Abstract
Introduction
A 61-year-old woman with a five-year history of progressive muscle weakness and atrophy had a muscle biopsy characterized by a combination of dystrophic features (necrotic fibers and endomysial fibrosis) and mitochondrial alterations [ragged-red cytochrome c oxidase (COX)-negative fibers].
Methods
Sequencing of the whole mtDNA, assessment of the mutation load in muscle and in accessible non-muscle tissues, and single fiber polymerase chain reaction (PCR).
Results
Muscle mitochondrial DNA (mtDNA) sequencing revealed a novel heteroplasmic mutation (m.4403G>A) in the gene (MTTM) that encodes tRNAMet. The mutation was not present in accessible non-muscle tissues from the patient or 2 asymptomatic sisters.
Discussion
The clinical features and muscle morphology in this patient are very similar to those described in a previous patient with a different mutation, also in MTTM, which suggests that mutations in this gene confer a distinctive “dystrophic” quality. This may be a diagnostic clue in patients with isolated mitochondrial myopathy.
Keywords: mtDNA, tRNAMet, de novo mutation, mitochondrial myopathy, dystrophic features, late-onset weakness
Mitochondrial DNA (mtDNA) is a circular molecule containing 13 genes that encode subunits of 4 complexes of the respiratory chain (I, III, IV, and V), 2 rRNA genes, and 22 tRNA genes.1 The tRNAs are essential for transcription and translation of the 13 protein-coding genes and are complemented by a number of imported nuclear-encoded factors.2
The number of pathogenic point mutations in tRNA genes has been increasing steadily since 1990, when mutations in tRNALeu(UUR) and tRNALys were associated with mitochonfrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS)3 and myoclonus epilepsy with ragged-red fibers (MERRF).4 To date, 261 mtDNA pathogenic point mutations have been listed in the mitomap database.5 Although tRNA (MTT) genes comprise only 9% of the mitochondrial genome, they are responsible for more than half of mtDNA-related diseases, which makes them pathogenic hotspots.1 Mutations in MTT genes usually cause multisystem disorders, including MELAS and MERRF, which are among the most common maternally-inherited mitochondrial diseases.
These mutations cause transcriptional and translational defects, mitochondrial respiratory chain dysfunction, and are associated with a striking variety of clinical features.2 The genotype-phenotype correlation is notoriously poor. Individuals who carry the same mutation often have diverse disease presentations, and different mutations within the same MTT gene can differ in both clinical symptoms and severity.6
We report a patient who had unusual dystrophic features on muscle biopsy and harbored a novel pathogenic m.4403G>A point mutation in the MTTM gene encoding tRNAMet. Notably, another patient with similar clinical and muscle pathology features harbored a different mutation in the same MTTM gene.7 Thus, mutations in this rarely affected gene seem to share a dystrophic phenotype, which may offer a useful diagnostic clue.
CASE REPORT
A 61-year-old Filipino woman presented with a five-year history of progressive symmetric weakness and atrophy involving proximal and distal muscles of the arms and legs. Due to difficulty lifting her feet when walking, she had fallen repeatedly in the past 3 years, resulting in elbow and ankle fractures. She could not walk more than 100 meters without resting and preferred to stand when in public due to significant difficulty rising from a seated position. Recent ophthalmological exam had revealed bilateral nonpolychromatic cataracts that were first diagnosed at age 60 and a normal retinal appearance. There was no family history of neuromuscular disease. Two of her sisters were examined and were found to be neurologically normal. By report, her adult daughter has no neurological impairments.
Physical exam revealed a thin woman with marked muscle atrophy, worst in bilateral intrinsic hand and forearm muscles. She was able to move against gravity in all muscles tested, but she could not resist the examiner in distal arm and leg muscles. There was no evidence of spasticity, clinical myotonia, or involuntary movements. Neck flexor, neck extensor, and trapezius muscles had normal strength. She had mild left ptosis but no diplopia, nystagmus, hearing impairment, dysphagia, or dysphonia. Gait was wide-based and waddling. Deep tendon reflexes were diminished in the arms but normal at the knees and ankles. Sensory exam and coordination were normal.
