Abstract
Cholesterol synthesis in fetal brain is inhibited because activity of DHCR24 (24-dehydrocholesterol reductase) is insufficient causing concentrations of the precursor desmosterol to increase temporarily to 15-25% of total sterols at birth. We demonstrate that failure of DHCR24 to be adequately upregulated during periods of elevated cholesterol synthesis in brain results from the presence in its promoter of the RE1 nucleotide sequence that binds the RE1-Silencing Transcription Factor (REST) and that REST, generally reduced in neural tissues, uncharacteristically but not without precedent, enhances DHCR24 transcription. DHCR24 and REST mRNA levels are reduced 3- to 4-fold in fetal mouse brain compared to liver, (P<0.001). Chromatin immunoprecipitation assays suggested that REST binds to the human DHCR24 promoter in the vicinity of the predicted human RE1 sequence. Luminescent emission from a human DHCR24 promoter construct with a mutated RE1 sequence was reduced 2-fold compared to output from a reporter with wild type RE1 (P<0.005). Silencing REST in HeLa cells resulted in significant reductions of DHCR24 mRNA (2-fold) and DHCR24 protein (4-fold). As expected, relative concentrations of Δ24-cholesterol precursor sterols increased 3- to 4-fold reflecting the inhibition of DHCR24 enzyme activity. In contrast, mRNA levels of DHCR7 (sterol 7-dehydrocholesterol reductase), a gene essential for cholesterol synthesis lacking a RE1 sequence, and concentrations of HMGR (3-hydroxy-3-methyl-glutaryl-CoA reductase) enzyme protein, were both unaffected. Surprisingly, a dominant-negative fragment of REST consisting of just the DNA binding domain (about 20% of the protein) and full length REST enhanced DHCR24 expression equally well. Furthermore, RE1 and SRE, the respective binding sites for REST and the sterol response element binding protein (SREBP), are contiguous. These observations led us to hypothesize that REST acts because it is bound in close proximity to SREBP amplifying its ability to upregulate DHCR24. Modulation of DHCR24 expression by REST likely persisted in the mammalian genome either because it does no harm or because suppressing metabolically active DHCR24 while providing abundant quantities of the multi-functional sterol desmosterol during neural development proved useful.
Keywords: Cholesterol, regulation, neural tissue, development, RE1, SRE
Introduction
Regulation of cholesterol metabolism in fetal mammalian brain is puzzling because, at a time when the entire cholesterol biosynthetic pathway is massively upregulated, the reaction that reduces the sterol C24,25-double bond (Supplementary Fig. S2), catalyzed by DHCR24 (24-dehydrocholesterol reductase), fails to keep pace.
Although the mature brain is our most cholesterol-rich organ [1], an efficient block forms early in gestation preventing mother from supplying more than about 25% of the cholesterol needed by her newborn fetus [2-4]. Despite the nearly complete bar to maternal cholesterol transfer imposed by the blood-brain barrier which forces the fetal brain to make most of its own cholesterol [4], activity of DHCR24 is inadequate [4;5]. As a result, desmosterol, as the ultimate cholesterol precursor, accumulates to 15-25% of total brain sterols at birth [4;6-8]. This unusually large pool of desmosterol does not persist, however, because an older brain (as well as peripheral nerves) make very little new cholesterol much preferring to recycle [9;10]. This enables most desmosterol to be converted to cholesterol, albeit somewhat slowly [5;8;11].
DHCR24, unique among the genes of cholesterol synthesis and metabolism in its failure to be expressed adequately, is a flavin adenine dinucleotide (FAD)-dependent oxidoreductase [12], thought capable of reducing the C24,25-double bond of most Δ24-cholesterol intermediates including desmosterol during cholesterol biosynthesis [13;14] (Supplementary Fig. S2). Here we demonstrate that expression of DHCR24 at levels sufficient to accommodate the high rates of cholesterol synthesis in the developing brain requires the presence of REST/NRSF (RE1-Silencing Transcription factor/Neuron-Restrictive Silencing Factor), a Krüppel-like zinc-finger protein whose expression is normally suppressed in neural tissues [15-18]. Most often REST, after recruiting corepressors and cofactors, initiates the essential process of transcriptionally silencing neuronal genes in non-neuronal cells. However, on rare occasions, and for a variety of reasons, REST can also function in the opposite role of enhancer [19-22] which, as we demonstrate, it does for DHCR24.
It is important to note that DHCR24 activity in fetal brain is sufficient to reduce the Δ24-double bonds of 70-75% of the cholesterol precursor molecules that the average fetal mouse brain needs to synthesize before birth [4]. As a consequence, large order of magnitude changes in DHCR24 ought not to be expected.
Several hundred-fold elevations of desmosterol and 2- to 4-fold reductions of DHCR24 are not likely to be without consequence to the fetal and embryonic brain. As will be discussed in detail below, both molecules are bioactive and are capable of altering sterol and triglyceride synthesis and transport, lipid raft formation, neural cell stability, etc. [14], processes known to be essential for proper brain development. So it may be that the reduced activity of DHCR24 has become essential for the complex mammalian brain and is now intentional and closely regulated.
Materials and Methods
Reagents
Dulbecco's MEM/F12 and bovine serum albumin (BSA IV) were obtained from Invitrogen (Grand Island, NY) while lipoprotein deficient bovine calf serum (LPDS) containing <25 μg/ml of cholesterol was purchased from Biomedical Technologies (Stoughton, MA). Sterols were purchased from Sigma Aldrich (St Louis, MO) or Steraloids (Newport, RI) and reagents for processing sterols for gas-chromatography (GC) from Sigma Aldrich or Fisher Scientific.
Animals
C57BL/6J mice were fed a commercial chow diet, bred and a successful mating identified by a vaginal plug. Pregnant mice were sacrificed following accepted humane protocols and fetal tissues harvested and placed in RNAlater (Ambion, Austin, TX) or else frozen in liquid nitrogen then stored at −70C for future sterol analysis [4;23]. All animal experiments complied with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and were performed with the approval of the Institutional Animal Care and Use Committee, Clement J. Zablocki Veterans Affairs Medical Center, Milwaukee, WI.
Quantitative RT-PCR
qRT-PCR was carried out with TaqMan® Genomic Assay kits designed by Applied Biosystems (Foster City, CA) for the appropriate genes as described previously [4]. The results as shown employed cyclophilin as the endogenous control. When we compared expression in brain and liver, measurements were repeated with β-actin and GAPDH which yielded similar results.
