WELGENE, Republic of Korea), according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1.0 μg of RNA using the ImProm II Reverse Transcriptase system kit (Promega, USA), followed by quantitative PCR (qPCR) analysis to determine gene expression levels (Bio-Rad, CFX96 Touch Real-Time PCR Detection System, USA). Primers used for qRT–PCR are listed in Supplementary Table S1 at JXB online. Transcript levels were calculated using the comparative threshold (CT) method, with ACT2 (At3g18780) as the internal control.
Yeast one-hybrid (Y1H) assays
The DupLEX-A system (OriGene Technologies, USA) was used with slight modifications for Y1H analysis of gene interactions. EIN3 full-length cDNA was cloned into the pJG4-5 prey vector, which includes a B42 transcriptional activation domain. Approximately 2kb of the ORE1 and AtNAP promoters were cloned separately into the lacZ (β-galactosidase) reporter plasmid pSH18-34. The yeast strain EGY48 (MATa, trp1, his3, ura3, leu2::6 LexAop-LEU2) was transformed with the indicated combinations of plasmids. Interactions were tested on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) medium (Ryu et al., 2005).
Transient expression assay in Arabidopsis protoplasts
For luciferase (LUC) reporter constructs, the promoters of ORE1, AtNAP, ANAC003, ANAC041, ANAC079, VND-INTERACTING2 (VNI2)/ANAC083, ANAC087, and ANAC102 were amplified from genomic DNA, cloned into pCR-CCD F (Kim and Somers, 2010), and recombined into the gateway version of the pGreen0800-LUC vector (Hellens et al., 2005), which contains 35Sp:RLuc (Renilla luciferase) as an internal control. Arabidopsis protoplasts were isolated and transfected as described (Hwang and Sheen, 2001; Yoo et al., 2007). Transfected protoplasts were incubated for 6h at 22 °C under dim light (5 μE m–2 s–1) and the luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega, USA), according to the manufacturer’s instructions.
Protein isolation and western blotting
To induce the expression of EIN3-FLAG, the 3- and 5-week-old iE/qm transgenic plants were sprayed with 20 μM and 100 μM estradiol for 6h, respectively. The third and fourth leaves were harvested, ground in liquid N2, and lysed with extraction buffer containing 50mM Tris-HCl (pH 7.5), 150mM NaCl, 10mM EDTA, 0.1% Nonidet P-40, 50 μM MG132, 1mM phenylmethylsulphonyl fluoride (PMSF), and a protease inhibitor cocktail. The protein extracts were heated at 95 °C for 5min in SDS–PAGE sample loading buffer, separated on 10% SDS–polyacrylamide gels, and transferred to polyvinylidene fluoride (PVDF) membranes (Gamble et al., 2002). The blot was probed with a monoclonal anti-FLAG antibody (Sigma-Aldrich, USA).
Chromatin immunoprecipitation (ChIP)-qPCR
Third and fourth leaves from 5-week-old iE/qm transgenic plants treated with 100 μM estradiol for 6h were harvested, and 2g was fixed in 1% formaldehyde solution and cross-linked under vacuum for 15min. A final concentration of 0.25M glycine was used to quench the formaldehyde for 5min under vacuum. After washing twice with cold deionized water, the tissue was ground in liquid N2 and extraction of chromatin was performed as described by Zhu et al. (2012). Prior to immunoprecipitation, 5 μg of anti-FLAG monoclonal antibody (Sigma-Aldrich, USA) was pre-incubated with 20 μl of protein A+G magnetic beads (Millipore, USA) at 4 °C on a rotator overnight. Sonicated chromatin supernatant (250 μl) was diluted to 500 μl and pre-cleared with 20 μl of protein A+G magnetic beads for 1h at 4 °C. Supernatants were incubated with the prepared antibody-bound beads at 4 °C for 2h, and beads were washed sequentially with low-salt wash buffer, high-salt wash buffer, and TE buffer. Elution and reverse cross-linking was performed as previously described (Zhu et al., 2012). The resulting immunoprecipitated DNA was purified with the Qiaquick PCR purification kit (Qiagen, USA), and used for qPCR to examine the enrichment of target genes using the primers listed in Supplementary Table S1 at JXB online.
Results
Identification of the senescence-associated NAC TFs acting downstream of EIN2
In an effort to expand the trifurcate pathway for leaf senescence, the aim was to identify molecular components functioning downstream of EIN2 in the regulation of leaf senescence. Genes whose expression is changed during leaf senescence and altered in the ein2 mutant would be good candidates as potential downstream targets of EIN2. NAC TF family proteins were the top candidates because publicly available microarray data revealed that NAC TF family genes are significantly up-regulated during leaf senescence in Arabidopsis, and the expression of some NAC TFs is altered in the ein2 mutant during leaf senescence (Schmid et al., 2005; Kim et al., 2009; Asahina et al., 2011; Breeze et al., 2011).
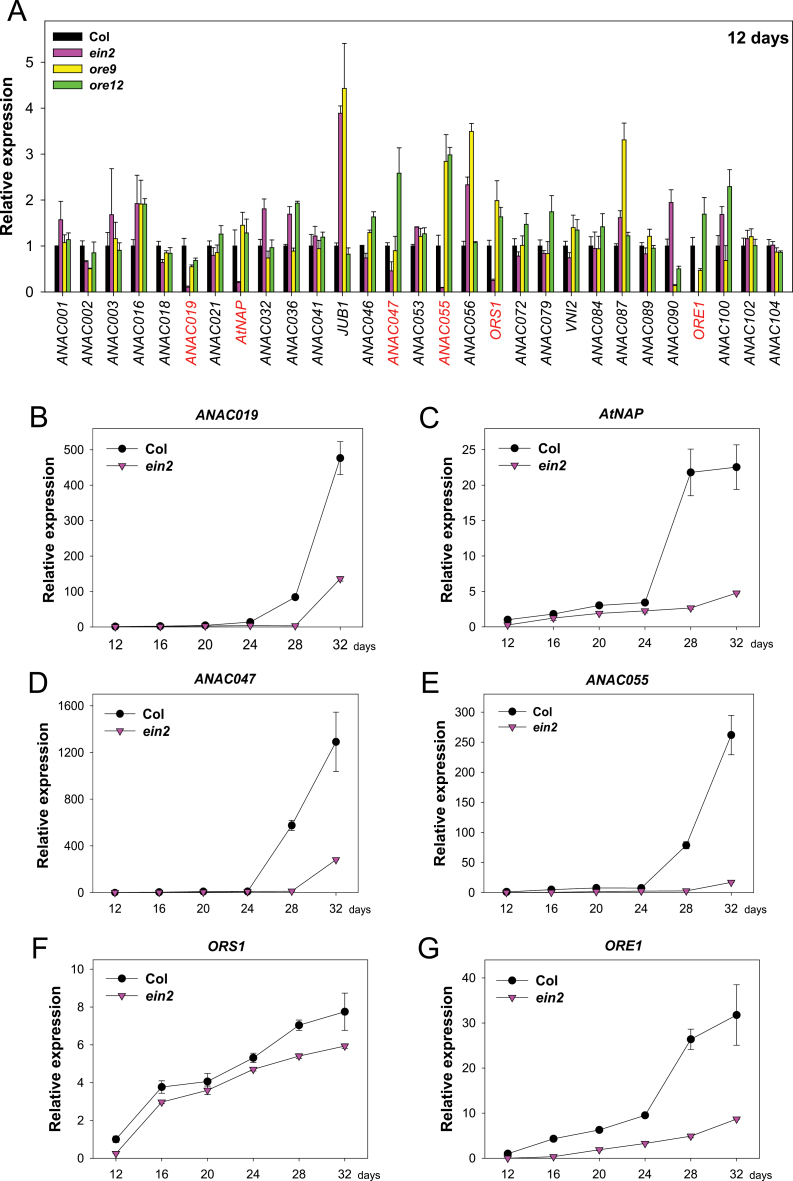
The focus of this strudy was the 29 NAC TF genes whose expression is increased at least 5-fold in senescent leaves, based on data from AtGenExpress (Schmid et al., 2005). The expression of the 29 senescence-induced NAC TF genes was examined in wild-type (Col) and ein2/ore3 mutant leaves at the mature stage (12-day-old third and fourth rosette leaves) by qRT–PCR (Fig. 1A). The expression analysis was also performed in the ore9 and ore12 mutants, which are well-known delayed leaf senescence mutants (Woo et al., 2001; Kim et al., 2006). Comparative expression analysis in these three mutants would allow the candidate downstream targets of EIN2 to be narrowed down by eliminating the NAC TFs whose expression is preferentially affected by delayed leaf senescence itself. As expected, the expression of ORE1 was strongly reduced in the mature leaves of ein2 (Fig. 1A). In addition to ORE1, ANAC019, AtNAP, ANAC047, ANAC055, and ORE1 SISTER1 (ORS1)/ANAC059 transcripts were significantly decreased in the mature leaves of ein2, compared with the reduction in the ore9 and ore12 mutants, implying that these six NAC TF genes are potential downstream targets of EIN2 in the control of the leaf senescence process.
