Abstract
Exosomes, secreted microvesicles transporting microRNAs (miRNAs), mRNAs, and proteins through bodily fluids, facilitate intercellular communication and elicit immune responses. Exosomal contents vary depending on the source and the physiological conditions of cells and can provide insights into how cells and systems cope with physiological perturbations. Previous analysis of circulating miRNAs in patients with complex regional pain syndrome (CRPS), a debilitating chronic pain disorder, revealed a subset of miRNAs in whole blood that are altered in the disease. To determine functional consequences of alterations in exosomal biomolecules in inflammation and pain, we investigated exosome-mediated information transfer in vitro, in a rodent model of inflammatory pain and in exosomes from patients with CRPS. Mouse macrophage cells stimulated with lipopolysaccharides (LPS) secrete exosomes containing elevated levels of cytokines and miRNAs that mediate inflammation. Transcriptome sequencing of exosomal RNA revealed global alterations in both innate and adaptive immune pathways. Exosomes from LPS-stimulated cells were sufficient to cause NF-kappaB activation in naïve cells, indicating functionality in recipient cells. A single injection of exosomes attenuated thermal hyperalgesia in a mouse model of inflammatory pain, suggesting an immunoprotective role for macrophage-derived exosomes. We also show that circulating miRNAs altered in patients with complex regional pain syndrome are trafficked by exosomes. Macrophage-derived exosomes carry a protective signature that is altered when secreting cells are exposed to an inflammatory stimulus. With their systemic signaling capabilities, exosomes can induce pleiotropic effects potentially mediating the multifactorial pathology underlying chronic pain and should be explored for their therapeutic utility.
Introduction
Chronic inflammatory pain ensues when the normal process of inflammation does not resolve, resulting in excess proinflammatory cytokines and chemoattractants that can eventually lead to central sensitization [13,15,49]. Small noncoding RNAs such as microRNAs (miRNAs) play a role in translational repression of mRNA and chromatin remodeling [5,29]. Analysis of tissue-specific miRNA levels indicates a correlation between the initiation and progression of inflammatory and neuropathic pain conditions [41]. Circulating miRNAs in bodily fluids are often present in small vesicles called exosomes. Exosomes, contain diverse classes of biomolecules including miRNAs, mRNAs, proteins, and lipids that are coexpressed, packaged, and secreted from cells into bodily fluids under normal and disease states [21,22,43,45,60,62]. Exosomes represent a novel pathway that allows a cohort of biomolecules to travel long distances and results in modulation of gene expression in recipient cells [36,57].
Complex regional pain syndrome (CRPS) is a severe neuropathic pain condition characterized by persistent inflammation [12,19,30,54]. Our previous study demonstrated that circulating miRNAs in whole blood can be useful in stratification of patients with CRPS and that dysregulation of specific miRNAs correlates to symptoms and comorbidities associated with the disease [51]. To elucidate the biological relevance of alterations in circulating miRNAs and cytokines, it is important to determine whether they are released in an uncontrolled manner upon injury or disease, or conversely, whether they are secreted in a regulated fashion as a compensatory mechanism in response to a stress signal. We hypothesized that delivery of a cohort of proteins, mRNAs, and miRNAs from one cell to another through the systemic circulation mediated via exosomal secretion would allow information transfer from one location to influence the entire body and is a mechanism that would have a profound effect if it is not properly regulated.
Macrophage-derived exosomes represent a large portion of circulating microvesicles in blood [35]. Exosomes from cells infected with intracellular pathogens stimulate a TLR-dependent inflammatory response in recipient cells [7,24] while dendritic cells (DC)-derived exosomes suppress the onset of murine collagen-induced arthritis and reduce its severity [10,38]. Here we show that LPS stimulation of RAW 264.7 cells leads to an increase in exosomal chemokines, and RNAs involved in regulating inflammation. Upon infection, NF-κB signaling cascade is activated and the pleiotropic nature of the NF-κB signaling allows for temporal regulation of inflammation, resulting in proinflammatory gene transcription early in the immune response and a transition to anti-inflammatory gene transcription later in the progression of inflammation [42]. We show that exosomes from LPS-stimulated cells can activate NF-κB in naïve macrophages. After a single intraplantar injection into complete Freund’s adjuvant (CFA) treated mice, exosomes from LPS-stimulated macrophages significantly reduced paw swelling. Additionally, macrophage-derived exosomes were able to relieve thermal hyperalgesia associated with CFA-induced inflammatory pain. Analyses of serum-derived exosomes from CRPS patients show that circulating miRNAs altered in patients with CRPS are trafficked by exosomes. These findings suggest a role for exosomes in dysregulated inflammation and chronic pain states.
Methods
Cell culture
RAW 264.7 cells (ATCC® TIB-71™) were maintained in complete media (1× DMEM, 10% heat-inactivated FBS). For exosome collection, RAW 264.7 cells (1×107) were plated in 150-mm dishes with complete culture media. At 24 h, media was replaced with exosome-depleted media (1× DMEM, 10% heat-inactivated FBS depleted of exosomes by ultracentrifugation) with or without 1 μg/ml LPS [Sigma, St. Louis, MO] and incubated overnight. Media was collected in 50-ml tubes at 24 h for exosome purification. Human monocytic THP-1 (ATCC® TIB-202™) cells were maintained in ATCC-formulated RPMI-1640 medium containing 2-mercaptoethanol (final concentration of 0.05 mM) and 10% FBS. For exosome purification, THP-1 cells were cultured in suspension at 2 × 105 viable cells/mL in culture media containing heat-inactivated FBS depleted of exosomes as mentioned above for RAW 264.7 cells.
Exosome purification
Exosome purification from cell culture media was performed as described previously [47]. Centrifugation was used to remove cell debris (500×g for 10 min); the supernatant was transferred and centrifuged (16,500×g for 20 min). Cell-free supernatants were filtered (0.22 μm; Merck Millipore Ltd., Cork, IRL) and exosomes were pelleted by ultracentrifugation (120,000×g for 70 min). The exosomal pellet was resuspended in buffer specific to downstream experiments and vortexed 2×15 seconds. For RNA purification, RNase inhibitors were added after the first centrifugation step at 1 U/ml (RNAsin Plus; Promega, Madison, WI) and at all subsequent steps at 1 U/μl. For purification from human samples, serum was diluted 1:1 with 1× PBS (−) Mg2+ (−) Ca2+ (Corning 21-031-CV; Corning, NY) and spun at 2000×g for 30 min at 4°C. The sample was transferred to a centrifuge tube and spun at 12,000×g for 45 min at 4°C, then transferred to an ultracentrifuge tube and spun at 110,000×g for 2 h at 4°C. The pellet was resuspended in 1×PBS (−) Mg2+ (−) Ca2+ and spun for an additional hour at 110,000×g before resuspension in RNA lysis buffer.
Human serum–derived exosomes
Patients with CRPS were recruited from the neurology pain clinic at Drexel University College of Medicine and met the clinical Budapest criteria for CRPS [31]. Healthy pain-free control subjects were recruited from the community’s general population. Blood samples were drawn from the cubital vein of subjects at rest, collected in serum-separating tubes and spun at 1940×g for 15 min at 4°C after 30 min incubation at room temperature.
Preparation of exosomes for TEM
Droplets of purified exosomes resuspended in 1% glutaraldehyde in 0.1 M sodium phosphate buffer were placed on 300-mesh carbon-coated polyvinyl formal copper grids (Formvar, Electron Microscopy Sciences Hatfield, PA) and left to adsorb for 30 min. After excess buffer was removed, dry grids were washed with deionized water and stained with 1% aqueous uranyl acetate before TEM analysis. For immunolabeling, exosomes were resuspended in 2% paraformaldehyde and droplets were left to adsorb on 300-mesh carbon-coated Formvar nickel grids for 20 min. After 2 washes in 1× PBS and 4 washes in 1× PBS/50 mM glycine, grids were incubated with blocking buffer (5% BSA/0.05% polysorbate 20/5% FBS in 1×PBS) for 10 min. The grids were immunolabeled with mouse anti-CD81 (1:100, Sigma) in 1:5 dilution of blocking buffer in 1×PBS for 30 min at room temperature. The unbound antibody was removed with 6 washes in 1:10 dilution of blocking buffer and then grids were incubated with 10 nm gold-labeled anti-rabbit IgG (1:25, Sigma) for 20 min at room temperature. After the unbound antibody was removed with 6 washes in 1:10 dilution of blocking buffer, grids were incubated in 1% glutaraldehyde for 5 min, washed with water, and stained with uranyl acetate as above.
Western blotting and cytokine array
Exosomes were resuspended in radioimmunoprecipitation assay buffer (Thermo Scientific, Waltham, MA) containing Halt protease inhibitor cocktail (Thermo Scientific) and the protein concentration was determined by Bradford analysis. For western blotting, the lysate was run on a 12% SDS-PAGE (NuPAGE, Novex/Life Technologies) for 1.5 h at 150 V. After 1 h transfer at 100 V, the nitrocellulose membrane was blocked with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween20 for 1 h, incubated with rabbit anti-HSP70 (Abcam, Cambridge, UK) or rabbit anti-TSG101 (Genetex, Irvine, CA) overnight and then with goat anti-rabbit IgG-HRP (System Biosciences, Mountain View, CA). LPS was detected after 1 h incubation with mouse anti-LPS (Abcam, ab35654) and goat antimouse IgG-HRP (Abcam, ab6789). Proteins were detected by Immobilon (Thermo Scientific) detection reagent and film exposure. For cytokine array, 100 μg of protein was incubated with the blots according to manufacturer’s specifications (R&D Systems, Minneapolis, MN).
