Abstract
Murine double minute 2 (MDM2) is a negative regulator of p53. A single-nucleotide polymorphism (SNP) (rs2279744: c.309T>G) in the promoter region of the MDM2 gene has been shown to result in higher levels of MDM2 RNA and protein. Regarding the contribution of c.309T>G in the MDM2 gene to the lung cancer risk, previous studies are conflicting. In order to evaluate the association between c.309T>G and the lung cancer risk, a case-control study was performed. The MDM2 genotypes were determined in 762 lung cancer patients and in 700 cancer-free control subjects using the Smart Amplification Process. Statistical adjustment was performed for gender, age and pack-years of smoking. The distributions of c.309T>G (T/T, T/G, G/G) were 20.1, 49.7, 30.2% in the case group and 21.7, 47.9, 30.4% in the healthy-control group. There were no overall associations between the MDM2 genotypes and the risk of lung cancer [T/G genotype: Adjusted odds ratio (AOR), 1.30; 95% confidence interval (CI), 0.88–1.93; and G/G genotype: AOR, 1.18; 95% CI, 0.78–1.80]. The subgroup analysis of gender, histology, smoking status and epidermal growth factor receptor mutation status also indicated that there was no association with lung cancer. Additionally, the genotypes did not have an effect on the age at the time of diagnosis of lung cancer (P=0.25). In conclusion, the G allele frequency in the lung cancer cases was 0.551, which was similar to other studies. The results of the present study suggest that the c.309T>G is not significantly associated with lung cancer.
Keywords: lung cancer, murine double minute 2, p53, single-nucleotide polymorphism 309, smart amplification process, smoking history, cancer susceptibility
Introduction
Lung cancer is the leading cause of cancer death worldwide (1), and evidence indicates that cigarette smoking is the major established risk factor (2). Additionally, exposure to environmental-chemical carcinogens are other associated risk factors (3). The mechanism of lung tumorigenesis is not fully understood. Epidemiological evidence indicates that complex interactions between numerous genetic and environmental factors are significant in the carcinogenesis of lung carcinoma (4).
The p53 gene is a well-known tumor suppressor gene that encodes a sequence-specific DNA-binding transcription factor that targets various genes that govern specific cellular processes (5). The murine double minute 2 (MDM2) protein plays an important role in regulating cell proliferation and apoptosis by transcriptional inhibition via direct physical binding to p53 and ubiquitination and proteasome-mediated degradation of p53 via its E3 ubiquitin ligase activity (6,7). A MDM2 single nucleotide polymorphism at the 309th nucleotide in the first intron (rs2279744), with a T to G change, could increase the affinity for stimulatory protein 1 (Sp1) binding and result in increased MDM2 expression (8) and subsequent attenuation of the P53 pathway. In humans, the SNP (c.309T>G) in the MDM2 gene has been correlated with the earlier onset of tumor formation in hereditary and sporadic cancers (9). A previous meta-analysis reported that the G/G genotype of c.309T>G was associated with a significantly increased risk for lung, colorectal, gastric and bladder cancers and a significantly decreased risk for prostate cancer (10). Molecular epidemiological studies of c.309T>G and lung cancer susceptibility (11–21) and survival rate (22–24) have reported disparate findings. It is noteworthy that there is a large difference among African-American, Caucasian and Asian populations with respect to the 309G allele frequency in lung cancer cases (25). The 309G allele is present at a higher frequency in the Asian population, including Japan, compared to Caucasians and African-Americans. Significant associations have been observed for c.309T>G in Japanese squamous cell lung cancer patients (20). Future studies analyzing the ethnic group-dependent susceptibility to cancer are required. Several genome-wide association studies (GWASs) on SNPs have identified the genomic loci associated with the risk of lung cancer in Caucasians (26–28), Europeans (29) and Asians (30–34). However, the associations of the MDM2 polymorphisms were not investigated in the aforementioned GWASs due to a lack of probes used to discriminate between the polymorphisms (20).