Serum CK ranged between 600 and 700 U/L (normal, 39-238 U/L). Venous lactate was elevated at 4.5mmol/L (normal <1.8mmol/L). Normal or negative laboratory studies included hemoglobin A1c, ANA, anti-SSA, anti-SSB, U1RNP, Scl-70, Jo-1, aldolase, mitochondrial M2 antibody, ESR, CRP, and serum protein electrophoresis. Nerve conduction studies were normal. EMG revealed changes compatible with myopathy, including complex repetitive discharges and positive sharp waves in all muscles tested. MRI of the cervical spine showed myelomalacia at the C3 level without degenerative spine disease or fatty replacement of paraspinal muscles. MRI of the brain was significant for moderate global cortical atrophy without evidence of leukoencephalopathy, brainstem lesions, or putaminal abnormalities.
METHODS
Histochemical studies of muscle were carried out as described.8 Biochemical analysis was performed in 10% muscle extracts.9
Total DNA was extracted from skeletal muscle, blood, saliva, and urinary sediment using the Purogen® DNA purification kit (Gentra System) and sequenced using Illumina TruSeq next generation sequencing methodology. The mutation was confirmed by Sanger methodology.
To assess quantitatively the level of the m.4403G>A mutation, restriction fragment length polymorphism (RFLP) was performed on DNA extracted from muscle, blood, saliva, urinary sediment, and skin fibroblasts. A 208 base-pair (bp) mtDNA fragment spanning nucleotides (nts) 4223–4430 was amplified from total DNA samples using a reverse mismatch primer (5′GCCCGATAGCTTATTTAGCTGACCTAA-3′) (nts 4404–4430, mismatch nucleotide underlined) and a forward primer (nts 4223–4249). The wild-type mtDNA was cut into 2 fragments (145 and 63 bp), and the mutant mtDNA was digested into 4 fragments (145, 63, 34 and 29 bp) by the restriction enzyme Tsp509I. The digestion products were run in a 12% non-denaturing acrylamide gel and subjected to SYBR Gold® Nucleic Acid Gel Stain (Invitrogen) to quantitate the percentage of the mutation.
RESULTS
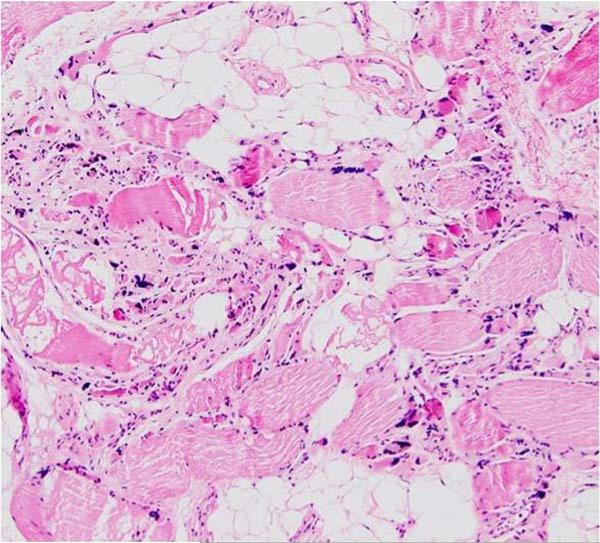
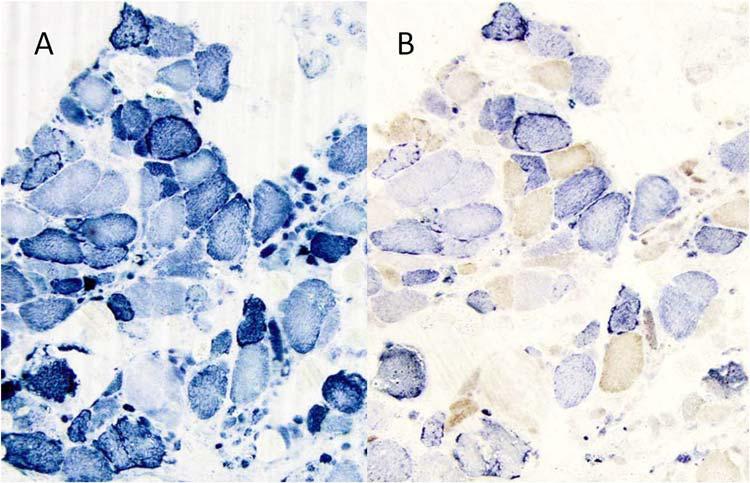
Muscle histochemistry showed numerous atrophic fibers dispersed among normal-sized or hypertrophic fibers. Generally moderate, but focally severe endomysial fibrosis was present, and perimysial adipose tissue was increased significantly (Figure 1). Rare rimmed vacuoles were seen within atrophic fibers, but no amyloid was detected. Roughly 40% of the fibers stained intensely with succinate dehydrogenase (SDH) to produce “ragged-blue fibers” (Figure 2A), but not all such fibers were COX-deficient (Figure 2B). Immunohistochemical stainings for dystrophin, caveolin 3, α-sarcoglycan, merosin, and dysferlin showed no specific deficiency.
