Abstract
SOX genes are developmental regulators with functions in the instruction of cell fate and maintenance of progenitor’s identity during embryogenesis. They play additional roles during tissue homeostasis and regeneration in adults particularly in the Central Nervous System (CNS). In the last years a growing number of evidences has shown that mutations and dysfunction of SOX factors are implicated in several human diseases, including a variety of cancers. In this review, we will summarize the current knowledge about SOX family in CNS tumors and their role in the origin and maintenance of the subpopulation of cancer stem cells in these tumors.
Keywords: SOX, CNS tumors, glioblastoma, glioma stem cell, cell of origin, oncogenic SOX2, therapy target
SOX family introduction
SOX (Sex-determining region Y (SRY)-box protein) family members are characterized by a conserved high mobility group (HMG) DNA-binding domain [1]. There are, at least, 20 members divided into 8 groups (from A to H), based on their HMG sequence identity in humans [2]. Members within a group preserve higher than 80% identity in their HMG-domain and share other well-conserved regions [3]. In addition, they share biochemical properties, have overlapping expression patterns and perform synergistic or redundant functions. In contrast, members from different groups usually perform different functions. SOX genes are developmental regulators with functions in sex determination, chondrogenesis, hematopoiesis, neural crest development and neurogenesis [4]. SOXB1, SOXB2, and SOXE members have a role in the instruction of cell fate and maintenance of progenitor’s identity during embryogenesis. They are also important for stem cell maintenance and play additional roles during tissue homeostasis and regeneration in adults particularly in the CNS [5]. In the last years a growing number of evidences have shown that mutations and dysfunction of SOX factors are implicated in several human diseases, including a variety of cancers [6]. These diseases are originated in tissues overlapping with their expression pattern during embryonic development. Since SOX factors play an integral role in the maintenance of neural stem cells and in the specification and differentiation of neurons, astrocytes and oligodendrocytes, it seems reasonable to surmise that aberrant expression of members of this family is implicated in the development and maintenance of CNS tumors.
Central nervous system tumors
Tumors of the CNS consist of a heterogeneous group of neoplasias accounting for around 3% of the total number but representing 7% of deaths caused by cancer. Every year in the world, approximately 350.000 people are diagnosed with gliomas, making it the most common primary brain tumor (IARC http://globocan.iarc.fr, accessed on day/month/year). Gliomas display histological similarities to glial cells and according to which cell they most resemble, the World Health Organization (WHO) classifies them into astrocytoma, oligodendroglioma, ependymoma or mixed oligoastrocytoma. This classification is based solely on morphology. Based on histopathological and clinical criteria they are classified into four classes of malignancy [7].
Glioblastoma multiforme (GBM) belongs to grade IV and accounts for 80% of the total primary malignant brain tumors in adults. The incidence ranges from 5 to 20 cases per 100,000 people per year [8] with an associated median survival of 15 months [9]. This survival identifies GBM as one of the most aggressive and fatal cancer overall.
The clinical hallmarks of GBM are its aggressive growth and inexorable recurrence as a consequence of the resistance to apoptosis, genomic instability and poor response to therapy [10]. It is also characterized for the presence of necrotic areas, for being highly invasive, infiltrative and with intense angiogenesis. In the last years different GBM characterizations have emerged based on the molecular knowledge of the genome [11-14] and transcriptome [15,16]. These studies have provided a high-resolution picture of the GBM landscape uncovering the major structural genomic and expression alterations that may drive disease pathogenesis and biology. These comprehensive data sets reveal GBM as a heterogeneous collection of distinct diseases with multiple dependencies both within and across each particular subtype. Contributing to this grim picture is the fact that despite a huge effort to understand this disease and to develop effective therapies over the past few decades, there are still no such agents [17].
CNS tumors constitute the largest group of solid neoplasms of childhood and the ones that cause the highest mortality rates in this group age [18]. Because the developing brain is highly vulnerable to treatment-induced cognitive and endocrine sequelae, particularly from radiotherapy, ongoing studies are focusing on improving the duration and the quality of survival, in affected patients [18]. Medulloblastoma (MB) is the most common pediatric brain cancer and the treatment of patients with this disease poses an additional problem. Current therapies for MB cause dramatic impairment of cognitive function and predispose patients to future treatment-associated neoplasms [19]. Pediatric high-grade gliomas (pHGG) including diffuse intrinsic pontine glioma (DIPGs) comprise 15% to 20% of all childhood tumors of the CNS, and more than 70% of patients die within 2 years of diagnosis. Consequently, understanding the molecular circuitries underlying the development of pHGG is crucial to identify relevant therapeutic targets [20] for these neoplasms.
The cellular origin of CNS tumors
For many years, cancer has been based on a stochastic model, which considers that all cells within the tumor are highly proliferative, possess tumorigenic potential and are capable of tumor progression and repopulation. In the last decade, the demonstration that cancers are heterogeneous masses containing a hierarchy of cells has modified the original model [21]. This new theory postulates that a small subpopulation of cancer cells (called cancer stem cells, CSCs) drives tumor formation, growth, metastasis and resistance to therapeutic treatments [22]. There is compelling evidence in support of its existence in hematological malignancies and in numerous solid epithelial types of cancer including GBM and medulloblastoma [23]. In the brain, the CSCs population display Neural Stem Cells (NSCs) characteristics; unlimited proliferation, self-renewal potential and multipotency to differentiate into astrocytes, oligodendrocytes and neurons (Figure 1). Furthermore, these cells form tumors phenotypically similar to the original human ones when injected into the brain of immunodeficient mice, indicative of being responsible for the initiation and maintenance of adult and pediatric brain tumors [24-26]. CSCs display much greater tumorigenic potential than matched non-stem tumor cells, have the ability to migrate and are more resistant to apoptosis and therapy suggesting that these cells are also responsible for tumor relapses particularly in GBM cases [27].