Chromatin immunoprecipitation (ChIP) Assay
The assay was carried out in HeLa cells grown in DMEM plus FBS using the EZ Chip Chromatin Immunoprecipitation Kit (Upstate/Millipore, Billerica, MA) according to the manufacturer's instructions. REST bound to chromatin was immunoprecipitated with antibody supplied by Upstate. After reversing cross-linking, the precipitated DNA was purified and quantified by PCR using primers 5’-AGT ATT CGC AGC GGA CCC CAC and 3’-GGT TCG CGC CTC CTG TCA CT, F and R, respectively, (Fig. 2) covering a 292 nt region spanning the putative REST binding sites. Positive controls were provided by the input sample; by precipitation with antibody to Polymerase II (Poly II) followed by amplification with primers F and R; and by precipitation with REST antibody followed by PCR with primers 5’-TGC ATT GCT GAT GCT GCT GCT and 3’-TGG CCA CCA CCT CCC AGA GTC (137 bp) for Synapsin 1 (SYN1) [24]. Negative controls included precipitation by mouse IgG; amplification of the human GAPDH promoter with the primer pair 5’-TAC TAG CGG TTT TAC GGG CG, 3’-TCG AAC ACG AGG AGC AGA GAG CGA (165 bp) after precipitation with REST antibody; and, after precipitation with REST antibody, with primers 5’-AGG CAG CTG GAG AAG TTT GT and 3’-CTT GCA GAT CTT GTC GTA CA that amplified a 1221 bp segment (3’-DHCR24) spanning the final DHCR24 intron/exon boundary excluding nonspecific REST binding to DHCR24. The sizes of the PCR products were confirmed by comparing their mobilities with those of molecular weight standards.
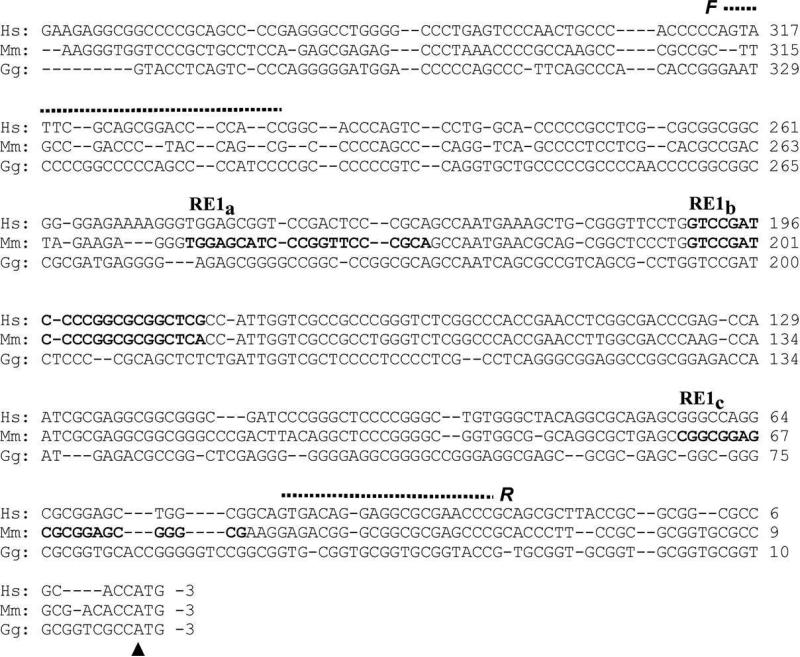
Fig 2.
The putative DHCR24 promoter is well conserved in the chicken (Gg), mouse (Mm) and human (Hs) genomes. The sequences in bold type represent the three REST/NSRF binding sites in the mouse (RE1a, RE1b and RE1c) and the one in the human (RE1b) gene predicted by MatInspector. F and R and the dashed lines indicate the sequences of the respective forward and reverse primers used to amplify the region bracketing the three suspected REST binding domain(s) for the ChIP assay. The vertical arrowhead marks the likely translation start site. In particular, the RE1b sequence, although not suggested by MatInspector as a REST/NSRF binding sequence in the chicken, is remarkably similar to both mouse and human.
Luciferase Assay
The gene encoding human REST, CloneID:40146881, obtained from Open Biosystems (now Fisher Scientific) was subcloned into pcDNA4/mycHisB (Life Technologies). A dominant negative version of REST (DN-REST), containing only the REST DNA binding domain, aa 234-437, about 20% of the native protein [18], was also subcloned into the same vector.
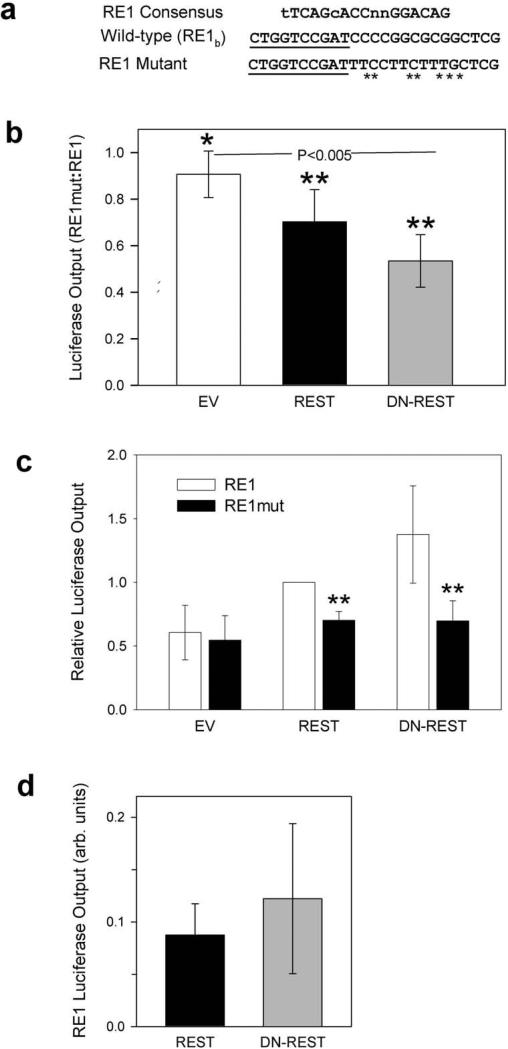
The Renilla luciferase construct used in these studies driven by the porphobilinogen deaminase promoter (phRL-PBGD), and the Firefly luciferase construct driven by 300 bp of the human DHCR24 promoter (pGL3-basic/300luc) have been described previously [25]. The predicted RE1-binding site RE1b (Fig. 2) was mutated (RE1mut, Fig. 5a, with altered bases indicated by an asterisk) in pGL3-basic/300luc using megaprimed PCR [26].
