Abstract
Background & Aims
All-oral regimens combining different classes of direct-acting antivirals (DAA) are highly effective for treatment of patients with chronic hepatitis C. NS5A inhibitors will likely form a component of future interferon-sparing treatment regimens. However, despite their potential, the detailed mechanism of action of NS5A inhibitors is unclear. To study their mechanisms, we compared their kinetics of antiviral suppression with those of other classes of DAA, using the hepatitis C virus (HCV) genotype 1a cell culture-infectious virus H77S.3.
Methods
We performed detailed kinetic analyses of specific steps in the HCV life cycle using cell cultures incubated with protease inhibitors, polymerase inhibitors, or NS5A inhibitors. Assays were designed to measure active viral RNA synthesis and steady-state RNA abundance, polyprotein synthesis, virion assembly, and infectious virus production.
Results
Despite their high potency, NS5A inhibitors were slow to inhibit viral RNA synthesis compared to protease or polymerase inhibitors. By 24 hrs after addition of an NS5A inhibitor, polyprotein synthesis was reduced less than 50%, even at micromolar concentrations. In contrast, inhibition of virus release by NS5A inhibitors was potent and rapid, with onset of inhibition as early as 2 hrs. Cells incubated with NS5A inhibitors were rapidly depleted of intracellular infectious virus and RNA-containing HCV particles, indicating a block in virus assembly.
Conclusions
DAAs that target NS5A rapidly inhibit intracellular assembly of gentoype 1a virions. They also inhibit formation of functional replicase complexes, but have no activity against pre-formed replicase, thereby resulting in slow shut-off of viral RNA synthesis.
Keywords: antiviral agent, replication, therapy, RNA-dependent RNA polymerase
BACKGROUND
The treatment of chronic hepatitis C is being revolutionized by the availability of direct acting antivirals (DAAs) that target viral proteins essential for virus replication and that offer the potential for all oral therapies and cure rates approaching 100%1. The first of these DAAs, boceprevir (BOC) and telaprevir, were approved for treatment of HCV in 20112. These linear ketoamides inhibit the major viral protease, NS3/4A, and their addition to interferon-based therapy increased sustained virologic response rates to 60–80% in treatment-naïve patients. More recently, a third NS3/4A protease inhibitor, the macrocyclic compound simeprevir, and a potent nucleotide inhibitor of the viral NS5B RNA-dependent RNA polymerase, sofosbuvir, have achieved regulatory approval.
DAAs that target NS3/4A (protease) and NS5B (RNA-dependent RNA polymerase) inhibit the enzymatic activity of these proteins. Detailed knowledge of the structure of these enzymes and their mechanisms of action greatly facilitated the development of these compounds. In contrast, small molecule inhibitors that target the viral NS5A protein were identified by screening libraries for compounds that block amplification of HCV RNA replicons in cell-based assays3–5. Sequencing of replicons selected for resistance to these compounds identified NS5A as their target, and more recent data confirm that they directly bind this viral nonstructural protein4–6. However, the mechanism of action of these NS5A inhibitors remains unclear. NS5A has no known enzymatic activity, and no cellular orthologs or viral homologs other than NS5A proteins of other hepaciviruses. Furthermore, NS5A is known to function in multiple aspects of the HCV life cycle as well as to modulate aspects of host cell signaling pathways that may favor virus replication7–10.
Although none have yet achieved regulatory approval, multiple NS5A inhibitors are currently in clinical development. These include, among others, daclatasvir (DCV, previously BMS-790052), ledipasvir (LDV, previously GS-5885), and MK-87423, 11, 12. As a class, these inhibitors show substantial promise as anti-HCV therapeutics. With 50% effective concentrations (EC50) in replicon assays in the low picomolar range, they are the most potent class of DAA to target HCV. Although NS5A inhibitors appear to have a low barrier to resistance, they are likely to be a key component of successful interferon-sparing DAA combinations13, 14.
To better understand the mechanism of action of NS5A inhibitors, we compared the activities of different classes of DAAs in multiple assays of antiviral activity that specifically examine different steps in the viral life cycle, including polyprotein synthesis, RNA synthesis, and the intracellular assembly and release of infectious genotype 1a virus. Our results complement recent efforts to develop multi-scale mathematical models of the clinical response to DAAs15, point to the need for greater attention to the full viral life cycle in evaluating both antiviral activity and antiviral resistance, and provide fresh insight into how these emerging therapeutics impact growth of the virus.
MATERIALS AND METHODS
Cell lines and plasmids
Details of cells used in these studies are provided in Supplementary Materials and Methods. pH77S.3 is an infectious molecular clone of a cell-culture adapted genotype 1a HCV16. pH77S.3/GLuc2A is a derivative of it that expresses Gaussia princeps luciferase (GLuc) from sequence inserted between p7 and NS216. L31V, Y93H, and Q30R resistance variants were constructed by site-directed mutagenesis or custom DNA synthesis. Final plasmid constructs were verified by sequence analysis.
Virus infections and antiviral assays
HCV RNA was transcribed from XbaI-linearized plasmid DNA using the T7 Megascript kit (Ambion) from pH77S.3, purified using the RNeasy Mini kit (Qiagen), and electroporated into Huh-7 cells as described previously17. Cells were cultured for 7 days to allow for HCV replication levels to reach a stable maximum prior to seeding to different plate formats for specific antiviral assays. Stock solutions of LDV, DCV, MK-8742, BOC, VPV, HCV-796, MK-0608 and compound 23 were prepared in DMSO. Additional details can be found in Supplementary Materials and Methods.
Statistical methods
For determination of 50% (EC50) and 90% (EC90) effective concentrations of DAAs, data were fit to a four-parameter dose response curve with variable slope using Prism 5.0c for Mac OS X software (GraphPad Software, Inc.). Results are reported as the estimated concentration ± 95% confidence interval.