Figure 1.
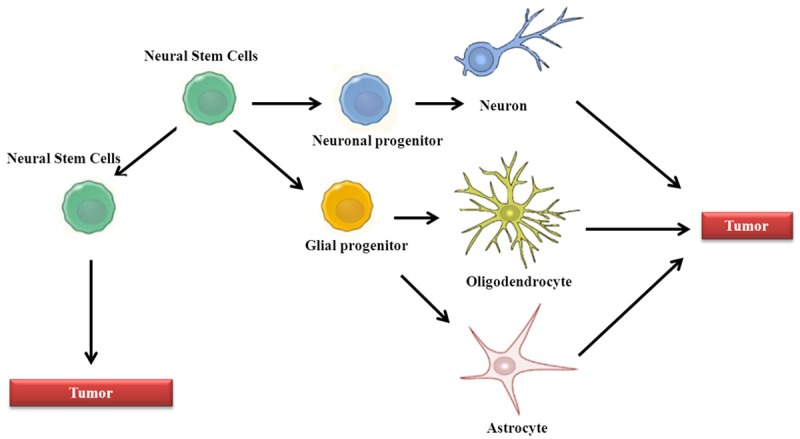
Glioma cell of origin and glioma stem cell (GSC) evolution. Normal cellular hierarchy comprises neural stem cells that progressively produce new stem cells and more restricted progenitor cells, finally yielding oligodendrocytes, astrocytes and neurons. Accumulation of genetic mutations in different cell types is sufficient to induce gliomas. These gliomas contain a population of GSCs with self-renewal capacity and ability to differentiate to all the lineages.
The demonstration of functional neurogenesis in the adult brain [28,29], the observation of a population with stem cells properties within the tumor bulk, opened the debate regarding the putative cell of origin of CNS tumors and postulated NSCs as the putative cell of origin for CNS tumors, mainly gliomas. Indeed, different genetic models have revealed that inactivation of p53/Rb/Pten/Nf1 tumor suppressors or enhancement in EGFR/PDGFR/PI3K oncogenic pathways in NSCs serve as glioma source [30,31]. Intriguingly, different studies have demonstrated that targeting the same pathways in astrocytes, oligodendrocyte progenitors and neurons is sufficient to undergo oncogenic transformation and form malignant gliomas [32-34]. These evidences strongly indicate that glioma cell of origin is diverse probably explaining the complex and heterogeneous pathology, morphology and clinic that characterize this type of tumor (Figure 1).
The concept of CSC may have profound implications from the point of view of therapy in that expose this population as a crucial target [35]. Therefore dysregulation of pathways controlling normal NSCs could constitute a requirement for cancer development and might play predominant roles in CSCs too. For example Notch is required from the transition from primitive to definitive NSC and their maintenance. Aberration in Notch pathway results in tumor formation and its expression is deregulated in GBM [36,37]. Sonic Hedgehog (SHH) pathway is required for neural stem/progenitor cell maintenance promoting their proliferation and self-renewal [38] and this pathway is also found deregulated in GBM [39]. Furthermore, a recent elegant study has demonstrated how developmental and regional differences influence neoplastic transformation in the CNS [40]. Thus, an active N-Myc mutant (T58A) generates medulloblastoma/primitive neuroectodermal tumors when transduced in cerebellar and brain stem NSCs, whereas develops diffuse glioma in forebrain NSCs. Tumors generated from diverse regions display different gene expression pattern including SHH dependence and independence within tumors from embryonic versus postnatal cerebellar NSCs [40]. Sox family members are critical transcription regulators of embryonic and neural stem cells, which are aberrantly expressed in several human cancers including GBMs. We plan to discuss, further down, their role in the development of the CNS and the implication of its deregulation in CNS tumors with particular attention in glioblastoma (Figure 2).
Figure 2.
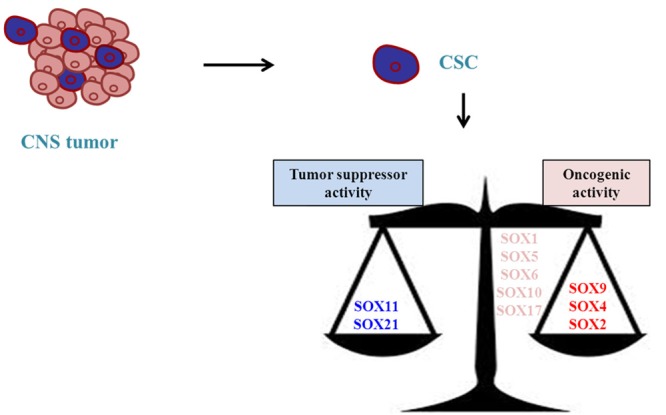
Balance between SOX members in the regulation of GSCs. Different SOX members display opposite functions in the regulation of GSCs. However, it remains elusive whether they are involved in the initiation of the glioma.
