Abstract
Transcription factors Pax3 and Zic1 are two important regulators of cell fate decision at the neural plate border, where they act synergistically to promote neural crest (NC) formation. To understand the role of these factors in NC development we performed a microarray analysis to identify downstream targets of Pax3 and Zic1 in Xenopus embryos. Among the genes identified was a member of transcription factor activator protein 2 (Tfap2) family, Tfap2 epsilon (Tfap2e). Tfap2e is first expressed at early neurula stage in NC progenitors and Rohon-Beard sensory neurons, and persists in a subset of migrating cranial NC cells as they populate the pharyngeal arches. This is in contrast to other species in which Tfap2e is not detected in the early NC lineage. Tfap2e morpholino-mediated knockdown results in a loss of NC progenitors and an expansion of the neural plate. Tfap2e is also sufficient to activate NC-specific genes in animal cap explants, and gain-of-function experiments in the whole embryo indicate that Tfap2e can promote NC formation. We propose that Tfap2e is a novel player in the gene regulatory network controlling NC specification in Xenopus downstream of Pax3 and Zic1.
Keywords: Neural crest, Sensory neuron, Tfap2, Pax3, Zic1, Xenopus
Introduction
The neural crest (NC) is one of the defining features of vertebrates. NC cells arise from the lateral edge of the neural plate, delaminate from the neuroepithelium and differentiate into a large repertoire of derivatives including the craniofacial skeleton, peripheral nervous system and portions of the cardiovascular system. Specification, maintenance and differentiation of NC cells depend on the activity of several classes signaling molecules and transcription factors that regulate these processes in time and space (reviewed in Stuhlmiller and Garcia-Castro, 2012; Bae and Saint-Jeannet, 2014).
It is now well accepted that, in response to signaling events mediated by Bmp, Wnt and Fgf, distinct sets of transcriptions factors are sequentially activated at the lateral edge of the neural plate. A first set of genes, known as neural plate border (NPB) specifiers, are initially broadly activated at the NPB, and their expression domain comprises the prospective NC tissue as well as other sub-domains of the NPB. These transcription factors include several homeobox-containing proteins, Pax3/7, Mxs1/2, Dlx5 and Tfap2a, as well as zinc finger-containing factors of the Zic family. These factors in turn activate a second set of genes more restricted to the NC territory, known as NC specifiers, which include among others, genes of the Snail, Sox, and Fox family of transcription factors. These NC specifiers are thought to regulate the expression of downstream NC effector genes implicated in the control of NC cells migration and differentiation. The proper expression of these three sets of genes in time and space is central to the specification of NC progenitors (reviewed in Meulemans and Bronner-Fraser, 2004; Sauka-Spengler and Bronner-Fraser, 2008; Betancur et al., 2010; Prasad et al., 2012).
Among these factors, Pax3 and Zic1 have been proposed as early regulators of NC fate (Monsoro-Burq et al., 2005; Sato et al., 2005, Hong and Saint-Jeannet, 2007; Garnett et al., 2012; Milet et al., 2013). Initially, Pax3 and Zic1 are broadly expressed at the NPB and become progressively restricted to different regions of the ectoderm. Pax3 is expressed in the presumptive hatching gland cells, and Zic1 marks the prospective pre-placodal ectoderm, while both factors are co-expressed in the NC forming region (Hong and Saint-Jeannet, 2007). Using gain of function and knockdown approaches in whole embryos we, and others, have shown that Pax3 and Zic1 are necessary and sufficient to promote hatching gland and pre-placodal fates, respectively, while their combined activity is essential to specify the NC in the whole embryo and in isolated explants (Monsoro-Burq et al., 2005; Sato et al., 2005, Hong and Saint-Jeannet, 2007, Milet et al., 2013).
