Abstract
Streptococcus suis (S. suis) type 2 is an extremely important Gram-positive bacterial pathogen that can cause human or swine endocarditis, meningitis, bronchopneumonia, arthritis and sepsis. Catabolite control protein A (CcpA) is a major transcriptional regulator in S. suis type 2 that functions in catabolite control, specifically during growth on glucose or galactose. The regulation of central metabolism can affect the virulence of bacteria. In the present study, a metabolomics approach was used along with principal components analysis (PCA) and partial least-squares-discriminant analysis (PLS-DA) models and 37 metabolites were found that differed substantially between native S. suis and a mutant lacking CcpA. These results showed that CcpA is an important protein in S. suis type 2 for studying bacterial protein function.
Keywords: Streptococcus suis type 2, carbon control protein A, metabolomics
Introduction
Streptococcus suis (S. suis) type 2 is an important Gram-positive bacterial pathogen of pigs that causes large economic losses in the meat industry, has a high rate of incidence and induces mortality worldwide (1,2).
Carbon catabolite repression (CCR) has a significant function in S. suis type 2 as it regulates the utilization of different carbon sources. CCR also affects the expression of numerous virulence factors that are dependent on the quantity and type of carbohydrates present (3,4). Catabolite control protein A (CcpA) is the major regulator of CCR in Gram-positive bacteria and is also involved in the regulation of virulence factors (5). Transcriptomic studies that compare native and mutant strains are a powerful approach for studying the regulation of carbohydrate metabolism and virulence (6). A previous study has shown that the mutation of CcpA leads to altered biofilm phenotypes in Streptococcus mutans (S. mutans) and other oral streptococci (7). CcpA was suggested to function in the regulation of virulence in numerous Gram-positive pathogens, particularly streptococci (8,9).
Our previous study systematically analyzed the virulence in S. suis type 2 using proteomics and found that CcpA is an important protein in this organism (10). In the present study, a metabolomics experiments was performed to identify the proteins and metabolites that differed in S. suis type 2 CcpA mutant strains. These results confirmed the importance of this protein in the organism.
Materials and methods
Bacterial strains and culture conditions
Native S. suis type 2 and CcpA mutant strains were cultured in Todd-Hewitt broth (Oxoid Ltd., Basingstoke, Hampshire, UK) with 5% CO2 at 37°C. This procedure was conducted to prepare cultures in the exponential growth phase for the subsequent experiments.
Metabolomics methodology and analysis
Quenching of the S. suis culture for the metabolomics experiments was performed by adding 60% MeOH and 0.85% (w/v) ammonium carbonate (pH 5.5). Centrifugation was performed at 10,000 × g for 2 min. The supernatants were then frozen in liquid nitrogen, freeze-dried and stored at −80°C until they were required for subsequent analysis. The regulation of amino acid metabolism by CcpA was detected using liquid chromatography-mass spectrometry (LC-MS)-based metabolomic methods. The results were analyzed using MassHunter software (version B.03.01) from Agilent Technologies, Inc. (Santa Clara, CA, USA). The principal components analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) were performed with the SIMCA-P software package (Umetrics, Inc., Umea, Sweden). The differentially expressed compounds were distinguished by P<0.05, which was considered to indicate a statistically significant difference (11,12). The metabolomics methods and analysis used in the study were carried out by Shanghai Sensichip Infotech Co., Ltd. (Shanghai, China).
Results
Differences in metabolites between the bacterial strains
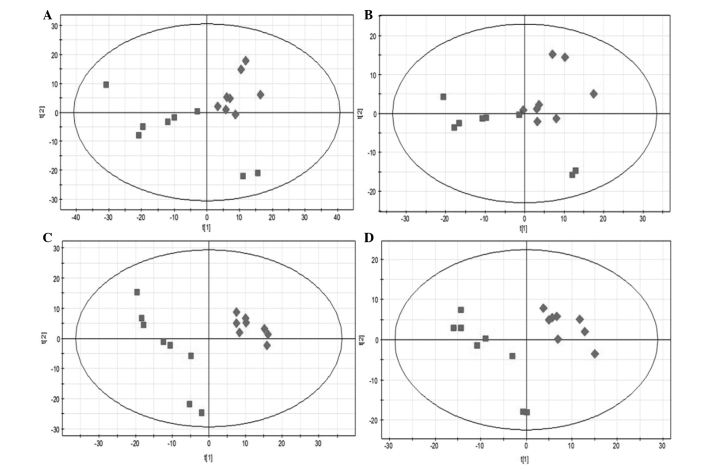
PCA and PLS-DA were used to analyze the metabolomics data. There was little overlap with the PCA model, suggesting this was appropriate for use in the present study. However, it was determined that PLS-DA should be used as the main model for analyzing the metabolomics samples as previously described (13). Based on the PLS-DA models, the native S. suis type 2 and CcpA mutant strains were successfully discriminated, with an R2X value of 0.439 and a Q2 value of 0.924 in the electrospray ionization (ESI)+ model using variable importance in the projection (VIP) of the first and second principal components (t[1] and t[2], respectively). An R2X value of 0.452 and a Q2 value of 0.878 were obtained with the ESI- model (Fig. 1). Using the T-test on the PLS-DA model, 37 differential metabolites were identified with a VIP score >1 (Table I). The model analysis suggested that the main differences were for the metabolites that were involved in amino acid, nucleic acid and fat metabolism, which all occur in the liver.