RESULTS
Kinetics of antiviral suppression by different classes of DAAs
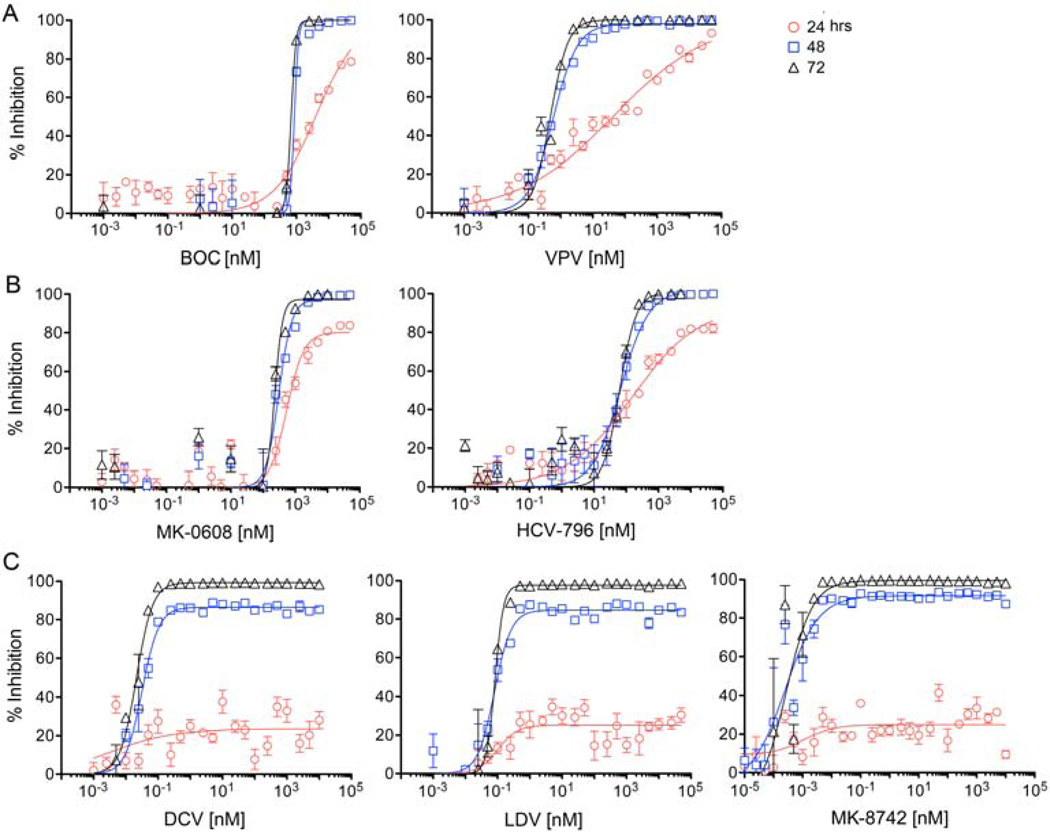
We compared the kinetics of antiviral suppression by the NS5A inhibitors, ledipasvir LDV,18, daclatasvir DCV,3, and MK-8742 with representative inhibitors of other DAA classes. For inhibitors of the NS3/4A protease, we studied BOC19, a linear ketoamide, and vaniprevir VPV,20, a macrocyclic protease inhibitor. We also studied MK-060821 a nucleoside analog inhibitor of the NS5B polymerase, and HCV-79622, a non-nucleoside NS5B inhibitor. Antiviral activity was assessed by monitoring GLuc secreted from cells that were stably infected with a cell culture-adapted virus, H77S.3/GLuc2A, that expresses GLuc as a fusion with its polyprotein (see Methods)16. EC50 and EC90 concentrations of each drug were estimated at 24, 48, and 72 hrs after the start of treatment based on the quantity of GLuc secreted over the preceding 24 hrs (Table S1). GLuc secretion is real-time measure of viral polyprotein synthesis, and correlates well with intracellular RNA abundance16. For all classes of DAAs tested, maximum antiviral activity (lowest EC50 and EC90) was not observed until 48–72 hrs after addition of the compound (Fig. 1). There were substantial differences in the response kinetics between inhibitor classes, however. While the slopes of the response curves for both NS3/4A protease (Fig. 1A) and NS5B polymerase (Fig. 1B) inhibitors were relatively shallow at 24 hrs compared to 48 hrs or 72 hrs, each of these compounds achieved nearly complete suppression of GLuc secretion at very high concentrations by 24 hrs (maximum inhibition, Emax = 80–100% at 24 hr, Table S1). In contrast, the Emax of the NS5A inhibitors, LDV, DCV, and MK-8742 (Fig. 1C), reached a plateau of 23–26% at low concentrations, and was not increased with higher concentrations of these DAAs at 24 hrs. This effect remained evident at 48 hrs, when the NS5A inhibitor Emax ranged from 85–91%, while that of the other compounds was 97–100% (Fig. 1, Table S1). Thus, despite their high potency (EC50 2–85 pM) at 72 hr, the NS5A inhibitors have only a limited ability to reduce GLuc secretion (polyprotein synthesis) at 24 hr, even at concentrations as high as 50 µM. This precluded estimation of the EC50 at 24 hrs for all three compounds.
Figure 1.
Kinetic analysis of the suppression of H77S.3/GLuc2A virus replication by different classes of DAAs: (A) NS3/4A protease inhibitors, boceprevir (BOC, linear ketoamide) and vaniprevir (VPV, macrocyclic); (B) NS5B inhibitors, MK-0608 (nucleoside) and HCV-796 (non-nucleoside); and (C) NS5A inhibitors, daclatasvir (DCV), ledipasvir (LDV), and MK-8742. FT3-7 cells were electroporated with H77S.3/GLuc2A RNA and passaged for one week before treatment with a range of DAA concentrations. Supernatant fluids were harvested from cultures daily and replaced with fresh medium containing DAAs. Results shown represent mean ± s.e.m. percent reduction in GLuc activity in supernatant fluids harvested at 24, 48 and 72 hrs after initiation of treatment, where 0% is GLuc activity in the absence of DAA and 100% is a reduction to background levels (cells transfected with replication-incompetent H77S-AAG/GLuc2A RNA). Data were fit to a four-parameter dose response curve.