Fig. 1.
Identification of the six senescence-associated NAC TFs as potential downstream components of EIN2. (A) Expression of 29 senescence-associated NAC TF genes in Col, ein2, ore9, and ore12 mutant leaves at the mature stage (12-day-old third and fourth rosette leaves). Transcript levels of each TF gene were examined by qRT–PCR. For qRT–PCR, ACT2 was used as an internal control. Transcript abundance of the NAC TF genes in each mutant was determined relative to that in wild-type leaves. The error bars represent the standard deviation (SD; n=4). The six NAC TF genes whose expression was decreased by >50% in the mature leaves of ein2 mutants compared with the wild-type are highlighted by grey text. (B–G) Age-dependent changes in the expression of candidate NAC TF genes downstream of EIN2. Transcript levels of ANAC019 (B), AtNAP (C), ANAC047 (D), ANAC055 (E), ORS1 (F), and ORE1 (G) were analysed by qRT–PCR in the third and fourth rosette leaves from wild-type and ein2 plants at the indicated ages. Transcript levels of each gene during leaf ageing were determined relative to levels in wild-type 12-day-old leaves. Error bars represent the SD (n=4).
Expression changes in the six NAC TF genes in wild-type and ein2 mutant leaves were further compared at 4 d intervals during leaf ageing (Fig. 1B–G). Consistent with previous findings (Schmid et al., 2005), the expression of the six NAC TF genes increases as a leaf gets older in wild-type leaves. Overall, these NAC TF genes were expressed at lower levels in ein2 leaves compared with wild-type leaves throughout the developmental stages of the leaf examined. The expression kinetics of each NAC TF gene in the ein2 mutant, however, were distinct during leaf ageing. For example, the abundance of ANAC019 and ANAC047 transcripts was dramatically increased in 28-day-old wild-type leaves, but transcripts of these genes in the ein2 leaves reached similar levels to those in 28-day-old wild-type leaves ~4 d later (Fig. 1B, D). In the case of AtNAP, ANAC055, and ORE1, transcript levels were also strongly induced in 28-day-old wild-type leaves, but did not significantly change in the ein2 mutant leaves until 32 d (Fig. 1C, E, G). These data indicate that ANAC019, AtNAP, ANAC047, ANAC055, and ORE1 are preferentially under the control of EIN2 during leaf ageing. In contrast, the expression kinetics of ORS1 during leaf ageing were similar in the wild-type and ein2 mutants, although its expression was lower in the ein2 leaves at all ages examined (Fig. 1F). This implies that ORS1 expression might be controlled by EIN2-independent as well as EIN2-dependent senescence signals. Collectively, these results suggest that at least six NAC TFs, including ORE1, act downstream of EIN2, which may be new components in the gene regulatory network governed by EIN2-mediated senescence signals.
EIN3 promotes leaf senescence through the activation of the two master NAC TFs, ORE1 and AtNAP
Next, experiments were carried out to examine how the EIN2-mediated senescence signal is transferred into the six NAC TFs. EIN3 has been long known as a key TF in EIN2-mediated ethylene signalling (Chao et al., 1997). Recent evidence indicates that EIN3 might function as an upstream regulator of these NAC TF genes during leaf senescence. First, EIN3 expression is induced durng leaf ageing, and the double mutant of EIN3 and its close homologue EIL1, ein3 eil1, exhibits delayed age-dependent leaf senescence (Li et al., 2013). Secondly, a previous ChIP-seq analysis revealed that EIN3 binds to the promoter of ORE1 after ethylene treatment in Arabidopsis seedlings (Chang et al., 2013). Thus, it was investigated whether EIN3 indeed acts upstream of the NAC TF genes whose expression was preferentially controlled by EIN2.
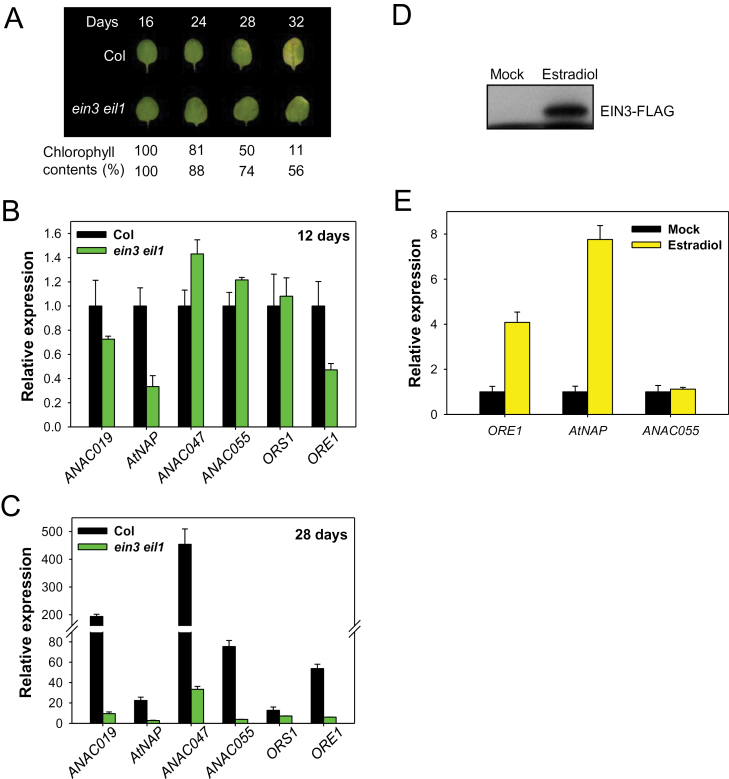
The effects of the ein3 mutation on the expression of the six NAC TF genes (ANAC019, AtNAP, ANAC047, ANAC055, ORS1, and ORE1) were first monitored (Fig. 2A–C). The ein3 eil1 double mutant was used instead of the ein3 single mutant due to the functional redundancy of EIN3 and EIL1 (Alonso et al., 2003; Binder et al., 2004). In mature (12-day-old) leaves, only ORE1 and AtNAP were down-regulated by >50% in the ein3 eil1 mutant compared with the wild type (Fig. 2B). In contrast, in 28-day-old leaves, all six NAC TF genes were expressed at significantly lower levels in the ein3 eil1 mutant, probably because of the delayed senescence phenotype of the ein3 eil1 mutant (Fig. 2A, C). These results imply that EIN3 might play a positive role in controlling leaf senescence through the activation of ORE1 and AtNAP.