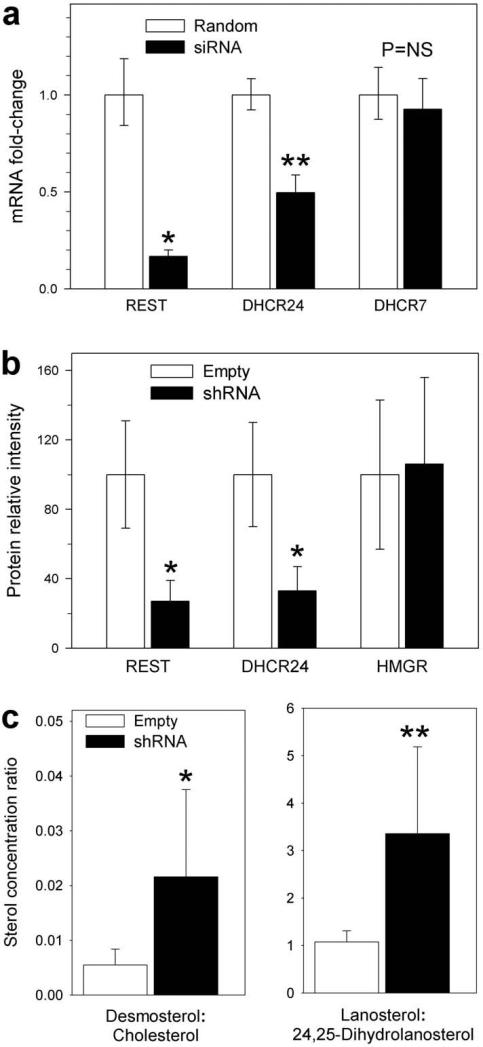
Fig 5.
Silencing REST in HeLa cells inhibits expression, protein and activity of DHCR24 but does not alter DHCR7 mRNA or HMGR protein levels. All data presented as mean ± 95% CI. (a) qRT-PCR of REST, DHCR24 and a differently structured but essential sterol reductase, DHCR7, that appears to lack RE1 sequences. HeLa cells were transfected for four hours with pooled REST siRNA, then washed, grown in normal medium and harvested 2 days later (black bars). Controls (white bars, n=12) are cells transfected with random RNA. REST expression is reduced 10-fold and DHCR24 expression 2-fold but DHCR7 transcription remains unchanged; *P < 0.001, **P < 0.025 pre- vs. post-silencing, n=18, two-tailed t-test. (b) Relative REST, DHCR24 and HMGR protein levels (normalized to mean GAPDH) from cells incubated for 6 days with a stable lentivirus construct containing a REST targeting vector, shRNA (black bar), or an empty vector (white bar); *P < 0.0025, two-tailed t-test, n=6. HMGR protein is unchanged. (c) Ratios of two of the most abundant Δ24-sterol precursors to their respective C24,25-saturated products after a 6 day incubation in lipoprotein deficient medium in cells treated with vectors containing (black bars) or empty of (white bars) REST silencing shRNA (lanosterol:24,25-dihydrolanosterol = 1.08 ± 0.23 vs. 3.36 ± 1.83, and desmosterol:cholesterol = 0.0055 ± 0.0029 vs. 0.022 ± 0.016, all n=9). *P = 0.068, **P = 0.028, two-tailed t-test.
HelaT cells were grown in triplicate in 24-well plates and transfected with 250 ng of Firefly luciferase plasmid, 25 ng phRL-PBGD, and 25 ng pcDNA4/REST or pcDNA4/DN-REST plasmid, using TransIT-2020 (Mirus Bio, Madison, WI) for 24 h. Cells were then lysed and assayed for Firefly and Renilla luciferase activities using the Dual Luciferase Reporter Assay System (Promega, Fitchburg, WI). Relative luciferase activity was calculated as the ratio of Firefly to Renilla activity for each sample.
Transient REST silencing
Eighty percent confluent HeLa cells in 8-well plates were incubated with a pool containing the four REST silencing double stranded 21 nucleotide (nt) RNAs designed and supplied by Dharmacon (Pittsburgh, PA): CAA CGA AUC UAC CCA UAU UUU; GGU GAA ACU UUA AAU GGU AUU; UGA CUU GCC UGA UAA UAU UUU; and CAU CCU ACU UGU CCU AAU AUU together with DHARMAfect transfection reagent according to the manufacturer's protocol. After four hours the transfection reagent was replaced by normal culture medium (DMEM plus 20% FBS) and the cells allowed to grow for an additional 44 h. Following reverse transcription the relative concentrations of REST, DHCR24 and DHCR7 mRNA were measured by qRT-PCR as described previously [4]. Controls were provided by cells incubated with a random double stranded RNA: UAA GGC UAU GAA GAG AUA C.
Stable REST silencing
HeLa cells were cultured in DMEM containing 10% FBS plus 100 U/ml penicillin and 100 μg/ml streptomycin. One day before transfection, the cells were seeded in 6-well cell culture plates to a final density of 60-70%. Lentiviral shRNAmir vectors (pGIPZ) containing the Hs REST hairpin targeting sequence TGC TGT TGA CAG TGA GCG CGC GGC TAC AAT ACT AAT CGA TTA GTG AAG CCA CAG ATG TAA TCG ATT AGT ATT GTA GCC GCA TGC CTA CTG CCT CGG A or a non-silencing control sequence (Open Biosystems, Pittsburgh, PA) were transfected into the cells with Arrest-In transfection reagent according to the manufacturer's protocol. After 48 h, cells were cultured in 5 μg/ml puromycin-containing media to select pGIPZ vector-expressing cells. Gradually increasing the concentration of puromycin to 30 μg/ml, after 3~4 weeksof culture, vector-encoded independent GFP expression was observed in all cells and confirmed by fluorescent-activated cell sorting. REST expression in the plasmidtransfected cells was characterized by real-time qPCR and immunoblotting. For sterol measurements, cells stably transfected with the plasmid containing silencing or non-silencing (control) sequences were cultured in DMEM/F12 supplemented with 1% LPDS and 0.5% BSA for 6 to 10 days (medium changed every day) then extracted and analyzed as described below.
Sterol measurements
Sterols, were extracted, hydrolyzed and measured as the trimethylsilyl ether derivatives by capillary column gas-chromatography (tissues) and GC/MS selected ion monitoring (cultured cells) as described previously [4]. For selected ion monitoring m/z was set to 393 (lanosterol), 395 (24,25-dihydrolanosterol), 343 (desmosterol) and 329 (cholesterol).
Immunoblotting
Rabbit anti-REST, anti-HMGR (HMG-CoA reductase) antibodies were obtained from Millipore (Billerica, MA), rabbit anti-DHCR24 antibody against an N-terminal peptide corresponding to amino acids 68-85 of the Hs DHCR24 protein were supplied by Sigma-Aldrich (St. Louis, MO), and a mouse monoclonal anti-GAPDH antibody by Abcam (Cambridge, MA). Whole cell lysates and nuclear proteins were prepared from ~107 cultured cells using a NE-PER kit (Pierce, Waltham, MA) and protein concentrations determined using a BCA protein assay kit (Fisher Scientific). Forty μg protein aliquots from each sample were separated on SDS–polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk and incubated with a REST (1:500), a DHCR24 (1:4000) and a GAPDH antibody (1:10,000) followed by incubation with HRP-conjugated second antibodies. Specific interactions were revealed using the ECL (Enhanced Chemiluminescence) detection system (Amersham, Pittsburgh, PA). Densitometric quantification of the immunoblot bands from controls (empty vector) or REST knock-down samples were quantified using NIH image 1.63 software.