SOX2
SOX2 is a member of the SOXB1 (together with SOX1 and SOX3) required for the maintenance of the early embryo, before implantation [41]. SOXB1 group members are coexpressed in the neuroepithelium and show certain degree of functional redundancy in the developing CNS [41]. In particular, SOX2 is one of the four essential factors required for induced pluripotent stem (iPS) cell induction [42]. It is widely expressed in the embryo, in particular in the developing CNS where its expression is initiated concomitant with the acquisition of neural progenitor identity and it functions to maintain it [1,43,44]. In the adult, its expression is maintained in different populations of stem cells [45-48], acting intrinsically to confer stem cell properties, but also more broadly by regulating the expression of critical niche factors as observed in the CNS [49].
SOX2 and GBM
SOX2 is highly expressed in several human cancers [50], including GBM [51-53]. Interestingly, the expression of SOX2 and other stem cell markers identifies a subset of patients with the poorest clinical outcome highlighting the clinical relevance of SOX2 in GBM and in several other neoplasms [54].
Functionally, SOX2 is enriched in human-derived glioma stem cells (GSCs) where it sustains stemness properties and maintenance of tumorigenicity [55,56]. Indeed, siRNA-mediated downregulation of SOX2 in GSCs impaired proliferation and their ability to form tumors in vivo [55]. SOX2 maintains GSC stemness using the same molecular targets of normal NSCs [55], supporting a hierarchical model of GBM controlled by SOX2 and opening the approach to find downstream genes as therapeutic targets. Furthermore, elevated expression of SOX2 is essential but not sufficient for maintaining the self renewal of GSCs [53] indicating that other factors cooperate to activate stem cell-like properties. Supporting this notion, just recently Suva et al identified a core set of neurodevelopmental transcription factors (TFs) (POU3F2, SOX2, SALL2, and OLIG2) essential for GBM propagation. These TFs coordinately bind and activate stem-like tumor propagating cells (TPCs)-specific regulatory elements and are sufficient to fully reprogram differentiated GBM cells to “induced” TPCs, recapitulating the epigenetic landscape and phenotype of native TPCs [57]. In addition, SOX2 drives additional cancer-associated phenotypes and SOX2-driven malignant GSCs are highly invasive and have migratory characteristics [53], mimicking those of NSCs [58]. Indeed, SOX2 depletion induced attenuated cell proliferation is caused by decreased levels of Cyclin D1 [59], while the impaired invasive activity is mediated by inhibition of focal adhesion kinase (FAK) signaling and downstream proteins such as HEF1/NEDD9 and matrix metalloproteinases 1 and 2 [59].
In the last years the mechanism of SOX2 activation in GBM has started to be unraveled. Our group identified SOX2 gene amplification and promoter DNA hypomethylation in a set of GBM patients as the leading mechanism responsible for SOX2 aberrant expression [53]. SOX2 presents a high CpG density throughout the promoter that may poise the gene for repression upon differentiation [60], suggesting that SOX2 promoter hypomethylation in GBM might reflect a more primitive cellular state resembling that found in NSCs [60]. SOX2 is also regulated transcriptionally and acts downstream relevant pathways in GBM formation. TGF-β regulates GSCs through SOX2 [56]. PDGF also modulates SOX2 activity. In fact, transforming activity of PDGF in neural progenitors and PDGF-dependent tumors in mice triggered SOX2 expression [61]. In human GSCs, siRNA-induced downregulation of SOX2 confers sensitivity to treatment with PDGF and IGF1 receptor inhibitors [62] suggesting that resistance to PDGF and IGF1-receptor inhibitors in GBM are related to SOX2 expression. Moreover, SOX2 is activated at translational level by eukaryotic initiation factor 4E (eIF4E) [63]. Indeed, there is a positive correlation between SOX2 and eIF4E in GBM human samples and down-regulation of eIF4E decreases SOX2 protein level without altering its mRNA level in GSCs. Post-transcriptionally, different miRNAs including miR-9, miR-145, miR-21, miR-137 regulate GSCs and impart chemoresistance regulating SOX2 activity [64-67].
In order to address downstream targets of SOX2, microarray analyses identified 489 genes and 105 precursor microRNAs whose expression is altered in response to SOX2 silencing [68]. Among the relevant identified targets, BEX1 and BEX2 tumor suppressors and miR-143, miR-145, miR-253-5p and miR-452 are downregulated with SOX2 knockdown. Interestingly, in this study they found that SOX2 and miR-145 form a double negative feedback loop [68]. It is known that miR-145 acts to silence multiple pluripotency factors, including SOX2, [69] during the switch from self-renewal to lineage commitment. Therefore it might be a mechanism to regulate the balance between an undifferentiated and committed state. However, this regulation warrants further investigation to determine their putative function in GBM.
In a very elegant study conducted by the group of Dr. Silvia Nicolis they address the question whether Sox2 was required by oligodendroglioma stem cells, mirroring its requirement for normal NSCs. They used their Sox2flox conditional mutation [70], in combination with the pHGG mouse model [61], to address the effects of Sox2 ablation on tumor reinitiation following tumor cell transplantation into brain. As expected, mice transplanted with SOX2-deleted cells remained tumor-free throughout the time window in which controls developed lethal tumors. Moreover, they showed that loss of tumorigenesis of SOX2-ablated cells was prevented by transduction with a Sox2-expressing virus. From a more practical point of view they demonstrated that vaccination with Sox2 peptides elicited a response that significantly delayed tumor development, underscoring the feasibility of using SOX2 as a target [71].