Figure 1.
Clusters of highly atrophic fibers are interspersed among normal-sized or hypertrophic fibers. There is moderate to severe endomysial fibrosis, and perimysial adipose tissue is increased. H&E x40.
Figure 2.
A) The SDH histochemical stain shows an irregular interfibrillar network in many fibers. A few fibers are stained excessively with SDH (“ragged-blue” fibers), ×100. B. With the combined SDH/COX stain, about 40% of fibers in a cross section show a variable degree of cytochrome c oxidase (COX) deficiency. Most ragged-blue fibers retain COX activity. ×100.
Biochemical analysis of respiratory chain complexes was essentially normal, except for mildly increased activities of citrate synthase and succinate dehydrogenase and slightly decreased activity of complex I (data not shown).
Sequencing of the entire muscle mtDNA by next generation sequencing methodology revealed a heteroplasmic single-base mutation, m.4403G>A in the MTTM gene. This change is not reported in the URLs of MITOMAP and mtDBHuman Mitochondrial Genome Database5 and was not found in 100 mtDNAs from normal or disease controls. Mitosequence analysis also revealed the presence of a homoplasmic nucleotide change (G>A at nt 4491) very close to the MTND2 gene. This polymorphism is characteristic of haplotype M9, which is common in East Asia10 and can be explained by the Filipino ethnicity of our patient.
Quantitative PCR/RFLP analysis showed that the mutation was heteroplasmic in muscle (mutation load, 63%), but it was undetectable in blood, saliva, urinary sediment, and skin fibroblasts.
Single-fiber PCR analysis showed that the proportion of mutant mtDNAs was 66±6% in 7 COX-negative ragged-red fibers (RRFs) and 51±6% in 8 COX-positive non-RRFs. This difference was significant statistically (P<0.01, 2-tailed testing, with paired t-test). These results are shown as Appendix 1 in Supplementary Information, available online.
The mutation could not be detected in non-muscle tissues from the patient and 2 clinically asymptomatic sisters. These results are in agreement with those of Vissing et al.,7 who documented the mutation in muscle, but not in blood or urinary sediment.
DISCUSSION
Although most point mutations in mt tRNA genes cause multisystem disorders, some have been associated with pure myopathy7, 11-14. We report a patient with isolated myopathy due to a novel m. 4403 A>G transition in MTTM. This mutation involves the acceptor stem region of the tRNAMet, which is important for attachment of the amino acid.
We consider this amino acid change pathogenic for several reasons. First, it alters an evolutionarily highly conserved base pair (see Appendix 2 in Supplementary Information, available online) within the acceptor stem region of tRNAMet and probably damages the secondary structure of the tRNA. Second, it is not reported in different databases and was absent in 100 control DNA samples. Third, we found a statistically significant difference between the mutation loads of COX-negative and COX-positive fibers. That this difference is smaller than in typical mitochondrial myopathies is probably due to the diffuse fibrosis, which hindered “clean” picking of ragged-blue fibers.
This case has some additional interesting features, most notably the unusual muscle morphology and the similarity to a previously reported patient who also harbored an MTTM mutation. In 1998, Vissing et al.7 described a 28-year-old woman who had exercise intolerance since childhood. She had short stature, diffuse muscle atrophy and weakness, and mild steppage gait. Other organs were not affected. Serum CK was increased mildly after exercise. Resting plasma lactate was elevated and increased excessively after standardized cycle ergometry. A deltoid muscle biopsy showed fibrosis and fat infiltration, and the modified trichrome stain revealed typical ragged-red fibers that were COX-deficient. A heteroplasmic mutation (m.4409T>C) in MTTM was detected in muscle, but not in her blood or in the blood of 3 asymptomatic siblings.
It is very uncommon in mitochondrial disease to see necrotic fibers in muscle. The fact that these were abundant in our patient and in the patient reported by Vissing et al, both of whom harbored mutations in MTTM, suggests that this is a distinctive feature of changes in this gene. The higher mutation load (77%) in muscle from the previous patient7 may explain why her myopathy started earlier and was more severe than in our patient. However, the remarkable overlap between these 2 patients, both in clinical presentation and muscle pathology, suggests a close genotype-phenotype correlation, which is most unusual in mtDNA-related diseases. Obviously, more than 2 patients are needed to verify these conclusions.