Fig. 2.
EIN3 is necessary and sufficient to induce the expression of the ORE1 and AtNAP genes. (A) Changes in chlorophyll content in the third and fourth rosette leaves of Col and ein3 eil1 mutant plants during leaf ageing. The photographs show representative leaves at the indicated age. Chlorophyll content is compared with the values from each genotype at day 12. (B and C) Expression of the six NAC TF genes in the third and fourth leaves of wild-type and ein3 eil1 mutants at 12 d (B) and 28 d (C) of leaf age. For qRT–PCR, ACT2 was used as an internal control. Transcript levels of each gene were determined relative to levels in wild-type 12-day-old leaves. Error bars represent the SD (n=4). (D) Expression of ORE1, AtNAP, and ANAC055 in transgenic ein3 eil1 ebf1 ebf2 plants overexpressing estradiol-inducible EIN3 (iE/qm). Three-week-old iE/qm transgenic plants were treated with 20 μM estradiol for 6h, and protein and RNA were isolated from the third and fourth leaves. The tagged EIN3 protein was visualized by immunoblot analysis using an anti-FLAG antibody. (E) For qRT–PCR, ACT2 was used as an internal control. Transcript levels of each gene after estradiol treatment were determined relative to the mock treatment. The error bars represent the SD (n=4).
The expression of the ORE1 and AtNAP genes was then examined in transgenic plants expressing estradiol-inducible EIN3-FLAG in the ein3 eil1 ebf1 ebf2 quadruple mutant background (iE/qm). Three-week-old iE/qm plants were sprayed with 20 μM estradiol and the third and fourth leaves were harvested 6h after treatment. As shown in Fig. 2D, EIN3 protein efficiently accumulated in the iE/qm transgenic plants following estradiol treatment. ORE1 and AtNAP transcripts were significantly induced in the iE/qm transgenic plants by estradiol treatment, while ANAC055 transcript did not change (Fig. 2E). Overall, gene expression analysis of the NAC TF genes in the mutant and EIN3-inducible lines demonstrate that EIN3 is necessary and sufficient to induce the expression of ORE1 and AtNAP to promote leaf senescence.
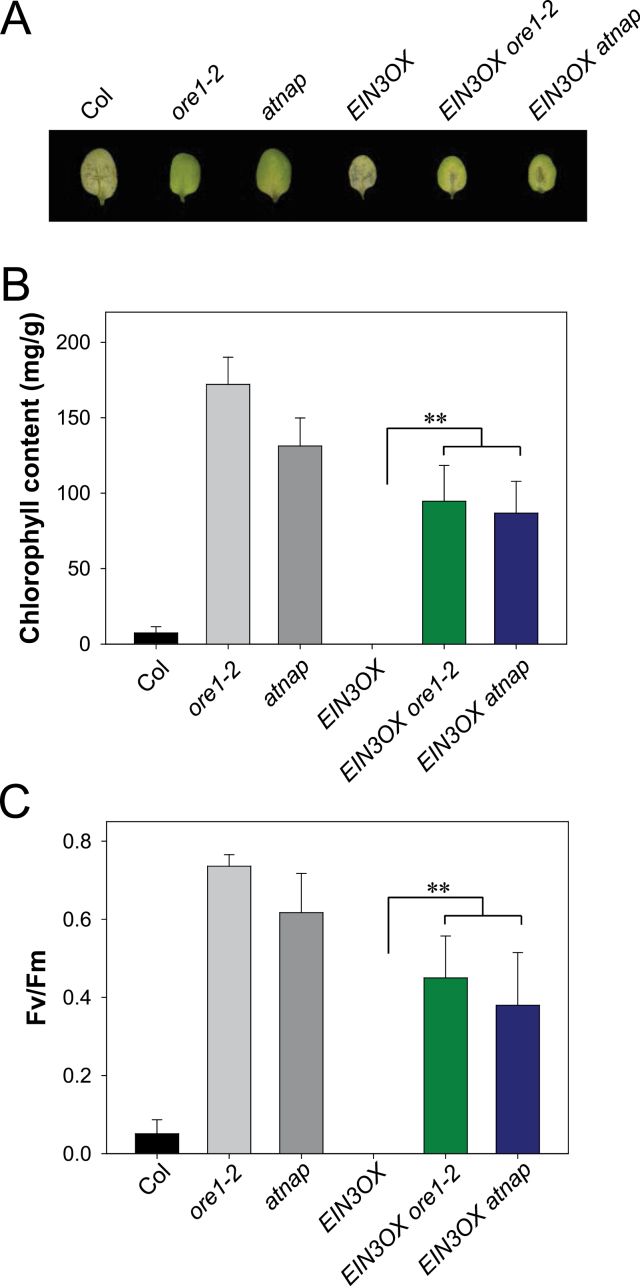
It is well known that ORE1 and AtNAP are master positive regulators of leaf senescence (Guo and Gan, 2006; Kim et al., 2009). Given that ORE1 and AtNAP function downstream of EIN3 in leaf senescence (Fig. 2), ORE1 and AtNAP loss-of-function mutations would be expected to repress EIN3-induced early senescence. A transgenic line expressing EIN3-FLAG driven from the 35S promoter (EIN3OX) was crossed with the ore1 or atnap mutants, and the dark-induced leaf senescence phenotype was examined in the double homozygous lines (Fig. 3). As expected, EIN3OX leaves exhibited early senescence phenotypes during dark incubation (Fig. 3). Mutation of ORE1 partially suppressed the EIN3-induced early leaf senescence phenotype (Fig. 3A–C). Similarly, the loss of chlorophyll content and photochemical efficiency during dark-induced leaf senescence was also delayed in both the atnap and EIN3OX atnap mutants, compared with wild-type and EIN3OX mutant leaves (Fig. 3A–C). The data demonstrated that EIN3 requires both ORE1 and AtNAP to induce leaf senescence. Taken together, these results revealed that EIN2-mediated senescence signalling induced the expression of the ORE1 and AtNAP genes through EIN3, and that this gene regulatory network played a major role in controlling leaf senescence.
Fig. 3.
The ORE1 or AtNAP loss-of-function mutant suppresses the early senescence phenotypes of an EIN3 overexpressor during dark-induced leaf senescence. (A) Representative leaves of Col, ore1, atnap, EIN3OX, EIN3OX ore1, and EIN3OX atnap plants after incubation in darkness for 6 d. (B and C) Analysis of chlorophyll content (B) and photochemical efficiency of PSII (C) of detached leaves of the indicated genotypes at 6 d after dark incubation (Student’s t-test, **P<0.01). Error bars indicate the SD (n >20).