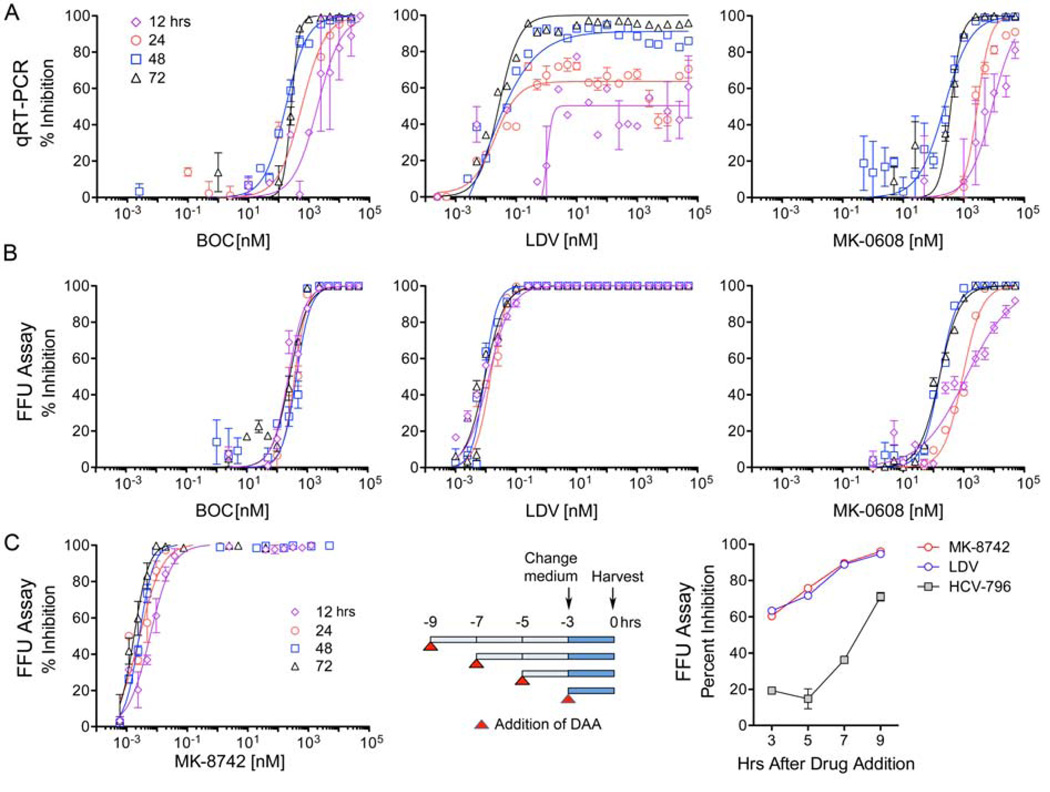
Next, we probed the kinetics of antiviral suppression using quantitative real-time qRT-PCR to assess residual intracellular HCV RNA abundance at various times after the start of treatment (Fig. 2A, Table S2). These experiments used cells infected with H77S.3 virus with no GLuc insertion. Results agreed closely with the GLuc assays for BOC, MK-0608 and LDV activities (compare Figs. 2A with Figs. 1A–C). Notably, LDV demonstrated less capacity to reduce viral RNA abundance at 12 and 24 hrs compared with BOC or MK-0608, even at very high concentrations (Fig. 2A, compare center with left and right panels). Similar results were obtained with the NS5A inhibitor, DCV (Fig. S1A in Supplementary Material).
Figure 2.
Kinetics of antiviral suppression by representatives of 3 different classes of DAAs: NS3/4A inhibitors (BOC), NS5A inhibitors (LDV), and NS5B inhibitors (MK-0608). (A) Antiviral suppression assessed by qRT-PCR measurements of residual RNA abundance at 12, 24, 48 and 72 hrs following initiation of treatment. (B) Antiviral effect of DAAs assessed by reductions in infectious virus released into supernatant fluids between 0–12, 12–24, 24–48, and 48–72 hrs after treatment initiation. Infectious virus release was quantified by FFU assay. Data shown represent mean ± s.e.m. of 2–3 independent results, and were fit to a dose-response curve as in Fig. 1. (C) Kinetics of inhibition of infectious viral release by MK-8742. (left panel) Concentration-related MK-8742 suppression of virus release at 12, 24, 48 and 72 hrs was determined as for other DAAs in panel B. (center panel) Experimental design for determining the impact of short-term DAA (MK-8742, LDV or HCV-796) treatment on infectious virus release. DAAs were added at 2 hr intervals to H77S.3-infected cells at concentrations equivalent to 5× EC90 in the 72 hr GLuc assay (Fig. 1). At 6 hrs, medium was removed from all cultures and replaced with fresh medium containing the compounds. Supernatant fluids were harvested 3 hrs later and infectious virus quantified by FFU assay. (right panel) Infectious virus titer of supernatant fluids at time of harvest determined by FFU assay. The experiment was done in triplicate and is representative of multiple experiments. DAA concentrations were: MK-8742 0.075 nM, LDV 0.86 nM, and HCV-796 1110 nM.
We next employed an assay for infectious focus-forming units (FFU) of virus to determine the quantity of infectious virus released from cells into supernatant fluids at 12–24 hr intervals following addition of the compound (Fig. 2B). These results were dramatically different, as both LDV and BOC rapidly inhibited infectious virus release with little if any difference in the EC50 at 12 vs. 72 hrs of treatment (Fig. 2B, center and left panels, Table S3). At 12 hrs, BOC was much more potent in the FFU assay (EC50 = 237 nM, Table S3) compared to the RT-PCR assay (EC50 = 1,930 nM, Table S2). For LDV, however, at 12 hrs the disparity in the kinetics of suppression of infectious virus release (EC50 = 0.012 nM, Table S3) versus viral RNA abundance (EC50 = 2,500 nM, Table S2) was much greater. MK-8742 (Fig. 2C, left) and DCV (Fig. S1B) also rapidly inhibited infectious virus release, indicating that this is a general property of this class of NS5A inhibitors. The inhibition of virus release was potent and immediate: 60–90% inhibition within 2–3 hrs of the addition of MK-8742 or LDV (Fig. 2C right) to the culture medium. Inhibition of infectious virus release was considerably slower with the non-nucleoside NS5B inhibitor, HCV-796 (Fig. 2C, right). Importantly, we confirmed that this was not due to carry-over of the DAA in viral titrations (Fig. S2).