To understand the role of these factors during NC development we performed a microarray analysis to specifically identify Pax3 and Zic1 downstream targets (Bae et al., 2014). Among the genes recovered was a member of transcription factor activator protein 2 (Tfap2) family, Tfap2 epsilon (Tfap2e). This group of transcription factors has highly conserved functions in the development of the NC and its derivatives in vertebrate embryos (Hilger-Eversheim et al., 2000; Hoffman et al., 2007). However, Tfap2e has not been associated with NC progenitors development in fish and mouse; in these species it is expressed in melanoblasts and olfactory bulb, respectively (Van Otterloo et al., 2010; Feng and Williams, 2003). Interestingly, we found that unlike other species, Xenopus Tfap2e is specifically expressed in NC progenitors and Rohon-Beard sensory neurons. Moreover, using gain and loss-of-function approaches we show that Tfap2e is both necessary and sufficient to promote NC formation in the embryo and in isolated explants. We propose that Tfap2e is a novel and essential component of the Xenopus NC gene regulatory network downstream of Pax3 and Zic1.
Materials and Methods
Plasmid constructs
Xenopus laevis Tfap2e (accession # BC111478) was purchased from Open Biosystems (Thermo Scientific, USA). A hormone-inducible version of Tfap2e was generated by sub-cloning the coding region of Tfap2e into pCS2+GR (Tfap2e-GR). The activity of the fusion proteins can be regulated by addition of dexamethasone to the culture medium of whole embryos or animal explants (Kolm and Sive, 1995).
Embryos, injections and explants culture
Xenopus laevis embryos were staged according to Nieuwkoop and Faber (1967) and raised in 0.1X NAM (Normal Amphibian Medium; Slack and Forman, 1980). Fgf8a (5 pg; Christen and Slack, 1997), and Tfap2e-GR mRNAs were synthesized in vitro using the Message Machine kit (Ambion, Austin, TX) and injected in the animal pole region of 2-cell stage embryos. Wnt8 plasmid DNA was injected to avoid axis duplication (100 pg; Wolda et al., 1993). Tfap2e (Tfap2eMO; 30-40 ng; GGGCACGATCCACAGAAGAAAAGCA), Fgf8 (Fgf8MO; 50 ng; Fletcher et al., 2006), Wnt8 (Wnt8MO; 40 ng; Park and Saint-Jeannet, 2008), Pax3 (Pax3MO; 60 ng; Monsoro-Burq et al., 2005) and Zic1 (Zic1MO; 45 ng; Sato et al., 2005) morpholino antisense oligonucleotides were purchased from GeneTools (Philomath, OR). The specificity of the Tfap2eMO was tested in an in vitro transcription/translation coupled rabbit reticulocyte lysate assay (Transcend, Promega). In whole embryos antisense oligonucleotides were injected in one blastomere at the 2-cell stage and embryos analyzed by in situ hybridization at stage 15. To identify the injected side, 500 pg of β-galactosidase mRNA was coinjected as a lineage tracer. For animal explant experiments, both blastomeres of 2-cell stage embryos were injected with Tfap2eGR mRNA, in the animal pole region, and explants were dissected at the late blastula stage and immediately cultured in vitro for several hours in NAM 0.5X plus 10 μM of dexamethasone (Dex; Sigma-Aldrich). Animal explants were subsequently analyzed by qRT-PCR as described (Hong and Saint-Jeannet, 2007).
Lineage tracing and in situ hybridization
Embryos at the appropriate stage were fixed in MEMFA, processed for Red-Gal (Research Organics) staining to visualize the lineage tracer (β-gal mRNA), and in situ hybridization. Antisense DIG-labeled probes (Genius kit; Roche) were synthesized using template cDNA encoding Tfap2e (pSPORT6-Tfap2e; OpenBiosystems), Tfap2a (Luo et al., 2003), Keratin (XK81; Jonas et al., 1985), Snail2 (Mayor et al. 1995), Sox10 (Aoki et al., 2003), Sox8 (O’Donnell et al., 2006), Sox2 (Mizuseki et al., 1998), Runx1 (Park et al., 2012) and Xhe (Katagiri et al., 1997). Whole-mount in situ hybridization was performed as previously described (Harland, 1991). For in situ hybridization on sections, embryos were fixed in 4% paraformaldehyde in phosphate buffer saline (PBS; Gibco) for 1 hour, embedded in Paraplast+ and 12 μm sections hybridized with the appropriate DIG-labeled probes as described (Henry et al. 1996). Sections were then briefly counter stained with eosin.