EIN3 activates the expression of the ORE1 and AtNAP genes by directly binding to their promoters
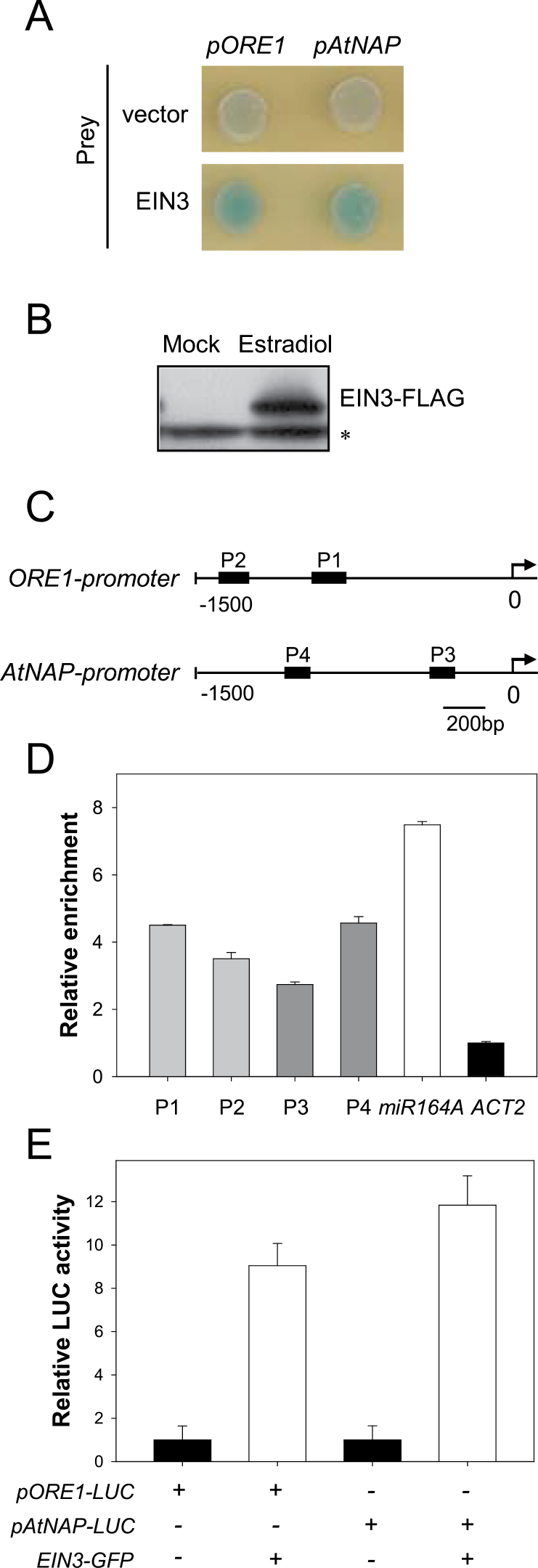
To examine whether the EIN3-mediated activation of the ORE1 and AtNAP genes is achieved by direct binding of EIN3 to their promoters, Y1H analysis was performed (Fig. 4A). The EGY48 yeast strain was co-transfected with an effector plasmid containing the full-length cDNA of EIN3 fused to the B42 transcriptional activation domain and a reporter vector containing either the ORE1 or the AtNAP promoter fused to the lacZ gene. Co-expression of EIN3 induced the expression of the lacZ reporter gene driven by the ORE1 or AtNAP promoter (Fig. 4A), indicating that EIN3 bound directly to the promoters of ORE1 and AtNAP in yeast.
Fig. 4.
EIN3 binds to the promoters of ORE1 and AtNAP, and induces their transcription. (A) Binding of EIN3 to the promoters of ORE1 and AtNAP in Y1H assay. An effector plasmid containing EIN3 and a reporter plasmid (pORE1-lacZ or pAtNAP-lacZ) were co-transformed into the EGY48 yeast strain. The growth of a blue yeast colony on selective medium containing X-gal indicates a positive interaction. The effector plasmid without EIN3 (vector alone) plus the reporter plasmid served as a negative control. (B) Protein levels of EIN3 in 5-week-old iE/qm transgenic plants treated or not with 100 μM estradiol for 6h. The tagged EIN3 protein was visualized by immunoblot analysis using an anti-FLAG antibody. An asterisk indicates non-specific bands detected by the anti-FLAG antibody. (C) Schematic diagram of the ORE1 and AtNAP promoters. P1–P4 represent the positions of amplicons used for ChIP-qPCR analysis. P1–P4 were chosen because these regions contain putative EIN3 binding sites (EBS, TACAT or TTCAAA). (D) Enrichment of EIN3-associated fragments after ChIP-qPCR. Chromatin from the leaves of 5-week-old iE/qm transgenic plants treated with 100 μM estradiol for 6h was immunoprecipitated with an anti-FLAG antibody. Enrichment was quantified by qPCR using specific primers. Fold changes in enrichment were normalized to ACT2. The promoter of miR164A was used as a positive control. The error bars represent the SD (n=4). (E) Transactivation of the ORE1 and AtNAP promoters by EIN3 in Arabidopsis protoplasts. Protoplasts were co-transfected with the pORE1-LUC or pAtNAP-LUC reporter and an effector plasmid expressing EIN3–GFP (green fluorescent protein). Luciferase activity was determined relative to that in protoplasts that were transfected with the reporter plasmid and an effector plasmid expressing GFP only. Relative expression of ORE1-LUC or AtNAP-LUC was normalized to that of 35Sp:RLuc (internal control). Error bars represent the SD (n=8).
ChIP-qPCR using iE/qm transgenic plants was carried out to determine whether EIN3 binds to the promoters of the ORE1 or AtNAP genes in plant cells. EIN3-FLAG protein strongly accumulated in the leaves of 5-week-old iE/qm transgenic plants treated with 100 μM estradiol for 6h (Fig. 4B). The miR164A promoter, which was shown to be a direct target of EIN3 (Li et al., 2013), was greatly enriched when a FLAG antibody was used to immunoprecipitate the FLAG-tagged EIN3 protein. In the same plants, a significant enrichment of EIN3 was also observed in the promoter regions of ORE1 (P1 and P2) and AtNAP (P3 and P4) (Fig. 4C, D), supporting the conclusion that ORE1 and AtNAP are direct downstream targets of EIN3 in vivo.
To investigate further whether EIN3 functions as a transcriptional activator of ORE1 and AtNAP in plant cells, luciferase-based transactivation assays were performed using Arabidopsis mesophyll protoplasts. A reporter construct containing the firefly luciferase (LUC) reporter gene under the control of either the ORE1 or the AtNAP promoter (pORE1-LUC or pAtNAP-LUC) was transfected into protoplasts with or without the 35Sp:EIN3-GFP effector plasmid. Luciferase activity was significantly increased when either the pORE1-LUC or the pAtNAP-LUC reporter construct was co-transfected with 35Sp:EIN3-GFP, compared with controls that were solely transfected with the reporter constructs (Fig. 4E), indicating that EIN3 transactivates the promoters of ORE1 and AtNAP in protoplasts. Taken together, these results demonstrate that EIN3 directly activates ORE1 and AtNAP transcription by binding to their promoter regions.