RNA sequencing
The SOLiD whole transcriptome analysis kit protocol with the fragmentation step omitted was used to generate a cDNA library for each sample. Total RNA, ranging in size from kilobases down to 10-mers, was purified from exosomes using the mirVana miRNA isolation kit (Life Technologies) following manufacturer’s protocol. RNA concentration was measured using Nanodrop 1000 (NanoDrop Technologies, Wilmington, DE). Total RNA from 3 independent exosome purifications was pooled to obtain 4 μg exosomal RNA per library (due to a limited amount of RNA in individual preparations), analyzed for integrity using the Agilent RNA 6000 Pico Kit (RNA integrity number between 1.6 and 2.1), and gel purified. Sequencing adapter ligation and cDNA reverse transcription were performed with SOLiD Total RNA-seq kit. DNA fragments in the target range of 150 to 500 bp were enriched using Agencourt AMPure XP PCR bead capture purification (Beckman Coulter; Brea, CA) before sequencing 50-bp pieces with no paired ends. The SOLiD 5500XL high-throughput sequencing platform (Applied Biosystems, Carlsbad, CA) was used for sequencing.
Sequencing reads were aligned to the mouse reference genome version mm9 (July, 2007) and transcripts were assembled based on refGene annotations (dated Dec 16th, 2012) obtained from the UCSC Genome Browser [37] and non-coding RNA transcript definitions (dated Dec 19th, 2012) from the fRNAdb database [39] at ncrna.org. Reads were mapped using the LifeScope Whole Transcriptome Pipeline with default parameters, which effectively maps RNA fragments down to 22 nucleotides in length. The Cufflinks algorithm [61] was used for transcript assembly, abundance estimation and differential expression analysis, using the reference transcript annotation as a guide. Results generated from Cufflinks were investigated using the CummeRbund package [1]. Following differential expression analysis, transcripts were annotated using information from the Molecular Signatures Database [44,58] for biological interpretation.
miRNA profiling
TLDA microfluidic cards (Life Technologies) were used for miRNA profiling as previously described. Thirty nanograms of total RNA was used for each cDNA synthesis reaction. Taqman preamplification reaction was performed before the samples were loaded into the TLDA cards as described previously (26). For miRNAs profiled from exosomes collected from RAW 264.7 cells, significance was determined by applying a P value cutoff of 0.05 to the results of a paired-samples t test. For human exosomal miRNAs, significance was determined by applying the Benjamini-Hochberg false discovery rate correction to the results of a 2-tailed t test. The miRNA nomenclature used is consistent with Life Technologies Assay ID convention as outlined in TaqMan MicroRNA Assay Index File (AIF) (Applied Biosystems). The assay name is the miRBase ID for the targeted miRNA sequence when the assay was first released.
qPCR validation of exosomal mRNAs
cDNA was synthesized from 5 ng purified exosomal RNA using the WT-Ovation RNA Amplification System from NuGEN (San Carlos, CA). Taqman assays were performed in a reaction volume of 20 μl containing 10 μl Taqman Fast Universal PCR master mix (2×) no AmpErase UNG, 1 μl Taqman gene expression assay mix (20×), 2 μl cDNA (100ng), and 7 μl RNase-free water. Gapdh was used as the normalizer and a t test was used to perform statistical analysis. Assay IDs: Mm00443111_m1 [Ccl4], Mm00436450_m1 [Cxcl2], Mm00441242_m1 [Ccl2], Mm00443260_g1 [Tnf]) Mm00501607_m1 [Creb1] and Mm00497193_m1 [Zeb2] (Applied Biosystems).
NF-κB reporter assay
RAW-Blue cells (InvivoGen, San Diego, CA), maintained in complete media (1× DMEM, 10% heat-inactivated FBS), were seeded into a 96-well plate in exosome-free media on the day of the assay. Exosomes purified from RAW 264.7 cells without or after LPS stimulation were added at 4 concentrations. After 24 h, QUANTI-Blue assay was performed with QUANTI-Blue media, prepared as described by the manufacturer (InvivoGen). To 150 μl QUANTI-Blue media, 50 μl conditioned media was added and incubated at 37°C for 1 h. Plates were read at 650 nm (Spectramax Plus, Molecular Devices, Sunnyvale, CA).
In vivo studies
All behavioral tests were performed using 8-week-old C57BL/6 male mice purchased from Taconic (Cranbury, NJ). Mice were housed in 12 h light/dark cycles. Behavioral assays were performed by researchers blinded to the treatment received. The CFA-induced inflammatory pain model was established and CFA-induced mechanical and thermal hypersensitivity was measured as described previously (70). Baseline measurements were obtained before initiation of treatment. Twenty microliters of 50% CFA was administered by intraplantar injection into the right hind paw. Mechanical sensitivity was measured using a series of von Frey filaments (North Coast Medical, Inc., San Jose, CA). The smallest monofilament that evoked paw withdrawal responses on 3 of 5 trials was taken as the mechanical threshold. Thermal sensitivity was measured using the Hargreaves method. The baseline latencies were set to approximately 10 seconds with a maximum of 20 seconds as the cutoff to prevent potential injury. The latencies were averaged over 3 trials separated by 15-min intervals. At 3 h post-CFA injection and after confirming that the animals were sensitive, 20 μl exosomes (0.5 μg) in PBS were injected intraplanar to the right hind paw. Paw thickness was recorded (3 h and 1, 2, and 5 d) and paw withdrawal was measured by the von Frey (1, 5, 10, 15, and 21 d) and Hargreaves methods (3 h and 1, 5, and 10 d) (n=9).
Data analysis
Data are presented as mean ± SEM. Treatment effects were statistically analyzed with a 1- or 2-way ANOVA. Pairwise comparisons between means were tested using the post hoc Bonferroni method. Error probabilities of P < 0.05 were considered statistically significant.
Study approval
All subjects were enrolled after giving informed consent as approved by the Drexel University College of Medicine Institutional Review Board. The care and use of all mice were approved by the Institutional Animal Care & Use Committee of Drexel University College of Medicine.
Results
Exosome characterization
As previously demonstrated by our laboratory and others, exosomes were purified from RAW 264.7 cell culture media and human serum [20,47,63]. After purification, we used transmission electron microscopy (TEM) in conjunction with immune-gold labeling to analyze the specificity and morphology of exosomes purified from naïve and LPS-stimulated RAW 264.7 cell culture media. Exosomes maintain a vesicular morphology with an approximate diameter of 100 nm and show immunoreactivity for CD81, a tetraspannin protein found in exosomal membranes (Figure 1, A and B). Specificity of exosome preparations from naïve or LPS-stimulated RAW 264.7 cells was additionally verified by western blotting for the presence of HSP70, TSG101 (tumor susceptibility gene), and LPS. All exosomal protein lysates showed specificity for HSP70 and TSG101 (Figure 1C). LPS was undetectable in exosomes after LPS stimulation for 24 h. We then analyzed the integrity of total exosomal RNA using the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) (Figure 1, D and E). The concentrations of exosomal RNA from macrophage cell culture media without and with LPS stimulation were 2.4 ± 0.3 and 2.0 ± 0.5 ng/ml, respectively. Consistent with previous reports, exosomal RNA is relatively low in concentration and does not contain a prominent 18S or 28S rRNA peak [18].
Figure 1.
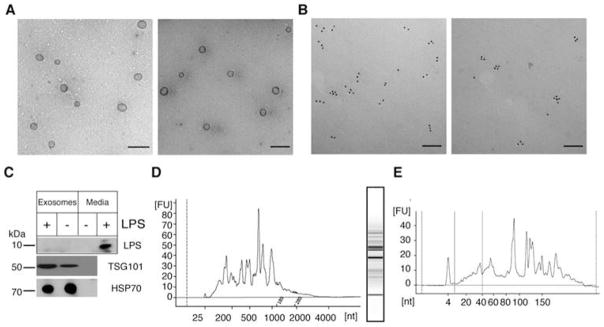
Characterization of exosome morphology and specificity. (A, B) TEM images of exosomes purified from RAW 264.7 cells before (left) and after (right) LPS treatment. (A) TEM after staining with uranyl acetate indicates RAW 264.7 cell–derived exosomes are intact after purification (scale=500 nm). (B) Specificity of exosomes was demonstrated by immune gold-labeling for CD81 (scale=100 nm). (C) Western blot analysis of exosomal lysate and media from stimulated (+) or unstimulated (−) RAW 264.7 cells indicates that 2 commonly detected exosomal proteins, HSP70 (mwpred= 70 kDa) and TSG101 (mwpred= 43 kDa, mwobs= 48 kDa), are present in exosomes and absent in exosome-free cell culture media (- lane). LPS is present in cell culture media but is undetectable in exosomes after LPS stimulation. (D) Bioanalyzer trace and gel showing integrity of total exosomal RNA from LPS-stimulated RAW 264.7 cells run on the eukaryotic total RNA pico chip. (E) Bioanalyzer trace of small exosomal RNAs from LPS-stimulated RAW 264.7 cells.
LPS stimulation alters exosomal RNA populations
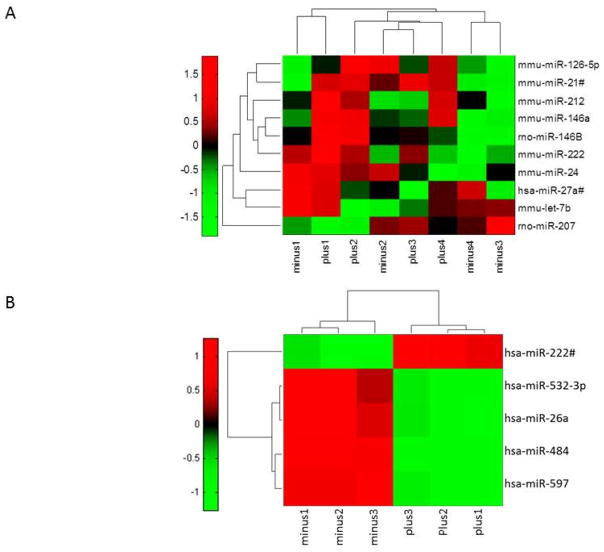
Exosomes contain a variety of coding and noncoding RNAs, but a comprehensive analysis of the total RNA population before and after an inflammatory stimulus has not been undertaken. In this study, we performed quantitative PCR (qPCR) on exosomal miRNA before and after LPS stimulation using Taqman low-density array (TLDA) cards to detect and quantitate up to 758 miRNAs. Our studies detected 433 miRNAs in exosomes derived from naïve and LPS-stimulated RAW 264.7 cells (Supplementary Table 1). We focused on 10 that were significantly altered after LPS treatment (Figure 2, Table 1). This subset of LPS-responsive miRNAs includes 2 miRNAs that are dysregulated in CRPS (miR-126-5p and miR-let7b) and 3 miRNAs previously reported to be upregulated in cells after LPS treatment (miR-146a, miR-146b, and miR-21-3p) [9,59]. Many of the LPS-responsive miRNAs that we detected in exosomes have validated mRNA targets that encode proteins involved in TLR signaling, chemokine signaling, and the TGF-β pathway [34]. We also performed a miRNA profiling study using exosomes purified from THP-1 cells, a human derived monocytic cell line, with and without LPS stimulation. Five significantly altered miRNAs and their validated target mRNAs [34,64] are shown in Table 2. The heat map is shown in Figure 2B. Hsa-miR-532-3p was previously reported to be downregulated in whole blood from CRPS. miR-532-3p also had a significant positive correlation with CRPS type, pain level, IL1Ra, and VEGF [51]. Our data provides evidence indicating that the exosomal biomolecular signature will differ between cell types and is reflective of the cellular and physiological state of the cells secreting the exosomes.