qRT-PCR analysis
For each sample, total RNAs were extracted from 10 animal explants using an RNeasy micro RNA isolation kit (Qiagen) according to the manufacturer’s directions. During the extraction procedure the samples were treated with DNase I, to eliminate possible contamination by genomic DNA. The amount of RNA isolated from tissues was quantified by measuring the optical density using a spectrophotometer. qRT-PCR was performed using primers for Snail2, Sox2, Keratin, Msx1, Ef1a (Hong and Saint-Jeannet, 2007), Sox10 (F:CTGTGAACAC AGCATGCAAA; R:TGGCCAACTGACCATGTAAA), Runx1 (F:ACTCTGAGTC CGGGGAAGAT; R:CCATATTCCGGTCTGTGCTT) and Tfap2a (F:GGGACAGA GACAGAGCCAAG; R:ATACTCGGGTCCTCAACGTG), and the QuantiTect SYBR green RT-PCR kit (Qiagen) on LightCycler (Roche). The reaction mixture consisted of 10 μl of QuantiTect SYBR Green RT-PCR Master Mix, 500 nM of forward and reverse primers and 60 ng of template RNA in a total volume of 20 μl. Cycling conditions were as follows: denaturation at 95°C (3 sec), annealing at 55°C (4 sec) and extension at 72°C (12 sec). By optimizing primers and reaction conditions a single specific product was amplified as confirmed by melting curve analysis. Each reaction included a control without template and a standard curve of serial dilution (in 10-fold increments) of test RNAs. In each case, Ef1a was used as an internal reference, and each bar on the histograms has been normalized to the level of EF1α expression (Hong et al, 2008). The histograms in each figure are presented as mean ± s.e.m of four independent experiments. A Student’s t-test was used to define statistically significant values in each group.
Results
Xenopus laevis Tfap2e
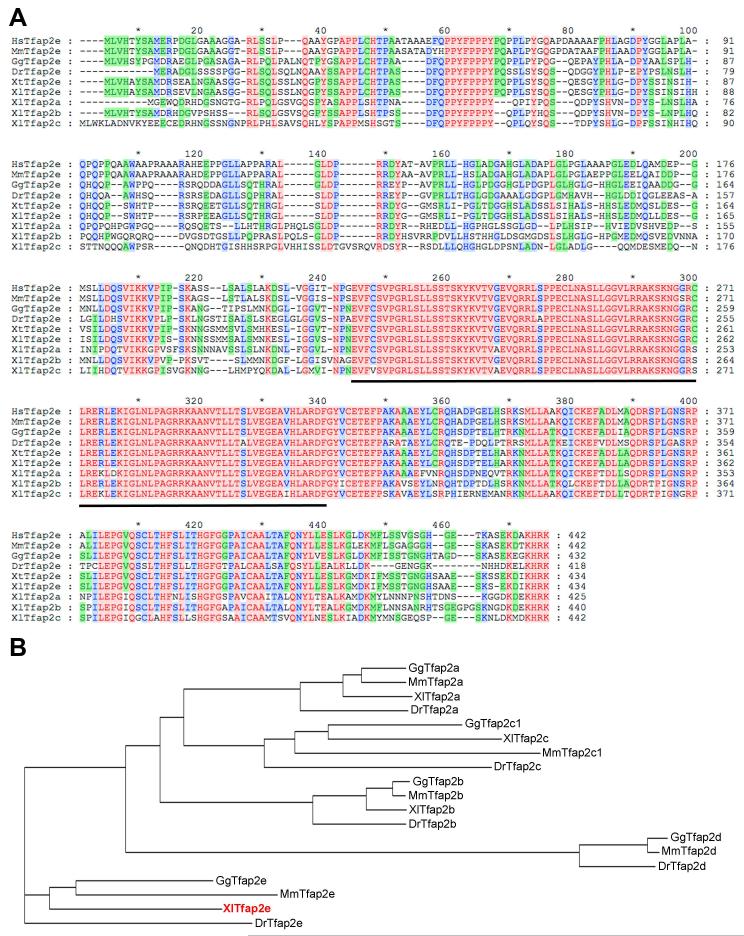