Among the other SOXB1 members, the role of SOX1 and SOX3 in GBM has not yet been studied but the knockdown of SOX2 inhibits the expression of SOX1 suggesting that this member might also display a role in this type of neoplasia [68]. Further studies will be necessary to clarify the role of SOX1 and SOX3 in GBM.
Together, all these results underscore the major role that SOX2 displays in the malignant phenotype of GBM.
SOX2 and pediatric tumors
Pathways essential for promoting neural precursor proliferation or growth arrest and differentiation have been implicated in CNS cancers and specifically in pediatric brain tumors; such as Sonic Hedgehog in medulloblastoma [72]. In agreement with this notion, Sox2 is upregulated in pHGG [75] and amplified in several pediatric cell lines [76]. Moreover, high levels of SOX2 are detected in a tissue array of DIPGs, consistent with a role for tumor stem cells in the origin and maintenance of these tumors [77]. SOX2 is also expressed in SHH-associated medulloblastoma with preponderance in adolescent and adult cases [78]. Deciphering the molecular circuitries controlled by Sox2 in pediatric brain tumors could provide insights into these neoplasm development, biology and possible novel molecular targeted therapies.
SOXB2
SOXB2 group (comprised by SOX14 and SOX21) is closely related to SOXB1. However, SOXB2 factors possess a repression instead of a C-terminal transactivation domain [79] and functionally Sox21 promotes neurogenesis by counteracting the activities of SOXB1 proteins in the developing CNS [80]. The decision of neural precursors to self-renew or to undergo neuronal differentiation therefore depends on the balance of SOXB1 and SOXB2 factors.
Analogous to their opposite roles in development and differentiation, forced expression of SOX21 inhibits SOX2 and induces apoptosis in human glioma cells [81]. Moreover, SOX21 inhibits gliomas progression in vivo by forming complexes with SOX2 and stimulating aberrant differentiation [82]. These results imply that SOX21 acts as a tumor suppressor negatively regulating SOX2. They further demonstrate the relevance of the balance between SOXB1 and SOXB2 in tissue homeostasis and disease in the CNS.
SOXC
SOXC proteins are implicated in the biology of different brain tumors [83,84] with SOX4 and SOX11 exhibiting opposing activities in GBM. On one hand, SOX4 is upregulated in human samples, where it is associated with TGF-β [85], an important signaling pathway in GBM formation and progression [86]. Functionally, the activation of canonical and non-canonical TGF-β signaling enhances GSCs tumor activity through SOX4 protein and consequent boost of SOX2 [56]. Further supporting this axis, inhibition of TGF-β signaling drastically decreases the tumorigenicity of GSCs by promoting their differentiation, and these effects are restored by SOX2 or SOX4 re-activation [56]. SOX4 induces the expression of SOX2 forming cooperative complexes with OCT-4 that bind to the SOX2 promoter [56]. In addition to their function regulating GSCs, combined high expression of OCT-4, SOX4 and SOX2 confers lower patient survival and correlates with p53-mutated status in GBM cases [87], highlighting the clinical relevance of this axis. Further investigations have revealed that SOX4 acts downstream of miR-204 to suppress GSCs self-renewal [88]. In summary, these findings indicate that SOX4 is a master regulator of GSCs although it remains unresolved whether Sox4 positive cells are the cells of origin of GBM. They also highlight that SOX transcription factors can act sequentially in tumor development, mimicking the action of those in neural lineage development [80].
The role of SOX11 in GBM is less clear. On one hand, SOX11 is transcriptionally overexpressed in GBM [89]; however, low levels correlate with a significant decrease in patient survival [90]. Agreeing with this notion, GSCs have lost SOX11 expression, and its ectopic restoration prevents their tumorigenesis in vivo blocking the expression of oncogenic Plagl1 [90]. Moreover, the identification of an immunogenic CD8+ T cell epitope derived from SOX11, which is abundantly and specifically overexpressed in malignant glioma, emphasizes the suitability of this protein for a T cell-based immunotherapy for these patients [91].
SOXE
SOX9 belongs to the related SOXE family and its presence during embryonic development and in adulthood has been associated with stem cell maintenance in the pancreas, hair follicle, breast intestine and CNS [92,93-95]. In the CNS, Sox9 is essential for gliogenesis and, in conjunction with Sox10, also maintains the multipotency of neural crest stem cells as well as directing differentiating cells to non-neuronal fates [96]. It acts as a downstream effector of the Shh and Notch pathways [93,97].
SOX9 expression levels are significantly higher in gliomas than in brain control tissue and increasing WHO grade gliomas display stronger SOX9 staining, together with higher SOX10 [98]. From the clinical point of view, the increased expression of SOX9 in GBM significantly correlates with a lower Karnofsky performance score. In addition, patients with high SOX9 expression present lower disease-free and overall survival rates than those with low SOX9 [99]. Thus, SOX9 expression might be a relevant independent prognostic factor for GBM patients. Other brain tumors such as medulloblastomas and ependymomas also display robust SOX9 expression [84].