ORE1 and AtNAP have partially additive functions in regulating age-dependent and artificially induced leaf senescence
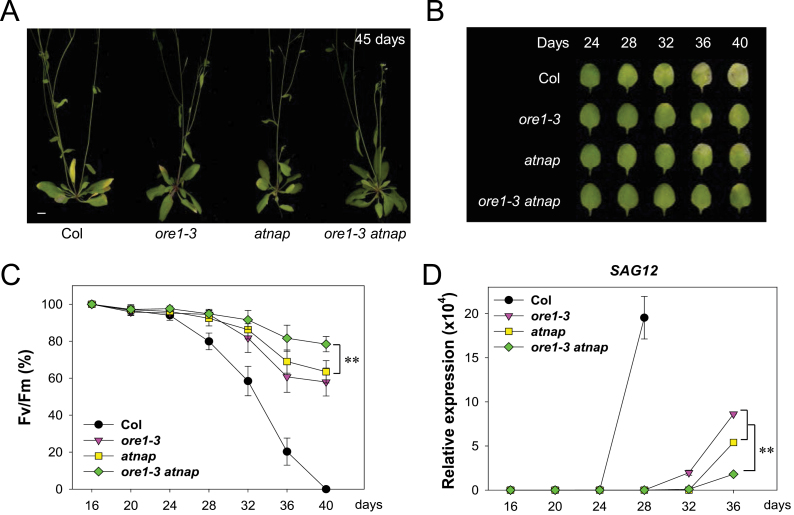
The genetic relationship between ORE1 and AtNAP in leaf senescence was explored by generating an ore1 atnap double mutant and analysing the leaf senescence phenotype during leaf ageing. Leaf senescence symptoms were first examined in detail during age-dependent in planta senescence. As previously reported (Guo and Gan, 2006; Kim et al., 2009), delayed loss of chlorophyll content with leaf ageing was observed in the ore1 and atnap single mutants (Fig. 5A, B). The 36-day-old leaves from wild-type plants lost 79.7% of the initial photochemical efficiency (F v/Fm) of PSII, while leaves from the ore1 and atnap mutants lost 39.3% and 31.0% of their initial PSII activity, respectively (Fig. 5C). Notably, 81.4% of the photochemical efficiency of PSII was retained in the leaves of ore1 atnap double mutant leaves at the same age. The expression of SAG12 also increased dramatically in 28-day-old wild-type leaves, but remained at a very low level until 28 d and 32 d in the ore1 and atnap single mutant leaves, respectively (Fig. 5D). In the ore1 atnap double mutant, induction of SAG12 was delayed even longer than in either of the single mutants (Fig. 5D).
Fig. 5.
The ore1 atnap double mutant exhibited a stronger delay in age-dependent leaf senescence than either mutant alone. (A) Whole-plant phenotypes of Col, ore1, atnap, and ore1 atnap mutant plants at 45 d after germination. The scale bar represents 1cm. (B) Age-dependent senescence phenotype of the third and fourth rosette leaves of Col, ore1, atnap, and ore1 atnap mutant plants at different ages. (C) The photochemical efficiency (F v/F m) of PSII was measured from the third and fourth leaves starting at 16 d of leaf age (Student’s t-test, **P<0.01). Error bars indicate the SD (n=12). (D) Age-dependent changes in SAG12 gene expression by qRT–PCR analysis. ACT2 was used as an internal control for qRT–PCR. The transcript level of SAG12 in wild-type at 12-day-old leaves was set at 1 (Student’s t-test, **P<0.01). The error bars represent the SD (n=4).
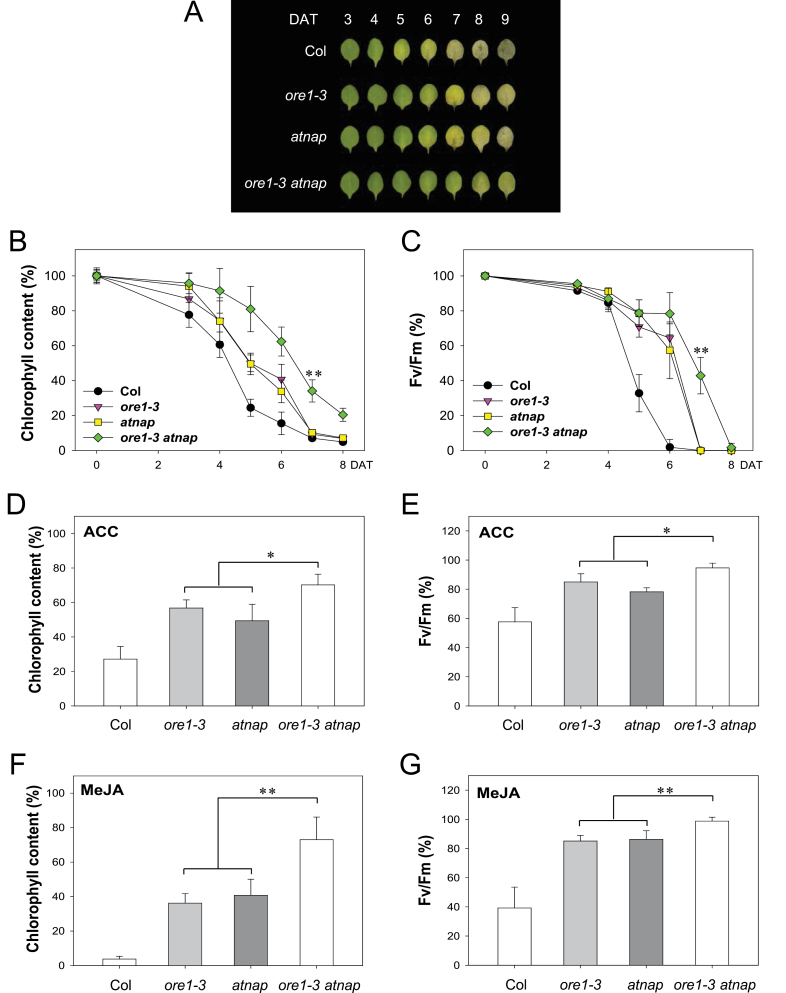
In artificially induced leaf senescence, the genetic relationship between ORE1 and AtNAP was further evaluated. Leaf senescence phenotypes were examined in wild-type, ore1, atnap, and ore1 atnap mutants during dark incubation. The leaves from the wild-type plants lost 84.4% of their chlorophyll after 6 d of dark incubation (Fig. 6A, B). However, for the ore1 and atnap single mutants, the chlorophyll content declined more slowly; even after 6 d, ~35% of the chlorophyll was retained (Fig. 6A, B). Measurement of the photochemical efficiency of PSII showed that, after 6 d of dark incubation, the leaves of ore1 and atnap still maintained >55% of their initial PSII activity, whereas wild-type leaves had lost their PSII activity almost entirely (Fig. 6C). In the ore1 atnap double mutant, 62.4% of the chlorophyll and 78.3% of the photochemical efficiency was retained even after 6 d of dark incubation (Fig. 6B, C). The leaf senescence symptoms of wild-type, ore1, atnap, and ore1 atnap mutants were assessed after treatment with two senescence-accelerating hormones, the ethylene precursor ACC, and MeJA. Not surprisingly, the leaves of the ore1 atnap mutant retained >70% of their chlorophyll following treatment with these hormones at 5 d after incubation, while ~50% and 40% of the chlorophyll was retained in each of the single mutant leaves treated with ACC and MeJA, respectively (Fig. 6D, F). A similar pattern was observed when photochemical efficiency was measured (Fig. 6E, G). Taken together, the ore1 atnap double mutant exhibited stronger senescence phenotypes than either of the single mutants. These results demonstrate that ORE1 and AtNAP might function in part additively in the regulation of leaf senescence and suggest that the two NAC TFs acting downstream of EIN3 independently regulate leaf senescence, but partially compensate for each other’s function.
Fig. 6.
ORE1 and AtNAP play partially additive roles in regulating artificially induced leaf senescence. (A) Phenotypes of Col, ore1, atnap, and ore1 atnap leaves after dark incubation for the indicated times. DAT, days after treatment. (B and C) Changes in photochemical efficiency (F v/F m) of PSII (B) and chlorophyll content (C) during dark-induced leaf senescence. Levels of photochemical efficiency and chlorophyll content on the days indicated were determined relative to those before dark incubation (Student’s t-test, **P<0.01). Error bars indicate the SD (n=6). (D and E) Changes in the chlorophyll content (D) and photochemical efficiency of PSII (E) during ACC-induced leaf senescence. (F and G) Changes in the chlorophyll content (F) and photochemical efficiency of PSII (G) during MeJA-induced leaf senescence. Levels of two senescence markers on the days indicated were determined relative to those before ACC or MeJA treatment (Student’s t test, *P<0.05 and **P<0.01). Error bars indicate the SD (n=6).