The aim of the present study was to investigate whether a functionally important SNP, MDM2 SNP309, was associated with the risk of lung cancer in a Japanese population. The genomic DNA was examined from blood samples for c.309T>G using the Smart Amplification Process (SmartAmp), which is a rapid, sensitive and simple mutation-detection assay that has been described previously (35,36). Subsequently, the association between c.309T>G and the lung cancer risk was examined. To the best of our knowledge, this is the first study to investigate the role of c.309T>G in the MDM2 gene in all the types of lung cancer in a Japanese population.
Materials and methods
Subjects
The study included 762 patients with lung cancer and 700 cancer-free controls. All the cases were histologically confirmed primary lung cancer patients who were diagnosed and recruited between January 2003 and June 2013 at Gunma University Hospital, Maebashi Red Cross Hospital and National Nishi-Gunma Hospital (Gunma, Japan). The control group consisted of hospital and population controls. The hospital controls were recruited from the same hospital as the patients. The population controls were recruited from a health survey for the evaluation of metabolic syndrome in Gunma. The potential controls that had a previous diagnosis of malignancy were excluded from the study. The protocols for sample collection, sample anonymity, storage and genomic DNA analysis were approved by the Institutional Review Board for the Ethics Committee for Human Genome Analysis at each hospital. Written informed consent from all the participants was obtained. The clinical study was conducted according to the Declaration of Helsinki Principles.
Patient demographics
The demographics, cancer history and smoking history of the subjects were documented using a structured chart review. The patients were categorized based on the smoking status. The cumulative cigarette dose [pack-years (PY)] was calculated using the following formula: PY = packs per day × years smoked. Never-smokers were defined as individuals with PY=0, mild smokers were PY<50 and heavy smokers were PY≥50.
Genotyping for polymorphisms and gene mutation analysis
Peripheral venous blood samples were collected and DNA was extracted using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Genotyping of c.309T>G in the MDM2 gene was performed by the Duplex SmartAmp method as described previously (36) with an Mx3000P PCR system (Agilent Technologies, Santa Clara, CA, USA). The lung cancer tissues were immediately frozen following surgical removal and stored at −80°C until DNA extraction using a Wizard Genomic DNA purification kit (Promega Corporation, Madison, WI, USA). The epidermal growth factor receptor (EGFR) mutations were analyzed by peptide nucleic acid-enriched sequencing, as described previously (37).
Statistical analyses
The demographic and clinical information were compared across genotypes and cancer stages using Pearson’s χ2 tests (for categorical variables) and Kruskal-Wallis tests (for continuous variables) where appropriate. The strengths of the associations between the genotypes and risk were measured as odds ratios (ORs) adjusted for gender, age and PY of smoking with 95% confidence intervals (CIs) by unconditional logistic regression analysis. All the statistical analyses were conducted at the two-sided P=0.05 level and SPSS Statistics version 20 (SPSS, Inc., Chicago, IL, USA) was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Demographics
A total of 762 primary lung cancer patients and 700 controls were included in the analysis and Table I shows the associated demographics. Compared to the controls, the patients were older with a higher proportion of males. The mean PY value was higher in the cases compared to the controls (Table I). Complete matching was not achieved for gender, age and smoking history. Therefore, these factors were adjusted in the logistic model. The frequency of c.309T>G did not differ between the hospital and population controls (P=0.74; data not shown). Therefore, the controls were treated as a single group. The distributions of c.309T>G (T/T, T/G, G/G) were 20.1, 49.7, 30.2% in the patient group and 21.7, 47.9, 30.4% in the healthy-control group. There were no significant differences in the c.309T>G genotypes between cases and controls. The allele frequencies of the G allele in the case and control groups were 0.551 and 0.544, respectively.
Table I.