Functionally, SOX9 knockdown impairs cell proliferation in glioma cell lines [100], induces the cell arrest in G2/M phase of cell cycle and enhances the apoptosis in glioma cells [99]. The inhibition of its activity mediates the impaired cell cycle progression and reduced cell invasion induced by miR-145 tumor suppressor [101]. In contrast, ectopic expression of SOX9 cooperates to transform NSCs and form tumors with a primitive neuroectodermal tumor profile [40], establishing his functional relevance in the regulation of GSCs (Figure 1). Beyond the CNS tumors, Sox9 interacts with pathways and genes also altered in GBM such as EGFR, BMI-1 and PTEN [102-105] and these connections might be interesting to be further investigated in GBM.
Other SOX members might have a prominent role in GBM as well. SOX5 and SOX6 are highly expressed in glioma while are not detected in non-neoplastic tissues [106,107]. Furthermore, there is a positive correlation between the presence of SOX5 and SOX6 IgGs from the sera of glioma patients and GBM patients survival suggesting they may be useful not only as diagnostic markers, but also as prognostic markers in glioma patients [107]. Finally, SOX17 expression in glioma endothelial cells is related to the angiogenic properties of tumor vessels, suggesting that SOX17 might play a relevant function in GBM promoting tumor angiogenesis and vascular abnormalities [65].
In summary, SOX proteins are differently expressed in GBM with the majority of them inducing aberrant cell growth and promoting tumorigenic activities at various levels. However, their function needs to be further investigated in order to determine whether targeting SOX proteins is a promising therapeutic strategy for GBM in humans.
Future perspectives
SOX factors are critical regulators during embryogenesis playing an integral role in the maintenance of NSCs and lineage differentiation both during embryo development and adult stage. Furthermore, their aberrant expression is observed in malignant gliomas where they exhibit various features of tumorigenesis and tumor progression. SOX2, SOX4 and SOX9 have been consistently shown to act as oncogenes while SOX11 and SOX21 behave as tumor suppressors (Figure 2). We have summarized the main role of the different SOX in CNS tumors in Table 1. Other members have not displayed a clear function yet.
Table 1.
Major discoveries associated to SOX2 protein in CNS tumors
| Sox member | Finding | Reference |
|---|---|---|
| SOX2-Glioblastoma | sustains stemness properties and tumorigenicity | [55] |
| Transcriptional regulation mediated by TGF-β | [56] | |
| Genetic and Epigenetic modifications | [53] | |
| Factor responsible for glioblastoma stem cells reprogramming | [57] | |
| SOX2-medulloblastoma | Sustains stemness properties but not involved in tumor survival | [78] |
| SOX2-oligodendroglioma | Required to maintain stemness properties and tumorigenicity | [71] |
| SOX4-Glioblastoma | Sustains stemness regulated by TGF-β and modulating SOX2 | [56] |
The outcome of SOX factors activation seems to depend on the tumor origin and cellular context reflecting their roles in different territories during development. They regulate key processes related to tumor biology, including cell proliferation, migration, epithelial mesenchymal transition, angiogenesis, apoptosis, and regulation of GSCs and their opposite roles in these processes could be related to, yet unexplored, regulation of protein activity through transcriptional, post-transcriptional and post-translational events. Future insight into the identification and functional characterization of their downstream target genes in GBM maintenance and progression are needed to determine which SOX may be targeted as a therapeutic strategy for GBM.
Among all the SOX factors, SOX2 is emerging as a very complex factor with multiple functions (Figure 3) not only in transcription but also in chromatin remodelling possibly through its association with the swi/snf complex, NuRD complex and others [108]. Moreover, it has been suggested a role of SOX2 in post-transcription regulation through its association with RNA binding proteins [109] and a putative role as a RNA splicer [110]. These functions have been previously ascribed to central regulator such as p53 [111]. Nevertheless, further experiments will be necessary to clarify the functional role and the mechanisms of these interactions to understand the complexity of SOX2 networks. This information might indicate that Sox2 targeting should be considered in ongoing efforts to develop novel stem cell targeting therapies. We have ahead of us a very interesting horizon that comes with sophisticated tools such as RNA deep sequencing, ChIRP, RIP-seq, among others, that will allow us to study and decipher the secrets that SOX family members are still hiding. Understanding the underpinnings of these molecular networks would allow proposing tailored therapies against SOX deregulation.
Figure 3.
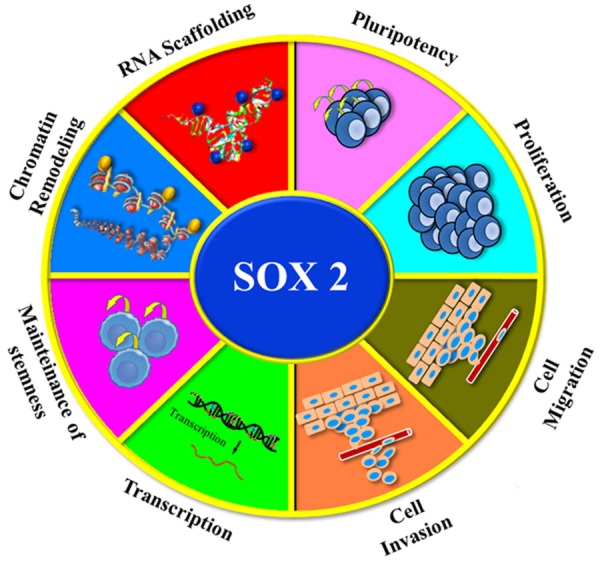
SOX2 functions in tissue homeostasis and cancer. SOX2 is emerging as a very complex factor with multiple functions. Here we include the most relevant for glioma.