Thus NS5A inhibitors, and to a lesser extent BOC and possibly other NS3/4A inhibitors, have dual effects on the viral life cycle: rapidly suppressing virus release and more slowly reducing RNA abundance. However, each class of DAA tested strongly inhibited both RNA synthesis and virus release by 72 hrs of treatment.
NS5A inhibitors cause an immediate but only partial block in viral RNA synthesis
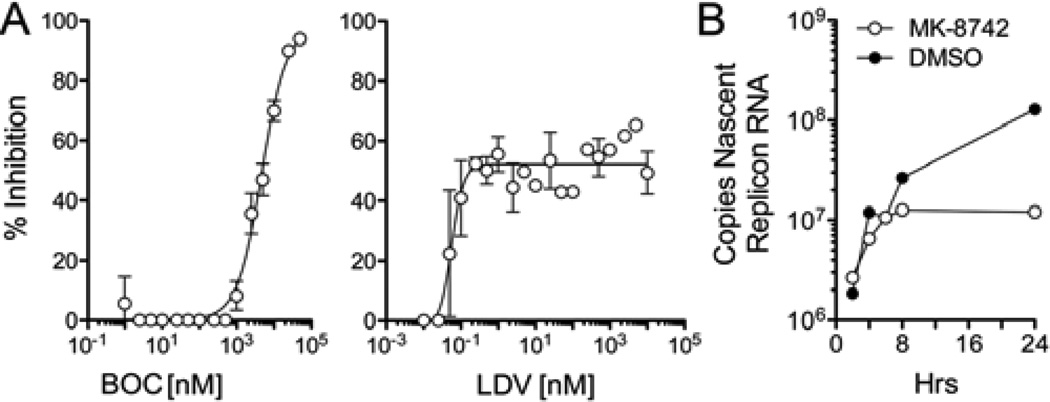
The data shown in Fig. 2 suggest a delay in inhibition of viral RNA synthesis by BOC and LDV. To directly assess this, we measured incorporation of 5-ethynyl uridine into newly synthesized viral RNA during the first 12 hrs of treatment with these compounds (see Supplementary Methods). These results revealed a striking difference in the effects of these two compounds on HCV RNA synthesis (Fig. 3A). High concentrations of BOC effectively ablated new viral RNA synthesis with an Emax approaching 98% (Fig. 3A, left). The BOC EC50 in this assay was 4670 nM (95% C.I. 3261–6690 nM), approximating the EC50 determined in the RT-PCR assay at 12 hrs (2500 nM) but well above the RT-PCR EC50 at 72 hrs (261 nM) (Fig. 2A, left panel; Table S2). In contrast, very high concentrations of LDV (>1000 nM) achieved an Emax of only 52% (Fig. 3A, right). However, this degree of inhibition was reached at very low concentrations. The 50% maximal inhibitory concentration for this partial suppression of RNA synthesis was 0.059 nM (95% C.I. 0.42–0.82 nM), which is comparable to the RT-PCR EC50 at 72 hrs (0.027 nM) (Fig. 2A, center panel; Table S2). Thus, while the NS3/4A inhibitor, BOC, causes an early and complete suppression of RNA synthesis at very high concentrations, low concentrations of the NS5A inhibitor, LDV, potently but only partially inhibit RNA synthesis. These data are consistent with the differences observed in the Emax of these compounds at 24 hrs in the GLuc assays (Fig. 1), and suggest that NS5A inhibitors may block assembly or otherwise inhibit newly forming viral replicase complexes, but have no effect upon those already assembled (see Discussion). Similar studies with MK-8742 suggested that there was little change in the rate of synthesis of a genotype 1b subgenomic HCV RNA replicon19 until 8 hrs after addition of the compound (Fig. 3B).
Figure 3.
Inhibition of HCV RNA synthesis by NS5A inhibitors. (A) Inhibition of viral RNA synthesis in H77S.3-infected cells by (left) BOC or (right) LDV. RNA synthesis was measured by incorporation of 5-ethynyl uridine into nascent HCV RNA between 2–12 hrs after addition of the DAAs to the culture medium (see Methods). (left panel) BOC treatment resulted in a 97% reduction of RNA synthesis at the highest concentrations tested (EC50 = 4688 nM, 95% C.I. = 3279–6742 nM). (right panel) The maximal inhibition of RNA synthesis by LDV was 52% and was achieved at low concentrations of the drug (50% maximal suppression at 0.056 nM, 95% C.I.=0.036–0.086 nM) and not increased at much higher concentrations. (B) Kinetic analysis of the inhibition of RNA synthesis by MK-8742. Genotype 1b subgenomic RNA replicon cells were studied as in panel A, with cells harvested at intervals following addition of the compound. Results shown represent the number of copies of newly synthesized replicon RNAs at each time point, and reveal a shut-off of RNA synthesis at ~8 hrs in MK-8742 cells.
NS5A inhibitors disrupt HCV assembly
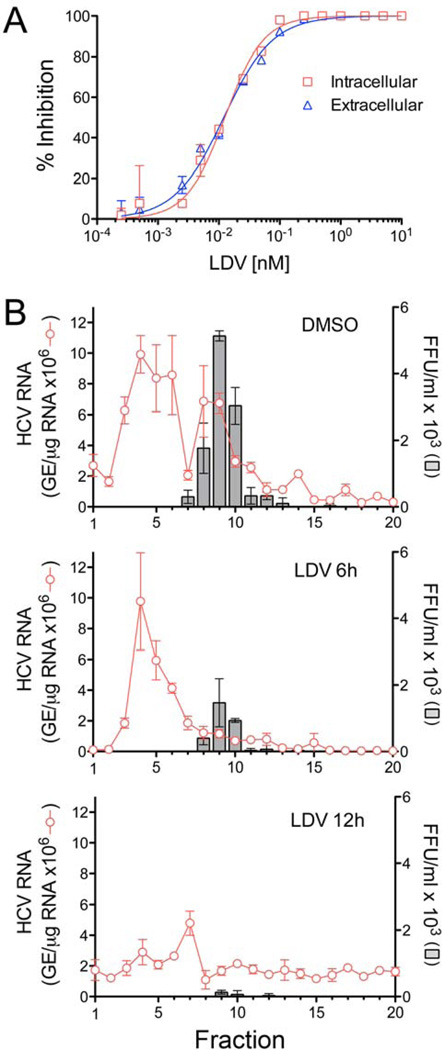
The nearly complete inhibition of viral release by NS5A inhibitors prior to shut-down of viral RNA synthesis and major reductions in viral RNA abundance (Fig. 2A and B, center panels) could reflect a blockade of cellular egress, or an upstream inhibition of viral assembly. To distinguish between these possibilities, we measured the impact of LDV on accumulation of infectious intracellular virus. H77S.3-infected cells were treated with a range of concentrations of LDV for 24 hrs prior to harvest. Extracellular virus released into cell culture medium and intracellular infectious virus were quantified by FFU assay (see Supplementary Methods). These experiments revealed equivalent reductions in intracellular and extracellular virus titers, with EC50 values ~0.012 nM (Fig. 4A). Thus, LDV disrupts a step in viral assembly that precedes the release of virus. We confirmed this by analyzing the sedimentation profile of viral RNA present in lysates of cells treated with LDV at 3× the EC90 for 6 or 12 hrs (Fig. 4B). Infectious viral particles sedimented as a discrete peak (Fig. 4B, grey bars) in fractions 8–10 of rate-zonal gradients loaded with lysate from untreated cells, and this was associated with a co-sedimenting peak of viral RNA. After 6 hrs LDV treatment, this peak was sharply reduced and most HCV RNA had shifted to a more slowly sedimenting species present in fractions 3–5 (Fig. 4B, red lines). By 12 hrs, the amount of viral RNA in this slowly sedimenting peak was sharply reduced.