Characteristics of lung cancer cases and healthy controls.
| Cases (n=762) | Controls (n=700) | ||
|---|---|---|---|
|
|
|
||
| Variable | n (%) | n (%) | P-value |
| Gender | <0.001a | ||
| Male | 472 (61.9) | 326 (46.6) | |
| Female | 290 (38.1) | 374 (53.4) | |
| Age, years | <0.001b | ||
| Mean ± SD | 68.4±9.8 | 64.1±12.7 | |
| Range | 31–95 | 20–93 | |
| MDM2 polymorphism | 0.69a | ||
| T/T | 153 (20.1) | 152 (21.7) | |
| T/G | 379 (49.7) | 335 (47.9) | |
| G/G | 230 (30.2) | 213 (30.4) | |
| G allele frequency | 0.551 | 0.544 | |
| PY | |||
| Mean ± SD | 36.4±37.2 | 26.6±32.7 | 0.001b |
| Range | 0–265 | 0–184 | |
| Histology | |||
| AD | 487 (63.9) | ||
| SQ | 182 (23.9) | ||
| Others | 82 (10.8) | ||
| Un-differentiated | 11 (1.4) | ||
Pearson’s χ2 test.
Kruskal-Wallis test.
SD, standard deviation; MDM2, murine double minute 2; PY, pack-years; AD, adenocarcinoma; SQ, squamous cell carcinoma.
MDM2 and lung cancer risk
There was no overall association between c.309T>G and the risk of lung cancer (Table II). The adjusted OR for developing lung cancer was 1.18 (95% CI, 0.76–1.80) for the G/G, compared to the T/T. The data was further stratified by gender, cancer histology, EGFR-mutation status of adenocarcinoma (AD) and smoking-status subgroups. There were no associations among any subgroup (Table II).
Table II.
Genotype frequencies of the MDM2 309T>G polymorphism among controls and cases and their association with lung cancer risk.
| Adjusted OR (95% CI)a | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Variable | No. | T/T (%) | T/G (%) | G/G (%) | T/T vs. G/G | T/T vs. T/G+G/G | T/T+T/G vs. G/G |
| Control | 700 | 152 (21.7) | 335 (47.9) | 213 (30.4) | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) |
| Case | 762 | 153 (20.1) | 379 (49.7) | 230 (30.2) | 1.18 (0.76–1.80) | 1.25 (0.87–1.81) | 0.98 (0.71–1.37) |
| Femaleb | 290 | 53 (18.3) | 143 (49.3) | 94 (32.4) | 1.31 (0.91–1.87) | 1.51 (0.83–2.75) | 1.34 (0.76–2.34) |
| Maleb | 472 | 100 (21.2) | 236 (50.0) | 136 (28.8) | 0.94 (0.55–1.60) | 1.13 (0.72–1.79) | 0.83 (0.55–1.24) |
| AD | 487 | 101 (20.7) | 233 (47.8) | 153 (31.4) | 1.05 (0.84–1.31) | 1.14 (0.77–1.67) | 1.02 (0.72–1.44) |
| SQ | 182 | 35 (19.2) | 98 (53.8) | 49 (26.9) | 1.06 (0.77–1.46) | 1.42 (0.80–2.49) | 0.87 (0.52–1.43) |
| EGFR-WT-AD | 207 | 41 (19.8) | 102 (49.3) | 64 (30.9) | 1.09 (0.83–1.42) | 1.28 (0.80–2.05) | 1.01 (0.67–1.52) |
| EGFR-Mt-AD | 175 | 39 (22.3) | 78 (44.6) | 58 (33.1) | 1.00 (0.75–1.35) | 0.97 (0.58–1.61) | 1.04 (0.66–1.64) |
| Never-smoker | 251 | 43 (17.1) | 129 (51.4) | 79 (31.5) | 1.07 (0.75–1.55) | 1.46 (0.78–2.73) | 0.89 (0.52–1.54) |
| Light smoker | 258 | 49 (19.0) | 127 (49.2) | 82 (31.8) | 1.09 (0.85–1.41) | 1.30 (0.83–2.03) | 1.00 (0.68–1.49) |
| Heavy smoker | 246 | 57 (23.2) | 120 (48.8) | 69 (28.0) | 1.00 (0.71–1.41) | 1.07 (0.60–1.92) | 0.95 (0.55–1.64) |
Adjusted for gender, age and smoking status.