Statistical Analysis
Most results are presented as mean ± 95% confidence interval (95% CI) with differences between groups analyzed by the two-tailed t-test. Ratios of luminescent outputs obtained from reporter constructs containing a mutated RE1 sequence (RE1mut) divided by the output from wild type RE1 reporters and ratios of C24,25 unsaturated to saturated sterols were employed to calculate differences by the two-tailed single sample t-test with the null hypothesis that mean = 1; i.e. that results before and after mutating RE1 or sterol ratios before and after silencing REST are equal (GraphPad, Systat, Chicago, IL).
Results
Quantitative RT-PCR (qRT-PCR) confirmed that mRNA levels of both Rest and Dhcr24 are reduced 3- to 4-fold (p<0.005) in fetal and newborn mouse brain compared to liver (Fig. 1). Fig. 1 also includes a plot demonstrating massively increased desmosterol concentrations in brain compared to liver.
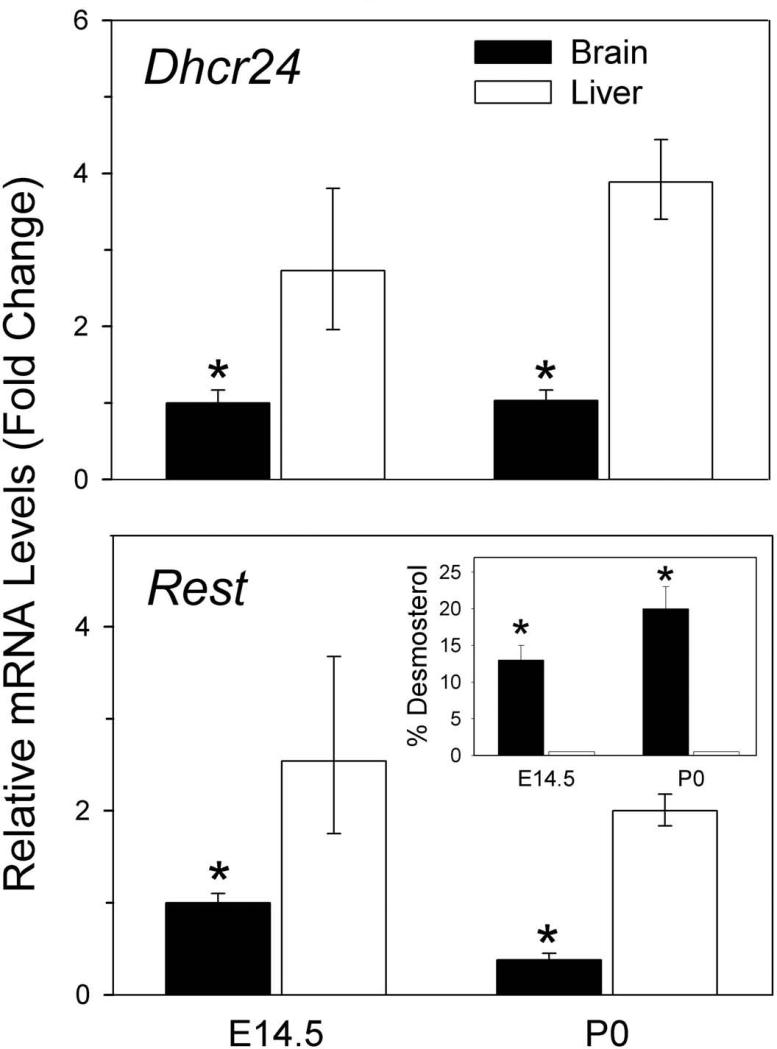
Fig 1.
Rest and Dhcr24 expression are markedly reduced in mouse brain compared to liver. qRT-PCR analysis of Dhcr24 (Top) and Rest (Bottom) from fetal and newborn mouse brain (black bars) and liver (white bars) demonstrating reduced expression of both in brain; mean ± 95% confidence interval (95% CI). *P < 0.001 brain vs. liver, n=12, two-tailed t-test. These results employed β-actin as the endogenous control but cyclophilin and GAPDH yielded very similar results (data not shown) confirming that the differences between brain and liver are real. Insert: Desmosterol as percent of total sterols vs. age illustrating markedly elevated desmosterol levels in fetal brain (black bars) compared to liver (white bars) at E14.5 and at birth (P0); mean + SD, *P < 0.005 brain vs. liver, n=3, two-tailed t-test.
A detailed analysis by MatInspector [27] of 2500 bp of the predicted, and recently confirmed [25;28;29], mouse and human DHCR24 promoters, well conserved across the vertebrate genome, suggested potential binding sequences for the RE1-silencing transcription factor REST, with three in the mouse, RE1a-c, and one, RE1b, in the human gene (Fig. 2). The mammalian RE1b sequence, especially, is very similar to the sequence observed in the chicken. These sites, all within 260 bp of the putative translation start site pointed to REST, a gene poorly expressed in neural tissues, as a possible modulator of DHCR24 transcription. However, since expression and activity of DHCR24 are inhibited in brain (Fig. 1), REST would not be acting here in its usual role of suppressor but instead is functioning in its more uncommon one of transcriptional enhancer. We focused our investigation here on the human gene Chromatin immunoprecipitation (ChIP) assays (Fig. 3) demonstrated that REST binds to the human DHCR24 promoter within the 292 nt region containing the predicted REST sequences. Positive controls polymerase II and Synapsin 1 elicited strong signals for REST while there was little or no response from negative controls including the amplification of a large segment of the final DHCR24 intron/exon boundary.
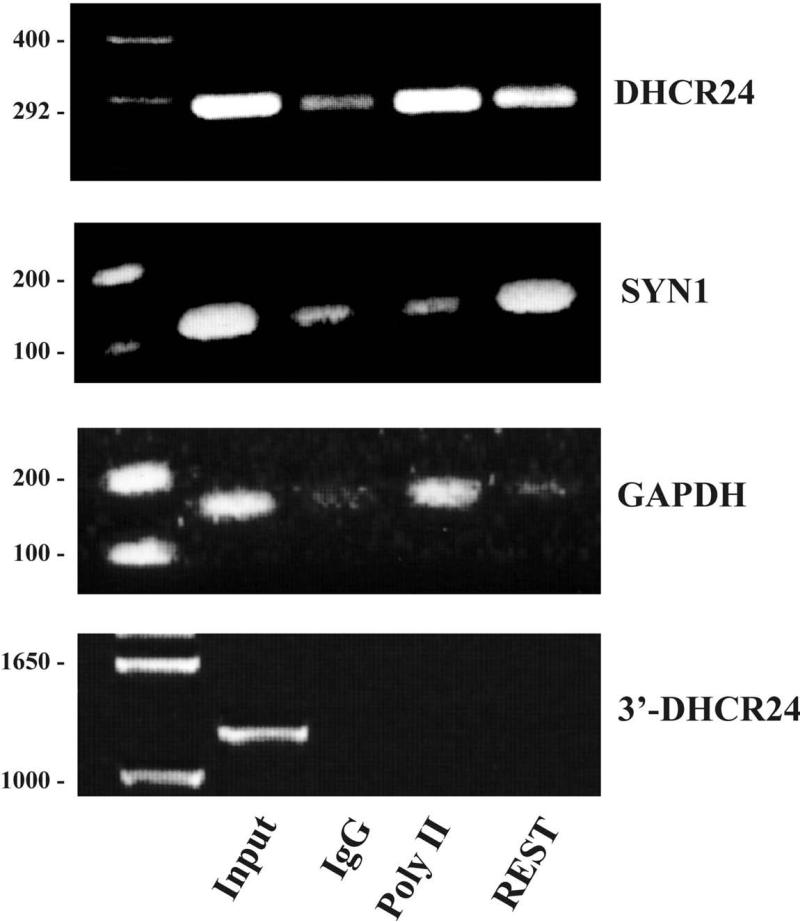
Fig 3.