Acknowledgements
This work is supported by grants from Spanish Ministry of Economy and Competition (MINECO) (Miguel Servet contract CP10/00539 to AM, Ramón y Cajal contract RYC-2009-05571 to MMA and PI10/00399 to MMA), European Union (Marie Curie CIG 2012/712404 to AM and Marie Curie IRG270459 to MMA), Diputación Foral Gipuzkoa (DFG12/004 to AM) and Industry Department of Basque Government (SAIO12-PE12BNO13 to NS). AAR is the recipient of a predoctoral fellowship from the Foundation “Amigos de la Universidad de Navarra”.
Disclosure of conflict of interest
None.
References
- 1.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 2.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 3.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 4.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Castillo SD, Sanchez-Cespedes M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert Opin Ther Targets. 2012;16:903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 7.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumors of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crocetti E, Trama A, Stiller C, Caldarella A, Soffietti R, Jaal J, Weber DC, Ricardi U, Slowinski J, Brandes A RARECARE working group. Epidemiology of glial and non-glial brain tumors in Europe. Eur J Cancer. 2012;48:1532–1542. doi: 10.1016/j.ejca.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomized phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 11.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller , L Ding CR, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, Brennan C, Ligon KL, Furnari F, Cavenee WK, Depinho RA, Chin L, Hahn WC. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gajjar A, Packer RJ, Foreman NK, Cohen K, Haas-Kogan D, Merchant TE COG Brain Tumor Committee. Children’s Oncology Group’s 2013 blueprint for research: central nervous system tumors. Pediatr Blood Cancer. 2013;60:1022–1026. doi: 10.1002/pbc.24427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershon TR, Becher OJ. Medulloblastoma: therapy and biologic considerations. Curr Neurol Neurosci Rep. 2006;6:200–206. doi: 10.1007/s11910-006-0006-y. [DOI] [PubMed] [Google Scholar]
- 20.Broniscer A, Gajjar A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. Oncologist. 2004;9:197–206. doi: 10.1634/theoncologist.9-2-197. [DOI] [PubMed] [Google Scholar]
- 21.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 22.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 23.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 24.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 26.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 27.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 28.Rakic P. DNA synthesis and cell division in the adult primate brain. Ann N Y Acad Sci. 1985;457:193–211. doi: 10.1111/j.1749-6632.1985.tb20806.x. [DOI] [PubMed] [Google Scholar]
- 29.Gross CG. Neurogenesis in the adult brain: death of a dogma. Nat Rev Neurosci. 2000;1:67–73. doi: 10.1038/35036235. [DOI] [PubMed] [Google Scholar]
- 30.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li V, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S, Ayers-Ringler J, Nishiyama A, Stallcup WB, Berger MS, Bergers G, McKnight TR, Goldman SA, Weiss WA. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrasco-Garcia E, Sampron N, Aldaz P, Arrizabalaga O, Villanua J, Barrena C, Ruiz I, Arrazola M, Lawrie C, Matheu A. Therapeutic strategies targeting glioblastoma stem cells. Recent Pat Anticancer Drug Discov. 2013;8:216–227. doi: 10.2174/15748928113089990002. [DOI] [PubMed] [Google Scholar]
- 36.Kanamori M, Kawaguchi T, Nigro JM, Feuerstein BG, Berger MS, Miele L, Pieper RO. Contribution of Notch signaling activation to human glioblastoma multiforme. J Neurosurg. 2007;106:417–427. doi: 10.3171/jns.2007.106.3.417. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowitch DH, S-Jacques B, Lee SM, Flax JD, Snyder EY, McMahon AP. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J Neurosci. 1999;19:8954–8965. doi: 10.1523/JNEUROSCI.19-20-08954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 40.Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A, Phillips JJ, Taylor MD, Weiss WA. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 44.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 45.Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 46.Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fauquier T, Rizzoti K, Dattani M, Lovell-Badge R, Robinson IC. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc Natl Acad Sci U S A. 2008;105:2907–2912. doi: 10.1073/pnas.0707886105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. Int J Biochem Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 50.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O’Kelly M, Dutt A, Rozenblatt-Rosen O, Dziunycz P, Komisarof J, Chirieac LR, Lafargue CJ, Scheble V, Wilbertz T, Ma C, Rao S, Nakagawa H, Stairs DB, Lin L, Giordano TJ, Wagner P, Minna JD, Gazdar AF, Zhu CQ, Brose MS, Cecconello I, Jr UR, Marie SK, Dahl O, Shivdasani RA, Tsao MS, Rubin MA, Wong KK, Regev A, Hahn WC, Beer DG, Rustgi AK, Meyerson M. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genomics Proteomics. 2011;8:139–147. [PubMed] [Google Scholar]