Adjusted for age and smoking status.
MDM2, murine double minute 2; OR, odds ratio; CI, confidence interval; AD, adenocarcinoma; SQ, squamous cell carcinoma; EGFR, epidermal growth factor receptor; WT, wild-type; Mt; mutant.
MDM2 and cigarette smoking
As smoking is a stressor and an established cause of lung cancer, we evaluated whether an interaction existed between the MDM2 polymorphism and smoking among lung cancer patients. No significant associations were observed between the various c.309T>G genotypes and cumulative smoking. (Table III). However, the G/G genotype tended to be associated with a lower level of exposure to cigarette smoke compared to the other genotype (P=0.07) among squamous cell carcinoma (SQ) patients (Table III).
Table III.
Comparison by cumulative smoking.
| T/T | T/G | G/G | |||
|---|---|---|---|---|---|
|
|
|
|
|||
| Variable | n | Mean ± SD | Mean ± SD | Mean ± SD | P-valuea (T/T+T/G vs. G/G) |
| Total | 755 | 39.9±36.5 | 36.5±38.7 | 34.1±35.0 | 0.35 |
| Female | 286 | 8.2±18.7 | 8.2±18.6 | 7.6±18.4 | 0.88 |
| Male | 469 | 55.9±32.6 | 53.6±37.6 | 52.5±31.9 | 0.70 |
| AD | 482 | 26.3±30.1 | 20.6±28.8 | 23.1±31.4 | 0.99 |
| SQ | 182 | 70.8±33.9 | 62.0±32.9 | 57.2±34.5 | 0.07 |
| SCLC | 52 | 58.8±34.9 | 67.5±56.7 | 59.2±24.8 | 0.93 |
Kruskal-Wallis test.
SD, standard deviation; AD, adenocarcinoma; SQ, squamous cell carcinoma; SCLC, small cell lung cancer.
MDM2 and age at diagnosis
The association between age at diagnosis and c.309T>G in all the lung cancer patients was examined, as well as in the gender, histological-subtype and smoking-status subgroups. Kruskal-Wallis analysis indicated no association between the various c.309T>G genotypes and age at onset in the study population (Table IV).
Table IV.
Comparison by age at diagnosis.
| T/T | T/G | G/G | |||
|---|---|---|---|---|---|
|
|
|
|
|||
| Variable | n | Mean ± SD | Mean ± SD | Mean ± SD | P-valuea (T/T+T/G vs. G/G) |
| Total | 761 | 67.4±9.6 | 68.9±9.7 | 68.2±10.0 | 0.54 |
| Female | 289 | 66.3±9.7 | 67.2±10.2 | 66.7±10.4 | 0.82 |
| Male | 472 | 68.0±9.6 | 69.9±9.3 | 69.2±9.5 | 0.69 |
| AD | 487 | 66.7±10.3 | 67.2±10.3 | 66.9±10.2 | 0.67 |
| SQ | 182 | 71.0±6.3 | 72.8±7.9 | 72.0±9.1 | 0.86 |
| SCLC | 52 | 64.9±9.9 | 69.3±7.8 | 70.8±8.9 | 0.27 |
| Smoker | 507 | 68.3±9.5 | 69.7±9.7 | 69.0±10.0 | 0.60 |
| Never smoker | 251 | 65.6±9.8 | 67.4±9.6 | 66.7±9.9 | 0.82 |
Kruskal-Wallis test.