Chromatin immunoprecipitation (ChIP) assay in cultured HeLa cells demonstrates that REST binds to the DHCR24 promoter in the vicinity of the predicted RE1 binding sequences. PCR products following precipitation by antibodies for polymerase II (Poly II), REST and IgG are shown together with molecular weight standards on the extreme left: (a) DHCR24 amplified by the primers F and R shown in Fig. 2 and (b) positive control Synapsin 1 (SYN1), a neural specific gene known to bind REST [24]. GAPDH (c) and amplification with primers for a 1200 bp fragment spanning the final intron/exon boundary of DHCR24 (3’-DHCR24) (d) provide negative controls. Input is the total sample before treatment.
To confirm that it is the RE1b site (Fig. 2) in the human promoter that is essential for upregulating DHCR24 by REST, we carried out a series of studies employing a luciferase reporter system [25] constructed from either the wild type DHCR24 promoter (RE1) or the identical promoter with the RE1b-binding sequence mutated (RE1mut) as shown in Fig. 4a. As described above, in an effort to reduce scatter we express the results as luminescence ratios (Fig. 4b). But, in Fig. 4c we also present these data in their familiar normalized form so that the effects of mutating RE1 e more visually obvious. Relative luciferase outputs (RE1mut:RE1) obtained after incubating with either full length REST or DN-REST, a dominant negative REST fragment consisting of just the DNA binding region (about 20% of the native protein) were reduced by as much as 50% (P < 0.005) and demonstrate that mutating RE1b indeed suppresses DHCR24 expression (Fig. 4b and c). Mutating RE1b also tended to diminish the luminescence output of the RE1mut reporter compared to the output from the reporter constructed from wild type RE1 (P = 0.065, Fig. 4b) after both were cotransfected with an empty vector (EV) rather than with REST or DN-REST. This result suggests that, in the absence of other unrecognized co-factors or subtle experimental errors in the synthesis of the two reporters, it is likely that the pre-existing, endogenous cellular pool of REST, without being supplemented with exogenous protein, is of sufficient size and activity to amplify the expression of transfected luciferase DHCR24 promoter constructs sufficiently to uncover differences between the RE1 and RE1mut reporters.
Fig 4.
Luciferase reporter studies demonstrate that REST must bind to the RE1b site (Fig. 2) before full DHCR24 expression is achieved. (a) Human RE1 consensus [24] together with wild type DHCR24 (RE1) and mutated (RE1mut) RE1 sequences; mutated bases are indicated by asterisks; the underlined nucleotides mark -196 InvSRE one of the functional sterol response elements (SRE) recently identified by Zerenturk et al. [25] . (b) Relative outputs from luciferase reporter constructs with mutated RE1 depicted in Fig. 4a divided by outputs from reporters with wild type RE1 confirm that mutating RE1b suppresses DHCR24 expression, (all results given as mean ± 95% CI): In these studies the reporter constructs were cotransfected with a control empty vector (EV) or with vectors containing either wild type REST or a dominant negative fragment of REST (DN-REST). Mutating the RE1 sequence (RE1mut) significantly reduced luciferase output to 70 ± 15% (n=8) and 53 ± 11% (n=4) after co-transfection with vectors containing REST and DN-REST, respectively. Mutating RE1 also tended to reduce luminescence from these same promoters to 90 ± 10% (n=3) of their original value when co-transfected with an empty vector (EV) when only endogenous REST was available to upregulate DHCR24. One sample t-test with mean = 1; *P = 0.065, **P < 0.005 that RE1mut and wild type RE1 luminescence are equal (a 93.5% and a 99.5% probability, respectively, that they are different). (c) The same luminescence data shown normalized to the output from the wild type RE1 promoter (RE1) after cotransfection with full length REST. Differences between luminescence from wild type and mutated promoters cotransfected with REST or DN-REST retain their statistical significance but the more subtle changes in the empty vector (EV) results are now submerged in experimental noise. (**P<0.005) (d) The fragmentary polypeptide DN-REST tends to upregulate DHCR24 expression more than does full length REST: Luminescence outputs from wild type luciferase constructs after incubating with REST (0.088 ± 0.030, n=8) or DN-REST (0.12 ± 0.071, n=4), P = NS, REST vs. DN-REST.
It is also important to note that DHCR24 expression was enhanced at least as well by DN-REST as it was by REST and that total photon emission tends to be increased when the former is substituted for the latter (Fig. 4d).
If we are correct about the effect of REST on DHCR24, then silencing REST in any cell line in which DHCR24 is well expressed should suppress DHCR24 mRNA, protein levels and enzyme activity, the latter measured by quantifying increases in the concentrations of common C24,25-unsaturated sterols. A brief 4 h treatment of HeLa cells with REST siRNA followed by a further two days of growth in normal medium led to a 10-fold suppression of REST and a highly significant 2-fold inhibition of DHCR24 mRNA (Fig. 5a). In contrast, expression of DHCR7 (7-dehydrocholesterol Δ7-reductase), another enzyme essential for cholesterol biosynthesis, robustly active in all tissues including brain [11] but without identifiable RE1 sequences, remained unchanged. In a complementary experiment, HeLa cells were treated for 6-10 days with a stable REST hairpin silencing vector. As a result, levels of DHCR24 and REST protein declined 4-and 5-fold, respectively, (P < 0.0025, Fig. 5b) but, as expected, HMGR (3-hydroxy-3-methyl-glutaryl-CoA reductase) protein was unchanged. Concentrations of two of the most abundant Δ24-unsaturated sterols, lanosterol and desmosterol, (Fig. 5c) experienced respective 3- and 4-fold increases relative to their C24,25-saturated products (P = 0.028 and P = 0.068, respectively) reflecting the expected decrease in DHCR24 protein enzyme activity when REST is suppressed.
Discussion
Markedly increased concentrations of cholesterol precursors engendered by diseases of abnormal cholesterol biosynthesis, is well understood [30-32]. But, the massive, somewhat transient, accumulation of desmosterol in healthy fetal mammalian brain [4;6;7] has always been confounding.