Figure 4.
NS5A inhibitors block intracellular assembly of HCV. (A) Inhibition of intracellular infectious virus (red) and cell-free virus released from infected cells (blue) following 24 hrs treatment with LDV. Intracellular and extracellular virus were quantified in FFU assays. Results are expressed as % inhibition and overlaid for comparison. (B) Rate-zonal centrifugation of cell lysates derived from H77S.3-transfected cells that were either mock-treated (0.0575% DMSO, top panel) or treated with LDV at ~3× the EC90 in the GLuc assay (575pM, bottom panel) for 6 or 12 hrs. Fractions collected from the top of the gradients were tested for HCV RNA by qRT-PCR and for infectivity by FFU assay.
Collectively, these data indicate that LDV has an immediate (<3 hrs), potent effect on the intracellular assembly of new virus particles that precedes reductions in viral RNA abundance. Previous studies demonstrated that NS5A inhibitors induce a redistribution of NS5A expressed by subgenomic HCV replicons from an ER-like pattern to lipid droplets23, suggesting a possible explanation for their effect on viral RNA synthesis. We observed the accretion of NS5A into large aggregates in H77S.3 virus-infected cells treated with LDV (Fig. S3). However, these changes were not evident until 12 hrs treatment with LDV, and thus did not correlate temporally with the inhibition of intracellular virus assembly. We did not observe a redistribution of NS5A to lipid droplets at either early or late time points (Fig. S3).
Mutations in NS5A confer resistance against disruption of assembly by LDV
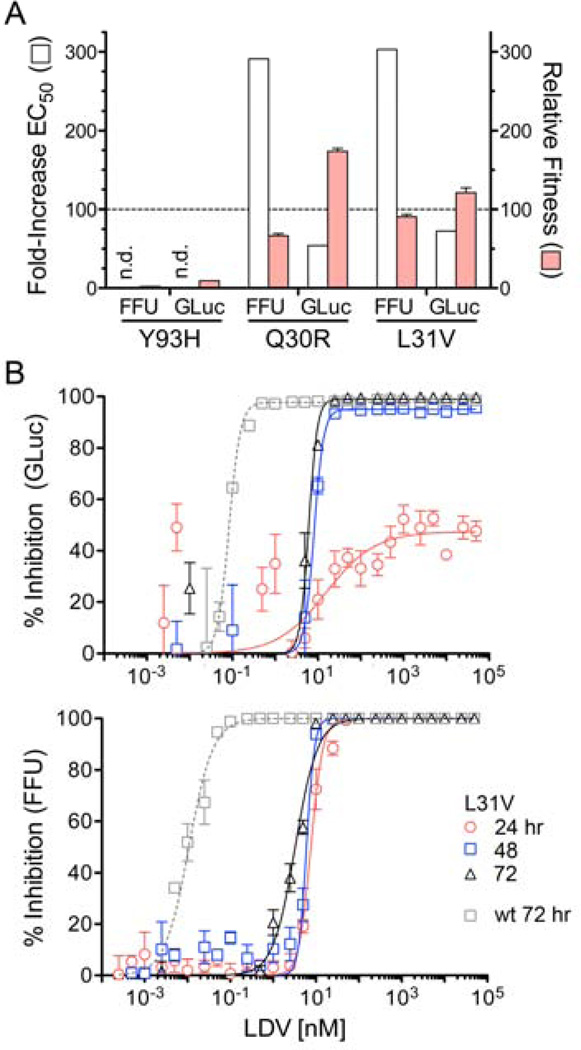
Mutations identified in HCV replicons24, and in genotype 1a viruses from patients treated with NS5A inhibitors25, confer resistance to the effects of these inhibitors on RNA replication. However, the impact of these resistance-associated variants (RAVs) on virus production and its ability to be disrupted by NS5A inhibitors are unknown. We thus studied three commonly reported RAVs: Q30R, L31V and Y93H, each constructed within the background of H77S.3 virus. Of these, Y93H was the least fit with replication levels (assessed by GLuc expression) and infectious virus yields reduced by 10- and 100-fold, respectively, compared to parental virus (Fig. 5A). In contrast, Q30R and L31V were able to replicate RNA and produce virus at levels within 2-fold that of the parental H77S.3 virus. The high fitness of Q30R and L31V variants allowed kinetic assessment of the impact of LDV on replication and virus production. L31V conferred resistance to the immediate inhibition of virus release by LDV, as well as the inhibition of RNA replication reflected in the GLuc assay (Fig. 5B). Resistance (fold-increase in EC50), however, was greater for infectious virus release (Fig. 5B). Importantly, differences in the kinetics of these responses were unchanged from those observed with H77S.3 virus (Figs. 1C and 2B). Similar results were obtained with Q30R (Fig. S4). Collectively, these data suggest that interactions of LDV with amino acid residues 30 and 31 impact its capacity to inhibit both virus assembly and RNA replication.