SD, standard deviation; AD, adenocarcinoma; SQ, squamous cell carcinoma; SCLC, small cell lung cancer.
Discussion
In the present molecular epidemiological study, the association of the genetic polymorphisms in MDM2 with the risk of developing lung cancer in a Japanese population was examined. The results obtained by analyzing 762 lung cancer patients and 700 controls demonstrated that c.309T>G in the MDM2 gene is not associated with lung cancer risk.
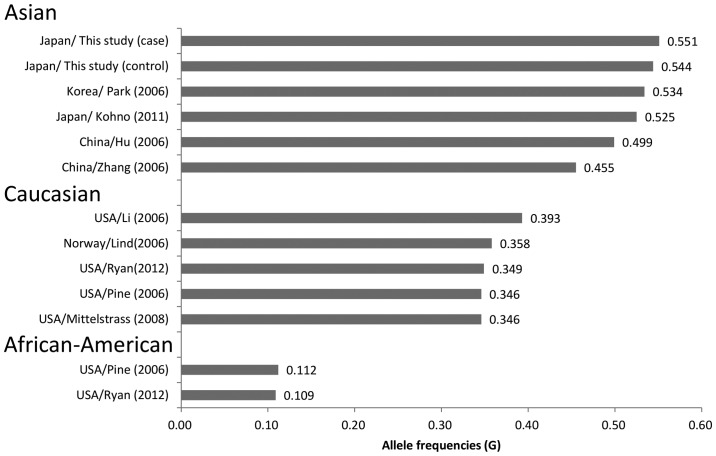
In the study, the frequency of the MDM2 309G alleles among the controls was 0.544, which is almost identical to the frequency observed in the Japanese (0.525) (20), Han Chinese of southeast China (0.499) (14) and Korean (0.534) (16) populations; but is significantly higher compared to the Han Chinese of northeast China (0.455) (15), Caucasian (0.346–0.393) (11–13,18,38) and African-American (0.109–0.112) (13,38) populations (Fig. 1). There are conflicting studies regarding the contribution of c.309T>G to the lung cancer risk (11–19,21). The differences in the allele frequencies among ethnic groups may have contributed to the disparities among the previous studies.
Figure 1.
Comparison of the allele frequencies of the 309 G allele.
Certain studies have linked the G allele with the lung cancer risk among Caucasians (11) and Asians (15). Conversely, only one study linked the 309T allele with the risk in Caucasians, notably in SQ patients and in males (12). The data of the present study suggests that c.309T>G is not associated with the lung cancer risk among the total population, which is in line with numerous epidemiological studies conducted in Caucasians (13,17,18), African-Americans (13) and Asians (14,16,20). A previous meta-analysis study (39) concluded that the association between c.309T>G and lung cancer was statistically significant in Asians, females and never-smokers, but not in Caucasians, males, ever-smokers or among any of the individual histological types.
Stratification by gender, histological subtype and smoking status revealed no association between c.309T>G and lung cancer susceptibility. Lind et al (11) reported a gender-specific risk-disposing effect of the 309T allele of MDM2 SNP309 for non-small cell lung cancer (NSCLC) patients. NSCLC patients were investigated and it was found that female carriers of G/G had an OR for lung cancer of 4.1, whereas male homozygotes had a non-significant OR of 1.3. The data of the present study showed corresponding ORs for females and males of 1.31 and 0.94, respectively, which tended to confirm this finding, although the ORs were much smaller and not significant. Bond et al (40) showed that the effect of the MDM2 G/G genotype was gender-specific and was increased in females with active estrogen-signaling pathways, which may explain the findings of the present study.
In terms of histological subtypes, Park et al (16) and Ren et al (21) reported that the T/T genotype was associated with a decreased risk of AD. However, the present study findings, as well as those of others (18,41), did not show an interaction between c.309T>G and the histological subtype.