Figure 2.
LPS-responsive exosomal miRNAs. (A) Heat map showing 10 of 433 detected miRNAs in RAW 264.7 cell–derived exosomes with significant alterations after LPS stimulation. Samples are labeled minus for control (exosomes from unstimulated RAW 264.7 cells) and plus for exosomes derived from LPS-treated RAW 264.7 cells (n=4). (B) Heat map showing 5 of 252 detected miRNAs in THP-1 cell–derived exosomes with significant alterations after LPS stimulation. Samples are labeled minus for control (exosomes from unstimulated THP-1 cells) and plus for exosomes derived from LPS-treated THP-1 cells (n=3) (log10fc red = high, black = average, green = low). Significance was determined by applying a P value cutoff of 0.05 to the results of a paired-samples t test.
Table 1.
LPS-responsive exosomal miRNAs and verified mRNA targets
| miRNA | Fold change | P value | Validated human mRNA targets |
|---|---|---|---|
| mmu-miR-21-3p | 114.067 | 0.0207 | Seed sequence is slightly different in human miR-21-3p |
| rno-miR-146b | 30.1762 | 0.0009 | CDKN1A, IRAK1, KIT, MMP16, NF-κB1, PDGFB, TRAF6 |
| mmu-miR-146a | 9.8140 | 0.0190 | BRCA1 & 2, CCNA2, CD40LG, CDKN1A, CDKN3, CFH, CXCR4, EGFR, ERBB4, FADD, FAS, FAF1, IL-8, IRAK 1 & 2, KIF22, MTA2, NF-κB1, PA2G4, ROCK1, SMAD4, TLR2, TLR4, TRAF6, WASF2 |
| mmu-miR-126-5p | 5.8577 | 0.0084 | ADAM9, CXCL12, MMP7, PTPN7, SLC45A3 |
| mmu-miR-212 | 3.7034 | 0.0229 | CCNA2, CCNB1, KCNJ2, MECP2, PEA15, PTCH1, RB1, TJP1 |
| mmu-miR-222 | 2.2474 | 0.0009 | BBC3, CDKN1B/C, CERS2, CORO1A, DICER, DIRAS3, ESR1, ETS1, FOXO3, ICAM1, KIT, MMP1, PPP2R2A, PTEN, RECK, SELE, SOD2, STAT5, TIMP3, TMED7, TNFSF10, TP53, TRPS1 |
| mmu-miR-24 | −1.3151 | 0.0486 | ACVR1B, AURKB, BRCA1, CCNA2, CDK4, CDKN1B, CDKN2A, DND1, DHFR, E2F2, FAF1, FEN1, FURIN, HNF4A, MAPK14, MYC, NOTCH1, POLD1, ST7L, TGFB1, TRIB3, VSIVgp5, ZNF217 |
| rno-miR-27a-5p | −2.2801 | 0.0272 | No validated targets for rare form |
| mmu-let-7b | −3.2575 | 0.0367 | ACTG1, CCNA1/2, CCND1/2, CDC25A, CDC34, CDK6, CYP2J2, HMGA2, IFNB1, LIN28A/B, MTPN, NR2E1, NRAS, PRDM1, RDH10, RPIA |
| rno-miR-207 | −5.4906 | 0.0462 | N/A No human homologue |
LPS-responsive exosomal miRNAs and verified mRNA targets. Statistical analysis of 433 detected miRNAs from LPS-treated and naïve macrophages revealed 10 miRNAs that were significantly altered after LPS stimulation (n=4), including two homologues that are also altered in patients with CRPS (bold). The fold change and P values are reported as well as the validated mRNA targets of these miRNAs from miRTarBase.
Table 2.
LPS-responsive exosomal miRNAs derived from THP-1 cells and their verified mRNA targets.
| miRNA | Fold change | P value | Validated human mRNA targets |
|---|---|---|---|
| hsa-miR-484 | −1.63636 | 6.78E-09 | VEGFB, VEGFR2 [64] FIS1 |
| hsa-miR-597 | −102.238 | 0.000983 | No validated targets |
| hsa-miR-222# | 36.49407 | 0.001038 | No validated targets for the rare form |
| hsa-miR-26a | −22.7778 | 0.001604 | ABCA1, ARL4C, CCND2, CCNE1/2, CDC6, CDK6, CDK8, CPEB2/3/4, CTGF, ESR1, EZH2, GDAP1, GSK3B, HMGA1/2, IFNB1, IL6, MAP3K2, MTDH, MYC, NOS2 PLAG1, PTEN, RB1, SERBP1, SMAD1/4, STRADB |
| hsa-miR-532-3p | −35.0463 | 0.002919 | No validated targets for rare form |
Statistical analysis of 252 detected miRNAs from LPS-treated and naïve THP-1 cells revealed 5 miRNAs that were significantly altered after LPS stimulation (n=3), including one miRNA that was also altered in whole blood from patients with CRPS (bold). The fold change, P values and validated mRNA targets of these miRNAs from miRTarBase and the reference [64] cited shown.
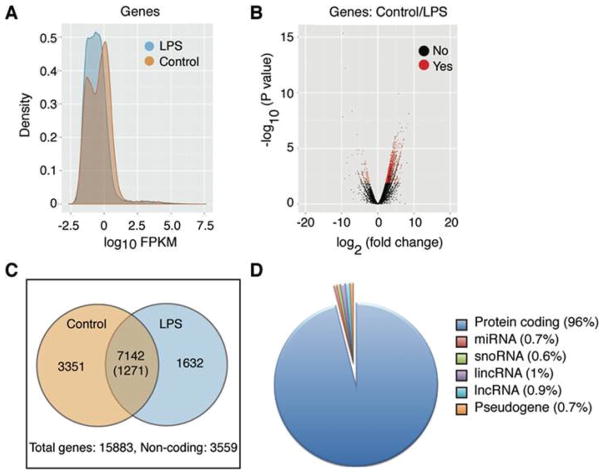
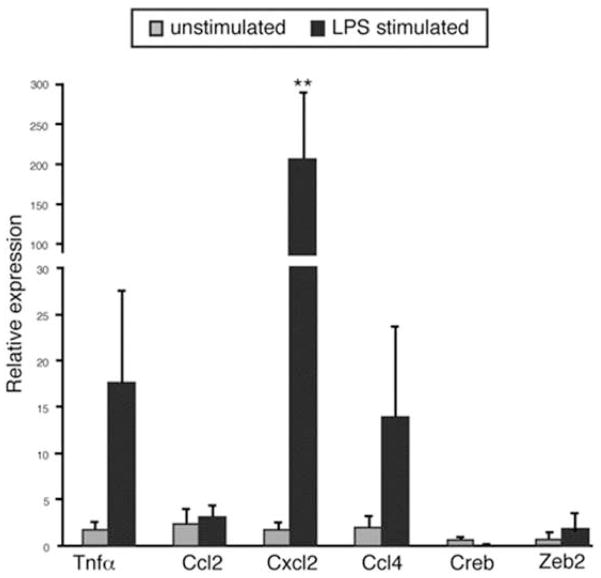
To identify the transcriptome secreted in exosomes from naïve or LPS-stimulated RAW 264.7 cells, we performed next-generation sequencing (NGS). Total reads were mapped from naïve (10323145 reads) and LPS treated (9418995 reads), respectively, and a total of 15883 genes were matched to the mouse genome. Significant differences were observed between naïve and LPS-stimulated exosomal RNA (Figure 3, A and B). Of the 15,883 unique exosomal transcripts, 3559 mapped to noncoding or unverified sequences and the remaining 12,324 were analyzed for differential regulation after LPS stimulation (Figure 3C). Of the 7142 transcripts encoding proteins found in exosomes, 3351 were specific to unstimulated exosomes. Additionally, 1632 genes were found only in LPS-stimulated exosomes and 1271 were differentially expressed after LPS stimulation (Supplementary Table 2). While the majority of the transcripts detected encode proteins, noncoding RNAs were also present (Figure 3D). Small nucleolar RNAs and various subunits of histone H1 were robustly represented in unstimulated exosomes, while reads that mapped to miRNAs were more abundant after LPS stimulation. Upon further investigation of sequences that encode LPS-responsive miRNAs and those altered in patients with CRPS, we found that at least three miRNAs (let-7b, let-7c and mmu-miR-24) were present as both pre-miRNA and mature miRNA forms. We used qPCR to validate 7 mRNAs including Gapdh; 4 mRNAs whose protein product was detected in exosomes (Cxcl2, Ccl2, Ccl4, and Tnfα, see below); and 2 mRNAs encoding transcription factors (Zeb2 and Creb). All transcripts were detected in both samples and Cxcl2 increased significantly after LPS stimulation as seen in the NGS data (Figure 4).