Tfap2e was recovered in a microarray screen designed to identify targets of Pax3 and Zic1 (Bae et al., 2014), two transcription factors that are necessary and sufficient to specify the NPB in Xenopus (Monsoro-Burq et al., 2005; Sato et al., 2005; Hong and Saint-Jeannet, 2007). Xenopus laevis Tfap2e possesses an open reading frame encoding 434 amino acids (Fig. 1A). At the amino acid level, Xenopus laevis Tfap2e shares 72% identity with human TFAP2E (NP_848643; Ebert et al., 2012), 70% identity with mouse Tfap2e (AAQ90059; Tummala et al., 2003), 79% identity with chicken Tfap2e (XP_417778), 70% identity with zebrafish Tfap2e (NP_957115; Van Otterloo et al., 2012) and 97% identity with Xenopus tropicalis Tfap2e (NP_001123400). When compared to other Xenopus laevis Tfap2 family members Tfap2e shows 63% identity with Tfap2a (NP_001081038; Winning et al., 1991), 65% identity with Tfap2b (NP_001087701; Zhang et al., 2006) and 56% identity with Tfap2c (NP_001083186; Zhang et al., 2006). Assignment of Xenopus laevis Tfap2e sequence to the Tfap2 family members was based on phylogenetic tree analysis of the predicted amino acid sequences compared to that of selected vertebrate species (Fig. 1B). This analysis indicates that Xenopus laevis Tfap2e represents the ortholog of mouse, chicken and zebrafish Tfap2e.
Figure 1. Sequence and structure comparison of Tfap2 proteins across species.
(A) The predicted amino acid sequences from human, mouse, chicken, zebrafish, Xenopus tropicalis, Xenopus laevis Tfap2e and Xenopus laevis Tfap2a, Tfap2c and Tfap2d genes were aligned using ClustalX2. Amino acid conservation among Tfap2 family members has been color-coded. Tfap2 DNA binding domain is underlined. (B) Phylogenetic tree analysis of Tfap2a, Tfap2b, Tfap2c, Tfap2d and Tfap2e proteins from Xenopus laevis (Xl), mouse (Mm), chicken (Gg) and zebrafish (Dr).