Figure 5.
Antiviral activity of LDV against resistance-associated variants (RAVs). (A) Antiviral resistance (fold-increase in EC50) and relative replication fitness of Y93H, Q30R, and L31V mutants compared with parental H77S.3 virus (‘wt’) were determined in GLuc and FFU reduction assays at 72 hrs (wt fitness set arbitrarily at 100). ‘n.d.’ = not determined (precluded by low fitness). Results shown represent means ± s.d. (B) LDV-mediated inhibition of (top panel) GLuc secretion and (lower panel) infectious virus release at 24, 48, and 72 hrs following addition of drug to L31V virus-infected cells. The antiviral effect against the parental virus (‘wt’) at 72 hrs is shown for comparison.
A selective PI4KIIIα quinazolinone inhibitor does not disrupt virus assembly
Both hypo- and hyper-phosphorylated forms of NS5A have been observed in many cell culture systems, and the phosphorylation of specific NS5A residues has been proposed to independently regulate virus assembly26, 27 or viral RNA replication8. Some results with genotype 2a JFH1 virus and the genotype 1b Con1 strain suggest that NS5A inhibitors act by blocking hyper- but not basal phosphorylation of NS5A28, a process that is negatively regulated by phosphatidylinositol-4 kinase III-α (PI4K-IIIα)29. We found no evidence that inhibitors alter the phosphorylation status of NS5A in H77S.3-infected cells. However, unlike NS5A expressed by JFH1 or Con1 that resolves as two bands in immunoblots, only a single NS5A band is detectable in lysates of H77S.3-infected cells27. Thus, this difference may be technical in nature, and not indicative of a difference in mechanism of action.
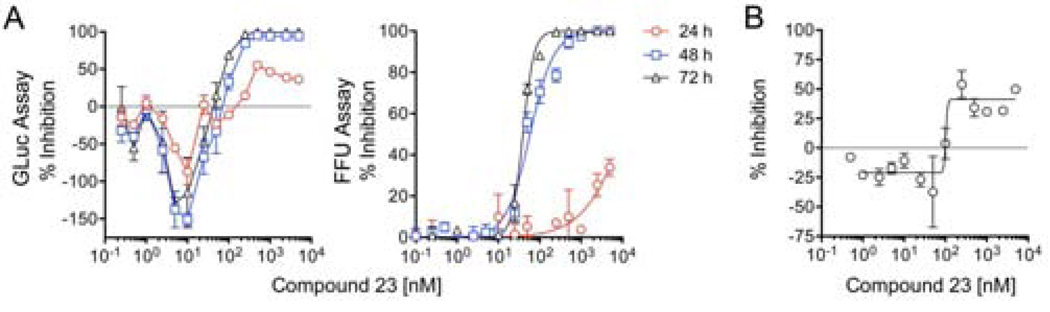
To gain insight into the role played by PI4KIIIα in the antiviral actions of NS5A inhibitors, we studied a recently described quinazolinone, i.e., ‘compound 23’ [5-(2-amino-4-oxo-3-(2-(trifluoromethyl)phenyl)-3,4-dihydroquinazolin-6-yl)-N-(2,4-difluorophenyl)-2-methoxypyridine-3-sulfonamide], a selective inhibitor of PI4KIIIα with substantial antiviral activity against HCV30. In the GLuc assay for polyprotein expression, compound 23 evoked a bi-phasic response. GLuc secretion was significantly enhanced at concentrations between 2.5–25 nM but strongly inhibited above 50 nM (Fig. 6A, left panel). Both positive and negative effects on GLuc secretion were relatively slow in onset. The inhibitory Emax was only 44% at 24 hrs, but nearly 100% by 48 hrs. The slow onset of GLuc inhibition was thus similar to what was observed with LDV and other NS5A inhibitors (Fig. 1C). The EC50 was 82.3 nM (95% C.I. 39.8–170) at 72 hr.
Figure 6.
Antiviral activity of compound 23, a selective PI4KIIIα inhibitor30. (A) Kinetic analysis showing (left panel) a concentration-dependent bi-phasic response to compound 23 in the GLuc assay that measures GLuc secreted by cells replicating H77S.3/GLuc2a virus and (right panel) compound 23 inhibition of release of infectious H77S.3 virus. Results shown represent the mean ± s.e.m. from triplicate cell cultures. (B) Compound 23 inhibition of H77S.3 RNA synthesis measured by incorporation of 5-EU into nascent RNA between 2–12 hrs after addition of the compound to the culture medium as in Fig 4. Results shown represent the mean ± s.e.m. from replicate independent experiments.
Compound 23 inhibited infectious virus release with an EC50 of 39.2 nM (95% C.I. 36.4–42.2) by 72 hr, but this effect was also very slow in onset with less than 50% inhibition by 24 hrs at 5000 nM, the highest concentration tested (Fig. 6A, right panel). This contrasts sharply with the rapid inhibition of infectious virus release with low concentrations of NS5A inhibitors such as LDV and MK-8742 (Figs. 2B–C and 4). Thus, the inhibition of virus release by compound 23 is likely related to shutdown of viral RNA synthesis. Taken collectively, these data indicate that PI4KIIIα activity is not essential for assembly and release of HCV and that the ability of NS5A inhibitors such as DCV, LDV, and MK-8742 to disrupt virion assembly is independent of any effect on the NS5A/PI4KIIIα complex.
We also measured 5-EU incorporation into viral RNA between 2–12 hrs after addition of compound 23 to cell cultures. As with the GLuc assay, low concentrations of the PI4KIIIα inhibitor caused a modest stimulation of viral RNA synthesis, while concentrations above 100 nM resulted in inhibition (Fig. 6B). The maximal inhibition (Emax) of 5-EU incorporation was only 41%, however. This was reached at 250 nM, with no greater inhibition at concentrations up to 5000 nM. Thus, in contrast to the striking difference between PI4KIIIα and NS5A inhibitors in the FFU assay (Fig. 2B, center vs. Fig. 6A, right), these different classes of inhibitors act similarly in their suppression of viral RNA synthesis. As discussed above, these results suggest that compound 23 inhibits new replicase complex formation but lacks activity against previously formed complexes. This suggests in turn that PI4KIIIα activity is required for formation of new replicase complexes, but not for ongoing RNA synthesis by mature complexes.