- 52.Holmberg J, He X, Peredo I, Orrego A, Hesselager G, Ericsson C, Hovatta O, Oba-Shinjo SM, Marie SK, Nister M, Muhr J. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS One. 2011;6:e18454. doi: 10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, Urquiza L, Jauregi P, Lopez de Munain A, Sampron N, Aramburu A, Tejada-Solís S, Vicente C, Odero MD, Bandrés E, García-Foncillas J, Idoate MA, Lang FF, Fueyo J, Gomez-Manzano C. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 56.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 57.Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cayre M, Canoll P, Goldman JE. Cell migration in the normal and pathological postnatal mammalian brain. Prog Neurobiol. 2009;88:41–63. doi: 10.1016/j.pneurobio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oppel F, Müller N, Schackert G, Hendruschk S, Martin D, Geiger KD, Temme A. SOX2-RNAi attenuates S-phase entry and induces RhoA-dependent switch to protease-independent amoeboid migration in human glioma cells. Mol Cancer. 2011;10:137. doi: 10.1186/1476-4598-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Appolloni I, Calzolari F, Tutucci E, Caviglia S, Terrile M, Corte G, Malatesta P. PDGF-B induces a homogeneous class of oligodendrogliomas from embryonic neural progenitors. Int J Cancer. 2009;124:2251–2259. doi: 10.1002/ijc.24206. [DOI] [PubMed] [Google Scholar]
- 62.Hägerstrand D, He X, Bradic Lindh M, Hoefs S, Hesselager G, Ostman A, Nistér M. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13:1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge Y, Zhou F, Chen H, Cui C, Liu D, Li Q, Yang Z, Wu G, Sun S, Gu J, Wei Y, Jiang J. Sox2 is translationally activated by eukaryotic initiation factor 4E in human glioma-initiating cells. Biochem Biophys Res Commun. 2010;397:711–717. doi: 10.1016/j.bbrc.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 64.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 65.Yang YP, Chien Y, Chiou GY, Cherng JY, Wang ML, Lo WL, Chang YL, Huang PI, Chen YW, Shih YH, Chen MT, Chiou SH. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 2012;33:1462–1476. doi: 10.1016/j.biomaterials.2011.10.071. [DOI] [PubMed] [Google Scholar]
- 66.Põlajeva J, Swartling FJ, Jiang Y, Singh U, Pietras K, Uhrbom L, Westermark B, Roswall P. miRNA-21 is developmentally regulated in mouse brain and is co-expressed with SOX2 in glioma. BMC Cancer. 2012;12:378. doi: 10.1186/1471-2407-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S, Xiang C, Poisson L, de Carvalho AC, Slavin S, Jacoby E, Yalon M, Toren A, Mikkelsen T, Brodie C. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4:665–676. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, Nicolis SK. Hippocampal development and neural stem cell maintenance require Sox2 dependent regulation of Shh. Nat Neurosci. 2009;12:1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- 71.Favaro R, Appolloni I, Pellegatta S, Sanga AB, Pagella P, Gambini E, Pisati F, Ottolenghi S, Foti M, Finocchiaro G, Malatesta P, Nicolis SK. Sox2 is required to maintain cancer stem cells in a mouse model of high-grade oligodendroglioma. Cancer Res. 2014;74:1833–1844. doi: 10.1158/0008-5472.CAN-13-1942. [DOI] [PubMed] [Google Scholar]
- 72.Roussel MF, Hatten ME. Cerebellum development and medulloblastoma. Curr Top Dev Biol. 2011;94:235–282. doi: 10.1016/B978-0-12-380916-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 74.Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 75.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, Bax DA, Coyle B, Barrow J, Hargrave D, Lowe J, Gajjar A, Zhao W, Broniscer A, Ellison DW, Grundy RG, Baker SJ. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J. Clin. Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bax DA, Mackay A, Little SE, Carvalho D, Viana-Pereira M, Tamber N, Grigoriadis AE, Ashworth A, Reis RM, Ellison DW, Al-Sarraj S, Hargrave D, Jones C. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16:3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ballester LY, Wang Z, Shandilya S, Miettinen M, Burger PC, Eberhart CG, Rodriguez FJ, Raabe E, Nazarian J, Warren K, Quezado MM. Morphologic characteristics and immunohistochemical profile of diffuse intrinsic pontine gliomas. Am J Surg Pathol. 2013;37:1357–1364. doi: 10.1097/PAS.0b013e318294e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahlfeld J, Favaro R, Pagella P, Kretzschmar HA, Nicolis S, Schüller U. Sox2 requirement in sonic hedgehog-associated medulloblastoma. Cancer Res. 2013;73:3796–3807. doi: 10.1158/0008-5472.CAN-13-0238. [DOI] [PubMed] [Google Scholar]
- 79.Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- 80.Bergsland M, Ramsköld D, Zaouter C, Klum S, Sandberg R, Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, Westermark B. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer. 2011;129:45–60. doi: 10.1002/ijc.25647. [DOI] [PubMed] [Google Scholar]
- 82.Caglayan D, Lundin E, Kastemar M, Westermark B, Ferletta M. Sox21 inhibits glioma progression in vivo by forming complexes with Sox2 and stimulating aberrant differentiation. Int J Cancer. 2013;133:1345–1356. doi: 10.1002/ijc.28147. [DOI] [PubMed] [Google Scholar]
- 83.Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57:201–214. doi: 10.1023/a:1015773818302. [DOI] [PubMed] [Google Scholar]