Developmental expression of Tfap2e
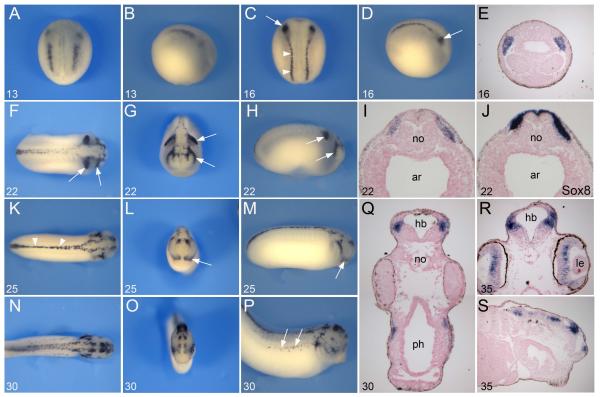
To evaluate the expression of Tfap2e, we performed whole-mount in situ hybridization on embryos at various stages of development. Tfap2e was first detected at stage 13 at the NPB. At this early stage Tfap2e expression was confined to two regions, a small anterior domain and an area that lines the posterior neural plate (Fig 2A, B). A few hours later (stage 16) the two domains appeared more continuous, extending along the entire length of the embryo, in a region corresponding to the prospective NC (Fig 2C, D). Interestingly, the most anterior domain of the cranial NC, the mandibular NC, exhibited a stronger signal than the posterior region of the cranial NC (Fig 2C-E). At stage 22 Tfap2e was primarily restricted dorsally to the trunk region and to the most anterior and the most posterior segments of the cranial NC, as these cells migrate to their respective branchial arches (Fig 2F-H). The Tfap2e expression domain completely overlapped with that of the NC-specific gene Sox8 (O’Donnell et al., 2006) in the stream of migrating cranial NC cells (Fig 2I-J). At stage 25 Tfap2e expression was down-regulated in the posterior stream of the cranial NC but persisted in the mandibular NC. Tfap2e was also detected in the dorsal aspect of the spinal cord as well as in the brain (Fig 2K-M). By stage 30/35 Tfap2e was detected in the dorsal neural tube, NC-derived melanocytes (Fig 2N-P), as well as in the retina and within discrete regions of the brain (Fig 2Q-S).
Figure 2. Developmental expression of Tfap2e by in situ hybridization.
(A-B) Tfap2e onset of expression at the NPB at stage 13. (C-D) At stage 16 Tfap2e expression extends to the entire length of the embryo, in region of the prospective trunk (arrowheads) and cranial NC. The most anterior segment of the cranial NC, the mandibular NC, shows stronger expression than the rest of the cranial NC (arrows). Panels (A, C), dorsal views, anterior to top. Panels (B, D), lateral views, anterior to right. (E) Transverse section of a stage 16 embryo, dorsal to top, showing Tfap2e expression in the mandibular NC. (F-H) At stage 22 as the neural tube closes, Tfap2e expression is restricted to the dorsal aspect of the spinal cord and in the most anterior and posterior segment of the migrating cranial NC (arrows). (I-J) On a transverse section of a stage 22 embryo Tfap2e and Sox8 have overlapping expression domains. (K-M) At stage 25 Tfap2e expression is detected in the mandibular NC (arrows), the dorsal aspect of the spinal cord (arrowheads) and in the brain. (N-P) At stage 30 Tfap2e expression is detected in the brain, dorsal neural tube and NC-derived melanocytes (arrows). (Q) Section of a stage 30 embryo at the level of the hindbrain. Transverse (R) and longitudinal (S) sections of a stage 35 embryo, showing expression in the brain and retina. Panels (F, K, N), dorsal views, anterior to right. Panels (G, L, O), frontal views, dorsal to top. Panels (H, M, P), lateral views, anterior to right. The stages are indicated in the lower left corner of each panel. ar, archenteron; hb, hindbrain; le, lens; no, notochord; ph, pharynx.