Figure 3.
Statistical analysis of exosomal RNA sequencing data from naïve and LPS-stimulated macrophages. (A) Density analysis of RNA sequencing data showing that the RNA population profiles differ after LPS treatment (FPKM, fragments per kilobase of exon per million). (B) Volcano plot showing the significantly different genes detected in exosomes after LPS stimulation compared with naïve. Red points indicate significantly differentially expressed transcripts (p <= 0.05). (C) Venn diagram of genes that are common and differentially expressed after LPS treatment. The numbers outside the circles indicate the total genes and noncoding RNAs that were detected. (D) Percentage of detected transcripts by RNA type showing that the majority of transcripts detected are protein coding. lincRNA, long intergenic noncoding RNA; lncRNA, long noncoding RNA; miRNA, microRNA; snoRNA, small nucleolar RNA.
Figure 4.
qPCR validation of mRNAs detected by NGS. The mRNA levels of 4 cytokines and 2 transcription factors from exosomes secreted by naïve and LPS-stimulated RAW 264.7 cells were normalized to Gapdh (n=3) **P < 0.01.
Pathway analysis of exosomal RNAs from LPS-stimulated cells compared to total transcripts detected reveals perturbations in multiple cellular pathways (Supplementary Table 3). The reactome database was used to analyze the gene ontology of total transcripts. The top 3 pathways that are represented by exosomal transcripts involve signaling, metabolism, and the immune system (Figure 5). Significantly altered inflammatory pathways representing the innate and adaptive immune systems are shown in Table 3.
Figure 5.
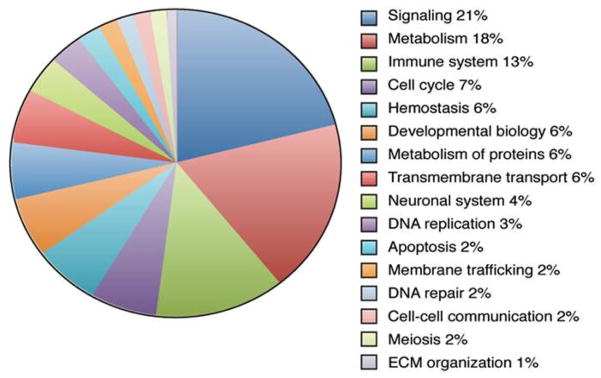
Gene ontology. The distribution of exosomal mRNAs categorized by cellular function based on global reactome pathways. Of the 15,883 genes detected, 5445 are represented in the pie chart. Read clockwise from top.
Table 3.
Immune pathways that are significantly altered after LPS treatment
| Pathway | Transcripts | Fold change | P value | |
|---|---|---|---|---|
| Total | Altered | |||
| Adaptive immune system | 425 | 52 | 1.40 | 0.0078 |
| Class 1 MHC-mediated antigen-processing presentation | 207 | 25 | 1.38 | 0.0596 |
| Antigen-processing cross-presentation | 67 | 11 | 1.88 | 0.0293 |
| Downstream signaling events of B-cell receptor | 88 | 17 | 2.22 | 0.0013 |
| Activation of NF-κB in B cells | 58 | 10 | 1.98 | 0.0271 |
| Antigen processing ubiquitination proteosome degradation | 172 | 23 | 1.53 | 0.0257 |
| Innate immune system | 171 | 19 | 1.27 | 0.1631 |
| Toll receptor cascades | 103 | 14 | 1.56 | 0.0632 |
| MAPK activation in TLR cascade | 49 | 8 | 1.87 | 0.0598 |
| TRAF6-mediated induction of NF-κB & MAPK upon TLR7/8 activation | 69 | 10 | 1.66 | 0.0751 |
| Cytokine signaling in immune system | 213 | 29 | 1.56 | 0.0107 |
Exosomal RNA datasets from naïve and LPS-treated samples were compared and significantly altered pathways involved in immune response are shown with corresponding fold change and P values. Global pathways are shown in bold. TRAF = TNF receptor associated factor.
LPS stimulation leads to increased exosomal cytokines
After LPS treatment, macrophages secrete a variety of chemokines and cytokines that induce the synthesis of additional pro- and anti-inflammatory mediators and act as homing signals for other immune cells. We hypothesized that stimulation of macrophages with LPS would lead to secretion of exosomes carrying a unique cytokine signature that could “prime” the recipient cell for an immune challenge. Consistent with previous studies, LPS stimulation of RAW 264.7 mouse macrophage cells led to the secretion of cytokines into culture media after 24 h (data not shown). Of the 16 cytokines secreted by RAW 264.7 cells after LPS stimulation, 10 were detected in RAW 264.7 cell–derived exosomes (Figure 6). Four proinflammatory cytokines were excluded from exosomes but present in the media (IL-1a, IL-1b, GM-CSF, and IL-6). Those present in LPS-treated exosomes include 2 anti-inflammatory mediators (G-CSF and IL-1Ra), as well as TNFα and a variety of chemokines. Both CCL3 and CCL4 were present in untreated exosomes as well as in LPS treated exosomes.
Figure 6.
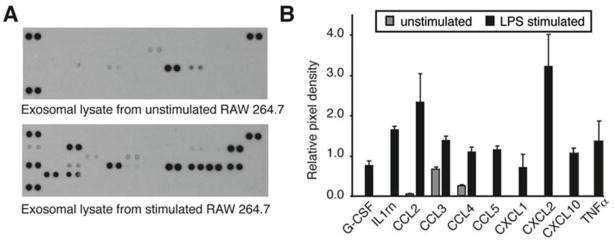
Differential transport of exosome-associated cytokines after LPS treatment. (A) Using an array of 40 cytokines, we quantified exosomal protein content before and after LPS stimulation. (B) Ten cytokines were upregulated in exosomes after LPS treatment. The values are the average and standard deviation from 3 independent experiments (n=3).
In vitro studies of exosomal function
Our analysis demonstrated that in addition to increased cytokines and LPS-responsive RNAs, exosomes from LPS-stimulated RAW 264.7 cells contained many miRNAs and mRNAs in common with those derived from naïve cells. This led us to investigate whether the LPS-induced signature transfers functionality to the recipient cell. Using RAW-Blue cells (InvivoGen; San Diego, CA), which have an inducible, chromosomally integrated secreted alkaline phosphatase (SEAP) gene downstream of the NF-κB promoter, we studied the dose-dependent activation of NF-κB induced by exosomes purified from culture media of naïve or LPS-stimulated RAW 264.7 cells (Figure 7). Using exosomal protein concentration to determine dose, we added 4 concentrations of exosomes from naïve or LPS-stimulated cells to RAW-Blue cells. After a 24 h incubation, we assayed the culture media and saw that exosomes from LPS-stimulated cells caused significantly more NF-κB activation at 50- and 100-μg/ml concentrations compared with exosomes from control cells. Our results demonstrate that purified exosomes are functional and that the exosomes derived from LPS-stimulated macrophages can induce dose-dependent activation of NF-κB.
Figure 7.
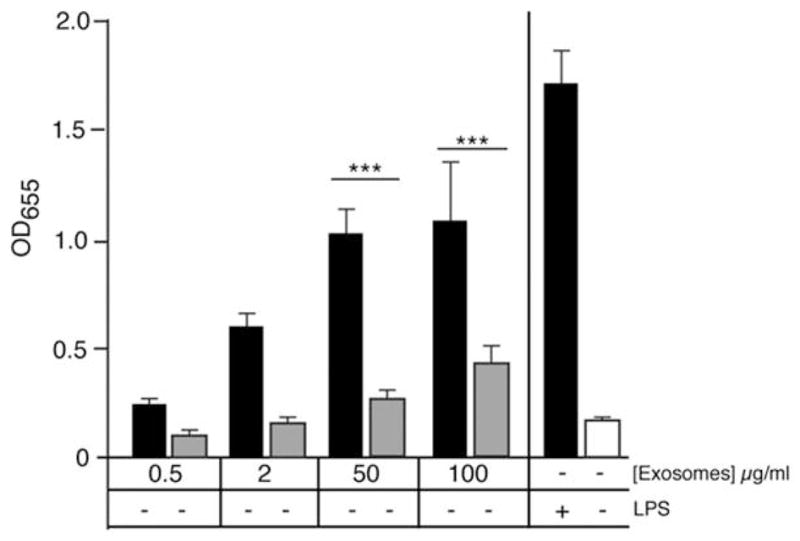
Exosomes purified from LPS-stimulated cells cause activation of NF-κB in naïve cells. Secreted exosomes were purified from RAW 264.7 cell media after 24 h incubation ± 1 μg/ml LPS. Exosomes were incubated at 4 concentrations (based on protein content) with RAW-blue reporter cells and QUANTI-Blue assay was performed at 24 h. Columns 1–8 are exosomes from (+) LPS-stimulated cells or (−) untreated cells and columns 9–10 are media control for NF-κB activation. The average of 3 experiments was analyzed by ANOVA and a Bonferroni posttest to determine P < 0.001.
In vivo studies on exosomal function
To determine the influence of macrophage-derived exosomes on inflammatory pain, we used a mouse model of inflammatory pain, induced by intraplantar injection of CFA suspension of heat-killed Mycobacterium tuberculosis into the hind paw (Figure 8A). We first measured baseline paw thickness, thermal and mechanical sensitivities in 8-week-old male C57BL/6 mice and then established the inflammatory pain model. CFA administration leads to paw swelling as well as mechanical and thermal hypersensitivity within 1 h, while saline treatment does not result in hypersensitivity or swelling. Three hours after the administration of CFA or saline, these mice were given another hind paw injection of 20 μl PBS or 0.5 μg of exosomes purified from either LPS-stimulated or untreated RAW 264.7 cells. This concentration was the maximum volume that could be administered to a mouse paw for the second injection while maintaining consistency of purified exosomes. No swelling was observed in saline-treated animals due to exosome injection (Figure 8B). A single injection of exosomes from LPS-stimulated macrophages resulted in a significant reduction in paw thickness at 24 h (Figure 8C). Exosomes from untreated RAW 264.7 cells did not alter CFA-induced paw swelling. Measurement of mechanical sensitivity using von Frey filaments, beginning 24 h after exosome injection, showed that exosomes did not have any effect on mechanical allodynia induced by CFA (Figure 9A). Exosomes did not induce significant changes in paw withdrawal threshold in saline-treated animals, indicating that an intraplantar injection of exosomes does not evoke mechanical hypersensitivity.