Cigarette smoking may induce DNA damage, initiating and promoting carcinogenesis, and is a major risk factor for NSCLC. Park et al (16) and Zhang et al (15) reported that the T/T genotype was associated with a decreased risk among ever-smokers and ever- and never-smokers, respectively. However, the present study, as well as others (18,41), did not show an interaction between c.309T>G in the MDM2 gene and smoking status. A tendency towards a lower level of exposure to cigarette smoking was observed among the G/G SQ patients compared with T allele carriers (T/T+T/G), but the difference was not statistically significant. However, no significant association between c.309T>G and the lung cancer risk among smokers or SQ patients was observed. Therefore, it was concluded that SNP309 did not contribute to smoking-related tumors.
c.309T>G was reported to be associated with an earlier onset of disease for Li-Fraumeni syndrome and sporadic sarcoma (8). The present study did not observe a lower age of diagnosis, and similar findings were described in previous studies (13,14,18).
Although the reason for the differences in MDM2 polymorphisms among different studies is unclear, it may be due to differences in subjects, genetic backgrounds and/or environmental factors (42) among various populations. As an example of population heterogeneity, an interaction of c.309T>G with c.285G>C in the MDM2 gene was questionable (43), as it has been previously reported to act as an antagonist by overriding the effect of c.309T>G on Sp1-mediated transcription. However, the study by Ryan et al (38) recently reported that c.285G>C did not antagonize the effect of c.309T>G in lung cancer. Furthermore, the differences in frequencies of driver mutations, such as EGFR mutations (44), may contribute to the different effects of c.309T>G in various ethnic groups.
EGFR mutations are predominantly found in female, non-smoking AD patients and in patients of East Asian origin (45). A recent meta-analysis study (39) reported that the association between c.309T>G and lung cancer was statistically significant, particularly in the Asian population, female and never-smokers. No statistically significant association was observed in males or ever-smokers. The AD patients were stratified according to EGFR-mutation status, however, no association was observed. To the best of our knowledge, there have been no report in terms of analyzing the association between c.309T>G in the MDM2 gene and lung cancer susceptibility based on EGFR mutation status.
The limitations of the study include the retrospective nature, and the patient populations may be biased. Furthermore, the study was a hospital- and community-based case-control study, which could not rule out possible selection bias. However, by matching age, gender and PY of smoking, the potential confounding effects should have been reduced. To further elucidate the impact of c.309T>G on lung cancer susceptibility, future investigations of large ethnically-diverse population-based studies are warranted.
In conclusion, the present study is the first to evaluate the overall lung cancer risk and c.309T>G in a Japanese population. Considering the contradictory results across multiple studies, conclusions from any single study must be interpreted with caution. The results of the present study should be confirmed by other prospective studies.
Acknowledgements
The present study was supported by Grant-in-Aid for Scientific Research (C) (grant no. 22590516) from the Japan Society for the Promotion of Science. The study was partially supported by i) research grants for the RIKEN Omics Science Center from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) to Y.H.; and ii) a research grant from the MEXT to the RIKEN Center for Life Science Technologies. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Abbreviations
- SNP
single-nucleotide polymorphism
- OR
odds ratio
- CI
confidence interval
- PY
pack-year
- AD
adenocarcinoma
- SQ
squamous cell carcinoma