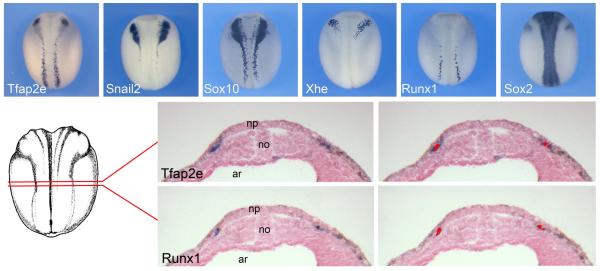
We also compared the expression of Tfap2e to that of genes with well-documented expression in various regions of the ectoderm at the neurula stage including Snail2 and Sox10 (NC), Xhe (hatching gland), Runx1 (Rohon-Beard sensory neurons) and Sox2 (neural plate). Based on this comparative analysis we can conclude that Tfap2e is expressed in cranial NC progenitor, in a pattern similar to that of snail2 and sox10, and possibly in Rohon Beard sensory neurons in the trunk (Fig 3A-F). Rohon Beard neurons arise from the posterior region of the NPB like the trunk neural crest. At the end of neurulation, these neurons are located in the dorsal spinal cord and innervate the skin to mediate the escape response to touch at the larval stages. Later in development Rohon Beard neurons undergo apoptosis, their function is then assumed by the NC-derived dorsal root ganglia neurons (Lamborghini, 1980; 1987). To further confirm that Tfap2e was also expressed in Rohon-Beard sensory neurons in the trunk, we performed in situ hybridization for Tfap2e and Runx1 on adjacent transverse sections of stage 15 embryos (Fig 3G). We found that Tfap2e was co-expressed with Runx1, however Tfap2e expression domain was also broader than Runx1 expression domain (Fig 3H-I’), suggesting that in the trunk region Tfap2e is expressed in both NC progenitors and Rohon-Beard cells.
Figure 3. Comparative expression of Tfap2e with other NPB genes at the neurula stage.
(A-F) Expression of Tfap2e, Snail2, Sox10, Xhe, Runx1 and Sox2 in stage-matched stage 16 embryos. Dorsal views, anterior to top. (G-I’) In situ hybridization on adjacent transverse sections of a stage 15 embryo (G), dorsal to top. Tfap2e (H) is co-expressed with Runx1 (I), as indicated by the red overlay in panels (H’) and (I’). ar, archenteron; no, notochord; np, neural plate.
Regulation of Tfap2e expression at the NPB
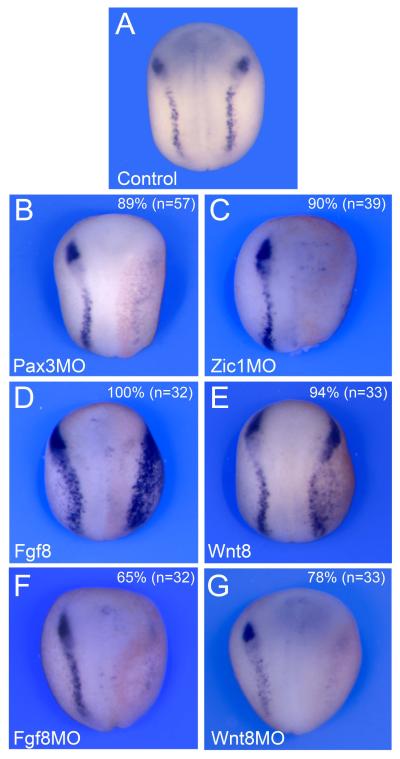
As a target of Pax3 and Zic1, Tfap2e expression is expected to depend on Pax3 and Zic1 activity at the NPB. To test this possibility we interfered with Pax3 or Zic1 function by injection of well-characterized morpholino antisense oligonucleotides at the 2-cell stage (Monsoro-Burq et al., 2005; Sato et al., 2005; Hong and Saint-Jeannet, 2007). In a large proportion of embryos injected with either Pax3MO or Zic1MO we observed a reduction of Tfap2e expression (Fig 4A-C). This result further demonstrates the position of Tfap2e downstream of Pax3 and Zic1 in the gene regulatory cascade leading to NC formation.
Figure 4. Regulation of Tfap2e expression at the NPB.
(A) Tfap2e expression at the NPB in a control embryo. (B-C) Unilateral injection of Pax3 (Pax3MO) or Zic1 (Zic1MO) morpholino antisense oligonucleotides at the 2-cell stage resulted in a strong reduction of Tfap2e expression at the neurula stage. (D-E) Misexpression of Wnt8 or Fgf8 in one blastomere at the 2-cell stage led to a lateral expansion of Tfap2e expression domain. (F-G) Conversely, morpholino-mediated knockdown of either Fgf8 (Fgf8MO) or Wnt8 (Wnt8MO) resulted in a loss of Tfap2e expression on the injected side. Embryos are viewed from the dorsal side, anterior to top. In all panels (B-G) the injected side is on the right, as indicated by the presence of the lineage tracer (Red-gal).