ORE1 and AtNAP control common as well as differential NAC TF genes
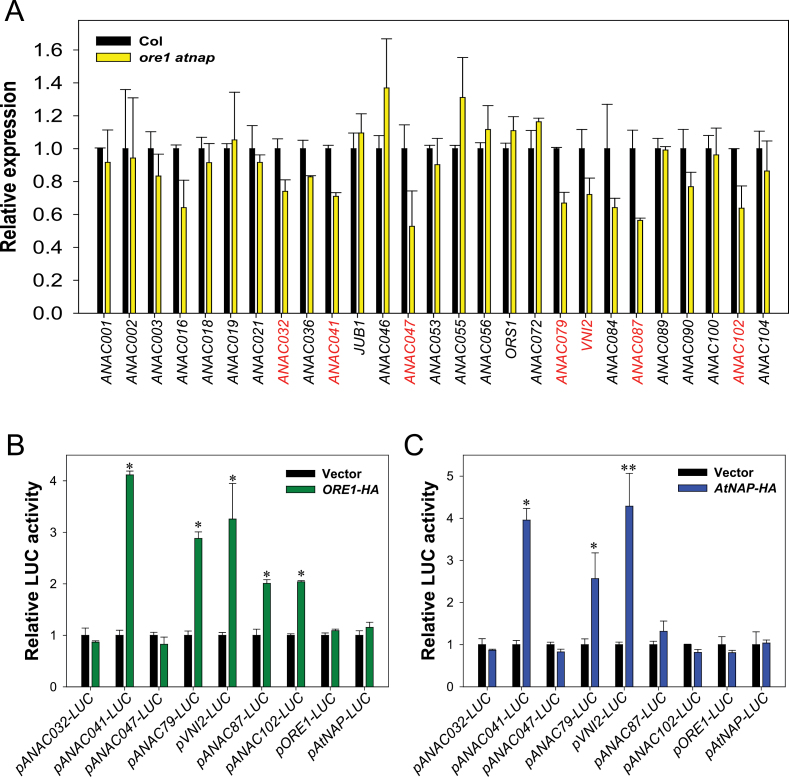
It has been shown that the promoters of many NAC TF genes contain consensus NAC-binding sites (Olsen et al., 2005; Balazadeh et al., 2010). In addition, ANAC016 binds to the promoter of AtNAP and ORS1 (Kim et al., 2013). Moreover, a recent global gene expression analysis in the ore1 mutant and inducible transgenic lines overexpressing ORE1 has revealed that expression of several NAC TF genes is affected by ORE1 (Balazadeh et al., 2010). Among them, ANAC041 and VNI2 have been predicted as direct targets of ORE1, because the promoters of the two genes have the ORE1 core binding site. These previous observations strongly support the possibility that ORE1 and AtNAP control leaf senescence, at least in part, through a NAC TF-mediated gene regulatory network. Therefore, qRT–PCR-based gene expression analysis was employed to identify the NAC TFs that lie downstream of ORE1 and AtNAP. Transcript levels of 27 senescence-associated NAC TF genes were evaluated in 16-day-old wild-type and ore1 atnap leaves (Fig. 7A). The expression of nine NAC TF genes was significantly reduced in ore1 atnap mutants, compared with wild-type leaves (Fig. 7A). Luciferase-based transactivation assays were performed using Arabidopsis protoplasts to determine the effect of ORE1 or AtNAP on the expression of seven NAC TF genes (Fig. 7B, C). The luciferase activity driven by the promoters of ANAC041, ANAC079, and VNI2 was increased at least 2-fold by transiently overexpressed ORE1 and AtNAP. In contrast, the luciferase activity of pANAC087-LUC and pANAC102-LUC was only increased by overexpression of ORE1-HA, not AtNAP-HA. Interestingly, neither ORE1 nor AtNAP activated their own promoters or each other’s promoters. Taken together, these results suggest that ORE1 and AtNAP serve as key regulators of leaf senescence by controlling common and differential downstream NAC TFs.
Fig. 7.
AtNAP and ORE1 control common as well as differential NAC TF genes. (A) Expression of 27 senescence-associated NAC TF genes in Col and ore1 atnap double mutant leaves at 16 d of leaf age. Transcript levels of each NAC TF gene were examined by qRT–PCR. ACT2 was used as an internal control for qRT–PCR. Transcript levels of the NAC TF genes in the ore1 atnap mutant were determined relative to levels in wild-type leaves. The error bars represent the SD (n=4). The nine NAC TF genes whose expression was significantly decreased in the mature leaves of the ore1 atnap mutant, compared wirh levels in wild-type leaves, are highlighted by grey text. (B and C) Transactivation of the promoters of the selected NAC TF genes by ORE1 (B) and AtNAP (C) in Arabidopsis protoplasts. Protoplasts were co-transfected with each NAC TF promoter-LUC reporter and an effector plasmid expressing ORE1-HA or AtNAP-HA. Luciferase activity was determined relative to that in protoplasts that were transfected with the reporter plasmid and an effector plasmid expressing HA only. The relative expression of each NAC TF promoter-LUC was normalized to 35Sp:RLuc (internal control) (Student’s t-test, *P<0.05 and **P<0.01). Error bars represent the SD (n=6).
Discussion
In a previous study, a gene regulatory network was proposed as underlying leaf senescence, the trifurcate feed-forward pathway which involves EIN2, miR164, and ORE1 (Kim et al., 2009). As a leaf ages, EIN2-mediated senescence signals induce the expression of ORE1, a positive regulator of leaf senescence, and simultaneously suppress the expression of miR164, which negatively regulates ORE1 at the post-transcriptional level. Mathematical modelling and genetic analysis results further suggested the existence of an ORE1-independent pathway activated by EIN2-mediated senescence signals. Recently, EIN3 has been shown to be involved in the trifurcate feed-forward pathway, by directly repressing miR164 expression (Li et al., 2013). However, of what the gene regulatory network activated by EIN2-mediated senescence signal is composed and how it manages the leaf senescence process remains to be elucidated.
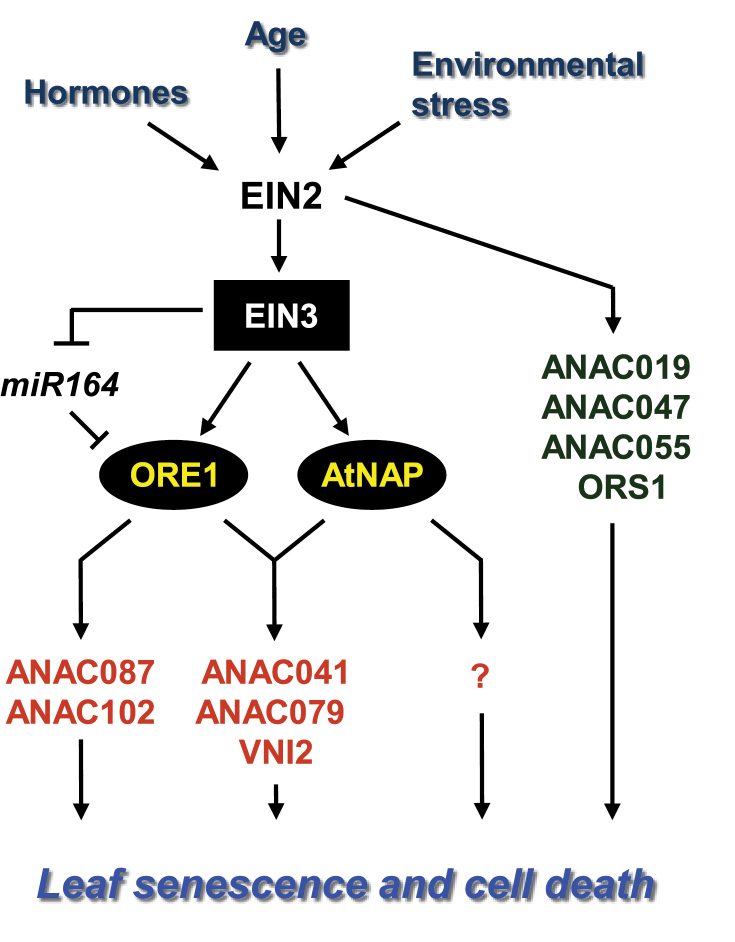
In this study, new molecular components of the trifurcate feed-forward pathway have been identified and characterized, enhancing understanding of the gene regulatory networks governing the leaf senescence process. New findings on the organization and function of the gene regulatory networks underlying leaf senescence included the following. First, six NAC TFs, including the two master regulators of leaf senescence, ORE1 and AtNAP, were found to participate in the gene regulatory network controlled by EIN2 (Fig. 1). Secondly, it was uncovered that EIN2-mediated senescence signal was transduced into ORE1 and AtNAP through the action of EIN3 (Figs 2–4). Furthermore, the data suggest that four additional NAC TFs seem to be mainly regulated by an EIN3-independent pathway(s) (Fig. 2). Thirdly, analysis of the downstream targets of ORE1 and AtNAP also provides new insights into how EIN2-mediated senescence signalling is differentially propagated through the key NAC TFs to execute the leaf senescence process (Fig. 7). Based on these data, a working model for an EIN2–EIN3–NAC TFs regulatory cascade with an important role in the control of leaf senescence was proposed (Fig. 8).