- NSCLC
non-small cell lung cancer
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Steliga MA, Dresler CM. Epidemiology of lung cancer: smoking, secondhand smoke, and genetics. Surg Oncol Clin N Am. 2011;20:605–618. doi: 10.1016/j.soc.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Fucic A, Gamulin M, Ferencic Z, et al. Lung cancer and environmental chemical exposure: a review of our current state of knowledge with reference to the role of hormones and hormone receptors as an increased risk factor for developing lung cancer in man. Toxicol Pathol. 2010;38:849–855. doi: 10.1177/0192623310378136. [DOI] [PubMed] [Google Scholar]
- 4.Yokota J, Shiraishi K, Kohno T. Genetic basis for susceptibility to lung cancer: Recent progress and future directions. Adv Cancer Res. 2010;109:51–72. doi: 10.1016/B978-0-12-380890-5.00002-8. [DOI] [PubMed] [Google Scholar]
- 5.Jin S, Levine AJ. The p53 functional circuit. J Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 6.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 7.Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 9.Bond GL, Hu W, Levine A. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 2005;65:5481–5484. doi: 10.1158/0008-5472.CAN-05-0825. [DOI] [PubMed] [Google Scholar]
- 10.Wo X, Han D, Sun H, et al. MDM2 SNP309 contributes to tumor susceptibility: a meta-analysis. J Genet Genomics. 2011;38:341–350. doi: 10.1016/j.jgg.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Lind H, Zienolddiny S, Ekstrøm PO, Skaug V, Haugen A. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer. 2006;119:718–721. doi: 10.1002/ijc.21872. [DOI] [PubMed] [Google Scholar]
- 12.Li G, Zhai X, Zhang Z, Chamberlain RM, Spitz MR, Wei Q. MDM2 gene promoter polymorphisms and risk of lung cancer: a case-control analysis. Carcinogenesis. 2006;27:2028–2033. doi: 10.1093/carcin/bgl047. [DOI] [PubMed] [Google Scholar]
- 13.Pine SR, Mechanic LE, Bowman ED, et al. MDM2 SNP309 and SNP354 are not associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:1559–1561. doi: 10.1158/1055-9965.EPI-06-0217. [DOI] [PubMed] [Google Scholar]
- 14.Hu Z, Ma H, Lu D, et al. Genetic variants in the MDM2 promoter and lung cancer risk in a Chinese population. Int J Cancer. 2006;118:1275–1278. doi: 10.1002/ijc.21463. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Miao X, Guo Y, et al. Genetic polymorphisms in cell cycle regulatory genes MDM2 and TP53 are associated with susceptibility to lung cancer. Hum Mutat. 2006;27:110–117. doi: 10.1002/humu.20277. [DOI] [PubMed] [Google Scholar]
- 16.Park SH, Choi JE, Kim EJ, et al. MDM2 309T>G polymorphism and risk of lung cancer in a Korean population. Lung Cancer. 2006;54:19–24. doi: 10.1016/j.lungcan.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Wheatley-Price P, Zhou W, et al. Genetic polymorphisms of MDM2, cumulative cigarette smoking and nonsmall cell lung cancer risk. Int J Cancer. 2008;122:915–918. doi: 10.1002/ijc.23178. [DOI] [PubMed] [Google Scholar]
- 18.Mittelstrass K, Sauter W, Rosenberger A, et al. Early onset lung cancer, cigarette smoking and the SNP309 of the murine double minute-2 (MDM2) gene. BMC Cancer. 2008;8:113. doi: 10.1186/1471-2407-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chua HW, Ng D, Choo S, et al. Effect of MDM2 SNP309 and p53 codon 72 polymorphisms on lung cancer risk and survival among non-smoking Chinese women in Singapore. BMC Cancer. 2010;10:88. doi: 10.1186/1471-2407-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohno T, Kunitoh H, Mimaki S, et al. Contribution of the TP53, OGG1, CHRNA3, and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. J Thorac Oncol. 2011;6:813–817. doi: 10.1097/JTO.0b013e3181ee80ef. [DOI] [PubMed] [Google Scholar]
- 21.Ren YW, Yin ZH, Wan Y, et al. P53 Arg72Pro and MDM2 SNP309 polymorphisms cooperate to increase lung adenocarcinoma risk in Chinese female non-smokers: a case control study. Asian Pac J Cancer Prev. 2013;14:5415–5420. doi: 10.7314/apjcp.2013.14.9.5415. [DOI] [PubMed] [Google Scholar]