Figure 8.
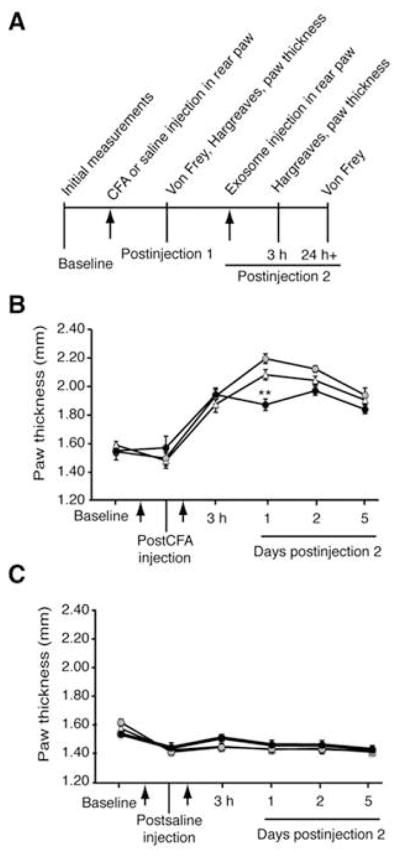
Exosomes from LPS-stimulated macrophages reduce paw thickness in a CFA model of inflammatory pain. (A) Paw thickness and pain behavior measurements before and after each injection. After baseline paw thickness was measured, the CFA model was established in 8-week-old male C57BL/6 mice by intraplantar injection (arrow) into the hind paw. Paw thickness was measured and then a second injection (arrow) of PBS or 20 μl of exosomes (0.5 μg) derived from LPS-stimulated or naïve RAW 264.7 cells were administered by intraplantar injection to the right hind paw 3 h after the CFA injection (n=9). (B) In the CFA-treated paw, one injection of exosomes from LPS-stimulated macrophages (black circle) decreased paw thickness at 24 h compared with exosomes from naïve RAW 264.7 cells (gray circles) or PBS injection (triangle). Statistical analysis performed was one-way ANOVA and Bonferroni posttest. (C) Animals treated with saline did not show any increase in paw thickness with a second injection of macrophage-derived exosomes. Data shown are mean ± SEM.
Figure 9.
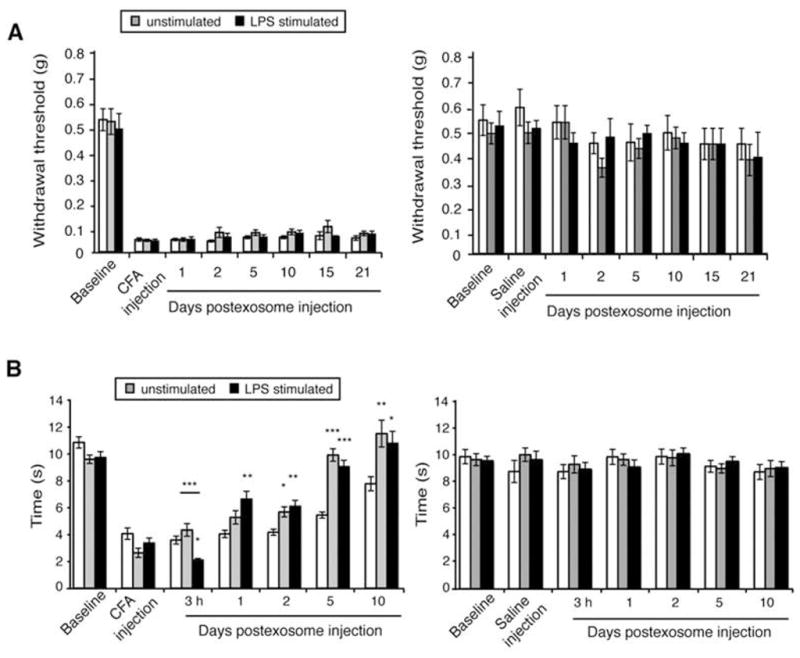
Macrophage-derived exosomes reduce thermal hyperalgesia induced by CFA. (A) Mechanical allodynia was measured by von Frey filaments before and after the CFA model was established in 8-week-old male C57BL/6 mice. A second injection of PBS (white) or exosomes from naïve (gray) or LPS-stimulated (black) RAW 264.7 cells was given 3 h after CFA treatment. Paw withdrawal thresholds were measured at 1, 2, and 5 days and then weekly for 21 d following exosome injection (n=9). Macrophage-derived exosomes had no effect on mechanical allodynia in the CFA-treated animals (right panel) and did not cause hypersensitivity in saline-treated animals as measured by either Hargreaves or von Frey filaments (right panels). (B) Thermal hyperalgesia was measured with Hargreaves direct heat source in CFA-treated (left panel) or saline-treated (right panel) animals. Although exosomes obtained from LPS-stimulated macrophages decreased the paw withdrawal latency immediately upon administration, the effect was transient. At 24 h, exosomes from LPS-stimulated cells reduced thermal hypersensitivity induced by CFA; by 48 h exosomes derived from untreated and LPS-stimulated RAW 264.7 cells reduced thermal hypersensitivity. This protective effect lasted for up to 10 days. Data shown are mean ± SEM. Statistical analysis was determined by ANOVA with Bonferroni posttest. *P<0.05. **P<0.01. ***P<0.001.
We also assessed thermal sensitivity to a radiant heat source using the Hargreaves method. CFA-treated animals became hypersensitive to heat within 1 h of CFA injection whereas saline-treated animals did not (Figure 9B). Saline-treated animals that received injections of exosomes had no hypersensitivity to heat (Figure 9B, right panel). In CFA-treated animals, injection of exosomes purified from LPS-stimulated macrophages (left panel) induced a transient decrease in paw withdrawal latency 3 h after exosome administration, indicating an increase in thermal hypersensitivity that was not observed in saline-treated animals. An injection of exosomes from untreated RAW 264.7 cells into either CFA- or saline-treated animals showed no effect on thermal hyperalgesia at 3 h. By 24 h, CFA-treated animals that received injections of exosomes from LPS-stimulated RAW 264.7 cells had increased paw withdrawal latency compared with CFA-treated animals that received PBS. At 48 h, CFA-treated animals displayed reduction in thermal hyperalgesia in response to exosome administration from both LPS-stimulated and naïve macrophages. Thus the reduction in thermal hypersensitivity observed after 48 h was independent of the inflammatory status of the macrophages from which these exosomes were derived. This attenuation of thermal hyperalgesia was specific to exosome injections and was not observed in CFA-treated animals that received an injection of PBS. One injection of exosomes was sufficient to specifically reduce CFA-induced thermal hypersensitivity within 24 h, accelerate the return to normal sensitivity, and reduce paw inflammation. These results indicate a protective role for exosomes derived from macrophages delivered to an inflamed paw.
Exosomal miRNA analysis in patients with CRPS
To determine whether exosomal miRNAs reflect the miRNA signature in whole blood of patients with CRPS identified in our previous study (26), we analyzed miRNAs in exosomes purified from the serum of 6 patients with CRPS and 6 healthy controls (Figure 10). Of the 12 subjects enrolled in this study, the patients with CRPS (5 women and 1 man) had a mean age of 37.8 years (range: 30–47 years), a mean disease duration of 7.7 years (range: 0.8–30 years), and reported median Numerical Rating Scale pain scores of 8.2 (range: 7–9). The 6 healthy control subjects (3 women and 3 men) had a mean age of 47.5 years (range: 32–69 years). Clinical characteristics of CRPS patients and controls used in exosomal miRNA profiling are shown in Supplementary Table 4.
Figure 10.
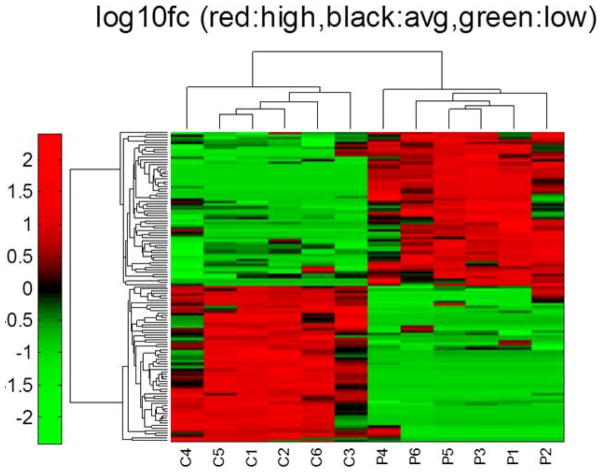
miRNA profiling of CRPS exosomes. Heat map showing 127 of 503 detected miRNAs in human serum exosomes with significant alterations in patients with CRPS. Samples are labeled “C” for control subject and “P” for patient with CRPS, (log10fc red = high, black = average, green = low) (n=6). Significance was determined by applying the Benjamini-Hochberg false discovery rate correction to the results of a 2-tailed t test.