Fig. 8.
A plausible model for the EIN2–EIN3–NAC TFs regulatory cascade in the control of leaf senescence. EIN2-mediated senescence signalling, triggered by various senescence-inducing factors including age, hormones, and environmental stresses, activates EIN3. EIN3 directly induces the expression of two key positive regulators of leaf senescence, ORE1 and AtNAP. Simultaneously, EIN3 directly suppresses the expression of miR164 (Li et al., 2013), which negatively regulates ORE1 at the post-transcriptional level. ORE1 and AtNAP activate the expression of common as well as distinct downstream NAC TF genes. In addition, EIN2-mediated senescence signal is transduced to four NAC TFs (ANAC019, ANAC047, ANAC055, and ORS1) via an EIN3-independent pathway.
NAC TFs as new components of the gene regulatory network activated by EIN2
EIN2, a central signalling component required for ethylene responses, has been long known as a master positive regulator of leaf senescence (Oh et al., 1997; Alonso et al., 1999), yet the gene regulatory network controlled by EIN2-mediated senescence signal is not fully understood. Here, an attempt was made to identify additional molecular components that act downstream of EIN2, as a first step towards understanding in detail the gene regulatory network activated by the EIN2-mediated senescence signal. In the present study, six NAC TF genes, including ORE1, AtNAP, and ANAC055, were found to be controlled by EIN2, based on gene expression analysis in the ein2 mutant (Fig. 1). It is intriguing that no significant reduction in the transcript levels of the six NAC TF genes was observed in mature leaves of the ore9 and ore12 mutants (Fig. 1A). These data further support the possibility that EIN2 is one of the key regulators controlling the expression of NAC TFs during leaf ageing. It is also likely that EIN2 might not be the only route capable of activating the six NAC TF genes because transcript levels of all six NAC TFs were eventually increased as a leaf gets older (Fig. 1B–G). This is further supported by the previous finding that ANAC016, which directly binds to the promoter of AtNAP in yeast (Kim et al., 2013), does not appear to be under the control of EIN2, based on the expression of ANAC016 in the ein2 mutant (Fig. 1A). Other TFs, including members of several TF families such as bZIP, bHLH, MYB, and AP2/ERF, were also identified as upstream molecules of ANAC019, ANAC055, and ANAC072 (Hickman et al., 2013), indicating the complexity of the gene regulatory network involving NAC TFs. Thus, further experiments for identifying EIN2-independent senescence signalling pathways will be needed to better understand the gene regulatory networks involving NAC TFs in the control of leaf senescence. Interestingly, levels of some NAC TF transcripts, including JUNGBRUNNEN1/ANAC042 reported as a negative regulator of leaf senescence (Wu et al., 2012), were even higher in the ein2 mutant. In the case of ANAC055, its transcript level was strongly reduced in the ein2 mutant but significantly increased in the ore9 and ore12 mutants (Fig. 1). This result indicates that gene regulatory networks involving NAC TFs might be complex and interconnected. Elucidation of components acting downstream of EIN2 through genome-wide screening will help to characterize the complex global gene regulatory network activated by EIN2 to control leaf senescence.
EIN3 directly activates the expression of ORE1 and AtNAP
In this study, several lines of evidence support the conclusion that EIN3 is an upstream TF controlling the expression of ORE1 and AtNAP. First, the expression of ORE1 and AtNAP was significantly altered in ein3 eil1 double mutants, even in young leaves (Fig. 2). Secondly, the ore1 and atnap mutations partially suppressed the EIN3-induced early leaf senescence phenotype (Fig. 3). Thirdly, EIN3 directly bound to the promoters of the ORE1 and AtNAP genes in Y1H and ChIP-PCR assays (Fig. 4A–C). Finally, transiently overexpressed EIN3 was sufficient to activate the expression of ORE1 and AtNAP (Fig. 4D).
It was notable that EIN3 functions as a transcriptional activator of ORE1 and AtNAP, which are known to be positive regulators of leaf senescence (Oh et al., 1997; Guo and Gan, 2006; Kim et al., 2009), whereas EIN3 is known to function as a direct repressor of miR164 (Li et al., 2013). This suggests that EIN3 can function as both a transcriptional activator and a repressor, depending on the target genes to regulate leaf senescence, in agreement with a previous study (Shi et al., 2012). This result further implies that EIN3 might simultaneously control the expression of ORE1 and its negative regulator, miR164, to regulate leaf senescence efficiently.
Chang et al. (2013) revealed that EIN3 binds to the promoters of the four NAC TF genes (ANAC019, ANAC047, ANAC055, and ORS1) after ethylene treatment in Arabidopsis seedlings, but the expression of these NAC genes was not altered in the mature leaves of the ein3 eil1 double mutant (Fig. 2B). This discrepancy implies that EIN3 and EIL1 might regulate different downstream targets under different physiological conditions or at different developmental stages. Thus, it is likely that the four NAC TF genes may not be direct targets of EIN3 and/or EIL1 in a mature leaf to trigger leaf senescence. Alternatively, it is equally possible that the four NAC TFs are targets of EIN3 and/or EIL1, but are regulated primarily by other TF(s) that act downstream of EIN2 in a mature leaf. In other words, the EIN2-mediated senescence signal seems to modulate leaf senescence through at least two independent pathways. These results imply that there are many routes to ensure leaf senescence and the associated cell death upon ageing. Future challenges will include identifying additional TFs that act downstream of the EIN2-mediated senescence signal, and determination of their molecular functions within the context of leaf senescence control.
The effects of mutations in ORE1 and AtNAP are partially additive in the regulation of leaf senescence
The functional relationship between ORE1 and AtNAP that act downstream of EIN3 was also investigated by analysing the leaf senescence phenotype of the ore1 atnap double mutant (Figs 5, 6). The partially additive phenotypes of the ore1 atnap double mutant imply that ORE1 and AtNAP have both overlapping and independent functions in the transmission of EIN3-mediated senescence signals. This finding suggests that EIN3-mediated senescence signals can be transmitted via two partially independent pathways, one involving ORE1, and a second involving AtNAP.