DISCUSSION
While previous modeling of the therapeutic response to DCV suggested a dual mode of action involving both inhibition of viral release and suppression of viral RNA synthesis15, the data presented here provide direct evidence for an immediate (< 3 hrs), effect of NS5A inhibitors on the intracellular assembly of HCV (Figs. 2 and 4). In the absence of inhibitors, NS5A is distributed between membrane-bound viral replicase complexes derived from endoplasmic reticulum where it functions in RNA replication, and lipid droplets to which it is recruited by core protein and functions in viral assembly26, 31, 32. NS5A inhibitors have been shown to induce a redistribution of NS5A within the cell23. We found LDV resulted in the accretion of NS5A into large, cytoplasmic aggregates (Fig. S3). However, this effect was not evident at early time points when viral assembly was blocked, and correlated more closely with inhibition of viral RNA synthesis (Fig. S3). LDV and other NS5A inhibitors are characterized by highly symmetrical structures, suggesting that they bind NS5A in a dimeric state6. Although we did not find that LDV enhanced co-localization of NS5A with lipid droplets as reported for NS5A inhibitors by others23, it seems likely that the inhibitors bind NS5A in proximity to the lipid droplet given their effect on virus assembly31. Binding of the inhibitor to NS5A could preclude its participation in protein-protein or protein-RNA interactions required for virus assembly. The rapidity with which NS5A inhibitors inhibit release of infectious virus (Fig. 2C) suggests that assembly and release is a very dynamic process, with newly assembled virus being rapidly exported from the cell.
In addition to the immediate effect on virus assembly, NS5A inhibitors potently induced an early (<12 hrs) but only partial block in viral RNA synthesis (Fig. 3A, right, and 3B). This occurred at low concentrations of LDV, only several fold above the EC50 in FFU assays. Unlike BOC, higher concentrations of LDV did not increase the magnitude of this early inhibition of RNA synthesis. These data best fit a model, previously proposed based on subcellular localization and biochemical fractionation analyses of NS5A33, in which NS5A inhibitors block the assembly of new replicase complexes but are unable to inhibit RNA synthesis in those that are already formed. Such a model is consistent with the low initial but increasing Emax observed over time in GLuc assays with NS5A inhibitors (Fig. 1C, Table S1). Consistent with this hypothesis, live cell imaging has shown that HCV replicase complexes persist for many hours in cultured cells, with new complexes replacing older ones at a relatively slow pace34. One possibility is that NS5A inhibitors block new replicase assembly by disrupting the interaction of NS5A with PI4KIIIα, a key host factor facilitating membrane recruitment or remodeling required for replicase assembly35, 36. Compound 23, a relatively specific PI4KIIIα inhibitor, similarly inhibits viral RNA synthesis. NS5A inhibitors could fail to block RNA synthesis in previously formed replicase complexes because the dimeric binding site no longer exists or is otherwise not accessible within the active complex, or possibly because NS5A is no longer required for RNA synthesis once the complex has assembled.
In contrast, BOC is capable of early, near complete inhibition of RNA synthesis at very high concentrations (Fig. 3A, left), suggesting it is able to penetrate and disrupt the activity of preformed replicase complexes. This suggests in turn the surprising conclusion that NS3 has a continuing function in RNA synthesis within fully assembled replicase complexes, separate and distinct from its role in polyprotein processing that must precede assembly of the complex.
Surprisingly, our data also suggest that the NS3/4A inhibitor, BOC, exerts an early blockade effect on infectious virus release (Fig. 2B). While surprising, NS3 is known to be recruited to lipid droplets together with NS5A and to function directly in viral assembly10, 37. Some RAVs associated with resistance to NS3/4A inhibitors also impair viral fitness by negatively impacting steps involved in the release of infectious virus16. Further experiments will be required to confirm whether NS3/4A inhibitors act as a class to block assembly or release of virus and to define how this occurs.
These studies provide novel insight into how NS5A inhibitors impact specific steps in the life cycle of HCV. Since all of the viral functions required for replication are imparted by only ten mature HCV proteins, these proteins are by necessity multi-functional. A more complete understanding of how DAAs impact these multiple functions should help to interpret the complex relationships between PK/PD, drug concentrations, and clinical responses. Our findings also point to the need for greater attention to be paid to the full viral life cycle in evaluating resistance to DAAs, as most studies of antiviral resistance have focused only on how resistance mutations influence the replication of subgenomic replicons. These replicons are incapable of assembly and release as infectious virus, processes that appear to play central roles in the antiviral response to NS5A inhibitors and some if not all NS3/4A protease inhibitors.
Supplementary Material
Acknowledgments
Grant Support: This work was supported in part by and by research grants from the National Institute of Allergy and Infectious Diseases (R01-AI095690) and the Investigator Initiated Studies Program (#40420) of Merck, Sharp and Dohme Corporation, and the University Cancer Research Fund of the University of North Carolina.
Abbreviations
- BOC
boceprevir
- DAA
direct-acting antiviral
- DCV
daclatasvir
- HCV
hepatitis C virus
- LDV
ledipasvir
- PEG-IFN
pegylated interferon α
- RBV
ribavirin
- SVR
Sustained Virologic Response
- VPV
vaniprevir
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: D.R.M., P.I., F.L., E.A-A., A.Y.H. and S.M.L. designed research; D.R.M., S.W., T.M., Z.F., P.I., F.L. and E.A-A. performed research and acquired the data; P.N. and R.D.F. contributed reagents; all authors analyzed the data and reviewed the manuscript for intellectual content; and D.R.M. and S.M.L. wrote the paper.
Author disclosures: S.M.L. is a consultant to Merck, Sharp & Dohme, Co., AbbVie, Inc., Gilead, Achillion Pharmaceuticals, Inc., Santaris, and Idenix. P.I., F.L., E.A-A. and A.Y.H. are employees of Merck, Sharp & Dohme, Co.