From a total of 503 miRNAs detected in at least one human serum–derived exosomal sample, we identified 127 miRNAs that were significantly different between CRPS and control-derived exosomes (Table 4 and Supplementary Table 5). Sixteen of the 18 miRNAs dysregulated in patients with CRPS from our previous study were detected in human serum–derived exosomes, but only 5 of these (miR-25-3p, miR-320B, miR-939, miR-126-3p, and RNU48) were significantly altered (Table 4). Additionally, the exosomal miRNA signature differed in the directionality of changes compared with that of the whole blood. In the exosomal fraction of patient blood, miR-320B, miR-939, miR-126-3p, and RNU48 were significantly upregulated, whereas they were downregulated in whole blood (26). Thus hsa-miR-25-3p was the only miRNA exhibiting the same trend in whole blood and exosomes in patients with CRPS. Three LPS-responsive miRNAs in exosomes from RAW 264.7 cells (miR-21-3p, miR-126-3p, and miR-212) were also significantly altered in patients with CRPS. These three exosomal miRNAs are increased both in patients with CRPS and in RAW cells after LPS stimulation. Analysis of exosomes from a larger sample of patients might yield more information on whether exosomal miRNA could be a useful biomarker or a secondary strategy for patient stratification.
Table 4.
Differentially expressed miRNAs in exosomes from patients with CRPS compared with control subjects
| miRNA | p-value | fold change | miRNA | p-value | fold change | miRNA | p-value | fold change | miRNA | p-value | fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-890 | 5.70E-08 | 1900.0 | hsa-miR-576-3p | 0.0004 | 14.1 | hsa-miR-628-3p | −0.003 | −3.7 | hsa-miR-190b | −0.0006 | −58.1 |
| hsa-miR-577 | 0.001 | 482.3 | hsa-miR-1208 | 1.50E-05 | 12.8 | hsa-miR-25# | −0.0006 | −3.8 | hsa-miR-650 | −0.007 | −63.6 |
| hsa-miR-15b# | 4.00E-08 | 284.7 | hsa-miR-320B | 0.002 | 12.6 | hsa-miR-30d# | −0.0006 | −4.1 | hsa-miR-639 | −0.002 | −64.1 |
| hsa-miR-422a | 2.30E-07 | 204.3 | hsa-miR-541# | 0.0007 | 11.2 | hsa-miR-29c | −0.007 | −6.8 | hsa-miR-379 | −6.40E-11 | −76.3 |
| hsa-miR-1290 | 0.003 | 178.6 | hsa-miR-22# | 0.001 | 10.3 | hsa-miR-646 | −0.0001 | −6.9 | hsa-miR-302c | −0.005 | −101.8 |
| hsa-miR-939 | 0.002 | 103.4 | hsa-miR-10b# | 0.003 | 10.1 | hsa-miR-26b# | −0.004 | −7.6 | hsa-miR-34a# | −6.50E-07 | −110.0 |
| hsa-miR-190 | 7.00E-08 | 77.9 | hsa-miR-15a | 0.0002 | 9.4 | hsa-miR-491-5p | −0.002 | −7.9 | hsa-miR-656 | −0.003 | −110.7 |
| hsa-miR-32 | 1.30E-06 | 75.2 | hsa-miR-660 | 0.007 | 8.2 | hsa-miR-758 | −0.0003 | −9.1 | hsa-miR-1259 | −0.0004 | −141.5 |
| hsa-miR-331-5p | 0.001 | 72.1 | hsa-miR-223# | 0.006 | 8.0 | hsa-miR-513C | −2.50E-06 | −10.2 | hsa-miR-518f | −7.40E-05 | −145.3 |
| hsa-miR-603 | 0.0003 | 71.7 | hsa-miR-93# | 0.008 | 7.5 | hsa-miR-1253 | −0.004 | −14.3 | hsa-miR-432 | −0.0001 | −155.5 |
| hsa-miR-129-5p | 2.70E-08 | 62.7 | hsa-miR-130b | 0.0003 | 7.5 | hsa-miR-636 | −0.0005 | −15.3 | hsa-miR-572 | −2.50E-05 | −161.9 |
| hsa-miR-126# | 0.004 | 62.3 | hsa-miR-625 | 0.004 | 6.8 | hsa-miR-1282 | −1.00E-06 | −15.4 | hsa-miR-615-3p | −2.80E-07 | −199.5 |
| hsa-miR-642 | 4.40E-07 | 57.5 | hsa-miR-330-3p | 0.0003 | 6.8 | RNU48 | −0.002 | −15.8 | hsa-miR-1294 | −7.00E-06 | −201.9 |
| hsa-miR-342-5p | 2.70E-09 | 51.6 | hsa-miR-212 | 0.008 | 6.6 | hsa-miR-433 | −0.009 | −11.9 | hsa-miR-154# | −3.80E-06 | −3341.1 |
| hsa-miR-125a-3p | 8.90E-08 | 46.6 | hsa-miR-9 | 0.0003 | 6.4 | hsa-miR-485-3p | −4.80E-05 | −12.3 | hsa-miR-1276 | −1.70E-07 | −268.6 |
| hsa-miR-590-3P | 8.00E-05 | 40.0 | hsa-miR-520c-3p | 0.002 | 6.4 | hsa-miR-200b | −0.002 | −12.7 | hsa-miR-519a | −0.004 | −279.6 |
| hsa-miR-301b | 9.70E-05 | 39.2 | hsa-miR-378 | 0.007 | 5.9 | hsa-miR-455-3p | −1.20E-06 | −16.2 | hsa-miR-324-3p | −4.70E-05 | −311.1 |
| hsa-miR-21# | 3.10E-06 | 38.2 | hsa-miR-886-5p | 0.002 | 5.7 | hsa-miR-1227 | −6.90E-06 | −21.6 | hsa-miR-872 | −3.00E-05 | −373.8 |
| hsa-miR-409-5p | 1.60E-09 | 37.3 | hsa-miR-200c | 0.006 | 4.7 | hsa-miR-184 | −1.00E-07 | −22.3 | hsa-miR-1233 | −4.60E-05 | −451.9 |
| hsa-miR-487b | 0.001 | 37.0 | hsa-miR-361-5p | 0.001 | 4.7 | hsa-miR-489 | −0.0001 | −22.9 | hsa-miR-302d | −3.20E-05 | −1332.2 |
| hsa-miR-218 | 0.003 | 32.3 | hsa-miR-98 | 0.003 | 4.4 | hsa-miR-512-3p | −1.60E-07 | −27.8 | hsa-miR-1276 | −1.70E-07 | −268.6 |
| hsa-miR-215 | 0.0001 | 31.0 | hsa-miR-509-5p | 0.001 | 4.2 | hsa-miR-34a | −2.50E-06 | −30.4 | hsa-miR-519a | −0.004 | −279.6 |
| hsa-miR-210 | 2.50E-07 | 25.0 | hsa-miR-654-5p | 0.0002 | 3.4 | hsa-miR-367 | −0.003 | −31.7 | hsa-miR-324-3p | −4.70E-05 | −311.1 |
| hsa-miR-545 | 0.001 | 24.8 | hsa-miR-140-3p | 0.009 | 3.3 | hsa-miR-645 | −0.007 | −33.2 | hsa-miR-872 | −3.00E-05 | −373.8 |
| hsa-miR-10b | 0.004 | 23.7 | hsa-miR-100 | 0.007 | 2.9 | hsa-miR-767-5p | −4.20E-06 | −34.8 | hsa-miR-1233 | −4.60E-05 | −451.9 |
| hsa-miR-372 | 0.003 | 22.6 | hsa-miR-502-5p | 0.0008 | 2.7 | hsa-miR-506 | −0.0004 | −37.6 | hsa-miR-302d | −3.20E-05 | −1332.2 |
| hsa-miR-551b | 0.0004 | 21.6 | hsa-miR-874 | 0.0006 | 2.4 | hsa-miR-522 | −0.0009 | −38.1 | hsa-miR-154# | −3.80E-06 | −3341.1 |
| hsa-miR-339-5p | 0.0008 | 21.4 | hsa-miR-497 | −0.005 | −1.7 | hsa-miR-20a# | −2.70E-05 | −39.2 | |||
| hsa-miR-501-5p | 0.0004 | 21.0 | hsa-miR-588 | −0.005 | −2.0 | hsa-miR-23a# | −5.30E-05 | −40.6 | |||
| hsa-miR-382 | 0.003 | 20.9 | hsa-miR-770-5p | −0.002 | −2.7 | hsa-miR-744# | −0.0003 | −40.7 | |||
| hsa-miR-1255B | 0.004 | 19.1 | hsa-miR-99b# | −0.0008 | −2.9 | hsa-miR-630 | −8.20E-05 | −42.4 | |||
| hsa-miR-144 | 0.002 | 18.9 | hsa-miR-1182 | −0.0002 | −3.3 | hsa-miR-518e | −1.80E-05 | −42.4 | |||
| hsa-miR-302b | 4.30E-06 | 17.4 | hsa-miR-1303 | −0.0004 | −3.5 | hsa-miR-520b | −1.40E-05 | −43.7 | |||
| hsa-miR-31 | 0.002 | 16.2 | hsa-miR-204 | −0.003 | −3.5 | hsa-miR-511 | −0.0008 | −44.6 | |||
| hsa-miR-601 | 0.0001 | 14.5 | hsa-miR-1269 | −0.0003 | −3.6 | hsa-miR-337-5p | −3.30E-05 | −50.4 | |||
miRNAs shown in bold were identified in our previous study to be differentially expressed in whole blood from patients with CRPS. Significance was determined by applying the Benjamini-Hochberg false discovery rate correction to the results of a 2-tailed t test.
Discussion
Homeostasis after injury or infection is re-established by temporally and spatially regulating both proinflammatory response to noxious conditions and protective mechanisms governing resolution of inflammation. Exosomes are a systemic form of intercellular communication recognized for their antigen-presenting capabilities and their ability to transfer pathogenic and nonpathogenic biomolecules [16,23,26–28,33,52,53]. We demonstrate that exosomes secreted by macrophages stimulated with LPS carry higher levels of 3 murine homologs for human miRNAs with known roles in preventing overactivation of the innate immune response (miR-21-3p, miR-146a, and miR-146b). NF-κB1 and other mRNAs involved in TLR signaling are repressed by these 3 miRNAs [34,59]. This leads to inhibition of proinflammatory cytokine transcription and translation, a necessary step in resolution of inflammation [3,11]. miR-21 is also an NF-κB-induced suppressor of inflammation and acts as a molecular switch regulating the synthesis of protective cytokines, IL-10 and IL-4 [3,11]. miR-21 has been proposed to be a hormone miRNA that influences the resolution of inflammation by binding TLR7/8 [25]. These data suggest that macrophages secrete exosomal miRNAs known to influence inflammatory pathways and that LPS stimulation leads to the secretion of exosomes that have increased levels of miRNAs that contribute to resolution of inflammation in the recipient cells. miRNA profile in THP-1-derived exosomes confirms that exosomal contents are reflective of the cellular environment of the secreting cells and will differ between cell types.