Figure 1.
(A and B) Principal components analysis (PCA) and (C and D) partial least squares-discriminant analysis (PLS-DA) score plots derived from liquid chromatography-mass spectrometry results. (A and C) Electrospray ionization (ESI)+ model and (B and D) ESI- model. Rhombuses, native Streptococcus suis type 2; squares, catabolite control protein A mutant strain.
Table I.
Metabolite differences between native Streptococcus suis type 2 and CcpA mutant strains.
| LC-MS | Retention time, min | Molecular mass | Metabolite | VIP scorea | Fold changeb |
|---|---|---|---|---|---|
| ESI+ | 0.66 | 103.0631 | γ-aminobutryic acid | 1.98 | 0.58 |
| 0.68 | 165.0456 | S-oxide | 1.55 | 0.29 | |
| 0.68 | 147.0527 | L-glutamate | 1.98 | 0.42 | |
| 0.74 | 159.1260 | DL-2-aminooctanoic acid | 1.28 | −0.45 | |
| 0.74 | 453.2959 | Glycerophospho-N-palmitoyl ethanolamine | 1.83 | −2.38 | |
| 0.82 | 149.0509 | L-methionine | 1.84 | 1.63 | |
| 1.02 | 244.0703 | Uridine | 1.91 | −1.56 | |
| 1.08 | 267.0971 | Deoxyguanosine | 2.29 | 3.20 | |
| 1.16 | 573.1530 | Biotinyl-5′-AMP | 1.74 | −2.28 | |
| 1.17 | 268.0795 | Inosine | 1.67 | −0.98 | |
| 3.72 | 441.2594 | Leukotriene E3 | 1.28 | −0.61 | |
| 10.14 | 255.2567 | Palmitic amide | 1.73 | −0.63 | |
| 10.65 | 329.2575 | 4,8-Dimethylnonanoyl-carnitine | 2.24 | 0.95 | |
| 13.44 | 281.2723 | Oleamide | 1.88 | 0.54 | |
| 13.93 | 160.0887 | Homoglutamine | 1.77 | 0.35 | |
| 15.26 | 384.2865 | 17,20-Dimethyl prostaglandin F1α | 1.84 | 0.76 | |
| 16.07 | 148.0376 | 2-Hydroxyglutarate | 2.21 | 0.88 | |
| ESI− | 0.67 | 133.0378 | L-aspartic acid | 1.76 | 0.32 |
| 0.68 | 147.0535 | L-glutamate | 1.63 | 0.44 | |
| 0.70 | 183.0656 | Phosphocholine | 1.82 | 1.19 | |
| 0.74 | 151.0504 | Guanine | 1.77 | 1.42 | |
| 0.75 | 324.0371 | Uridine monophosphate | 1.95 | 1.89 | |
| 1.00 | 168.0285 | Uric acid | 1.36 | −0.97 | |
| 1.00 | 152.0336 | Xanthine | 1.46 | 0.36 | |
| 1.02 | 244.0707 | Uridine | 1.56 | −0.58 | |
| 1.05 | 192.0272 | Citric acid | 1.64 | −2.85 | |
| 1.16 | 268.0820 | Inosine | 1.67 | −1.86 | |
| 1.17 | 118.0266 | Succinic acid | 1.52 | −0.15 | |
| 1.23 | 131.0948 | L-leucine | 1.47 | −0.82 | |
| 3.80 | 204.0905 | L-tryptophan | 1.53 | −0.25 | |
| 3.93 | 427.2449 | 20-Hydroxy N-arachidonoyl taurine | 1.33 | −0.22 | |
| 4.80 | 643.3191 | Gsh-prostaglandin A1 | 1.86 | −0.32 | |
| 4.93 | 723.3088 | Trypanothione | 1.52 | −0.71 | |
| 5.62 | 187.1212 | 8-Amino-7-oxononanoic acid | 1.35 | 0.68 | |
| 5.83 | 193.0742 | Phenylacetylglycine | 1.61 | −1.14 | |
| 11.72 | 450.1830 | Geranylgeranyl PP | 1.54 | 0.73 | |
| 14.44 | 166.0484 | 7-Methylxanthine | 1.53 | 1.53 |
VIP, variable importance in the projection (P<0.05).