DHCR24 appears to be unique among the genes of cholesterol biosynthesis in that it is the only one that is always “inadequately” upregulated in neural tissues. Here we demonstrate that the zinc-finger protein REST, expressed poorly in brain and other neural tissues (Fig. 1), and which binds strongly to the DHCR24 promoter at the RE1b sequence (Figs. 2 and 3) is needed to fully upregulate gene expression and yield enzyme abundant enough to permit all newly synthesized desmosterol to be promptly reduced to cholesterol in the fetal brain. We proved that REST is essential because mutating RE1b (Fig. 4a) significantly reduced the output of a DHCR24 promoter driven luciferase reporter (Fig. 4b); and silencing REST (Fig. 5) in cultured cells suppressed DHCR24 expression, protein levels and enzyme activity, the last signaled by increased concentrations of the C24,25-unsaturated cholesterol precursors desmosterol and lanosterol. In contrast, other enzyme proteins essential for cholesterol biosynthesis such as DHCR7, a sterol reductase lacking RE1 motifs that functions late in the pathway, and HMGR, an early pathway enzyme usually considered rate-limiting, were unaffected.
Generally, it is neural-specific genes that contain RE1 REST binding sequences but REST is poorly expressed in brain and peripheral nerves. This means that only non-neural tissues are likely to have sufficient REST protein to bind to these genes and assemble an activity complex capable of altering histone methylation and acetylation that leads to transcriptional suppression [33-36]. However, in rare instances REST can do the opposite and enhance gene expression, though with little commonality of mechanism or action [19-22]. Our observations that cotransfecting DHCR24 reporter constructs with DN-REST tended to increase luminescence output compared to cotransfection with full length REST (Fig. 4b-d) suggests that upregulation of DHCR24 by REST (UniProt Q13127) requires, at most, about 20% of the protein. DN-REST consists of six of the eight REST zinc fingers and little else except linkers. It thus seems highly unlikely that DN-REST could elicit an epigenetic response when it interacts with RE1 because this protein fragment consists of only the REST DNA-binding domain and lacks sequences able to recruit and bind the cofactors necessary to form a REST-like transcriptional suppression complex. It then follows that, unless DN-REST and REST accelerate DHCR24 expression by different mechanisms, REST does not upregulate DHCR24 by inhibiting the transcription of some, as yet, unknown DHCR24 repressor. Taken together, these results suggest that REST is most likely acting locally possibly stimulating DHCR24 expression by stabilizing and enhancing SREBP function when both join in tandem to their respective, neighboring nucleotide binding sequences (Fig. 4a). Admittedly this is somewhat speculative and, other scenarios are certainly possible. We also cannot rule out a contribution from post-translational inhibition of DHCR24 activity in brain by steroid hormones [8], an hypothesis well beyond the scope of the present study.
Furthermore, because DN-REST tends to be more efficient at activating DHCR24 expression than does REST (Fig. 4) is it possible that REST is also functioning traditionally as a transcriptional inhibitor, though a weak one, of DHCR24 providing for an additional level of regulation?
Because the reduced level of REST found in fetal brain leads to only partial DHCR24 inactivation, meaning that there is still sufficient enzyme activity to reduce 75-80% of the desmosterol in brain to cholesterol before the animal is born [4], we do not suggest that REST is the principle regulator of DHCR24 activity. Although 2- to 3-fold changes in DHCR24 expression, similar to those we have measured following REST suppression (Figs. 4 and 5), have also been reported in cultured cells following treatment with common inhibitors or promoters of cholesterol biosynthesis [25;29], Dhcr24 activity in rat liver can be upregulated by as much as 120-fold in response to cholesterol deprivation [13]. Furthermore, recent studies of the DHCR24 promoter have demonstrated at least two functional, sterol response elements (SRE) as well as the expected regulation of DHCR24 expression by sterols [25]. This means that primary regulation of DHCR24 expression, like most, if not all, genes involved in cholesterol synthesis and transport, appears to require binding of the active sterol response element binding protein (SREBP) cleavage fragment to SRE [37]. Our current study, in contrast, demonstrates that, because REST is essential for the full realization of DHCR24 expression, it serves as a secondary modulator or cofactor having its greatest effect in neural tissues in which REST expression is naturally reduced and then only when cholesterol synthesis is substantially elevated as it is in the brain of an embryo or fetus.
Nothing in our data, however, suggests that REST could not also serve as a cofactor for any or all genes regulated by SREBP. But, its absence impacts only DHCR24.
Similarly elevated desmosterol concentrations have also long been noted in the gonads [38] in which, at least in the mouse, REST appears to be well expressed [39]. Reduced DHCR24 activity in these tissues is generally thought to be caused by the naturally abundant progestins [40].
Despite being secondary and indirect, the effects of REST on lipid metabolism are not inconsequential when the cholesterol biosynthetic pathway is as active as it is in fetal brain. Although inactivating mutations of DHCR24 in humans lead to a rare, severe and often fatal birth defect [12;41] and one can build, at best, a viable but poorly functioning mouse from internally synthesized desmosterol and maternally supplied cholesterol [42], desmosterol itself under normal physiologic conditions has a number of non-trivial metabolic effects including an ability to reduce the formation of ordered lipid domains and perhaps impair raft-dependent signaling [43]. Desmosterol has also been reported to be a potent agonist for the liver X receptor (LXR) and to inhibit cholesterol biosynthesis by blocking SREBP cleavage [44;45]. Furthermore, DHCR24, in common with a number of other proteins associated with cholesterol metabolism, is multi-functional (recently reviewed by Zerenturk et al [14]) in that it appears to protect neuronal cells from degeneration [46;47]; to modulate levels of the tumor suppressor p53 by binding to its amino terminus enhancing survival of cells during oxidative stress [48;49]; and to inhibit Aβ cleavage of the amyloid precursor protein APP while assisting in lipid raft formation [50]. Thus, the 2- to 4-fold reduction in DHCR24 levels that we, for instance, observed in mouse brain (Fig. 2) combined with a several hundred-fold increase in the concentration of desmosterol, even if lasting for only 3-4 weeks in rodents or a few months to a year in humans, might be expected to have non-trivial effects on lipids, membranes and development.
How and when the DHCR24 promoter came to acquire an active REST binding sequence, we cannot say but its presence must have been either beneficial or indifferent to have been retained in the mammalian genome. The magnitude of the changes in desmosterol and DHCR24 concentrations in fetal brain are large and, although of somewhat limited temporal span, with sufficient metabolic effect to suggest the former. Perhaps it was advantageous to have desmosterol acting as an inhibitor keeping sterol biosynthesis [44;45] in check until parturition when massive quantities of cholesterol are needed very quickly for myelination. Or it may be that because desmosterol is a relatively poor substrate for esterification it is better at upregulating LXR and stimulating astrocyte sterol excretion [8;51]. So, it is possible that during the sculpting of the young brain, enhancing local sterol transport by increasing the supply of free sterol or modulating lipid raft formation by desmosterol and/or DHCR24 might prove useful. It is also not impossible that desmosterol and DHCR24, like 7-dehydrocholesterol and DHCR7 [52;53], have additional and, as yet undiscovered, signaling capabilities. And, it may well be that DHCR24 protein and desmosterol are so essential for brain development that we will find that their concentrations are far more closely managed than we had previously thought.