Previously, profiling and sequence analysis of exosomal RNA revealed both miRNAs and mRNAs in exosomes [32,40,48,50,55,65] and functionality in the recipient cell after uptake [56,62]. RNA sequencing of naïve and activated macrophages has identified an inflammation-induced transcriptome, which is temporally and spatially regulated by the timing of transcription factor activation and the localization of nascent transcripts [8,46]. To determine whether inflammation-induced alterations are reflected in exosomal transcriptome, we sequenced exosomal RNA from naïve and LPS-stimulated macrophages. Pathway analysis of exosomal mRNAs from naïve cells compared with those from LPS-stimulated cells indicates significant changes in both the adaptive and innate immune processes, specifically pathways related to NF-κB activation and Toll receptor cascades. We also observed a striking shift in the most abundant noncoding RNA populations with LPS stimulation. The presence of snoRNAs and pre-miRNAs suggest that exosomes deliver molecules that could induce temporal epigenetic regulation in recipient cells regulating the course of inflammatory gene expression [46]. The influx of inflammatory-relevant pre-miRNAs in LPS-stimulated cells could indicate the need for a rapid response to inflammation rather than regulation at the nuclear level. Mature miRNAs may fine-tune the regulation of inflammation by altering the mRNA levels of inflammatory proteins immediately, while pre-miRNAs offer a second wave of regulation at a later time point. Exosomes have the advantage of delivering proteins and miRNAs that are primed to act directly and immediately along with mRNAs that can be readily translated.
Here we show that exosomes derived from macrophages after LPS stimulation had increased cytokine content, predominantly chemokines. LPS induced secretion of interleukins and cytokines from macrophages; notably, TNFα, CCL3, CCL4, CCL5, G-CSF, IL-1RA, and CXCL2 were present in exosomes, but interleukin proteins were undetectable. Increase in exosomal cytokines after LPS stimulation of macrophages suggests that activated macrophages secrete exosomes that are primed to signal an immune response to recipient cells. Previous study on exosomes derived from cells infected with Mycobacterium bovis bacillus Calmette–Guérin or Salmonella indicates that, at 48 h after infection, secreted exosomes contain pathogen-associated molecular patterns that stimulate cytokine secretion in recipient cells, but the content of cytokines delivered in exosomes was not investigated [6,7]. We did not see detectable levels of LPS in exosomes secreted 24 h after LPS stimulation and the abundance of cytokines detected in exosomes after LPS stimulation of macrophages indicates that exosomal content can influence inflammatory response. This is further established by our finding that exosomes originating from macrophages stimulated with LPS lead to transcriptional activation of NF-κB, whereas exosomes from unstimulated cells were less effective in activating NF-κB even at the highest concentration tested. We suggest that LPS stimulation resulted in “primed” exosomes that not only caused robust NF-κB activation when added to RAW-Blue cells but also temporarily reduced paw swelling in CFA-treated animals 24 h after injection. The increase in immune-specific miRNAs and cytokines in exosomes from LPS-stimulated cells indicate that exosomal contents reflect the secreting cellular environment and can potentially mediate inflammatory pathways in a temporal manner.
While immune cells are known to secrete exosomes with source-specific immunoregulatory functions, an anti-inflammatory role for exosomes has recently been demonstrated. DC-derived exosomes protected mice from collagen-induced arthritis [14,38]. Here we show that immunosuppression can also be achieved by naïve exosomes derived from yet another kind of antigen-presenting cell, the macrophages. A direct injection of exosomes into CFA-treated paws attenuated thermal hypersensitivity as early as 24 h. At 48 h, CFA-treated animals displayed a significant reduction in thermal hyperalgesia in response to exosome administration derived from both naïve and LPS-stimulated macrophages. Interestingly, exosomes from LPS-stimulated macrophages showed an acute increase in thermal hypersensitivity at 3 h in CFA-treated, but not in saline-treated paws of control mice. We conclude that increased hypersensitivity seen only in CFA-treated paws at 3 h may be due to the unique inflammatory signature resulting from LPS stimulation, because exosomes were washed to remove all traces of media and LPS was undetected by immunoblotting. The fact that saline-treated animals did not show hypersensitivity at any point after the exosome injection may indicate a threshold that must first be overcome to induce hypersensitivity response seen at 3 h in CFA-treated animals injected with exosomes. The lack of pain and swelling in saline-treated paws after exosome injection also demonstrates that exosomal delivery does not produce a proinflammatory response. The lack of immune response caused by injection of exosomes and the fact that exosomes can be loaded with nucleic acids or drugs opens the possibility of exosomes as novel avenues for pain therapy [2,4,21].
It was recently shown that miRNAs are necessary to regulate the protein and mRNAs that induce inflammatory pain but do not affect acute pain responses in nociceptive neurons [66]. Attenuation of thermal hyperalgesia by macrophage-derived exosomes in CFA-treated animals could reflect the temporal regulation that exosomes can mediate by synergistically influencing multiple inflammatory pathways through delivery of immediate-acting biomolecules as well as those that are translation dependent or that cause changes in recipient cells by affecting gene transcription.
Although the mechanisms are unclear, chronic, nonresolving inflammation in CRPS is characterized by prolonged secretion of proinflammatory factors, disruption of the blood–brain barrier, and development of autoimmunity [17]. The pain and inflammation in CRPS are not alleviated by commonly used analgesics or anti-inflammatory drugs, and the development of persistent pain states and the maintenance of chronic inflammation point to systemic aberrations. Of the 18 circulating miRNAs that were shown to be predictive of CRPS [51], we detected 16 in exosomes derived from human serum. We identified a subset of exosomal miRNAs that showed significant alterations in patients with CRPS. miR-939, miR-320B, miR-126-5p, and miR-25-3p were previously shown to be significantly downregulated in whole blood. Three of these miRNAs, with the exception of miR-25-3p, were significantly upregulated in the exosomal fraction even with the small sample size. This negative correlation may indicate a protective role for exosomes in CRPS; by carrying increased amounts of miRNAs that can modulate proinflammatory mRNAs to recipient cells, exosome-mediated transport of miRNAs may be a systemic attempt to resolve the chronic condition. Three miRNAs that were significantly increased in exosomes from patients with CRPS were also upregulated in exosomes after LPS stimulation of macrophages (miR-126-5p, miR-21# or miR-21-3p, and miR-212). These three miRNAs have validated inflammatory-relevant mRNA targets which suggests that patient exosomes are delivering miRNAs in an attempt to reduce the availability of inflammatory mRNAs to recipient cells. The limited availability of patient blood prevented the complete characterization and further mechanistic studies of exosomal content, but these preliminary results point to a significant contribution of exosomes to CRPS pathology. Further studies are needed to investigate whether packaging of specific miRNAs into exosomes can be influenced by inflammatory stimulus.
Taken together, our data suggest that uptake of exosomes enables spatial and temporal regulation of molecular events in recipient cells, including the resolution of inflammation and the reduction in thermal hypersensitivity resulting from inflammatory pain. Though activated macrophages secrete a variety of cellular mediators into the local environment, exosomes can mediate both local and systemic intercellular communication. Because not all known secreted proteins detected in the medium are present in exosomes, the packaging of biomolecular cargo into exosomes can be thought of as a regulated and not a random process. The presence of a variety of noncoding RNAs and histone transcripts in exosomes indicates that the epigenetic regulation of cellular processes can be mediated via circulation; further studies are needed to confirm this. In a nonresolving inflammatory state, exosomes may be transporting biomolecules in an attempt to restore homeostasis. Exosomes derived from antigen-presenting cells, with their diverse array of biomolecules, can induce pleiotropic effects and should be further explored for their therapeutic utility in treating chronic pain.
Supplementary Material
Exosomal miRNA found in macrophage-derived exosomes. Purified exosomal RNA samples from RAW 264.7 cells (untreated and stimulated with LPS) were profiled for 758 miRNAs using TLDA cards. The ΔΔCt and P values of detected miRNAs for 4 separate treatments including the number of times each miRNA was detected are shown. Homologues of miRNAs identified in our previous study to be differentially expressed in whole blood from patients with CRPS shown in bold.
LPS-responsive exosomal mRNAs. Analysis of exosomal mRNA levels after LPS stimulation of RAW 264.7 cells compared with exosomes from naïve cells revealed that transcripts known to be involved in pain and inflammation are upregulated and transported in exosomes. All genes with greater than 2-fold increase after LPS stimulation are reported.
Pathway analysis of exosomal RNAs from LPS-stimulated cells compared with naïve cells. Cellular pathways that are altered by changes in exosomal content after LPS stimulation of cells are listed by fold change and significance.
Clinical characteristics of CRPS patients and control used in exosomal miRNA profiling.
All miRNAs detected in exosomes from human serum for control subjects and patients with CRPS. The ΔΔCt values and significance for the differentially regulated miRNAs are also shown. miRNAs that were previously reported to be significantly different in whole blood from CRPS and control samples are shown in bold at the top of the table.
Acknowledgments
This study was supported by grants from the NIH (1R21NS082991-01) and the Rita Allen Foundation and by a Professional Enrichment and Growth grant from Drexel University College of Medicine to Seena Ajit. The authors would like to acknowledge Erika Balogh and Dr. Soumitra Ghoshroy from the University of South Carolina Electron Microscopy Center for instrument use and for scientific and technical assistance. We would like to thank Marielle Perreault for her help in coordinating collection of blood samples and compiling clinical parameters. We thank Dr. Huijuan Hu, Dr. Paul McGonigle and Dr. Peter Katsikis for critical reading of the manuscript, Diana Winters for editing and Kathleen Delgrosso, Megan Musick, and Kathryn Scott from the Kimmel Cancer Center for the NGS technical support.