Fold change was calculated as binary logarithm of average mass response ratio between native Streptococcus suis type 2 and CcpA mutant strain, where the positive value means that the native Streptococcus suis type 2 of the metabolite is larger than the CcpA mutant strain.
CcpA, catabolite control protein A; LC-MS, liquid chromatography-mass spectrometry; ESI, electrospray ionization.
Discussion
S. suis type 2 is persistent in a wide variety of environmental conditions and this contributes to the serious economic losses endured by the swine industry when disease outbreaks occur. Another major concern to public health is the elusive mechanisms of invasion and infection, which remain to be clarified (14,15). CcpA-independent CCR appears to be important for the coordination of carbohydrate catabolism in this organism. The ability to integrate information regarding the metabolic status of the cells with a knowledge of the transcriptional responses would be advantageous. The mechanisms of catabolite control by CcpA have been studied in model bacteria previously (6). Transcriptomics and microarray studies have shown that this protein regulates carbohydrate metabolism in S. mutans (6). The deletion of CcpA substantially reduced the effects of repressing sugars on α- or β-glucosidase activity. CcpA regulates several proteins that are involved in metabolism, a number of which are important for bacterial colonization, survival or replication in vivo (16). CcpA is part of a conserved pathway in Gram-positive bacteria that controls sugar utilization, and targeting this protein or pathway is an attractive mechanism for diminishing virulence (17).
CcpA also affects sugar metabolism and virulence in S. suis type 2 and mutant strains lacking this protein exhibit a reduced expression of certain virulence genes, including sly and eno (1). CcpA is considered to regulate virulence factors through the expression of genes in the capsule locus (18). This is reminiscent of a functional study in Streptococcus pneumoniae, which showed that CcpA is a master regulator of catabolism and virulence (19). A notable impact on the capsule synthesis and virulence properties of S. suis has also been shown (20).
The LC-MS-based metabolomics experiments in the present study suggested that CcpA is involved in the regulation of amino acid metabolism, which may affect the metabolic fitness and is in accordance with the findings of other studies (21). The PLS-DA model showed that CcpA acts as a regulator of catabolite control, and has a particular influence on the metabolism of sugars, fats and amino acids.
The experiments also showed that CcpA is an important regulator of metabolism in S. suis type 2. Additionally, the analysis of the metabolomics results identified numerous metabolites, including ABC transporters, amino sugars, carbon metabolism, glycolysis and galactose metabolism. This result indicates that CcpA may be involved in mediating metabolic networks and coordinating the regulation of catabolism and anabolism to ensure optimum growth and propagation under particular growth conditions. Notably, Bacillus subtilis mutants lacking CcpA have a slower growth rate than native bacteria in minimal media containing glucose and ammonium as carbon and nitrogen sources (22,23). This result suggests that CcpA may regulate carbon and nitrogen metabolic pathways and function in connecting catabolism and anabolism.
From the data of the metabolomics analyses, it was found that glutamate, glutamine and other deviants and enzymes have different effects between the native S. suis type 2 and CcpA mutant strains. These, and other genes, were found to have extremely important effects for the methionine acquisition and synthesis of Streptococcus pneumoniae in the growth and virulence when using gene deletions methods (24). By contrast, the methionine aminopeptidase is a dinuclear metalloprotease, which is conserved in all forms of life in bacteria (25). Others, including the glutamine, glutamate and uridine biosynthesis and transport, are stringently controlled during bacteria metabolism (26–28). Thus, these results may provide a clear insight into the regulation of glutamine and glutamate metabolism or another mechanism of S. suis type 2 mediated by CcpA.
In conclusion, the metabolomics analyses performed in the present study proved useful for studying bacterial protein function. CcpA was found to affect numerous metabolic pathways in S. suis. The apparent associations with the metabolism of sugars, amino acids, amino sugars, nucleotides and ABC transporters are potentially noteworthy. However, more studies are required to confirm and understand these phenomena.