REFERENCES
- 1.Lange CM, Jacobson IM, Rice CM, et al. Emerging therapies for the treatment of hepatitis C. EMBO Mol Med. 2013;1:4–15. doi: 10.1002/emmm.201303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofmann WP, Zeuzem S. A new standard of care for the treatment of chronic HCV infection. Nat Rev Gastroenterol Hepatol. 2011;8:257–264. doi: 10.1038/nrgastro.2011.49. [DOI] [PubMed] [Google Scholar]
- 3.Gao M, Nettles RE, Belema M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conte I, Giuliano C, Ercolani C, et al. Synthesis and SAR of piperazinyl-N-phenylbenzamides as inhibitors of hepatitis C virus RNA replication in cell culture. Bioorg Med Chem Lett. 2009;19:1779–1783. doi: 10.1016/j.bmcl.2009.01.066. [DOI] [PubMed] [Google Scholar]
- 5.Lemm JA, O'Boyle D, 2nd, Liu M, et al. Identification of hepatitis C virus NS5A inhibitors. J Virol. 2010;84:482–491. doi: 10.1128/JVI.01360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Boyle Ii DR, Sun JH, Nower PT, et al. Characterizations of HCV NS5A replication complex inhibitors. Virology. 2013;444:343–354. doi: 10.1016/j.virol.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 7.Milward A, Mankouri J, Harris M. Hepatitis C virus NS5A protein interacts with bet-acatenin and stimulates its transcriptional activity in a phosphoinositide-3 kinase-dependent fashion. J Gen Virol. 2010;91:373–381. doi: 10.1099/vir.0.015305-0. [DOI] [PubMed] [Google Scholar]
- 8.Lemay KL, Treadaway J, Angulo I, et al. A hepatitis C virus NS5A phosphorylation site that regulates RNA replication. J Virol. 2013;87:1255–1260. doi: 10.1128/JVI.02154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobardt M, Hopkins S, Baugh J, et al. HCV NS5A and IRF9 compete for CypA binding. J Hepatol. 2013;58:16–23. doi: 10.1016/j.jhep.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyanari Y, Atsuzawa K, Usuda N, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 11.Link JO, Taylor JG, Xu L, et al. The Discovery of Ledipasvir (GS-5885), a Potent Once-Daily Oral NS5A Inhibitor for the Treatment of Hepatitis C Virus Infection. J Med Chem. 2013;57:2033–2046. doi: 10.1021/jm401499g. [DOI] [PubMed] [Google Scholar]
- 12.Coburn CA, Meinke PT, Chang W, et al. Discovery of MK-8742: An HCV NS5A Inhibitor with Broad Genotype Activity. Chem Med Chem. 2013;8:1930–1940. doi: 10.1002/cmdc.201300343. [DOI] [PubMed] [Google Scholar]
- 13.Lok AS, Gardiner DF, Lawitz E, et al. Preliminary study of two antiviral agents for hepatitis C genotype 1. N Engl J Med. 2012;366:216–224. doi: 10.1056/NEJMoa1104430. [DOI] [PubMed] [Google Scholar]
- 14.McPhee F, Hernandez D, Yu F, et al. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology. 2013;58:902–911. doi: 10.1002/hep.26388. [DOI] [PubMed] [Google Scholar]
- 15.Guedj J, Dahari H, Rong L, et al. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A. 2013;110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimakami T, Welsch C, Yamane D, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kannan RP, Hensley LL, Evers LE, et al. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol. 2011;85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawitz EJ, Gruener D, Hill JM, et al. A phase 1, randomized, placebo-controlled, 3-day, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis C. J Hepatol. 2012;57:24–31. doi: 10.1016/j.jhep.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 19.Malcolm BA, Liu R, Lahser F, et al. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother. 2006;50:1013–1020. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liverton NJ, Carroll SS, Dimuzio J, et al. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob Agents Chemother. 2010;54:305–311. doi: 10.1128/AAC.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll SS, Ludmerer S, Handt L, et al. Robust antiviral efficacy upon administration of a nucleoside analog to hepatitis C virus-infected chimpanzees. Antimicrob Agents Chemother. 2009;53:926–934. doi: 10.1128/AAC.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howe AY, Cheng H, Johann S, et al. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob Agents Chemother. 2008;52:3327–3338. doi: 10.1128/AAC.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Targett-Adams P, Graham EJ, Middleton J, et al. Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound modes of action. J Virol. 2011;85:6353–6368. doi: 10.1128/JVI.00215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fridell RA, Qiu D, Wang C, et al. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother. 2010;54:3641–3650. doi: 10.1128/AAC.00556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridell RA, Wang C, Sun JH, et al. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology. 2011;54:1924–1935. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 26.Tellinghuisen TL, Foss KL, Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Welsch C, Yi M, et al. Regulation of the production of infectious genotype 1a hepatitis C virus by NS5A domain III. J Virol. 2011;85:6645–6656. doi: 10.1128/JVI.02156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu D, Lemm JA, O'Boyle DR, 2nd, et al. The effects of NS5A inhibitors on NS5A phosphorylation, polyprotein processing and localization. J Gen Virol. 2011;92:2502–2511. doi: 10.1099/vir.0.034801-0. [DOI] [PubMed] [Google Scholar]
- 29.Reiss S, Harak C, Romero-Brey I, et al. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 2013;9:e1003359. doi: 10.1371/journal.ppat.1003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leivers AL, Tallant M, Shotwell JB, et al. Discovery of Selective Small Molecule Type III Phosphatidylinositol 4-Kinase Alpha (PI4KIIIalpha) Inhibitors as Anti Hepatitis C (HCV) Agents. J Med Chem. 2013;57:2091–2106. doi: 10.1021/jm400781h. [DOI] [PubMed] [Google Scholar]
- 31.Miyanari Y, Atsuzawa K, Usuda N, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 32.Appel N, Zayas M, Miller S, et al. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C, Ma H, Hang JQ, et al. The hepatitis C virus NS5A inhibitor (BMS-790052) alters the subcellular localization of the NS5A non-structural viral protein. Virology. 2011;414:10–18. doi: 10.1016/j.virol.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolk B, Buchele B, Moradpour D, et al. A dynamic view of hepatitis C virus replication complexes. J Virol. 2008;82:10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger KL, Cooper JD, Heaton NS, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger KL, Kelly SM, Jordan TX, et al. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, Yates J, Liang Y, et al. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.