Although we know a good deal about the neuropathology caused by mutated, nonfunctional DHCR24 [54], one wonders if a mammalian brain in which fetal DHCR24 protein levels and activity were not suppressed and desmosterol concentrations were more similar to those in liver or kidney might differ in any material way from a “normal” brain.
Acknowledgements
This work was supported in part by research grants from the U.S. Department of Veterans Affairs to G.S.T. and the National Health and the Medical Research Council (Australia) (1008081) and National Heart Foundation of Australia (G11S5757) to A.J.B. H.Y. was supported by National Institutes of Health grants HL68660 and HD062938. G.S.T. also wishes to thank Bibiana Pcolinsky for her excellent and dedicated technical support.
Reference List
- 1.Spady DK, Dietschy JM. Sterol synthesis in vivo in 18 tissues of the squirrel monkey, guinea pig, rabbit, hamster, and rat. J Lipid Res. 1983;24:303–315. [PubMed] [Google Scholar]
- 2.Chevallier F. Transferts et synthèse du cholesterol chez le rat au course de sa croissance. Biochim Biophys Acta. 1964;84:316–339. [PubMed] [Google Scholar]
- 3.Connor WE, Lin DS. Placental transfer of cholesterol-4-C14 into rabbit and guinea pig fetus. J Lipid Res. 1967;8:558–564. [PubMed] [Google Scholar]
- 4.Tint GS, Yu H, Shang Q, Xu G, Patel SB. The use of the Dhcr7 knockout mouse to accurately determine the origin of fetal sterols. J Lipid Res. 2006;47:1535–1541. doi: 10.1194/jlr.M600141-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinse CH, Shah SN. The desmosterol reductase activity of rat brain during development. J Neurochem. 1971;18:1989–1998. doi: 10.1111/j.1471-4159.1971.tb09604.x. [DOI] [PubMed] [Google Scholar]
- 6.Kritchevsky D, Holmes WL. Occurrence of desmosterol in developing rat brain. Biochem Biophys Res Comm. 1962;7:128–131. doi: 10.1016/0006-291x(62)90160-2. [DOI] [PubMed] [Google Scholar]
- 7.Fumagalli R, Paoletti R. The identification and significance of desmosterol in the developing human and animal brain. Life Sci. 1963;5:291–295. doi: 10.1016/0024-3205(63)90159-0. [DOI] [PubMed] [Google Scholar]
- 8.Jansen M, Wang W, Greco D, Bellenchi GC, di Porzio U, Brown AJ, Ikonen E. What dictates the accumulation of desmosterol in the developing brain? FASEB J. 2013;27:865–870. doi: 10.1096/fj.12-211235. [DOI] [PubMed] [Google Scholar]
- 9.Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr Opin Lipidol. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Goodrum JF. Cholesterol synthesis is down-regulated during regeneration of peripheral nerve. J Neurochem. 1990;54:1709–1715. doi: 10.1111/j.1471-4159.1990.tb01225.x. [DOI] [PubMed] [Google Scholar]
- 11.Fitzky BU, Moebius FF, Asaoka H, Waage-Baudet H, Xu L, Xu G, Maeda N, Kluckman K, Hiller S, Yu H, Batta AK, Shefer S, Chen T, Salen G, Sulik K, Simoni RD, Ness GC, Glossmann H, Patel SB, Tint GS. 7-Dehydrocholesterol-dependent proteolysis of HMG-CoA reductase suppresses sterol biosynthesis in a mouse model of Smith-Lemli- Opitz/RSH syndrome. J Clin Invest. 2001;108:905–915. doi: 10.1172/JCI12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterham HR, Koster J, Romeijn GJ, Hennekam RCM, Vreken P, Andersson HC, FitzPatrick DR, Kelley RI, Wanders RJA. Mutations in the 3β-hydroxysteroid Δ24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am J Hum Genet. 2001;69:685–694. doi: 10.1086/323473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae SH, Paik YK. Cholesterol biosynthesis from lanosterol: development of a novel assay method and characterization of rat liver microsomal lanosterol Δ24-reductase. Biochem J. 1997;326:609–616. doi: 10.1042/bj3260609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerenturk EJ, Sharpe LJ, Ilkonen E, Brown AJ. Desmosterol and DHCR24: Unexpected new directions for a terminal step in cholesterol synthesis. Prog Lipid Res. 2013;52:666–680. doi: 10.1016/j.plipres.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 16.Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 17.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood IC, Belyaev ND, Bruce AW, Jones C, Mistry M, Roopra A, Buckley NJ. Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J Mol Biol. 2003;334:863–874. doi: 10.1016/j.jmb.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Abramovitz L, Shapira T, Ben Dror I, Dror V, Granot L, Rousso T, Landoy E, Blau L, Thiel G, Vardimon L. Dual Role of NRSF/REST in Activation and Repression of the Glucocorticoid Response. J Biol Chem. 2008;283:110–119. doi: 10.1074/jbc.M707366200. [DOI] [PubMed] [Google Scholar]
- 20.Bessis A, Champtiaux N, Chatelin L, Changeux JP. The neuron-restrictive silencer element: A dual enhancer/silencer crucial for patterned expression of a nicotinic receptor gene in the brain. Proc Natl Acad Sci USA. 1997;94:5906–5911. doi: 10.1073/pnas.94.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kallunki P, Edelman GM, Jones FS. The neural restrictive silencer element can act as both a repressor and enhancer of L1 cell adhesion molecule gene expression during postnatal development. Proc Natl Acad Sci USA. 1998;95:3233–3238. doi: 10.1073/pnas.95.6.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seth KA, Majzoub JA. Repressor element silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) can act as an enhancer as well as a repressor of corticotropin-releasing hormone gene transcription. J Biol Chem. 2001;276:13917–13923. doi: 10.1074/jbc.M007745200. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Wessels A, Chen J, Phelps AL, Oatis J, Tint GS, Patel SB. Late gestational lung hypoplasia in a mouse model of the Smith-Lemli-Opitz syndrome. BMC Dev Biol. 2004;4:1. doi: 10.1186/1471-213X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerenturk EJ, Sharpe LJ, Brown AJ. Sterols regulate 3β-hydroxysterol Δ24-reductase (DHCR24) via dual sterol regulatory elements: cooperative induction of key enzymes in lipid synthesis by Sterol Regulatory Element Binding Proteins. Biochim Biophys Acta. 2012;1821:1350–1360. doi: 10.1016/j.bbalip.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Tseng WC, Lin JW, Wei TY, Fang TY. A novel megaprimed and ligase-free, PCR-based, site-directed mutagenesis method. Anal Biochem. 2008;375:376–378. doi: 10.1016/j.ab.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 28.Drzewinska J, Walczak-Drzewiecka A, Ratajewski M. Identification and analysis of the promoter region of the human DHCR24 gene: involvement of DNA methylation and histone acetylation. Mol Biol Rep. 2011;38:1091–1101. doi: 10.1007/s11033-010-0206-z. [DOI] [PubMed] [Google Scholar]