Acknowledgements
The metabolomics experiments and analysis were performed by the Shanghai Sensichip Infotech Co., Ltd. (Shanghai, China). The present study was funded by the National Natural Science Foundation of China (grant nos. 31101790/C1803 and 31172340/C180501)
References
- 1.Willenborg J, Fulde M, de Greeff A, Rohde M, Smith HE, Valentin-Weigand P, Goethe R. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology. 2011;157:1823–1833. doi: 10.1099/mic.0.046417-0. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Zhu J, Ren H, Zhu S, Zhao P, Zhang F, Lv H, Hu D, Hao L, Geng M, et al. Rapid visual detection of highly pathogenic Streptococcus suis serotype 2 isolates by use of loop-mediated isothermal amplification. J Clin Microbiol. 2013;51:3250–3256. doi: 10.1128/JCM.01183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev. 2003;67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng L, Chen Z, Itzek A, Ashby M, Kreth J. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J Bacteriol. 2011;193:516–526. doi: 10.1128/JB.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One. 2011;6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leiba J, Hartmann T, Cluzel ME, Cohen-Gonsaud M, Delolme F, Bischoff M, Molle V. A novel mode of regulation of the Staphylococcus aureus catabolite control protein A (CcpA) mediated by Stk1 protein phosphorylation. J Biol Chem. 2012;287:43607–43619. doi: 10.1074/jbc.M112.418913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Dang Y, Wang X, Lu H, Wang X, Lang X, Li X, Feng S, Zhang F, Ren L. Comparative proteomic analyses of Streptococcus suis serotype 2 cell wall-associated proteins. Curr Microbiol. 2011;62:578–588. doi: 10.1007/s00284-010-9747-6. [DOI] [PubMed] [Google Scholar]
- 11.Teng Q, Huang WL, Collette TW, Ekman DR, Tan C. A direct cell quenching method for cell-culture based metabolomics. Metabolomics. 2009;5:199–208. [Google Scholar]
- 12.Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal Bioanal Chem. 2011;401:1899–1904. doi: 10.1007/s00216-011-5245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrola J, Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho L, Duarte IF. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res. 2011;10:221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 14.Berthelot-Hérault F, Gottschalk M, Labbé A, Cariolet R, Kobisch M. Experimental airborne transmission of Streptococcus suis capsular type 2 in pigs. Vet Microbiol. 2001;82:69–80. doi: 10.1016/s0378-1135(01)00376-5. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Fan HJ, Lu CP. Identification and distribution of putative virulent genes in strains of Streptococcus suis serotype 2. Vet Microbiol. 2009;133:309–316. doi: 10.1016/j.vetmic.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y, Wu W, Zhang X, Lu Z, Chen J, Fang W. Catabolite control protein A of Streptococcus suis type 2 contributes to sugar metabolism and virulence. J Microbiol. 2012;50:994–1002. doi: 10.1007/s12275-012-2035-3. [DOI] [PubMed] [Google Scholar]
- 18.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bächi B, Bischoff M. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother. 2006;50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman GE, Yother J. CcpA-dependent and -independent control of beta-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J Bacteriol. 2007;189:5183–5192. doi: 10.1128/JB.00449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang A, Chen B, Yuan Z, Li R, Liu C, Zhou H, Chen H, Jin M. HP0197 contributes to CPS synthesis and the virulence of Streptococcus suis via CcpA. PLoS One. 2012;7:e50987. doi: 10.1371/journal.pone.0050987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Huang C, Zheng D, Wang Y, Yuan Z. CcpA-mediated enhancement of sugar and amino acid metabolism in Lysinibacillus sphaericus by NMR-based metabolomics. J Proteome Res. 2012;11:4654–4661. doi: 10.1021/pr300469v. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig H, Meinken C, Matin A, Stülke J. Insufficient expression of the ilv-lew operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J Bacteriol. 2002;184:5174–5178. doi: 10.1128/JB.184.18.5174-5178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 24.Basavanna S, Chimalapati S, Maqbool A, Rubbo B, Yuste J, Wilson RJ, Hosie A, Ogunniyi AD, Paton JC, Thomas G, Brown JS. The effects of methionine acquisition and synthesis on Streptococcus pneumoniae growth and virulence. PLoS One. 2013;8:e49638. doi: 10.1371/journal.pone.0049638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arya T, Kishor C, Saddanapu V, Reddi R, Addlagatta A. Discovery of a new genetic variant of methionine aminopeptidase from Streptococci with possible post-translational modifications: biochemical and structural characterization. PLoS One. 2013;8:e75207. doi: 10.1371/journal.pone.0075207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendriksen WT, Kloosterman TG, Bootsma HJ, Estevão S, de Groot R, Kuipers OP, Hermans PW. Site-specific contributions of glutamine-dependent regulator GlnR and GlnR-regulated genes to virulence of Streptococcus pneumoniae. Infect Immun. 2008;76:1230–1238. doi: 10.1128/IAI.01004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen PM, Chen YY, Yu SL, Sher S, Lai CH, Chia JS. Role of GlnR in acid-mediated repression of genes encoding proteins involved in glutamine and glutamate metabolism in Streptococcus mutans. Appl Environ Microbiol. 2010;76:2478–2486. doi: 10.1128/AEM.02622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloosterman TG, Hendriksen WT, Bijlsma JJ, Bootsma HJ, van Hijum SA, Kok J, Hermans PW, Kuipers OP. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J Biol Chem. 2006;281:25097–25109. doi: 10.1074/jbc.M601661200. [DOI] [PubMed] [Google Scholar]