Footnotes
Conflict of interest statement: Michael Gormley is an employee at Janssen Research and Development, Spring House, Pennsylvania.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Development Core Team R. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. http://www.R-project.org. [Google Scholar]
- 2.Ajit SK. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 2012;12(3):3359–3369. doi: 10.3390/s120303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alam MM, O’Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. European journal of immunology. 2011;41(9):2482–2485. doi: 10.1002/eji.201141740. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez-Erviti L, Seow Y, Yin HF, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatty WL, Rhoades ER, Ullrich HJ, Chatterjee D, Heuser JE, Russell DG. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1(3):235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 7.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110(9):3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150(2):279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianco NR, Kim SH, Ruffner MA, Robbins PD. Therapeutic effect of exosomes from indoleamine 2,3-dioxygenase-positive dendritic cells in collagen-induced arthritis and delayed-type hypersensitivity disease models. Arthritis and rheumatism. 2009;60(2):380–389. doi: 10.1002/art.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-kappaB. Immunological reviews. 2012;246(1):205–220. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 12.Bruehl S. An update on the pathophysiology of complex regional pain syndrome. Anesthesiology. 2010;113(3):713–725. doi: 10.1097/ALN.0b013e3181e3db38. [DOI] [PubMed] [Google Scholar]
- 13.Campbell JN, Meyer RA. Mechanisms of neuropathic pain. Neuron. 2006;52(1):77–92. doi: 10.1016/j.neuron.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charbonnier L-M, van Duivenvoorde LM, Apparailly F, Cantos C, Han WGH, Noël D, Duperray C, Huizinga TWJ, Toes REM, Jorgensen C, Louis-Plence P. Immature Dendritic Cells Suppress Collagen-Induced Arthritis by In Vivo Expansion of CD49b+ Regulatory T Cells. The Journal of Immunology. 2006;177(6):3806–3813. doi: 10.4049/jimmunol.177.6.3806. [DOI] [PubMed] [Google Scholar]
- 15.Chiu IM, von Hehn CA, Woolf CJ. Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nature neuroscience. 2012;15(8):1063–1067. doi: 10.1038/nn.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clayton A, Mason MD. Exosomes in tumour immunity. Curr Oncol. 2009;16(3):46–49. doi: 10.3747/co.v16i3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper MS, Clark VP. Neuroinflammation, Neuroautoimmunity, and the Co-Morbidities of Complex Regional Pain Syndrome. Journal of neuroimmune pharmacology: the official journal of the Society on Neuro Immune Pharmacology. 2012 doi: 10.1007/s11481-012-9392-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, Lotvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Mos M, Sturkenboom MC, Huygen FJ. Current understandings on complex regional pain syndrome. Pain practice: the official journal of World Institute of Pain. 2009;9(2):86–99. doi: 10.1111/j.1533-2500.2009.00262.x. [DOI] [PubMed] [Google Scholar]
- 20.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. Journal of cell science. 2000;113(Pt 19):3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 21.El Andaloussi S, Lakhal S, Mager I, Wood MJ. Exosomes for targeted siRNA delivery across biological barriers. Advanced drug delivery reviews. 2013;65(3):391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 22.El Andaloussi S, Mager I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 23.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30(20):6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabbri M, Paone A, Calore F, Galli R, Croce CM. A new role for microRNAs, as ligands of Toll-like receptors. RNA biology. 2013;10(2):169–174. doi: 10.4161/rna.23144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(26):9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fruhbeis C, Frohlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Mobius W, Goebbels S, Nave KA, Schneider A, Simons M, Klugmann M, Trotter J, Kramer-Albers EM. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS biology. 2013;11(7):e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomes C, Keller S, Altevogt P, Costa J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neuroscience letters. 2007;428(1):43–46. doi: 10.1016/j.neulet.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 29.Guil Sn, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. The international journal of biochemistry & cell biology. 2009;41(1):87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Harden RN, Bruehl S, Perez RS, Birklein F, Marinus J, Maihofner C, Lubenow T, Buvanendran A, Mackey S, Graciosa J, Mogilevski M, Ramsden C, Chont M, Vatine JJ. Validation of proposed diagnostic criteria (the “Budapest Criteria”) for Complex Regional Pain Syndrome. Pain. 2010;150(2):268–274. doi: 10.1016/j.pain.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harden RN, Bruehl S, Stanton-Hicks M, Wilson PR. Proposed new diagnostic criteria for complex regional pain syndrome. Pain Med. 2007;8(4):326–331. doi: 10.1111/j.1526-4637.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- 32.Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, Kim JH, Choi DS, Kim YK, Hwang D, Gho YS. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC genomics. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71(11):3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 34.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic acids research. 2011;39(Database issue):D163–169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, Paulaitis ME, Piper MG, Marsh CB. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121(6):984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Krämer-Albers E-M, Lim SK, Llorente A, Lötvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-’t Hoen ENM, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TSK, Rajendran L, Raposo G, Record M, Reid GE, Sánchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BWM, Vázquez J, Vidal M, Wauben MHM, Yáñez-Mó M, Zoeller M, Mathivanan S. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS biology. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC Genome Browser database: 2014 update. Nucleic acids research. 2014;42(1):D764–770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SH, Lechman ER, Bianco N, Menon R, Keravala A, Nash J, Mi Z, Watkins SC, Gambotto A, Robbins PD. Exosomes derived from IL-10-treated dendritic cells can suppress inflammation and collagen-induced arthritis. J Immunol. 2005;174(10):6440–6448. doi: 10.4049/jimmunol.174.10.6440. [DOI] [PubMed] [Google Scholar]
- 39.Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, Kojima A, Kimura Y, Komori T, Asai K. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic acids research. 2007;35(Database issue):D145–148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koh W, Sheng CT, Tan B, Lee QY, Kuznetsov V, Kiang LS, Tanavde V. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC genomics. 2010;11 (Suppl 1):S6. doi: 10.1186/1471-2164-11-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kynast KL, Russe OQ, Geisslinger G, Niederberger E. Novel findings in pain processing pathways: implications for miRNAs as future therapeutic targets. Expert Rev Neurother. 2013;13(5):515–525. doi: 10.1586/ern.13.34. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor perspectives in biology. 2009;1(6):a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. The international journal of biochemistry & cell biology. 2012;44(1):11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic acids research. 2012;40(1):D1241–1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 46.McCall CE, El Gazzar M, Liu T, Vachharajani V, Yoza B. Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J Leukoc Biol. 2011;90(3):439–446. doi: 10.1189/jlb.0211075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald MK, Capasso KE, Ajit SK. Purification and microRNA profiling of exosomes derived from blood and culture media. Journal of visualized experiments: JoVE. 2013;(76):e50294. doi: 10.3791/50294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature communications. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicotra L, Loram LC, Watkins LR, Hutchinson MR. Toll-like receptors in chronic pain. Experimental neurology. 2011 doi: 10.1016/j.expneurol.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic acids research. 2012;40(18):9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK. MicroRNA modulation in complex regional pain syndrome. J Transl Med. 2011;9(1):195. doi: 10.1186/1479-5876-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(30):11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rak J. Microparticles in cancer. Seminars in thrombosis and hemostasis. 2010;36(8):888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 54.Schwartzman RJ, Alexander GM, Grothusen J. Pathophysiology of complex regional pain syndrome. Expert Rev Neurother. 2006;6(5):669–681. doi: 10.1586/14737175.6.5.669. [DOI] [PubMed] [Google Scholar]
- 55.Skog J, Wurdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature cell biology. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119(3):646–648. doi: 10.1182/blood-2011-11-389478. [DOI] [PubMed] [Google Scholar]
- 58.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 61.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 63.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 64.Vecchione A, Belletti B, Lovat F, Volinia S, Chiappetta G, Giglio S, Sonego M, Cirombella R, Onesti EC, Pellegrini P, Califano D, Pignata S, Losito S, Canzonieri V, Sorio R, Alder H, Wernicke D, Stoppacciaro A, Baldassarre G, Croce CM. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(24):9845–9850. doi: 10.1073/pnas.1305472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, Zacharias W, Hao H, McMasters KM. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PloS one. 2012;7(10):e46874. doi: 10.1371/journal.pone.0046874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao J, Lee MC, Momin A, Cendan CM, Shepherd ST, Baker MD, Asante C, Bee L, Bethry A, Perkins JR, Nassar MA, Abrahamsen B, Dickenson A, Cobb BS, Merkenschlager M, Wood JN. Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J Neurosci. 2010;30(32):10860–10871. doi: 10.1523/JNEUROSCI.1980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exosomal miRNA found in macrophage-derived exosomes. Purified exosomal RNA samples from RAW 264.7 cells (untreated and stimulated with LPS) were profiled for 758 miRNAs using TLDA cards. The ΔΔCt and P values of detected miRNAs for 4 separate treatments including the number of times each miRNA was detected are shown. Homologues of miRNAs identified in our previous study to be differentially expressed in whole blood from patients with CRPS shown in bold.
LPS-responsive exosomal mRNAs. Analysis of exosomal mRNA levels after LPS stimulation of RAW 264.7 cells compared with exosomes from naïve cells revealed that transcripts known to be involved in pain and inflammation are upregulated and transported in exosomes. All genes with greater than 2-fold increase after LPS stimulation are reported.
Pathway analysis of exosomal RNAs from LPS-stimulated cells compared with naïve cells. Cellular pathways that are altered by changes in exosomal content after LPS stimulation of cells are listed by fold change and significance.
Clinical characteristics of CRPS patients and control used in exosomal miRNA profiling.
All miRNAs detected in exosomes from human serum for control subjects and patients with CRPS. The ΔΔCt values and significance for the differentially regulated miRNAs are also shown. miRNAs that were previously reported to be significantly different in whole blood from CRPS and control samples are shown in bold at the top of the table.