Finally, mutations in tRNAMet are exceedingly rare when compared with mutations in other mt tRNAs. In fact, besides our patient and that of Vissing et al, only 1 other heteroplasmic mutation in tRNAMet (m.4450G>A) has been reported in association with splenic lymphoma.15 Although the pathogenicity of the mutation was well documented, the link between the mutation and lymphoid proliferation remained unclear.
In conclusion, we describe a new MTTM mutation associated with a clinical and muscle histopathological phenotype that is remarkably similar the only previously described patient who harbored a mutation in the same gene. This correlation between genotype and phenotype, unusual for mtDNA mutations, may suggest the molecular diagnosis in patients who present with sporadic adult-onset isolated mitochondrial myopathy.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by grants from NICHD (P01HD032062), NINDS/NICHD (U54NS078059), and by the Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF).
LIST OF ABBREVIATIONS
- COX
cytochrome c oxidase
- MELAS
mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes
- MERRF
myoclonus epilepsy and ragged-red fibers
- mtDNA
mitochondrial DNA
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- RRF
ragged-red fibers
- SDH
succinate dehydrogenase
REFERENCES
- 1.Schon EA, DiMauro S, Hirano K-I. Human mitochondrial DNA: roles of inherited and somatic mutations. Nature Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiMauro S, Schon EA, Carelli V, Hirano M. The clinical maze of mitochondrial neurology. Nat Rev Neurol. 2013;9:429–444. doi: 10.1038/nrneurol.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 4.Shoffner JM, Lott MT, Lezza A, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 5.Mitomap MITOMAP: A human mitochondrial genome database. 2003 http://wwwmitomaporg.
- 6.Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J Pathol. 2012;226:274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- 7.Vissing J, Salamon MB, Arlien-Soborg P, Nœrby S, Manta P, DiMauro S, et al. A new mitochondrial tRNAMet gene mutation in a patient with dystrophic muscle and exercise intolerance. Neurology. 1998;50:1875–1878. doi: 10.1212/wnl.50.6.1875. [DOI] [PubMed] [Google Scholar]
- 8.Tanji K, Bonilla E. Optical imaging techniques (histochemical, immunohistochemical, and in situ hybridization staining methods) to visualize mitochondria. Meth Cell Biol. 2001;65:311–32. doi: 10.1016/s0091-679x(01)65019-2. [DOI] [PubMed] [Google Scholar]
- 9.DiMauro S, Servidei S, Zeviani M, Di Rocco M, De Vivo DC, DiDonato S, et al. Cytochrome c oxidase deficiency in Leigh syndrome. Ann Neurol. 1987;22:498–506. doi: 10.1002/ana.410220409. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M, Cabrera VM, Gonzalez AM, Larruga JM, Takeyasu T, Fuku N, et al. Mitochondrial genome variation in Eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moslemi A-R, Lindberg C, Toft J, Holme E, Kollberg G, Oldfors A. A novel mutation in the mitochondrial tRNAPhe gene associated with mitochondrial myopathy. Neuromusc Disord. 2004;14:46–50. doi: 10.1016/s0960-8966(03)00168-8. [DOI] [PubMed] [Google Scholar]
- 12.Jeppesen TD, Duno M, Risom L, Wibrand F, Rafiq J, Krag T, et al. A novel de novo mutation of the mitochondrial tRNALys gene mt.8340G>A associated with pure myopathy. Neuromusc Dis. 2014 doi: 10.1016/j.nmd.2013.08.004. in press. [DOI] [PubMed] [Google Scholar]
- 13.Baric I, Fumic K, Ramadza DP, Sperl W, Zimmermann FA, Muačević-Katanic D, et al. Mitochondrial myopathy associated with a novel 5522G>A mutation in the mitochondrial tRNATrp gene. Eur J Hum Genet. 2013;21:871–875. doi: 10.1038/ejhg.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silvestri G, Rana M, DiMuzio A, Uncini A, Tonali P, Servidei S. A late-onset mitochondrial myopathy is associated with a novel mitochondrial DNA (mtDNA) point mutation in the tRNATrp gene. Neuromusc Disord. 1998;8:291–295. doi: 10.1016/s0960-8966(98)00037-6. [DOI] [PubMed] [Google Scholar]
- 15.Lombes A, Bories D, Girodon E, Frachon P, Ngo MM, Breton-Gorius J, et al. The first pathogenic mitochondrial methionine tRNA point mutation is discovered in splenic lymphoma. Hum Mut. 1998;(Suppl. 1):S175–S183. doi: 10.1002/humu.1380110158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.