- 84.de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, Luider TM, den Boer ML, Pieters R. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10:648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin B, Madan A, Yoon JG, Fang X, Yan X, Kim TK, Hwang D, Hood L, Foltz G. Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One. 2010;5:e10210. doi: 10.1371/journal.pone.0010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 87.Galatro TF, Uno M, Oba-Shinjo SM, Almeida AN, Teixeira MJ, Rosemberg S, Marie SK. Differential expression of ID4 and its association with TP53 mutation, SOX2, SOX4 and OCT-4 expression levels. PLoS One. 2013;8:e61605. doi: 10.1371/journal.pone.0061605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weigle B, Ebner R, Temme A, Schwind S, Schmitz M, Kiessling A, Rieger MA, Schackert G, Schackert HK, Rieber EP. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol Rep. 2005;13:139–144. [PubMed] [Google Scholar]
- 90.Hide T, Takezaki T, Nakatani Y, Nakamura H, Kuratsu J, Kondo T. Sox11 prevents tumorigenesis of glioma-initiating cells by inducing neuronal differentiation. Cancer Res. 2009;69:7953–7959. doi: 10.1158/0008-5472.CAN-09-2006. [DOI] [PubMed] [Google Scholar]
- 91.Schmitz M, Wehner R, Stevanovic S, Kiessling A, Rieger MA, Temme A, Bachmann M, Rieber EP, Weigle B. Identification of a naturally processed T cell epitope derived from the glioma-associated protein SOX11. Cancer Lett. 2007;245:331–336. doi: 10.1016/j.canlet.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 92.Pritchett J, Athwal V, Roberts N, Hanley NA, Hanley KP. Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol Med. 2011;17:166–174. doi: 10.1016/j.molmed.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 93.Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- 94.Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 95.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chew LJ, Gallo V. The Yin and Yang of Sox proteins: Activation and repression in development and disease. J Neurosci Res. 2009;87:3277–3287. doi: 10.1002/jnr.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martini S, Bernoth K, Main H, Ortega GD, Lendahl U, Just U, Schwanbeck R. A critical role for Sox9 in notch-induced astrogliogenesis and stem cell maintenance. Stem Cells. 2013;31:741–751. doi: 10.1002/stem.1320. [DOI] [PubMed] [Google Scholar]
- 98.Kordes U, Hagel C. Expression of SOX9 and SOX10 in central neuroepithelial tumor. J Neurooncol. 2006;80:151–155. doi: 10.1007/s11060-006-9180-7. [DOI] [PubMed] [Google Scholar]
- 99.Wang L, He S, Yuan J, Mao X, Cao Y, Zong J, Tu Y, Zhang Y. Oncogenic role of SOX9 expression in human malignant glioma. Med Oncol. 2012;29:3484–3490. doi: 10.1007/s12032-012-0267-z. [DOI] [PubMed] [Google Scholar]
- 100.Swartling FJ, Ferletta M Kastemar M, Weiss WA, Westermark B. Cyclic GMP-dependent protein kinase II inhibits cell proliferation, Sox9 expression and Akt phosphorylation in human glioma cell lines. Oncogene. 2009;28:3121–3131. doi: 10.1038/onc.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013;15:1302–1316. doi: 10.1093/neuonc/not090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, Skotheim RI, Lothe RA, Lopez de Munain A, Briscoe J, Serrano M, Lovell-Badge R. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72:1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thomsen MK, Ambroisine L, Wynn S, Cheah KS, Foster CS, Fisher G, Berney DM, Moller H, Reuter VE, Scardino P, Cuzick J, Ragavan N, Singh PB, Martin FL, Butler CM, Cooper CS, Swain A Transatlantic Prostate Group. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010;70:979–987. doi: 10.1158/0008-5472.CAN-09-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eberl M, Klingler S, Mangelberger D, Loipetzberger A, Damhofer H, Zoidl K, Schnidar H, Hache H, Bauer HC, Solca F, Hauser-Kronberger C, Ermilov AN, Verhaegen ME, Bichakjian CK, Dlugosz AA, Nietfeld W, Sibilia M, Lehrach H, Wierling C, Aberger F. Hedgehog-EGFR cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumour-initiating pancreatic cancer cells. EMBO Mol Med. 2012;4:218–233. doi: 10.1002/emmm.201100201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ling S, Chang X, Schultz L, Lee TK, Chaux A, Marchionni L, Netto GJ, Sidransky D, Berman DM. An EGFR-ERK-SOX9 signaling cascade links urothelial development and regeneration to cancer. Cancer Res. 2011;71:3812–3821. doi: 10.1158/0008-5472.CAN-10-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ueda R, Yoshida K, Kawakami Y, Kawase T, Toda M. Expression of a transcriptional factor, SOX6, in human gliomas. Brain Tumor Pathol. 2004;21:35–38. doi: 10.1007/BF02482175. [DOI] [PubMed] [Google Scholar]
- 107.Ueda R, Yoshida K, Kawase T, Kawakami Y, Toda M. Preferential expression and frequent IgG responses of a tumor antigen, SOX5, in glioma patients. Int J Cancer. 2007;120:1704–1711. doi: 10.1002/ijc.22472. [DOI] [PubMed] [Google Scholar]
- 108.Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, van Ijcken W, Dekkers DH, Demmers J, Rijkers EJ, Bhattacharya S, Philipsen S, Pevny LH, Grosveld FG, Rottier RJ, Lenhard B, Poot RA. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 109.Fang X, Yoon JG, Li L, Tsai YS, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B. Landscape of the SOX2 protein-protein interactome. Proteomics. 2011;11:921–934. doi: 10.1002/pmic.201000419. [DOI] [PubMed] [Google Scholar]
- 110.Tung CL, Hou PH, Kao YL, Huang YW, Shen CC, Cheng YH, Wu SF, Lee MS, Li C. SOX2 modulates alternative splicing in transitional cell carcinoma. Biochem Biophys Res Commun. 2010;393:420–425. doi: 10.1016/j.bbrc.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 111.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]