- 29.Daimiel LA, Fernandez-Suarez ME, Rodriguez-Acebes S, Crespo L, Lasuncion MA, Gomez-Coronado D, Martinez-Botas J. Promoter analysis of the DHCR24 (3β-hydroxysterol Δ24-reductase) gene: characterization of SREBP (sterol-regulatory-element-binding protein)-mediated activation. Biosci Rep. 2013;33:57–69. doi: 10.1042/BSR20120095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tint GS, Salen G. Evidence for the early reduction of the 24,25 double bond in the conversion of lanosterol to cholesterol in cerebrotendinous xanthomatosis. Metabolism. 1977;26:721–729. doi: 10.1016/0026-0495(77)90059-2. [DOI] [PubMed] [Google Scholar]
- 31.Tint GS, Irons M, Elias ER, Batta AK, Frieden R, Chen TS, Salen G. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N Engl J Med. 1994;330:107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 32.Porter FD. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet. 2008;16:535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- 33.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-Dependent Silencing of Chromosomal Regions Encoding Neuronal Genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 35.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 37.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sion B, Grizard G, Boucher D. Quantitative analysis of desmosterol, cholesterol and cholesterol sulfate in semen by high-performance liquid chromatography. J Chromatogr A. 2001;935:259–265. doi: 10.1016/s0021-9673(01)01105-0. [DOI] [PubMed] [Google Scholar]
- 39.Rinn JL, Rozowsky JS, Laurenzi IJ, Petersen PH, Zou K, Zhong W, Gerstein M, Snyder M. Major molecular differences between mammalian sexes are involved in drug metabolism and renal function. Dev Cell. 2004;6:791–800. doi: 10.1016/j.devcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Lindenthal B, Holleran AL, Aldaghlas TA, Ruan B, Schroepfer GJ, Jr, Wilson WK, Kelleher JK. Progestins block cholesterol synthesis to produce meiosis-activating sterols. FASEB J. 2001;15:775–784. doi: 10.1096/fj.00-0214com. [DOI] [PubMed] [Google Scholar]
- 41.Clayton PT. Disorders of cholesterol biosynthesis. Arch Dis Child. 1998;78:185–189. doi: 10.1136/adc.78.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wechsler A, Brafman A, Shafir M, Heverin M, Gottlieb H, Damari G, Gozlan-Kelner S, Spivak I, Moshkin O, Fridman E, Becker Y, Skaliter R, Einat P, Faerman A, Bjorkhem I, Feinstein E. Generation of viable cholesterol-free mice. Science. 2003;302:2087. doi: 10.1126/science.1090776. [DOI] [PubMed] [Google Scholar]
- 43.Vainio S, Jansen M, Koivusalo M, Rog T, Karttunen M, Vattulainen I, Ikonen E. Significance of sterol structural specificity. Desmosterol cannot replace cholesterol in lipid rafts. J Biol Chem. 2006;281:348–355. doi: 10.1074/jbc.M509530200. [DOI] [PubMed] [Google Scholar]
- 44.Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, Hobbs HH. Sterol Intermediates from Cholesterol Biosynthetic Pathway as Liver X Receptor Ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Acebes S, de la CP, Fernandez-Hernando C, Ferruelo AJ, Lasuncion MA, Rawson RB, Martinez-Botas J, Gomez-Coronado D. Desmosterol can replace cholesterol in sustaining cell proliferation and regulating the SREBP pathway in a sterol-Δ24-reductase-deficient cell line. Biochem J. 2009;420:305–315. doi: 10.1042/BJ20081909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greeve I, Hermans-Borgmeyer I, Brellinger C, Kasper D, Gomez-Isla T, Behl C, Levkau B, Nitsch RM. The Human DIMINUTO/DWARF1 Homolog Seladin-1 Confers Resistance to Alzheimer's Disease-Associated Neurodegeneration and Oxidative Stress. J Neurosci. 2000;20:7345–7352. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iivonen S, Hiltunen M, Alafuzoff I, Mannermaa A, Kerokoski P, Puolivali J, Salminen A, Helisalmi S, Soininen H. Seladin-1 transcription is linked to neuronal degeneration in Alzheimer's disease. Neuroscience. 2002;113:301–310. doi: 10.1016/s0306-4522(02)00180-x. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Miloslavskaya I, Demontis S, Maestro R, Galaktionov K. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature. 2004;432:640–645. doi: 10.1038/nature03173. [DOI] [PubMed] [Google Scholar]
- 49.Kuehnle K, Crameri A, Kalin RE, Luciani P, Benvenuti S, Peri A, Ratti F, Rodolfo M, Kulic L, Heppner FL, Nitsch RM, Mohajeri MH. Prosurvival Effect of DHCR24/Seladin-1 in Acute and Chronic Responses to Oxidative Stress. Mol Cell Biol. 2008;28:539–550. doi: 10.1128/MCB.00584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crameri A, Biondi E, Kuehnle K, Lutjohann D, Thelen KM, Perga S, Dotti CG, Nitsch RM, Ledesma MD, Mohajeri MH. The role of seladin-1/DHCR24 in cholesterol biosynthesis, APP processing and Aβ generation in vivo. EMBO J. 2006;25:432–443. doi: 10.1038/sj.emboj.7600938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, Yvan-Charvet L, Lutjohann D, Mulder M, Vanmierlo T, Kim TW, Tall AR. ATP-binding cassette transporters G1 and G4 mediate cholesterol and desmosterol efflux to HDL and regulate sterol accumulation in the brain. FASEB J. 2008;22:1073–1082. doi: 10.1096/fj.07-9944com. [DOI] [PubMed] [Google Scholar]
- 52.Jiang XS, Wassif CA, Backlund PS, Song L, Holtzclaw LA, Li Z, Yergey AL, Porter FD. Activation of Rho GTPases in Smith-Lemli-Opitz Syndrome: Pathophysiological and Clinical Implications. Hum Mol Genet. 2010;19:1347–1357. doi: 10.1093/hmg/ddq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korade Z, Kenworthy AK, Mirnics K. Molecular consequences of altered neuronal cholesterol biosynthesis. J Neurosci Res. 2009;87:866–875. doi: 10.1002/jnr.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.FitzPatrick DR, Keeling JW, Evans MJ, Kan AE, Bell JE, Porteous MEM, Mills K, Winter RM, Clayton PT. Clinical phenotype of desmosterolosis. Am J Med Genet. 1998;75:145–152. [PubMed] [Google Scholar]