Further understanding of how the gene regulatory network involving ORE1 and AtNAP functions in the regulation of leaf senescence can be facilitated by the identification of downstream target genes. Recent studies utilizing microarray analysis in the ore1 mutant as well as inducible transgenic lines overexpressing ORE1 have identified potential targets of ORE1. Among them, BIFUNCTIONAL NUCLEASE1 has been characterized as a direct downstream target molecule of ORE1 (Balazadeh et al., 2010; Matallana-Ramirez et al., 2013). As a direct downstream target gene of AtNAP, SAG113, a gene encoding a protein phosphatase 2C family protein phosphatase, has been identified (Zhang and Gan, 2012). However, knowledge about the downstream gene regulatory network of ORE1 and AtNAP remains limited. In this study, downstream NAC TF genes of ORE1 and AtNAP were thus examined by performing transient transactivation assays in order to gain deeper insight into the gene regulatory networks involving ORE1 and AtNAP. It is intriguing that ORE1 and AtNAP did not activate each other, and ANAC087 and ANAC102 were preferentially activated by ORE1, not by AtNAP (Fig. 7B, C). The results further support the idea that ORE1 and AtNAP act independently in the regulation of leaf senescence by activating different NAC TFs, and they may differentially activate other downstream targets as well. ORE1 and AtNAP activated three common NAC TFs genes (ANAC041, ANAC079, and VNI2) (Fig. 7B, C), indicating that ORE1 and AtNAP might compensate for each other’s function by activating common downstream components.
It was notable that some of the NAC TFs activated by ORE1 in the transactivation assays were predicted as potential downstream targets of ORE1 in previous studies. For example, ANAC102 is also one of the downstream genes predicted to be activated by ORE1 in a recent network modelling analysis based on high-resolution time-course profiles of gene expression during leaf development (Breeze et al., 2011). It has also been known that ANAC041 and VNI2 might be downstream targets of ORE1 from a microarray analysis during leaf senescence (Balazadeh et al., 2010; Rauf et al., 2013). It is intriguing that ORE1 and AtNAP activate VNI2, a negative regulator of leaf senescence (Yang et al., 2011). These findings imply that ORE1 and AtNAP, in addition to acting as positive regulators of senescence-accelerating genes, may finely tune the progression rate of leaf senescence through activating genes involved in maintenance activity. Further identification of direct downstream targets of ORE1 and AtNAP is essential to provide new insights into how the gene regulatory network involving the EIN2-mediated senescence signal regulates the leaf senescence process. Further experiments combining genetic analysis, ChIP-seq, gene expression profiling, and computational analyses will contribute to the elucidation of complex gene regulatory networks involving EIN2, EIN3, and NAC TFs, and will help us to understand how these pathways are interconnected. Collectively, the present data provide insight into the global gene regulatory network involving EIN3 and NAC TFs, through which the EIN2-mediated senescence signalling pathway coordinates global gene expression during leaf senescence.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this work.
Acknowledgements
We thank K.H. Seo, J.E. Hyun, and J.Y. Kim for excellent technical assistance. We also thank H. Guo for ein3-1eil1-1 and iE/qm seeds, and S.D. Yoo for EIN3-FLAG overexpression seeds. This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the government of the Republic of Korea: the Research Center Program of the Institute for Basic Science (IBS) [grant no. CA1208] and the Basic Science Research Program [grant os 2010-0010915, 2012R1A1A3004599].
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. 1999. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis . Science 284, 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, et al. 2010. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis . The Plant Cell 22, 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M, Azuma K, Pitaksaringkarn W, et al. 2011. Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Riano-Pachon DM, Mueller-Roeber B. 2008. Transcription factors regulating leaf senescence in Arabidopsis thaliana . Plant Biology (Stuttgart) 10 Suppl 1, 63–75 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Kohler B, Mueller-Roeber B. 2010. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. The Plant Journal 62, 250–264 [DOI] [PubMed] [Google Scholar]
- Binder BM, O’Malley R C, Wang W, Moore JM, Parks BM, Spalding EP, Bleecker AB. 2004. Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiology 136, 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, et al. 2011. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. The Plant Cell 23, 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. 2003. The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnology Journal 1, 3–22 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis . The Plant Journal 42, 567–585 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, et al. 2013. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis . Elife 2, e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR, 1997. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144 [DOI] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shahak Y, Giller Y, Ratner K, Levine A, Granot D. 1999. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. The Plant Cell 11, 1253–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE. 2002. Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiology 128, 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. 2013. Towards systems biological understanding of leaf senescence. Plant Molecular Biology 82, 519–528 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R, Hill C, Penfold CA, et al. 2013. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. The Plant Journal 75, 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. 2001. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389 [DOI] [PubMed] [Google Scholar]
- John I, Drake R, Farrell A, Cooper W, Lee P, Horton P, Grierson D. 1995. Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants—molecular and physiological analysis. The Plant Journal 7, 483–490 [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. 2006. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis . Proceedings of the National Academy of Sciences, USA 103, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Somers DE. 2010. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiology 154, 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis . Science 323, 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC. 2013. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant and Cell Physiology 54, 1660–1672 [DOI] [PubMed] [Google Scholar]
- Li B, Wang Y, Zhang Z, Wang B, Eneji AE, Duan L, Li Z, Tian X. 2012. Cotton shoot plays a major role in mediating senescence induced by potassium deficiency. Journal of Plant Physiology 169, 327–335 [DOI] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H. 2013. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis . The Plant Cell 25, 3311–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids—pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382 [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136 [DOI] [PubMed] [Google Scholar]
- Liu X, Li Z, Jiang Z, Zhao Y, Peng J, Jin J, Guo H, Luo J. 2011. LSD: a leaf senescence database. Nucleic Acids Research 39, D1103–D1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue GP, Droge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B. 2013. NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis . Molecular Plant 6, 1432–1452 [DOI] [PubMed] [Google Scholar]
- Nam HG. 1997. The molecular genetic analysis of leaf senescence. Current Opinion in Biotechnology 8, 200–207 [DOI] [PubMed] [Google Scholar]
- Nooden LD. 1988. The phenomenon of senescence and aging. In: Nooden LD, Leopold AC, eds Senescence and aging in plants. London: Academic Press, 1–50 [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG. 1996. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Molecular Biology 30, 739–754 [DOI] [PubMed] [Google Scholar]
- Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. 1997. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana . The Plant Journal 12, 527–535 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. 2005. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science 10, 79–87 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM. 2000. Molecular aspects of leaf senescence. Trends in Plant Science 5, 278–282 [DOI] [PubMed] [Google Scholar]
- Raggi V. 1995. CO2 assimilation, respiration and chlorophyll fluorescence in peach leaves infected by Taphrina deformans. Physiologia Plantarum 93, 540–544 [Google Scholar]
- Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B. 2013. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis . The Plant Cell 25, 4941–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JS, Kim JI, Kunkel T, et al. 2005. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120, 395–406 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506 [DOI] [PubMed] [Google Scholar]
- Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S. 2012. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis . The Plant Cell 24, 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Stoddart JL. 1980. Leaf senescence. Annual Review of Plant Physiology and Plant Molecular Biology 31, 83–111 [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R. 2006. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology 141, 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. 1998. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Molecular Biology 37, 455–469 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. 2001. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis . The Plant Cell 13, 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, et al. 2012. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. The Plant Cell 24, 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM. 2011. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. The Plant Cell 23, 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang K, Gan SS. 2012. An abscisic acid–AtNAP transcription factor–SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiology 158, 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dong A, Shen WH. 2012. Histone variants and chromatin assembly in plant abiotic stress responses. Biochimica et Biophysica Acta 1819, 343–348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.