- 22.Heist RS, Zhou W, Chirieac LR, et al. MDM2 polymorphism, survival, and histology in early-stage non-small-cell lung cancer. J Clin Oncol. 2007;25:2243–2247. doi: 10.1200/JCO.2006.08.8914. [DOI] [PubMed] [Google Scholar]
- 23.Han JY, Lee GK, Jang DH, Lee SY, Lee JS. Association of p53 codon 72 polymorphism and MDM2 SNP309 with clinical outcome of advanced nonsmall cell lung cancer. Cancer. 2008;113:799–807. doi: 10.1002/cncr.23668. [DOI] [PubMed] [Google Scholar]
- 24.Dong J, Ren B, Hu Z, et al. MDM2 SNP309 contributes to non-small cell lung cancer survival in Chinese. Mol Carcinog. 2011;50:433–438. doi: 10.1002/mc.20727. [DOI] [PubMed] [Google Scholar]
- 25.Zhuo W, Zhang L, Zhu B, Ling J, Chen Z. Association of MDM2 SNP309 variation with lung cancer risk: evidence from 7196 cases and 8456 controls. PLoS One. 2012;7:e41546. doi: 10.1371/journal.pone.0041546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J, Chatterjee N, Rotunno M, et al. Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2012;2:131–139. doi: 10.1158/2159-8290.CD-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosgood HD, III, Wang WC, Hong YC, et al. Genetic variant in TP63 on locus 3q28 is associated with risk of lung adenocarcinoma among never-smoking females in Asia. Hum Genet. 2012;131:1197–1203. doi: 10.1007/s00439-012-1144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn MJ, Won HH, Lee J, et al. The 18p11.22 locus is associated with never smoker non-small cell lung cancer susceptibility in Korean populations. Hum Genet. 2012;131:365–372. doi: 10.1007/s00439-011-1080-z. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z, Wu C, Shi Y, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 33.Shiraishi K, Kunitoh H, Daigo Y, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–903. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 34.Lan Q, Hsiung CA, Matsuo K, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44:1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitani Y, Lezhava A, Kawai Y, et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 36.Enokida Y, Shimizu K, Atsumi J, et al. Rapid detection of SNP (c.309T>G) in the MDM2 gene by the Duplex SmartAmp method. PLoS One. 2013;8:e60151. doi: 10.1371/journal.pone.0060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyamae Y, Shimizu K, Mitani Y, et al. Mutation detection of epidermal growth factor receptor and KRAS genes using the smart amplification process version 2 from formalin-fixed, paraffin-embedded lung cancer tissue. J Mol Diagn. 2010;12:257–264. doi: 10.2353/jmoldx.2010.090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan BM, Calhoun KM, Pine SR, et al. MDM2 SNP285 does not antagonize the effect of SNP309 in lung cancer. Int J Cancer. 2012;131:2710–2716. doi: 10.1002/ijc.27573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He W, Long J, Xian L, et al. MDM2 SNP309 polymorphism is associated with lung cancer risk in women: A meta-analysis using METAGEN. Exp Ther Med. 2012;4:569–576. doi: 10.3892/etm.2012.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bond GL, Hirshfield KM, Kirchhoff T, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 41.Liu JN, Zhang XM, Guo YL, Sun T, Lin DX, Wen T. Genetic polymorphism in MDM2 is associated with susceptibility to colorectal cancer in a Chinese population. Zhonghua Zhong Liu Za Zhi. 2008;30:335–338. (In Chinese) [PubMed] [Google Scholar]
- 42.Alberg AJ, Ford JG, Samet JM American College of Chest Physicians. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132(Suppl 3):29S–55S. doi: 10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 43.Knappskog S, Bjørnslett M, Myklebust LM, et al. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell. 2011;19:273–282. doi: 10.1016/j.ccr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Nana-Sinkam SP, Powell CA. Molecular biology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e30S–e39S. doi: 10.1378/chest.12-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]