Pax3 and Zic1 are activated at the NPB in response to a specific set of inductive signals: first a Bmp signal, which must be partially attenuated by Bmp antagonists, and then a separate signal mediated by either canonical Wnt or Fgf signaling (reviewed in Stuhlmiller and Garcia-Castro, 2012; Bae and Saint-Jeannet, 2014). We evaluated the dependence of Tfap2e expression on both Wnt and Fgf signaling pathways. Embryos with increased canonical Wnt and Fgf signaling, by injection of the Wnt8 plasmid DNA or Fgf8a mRNA showed increased Tfap2e expression, while knockdown of either molecule with the corresponding morpholino antisense oligonucleotide resulted in a loss of Tfap2e expression on the injected side (Fig 4D-G). Altogether these results indicate that Tfap2e fulfills the criteria of a genuine NC specifiers downstream of the classical NC inducing signals, Wnt and Fgf, and a target of the two NPB specifiers Pax3 and Zic1.
Tfap2e is required for NC formation
To evaluate Tfap2e function during early NC development we performed knockdown of Tfap2e protein using morpholino antisense oligonucleotides. A Tfap2e morpholino (Tfap2eMO) was designed to specifically interfere with translation of Tfap2e mRNA (Fig 5A). In an in vitro transcription/translation assay Tfap2eMO blocked Tfap2e protein production (Fig 5B). Unilateral injection of Tfap2eMO (30 ng) in the animal region of 2-cell stage embryos resulted in a marked decrease in Snail2, Sox10 and Runx1 expression at stage 14 in a large proportion of injected embryos (Fig. 5C-E). Concomitant with the loss of these genes an expansion of Sox2 (neural plate) expression domain (Fig. 5F), and a repositioning of the medial boundary of Keratin (Fig. 5G) were observed on the injected side. Interestingly, while Tfap2a was largely unaffected in morphant embryos, its expression domain was shifted laterally, following the position of the expanded neural plate (Fig. 5H). At later stages the embryos had a severe reduction in the number of migratory cranial NC cells (Fig. 5F-G) consistent with an early loss of NC progenitors.
Figure 5. Tfap2e-depletion blocks NC formation and expands neural plate.
(A) Target sequence (red) of Tfap2e morpholino antisense oligonucleotide (Tfap2eMO). The position of the start codon (ATG) is indicated. (B) In vitro coupled transcription/translation reactions with plasmid encoding Tfap2e. Increasing amounts of Tfap2eMO, 10 ng (+), 100 ng (++) and 1000 ng (+++) blocks translation directed by Tfap2e mRNA. (C-H) Embryos injected with 30ng of Tfap2eMO in one blastomere at the 2-cell stage displayed a reduction or loss of Snail2 (C), Sox10 (D) and Runx1 (E) expression, and a lateral expansion of Sox2 (F) and reduction of Keratin (G) expression domains. Tfap2a expression levels were largely unaffected in the morphant embryos, however its domain of expression was shifted laterally (H). Dorsal views, anterior to top. Injected side (right side) is identified by the presence of the lineage tracer (Red-gal). (I) At tailbud stage these embryos showed a severe reduction in the number of migratory cranial NC cells on the injected side as compared to the uninjected side (right panel). Embryo viewed from the lateral side, dorsal to top, anterior to right (left panel), or to left (right panel). (J) Snail2 expression in Tfap2e morphants can be efficiently rescued by coinjection of mRNA encoding a hormone inducible version of Tfap2e (Tfap2eGR). Dorsal views, anterior to top.
To assess the specificity of the morphant’s phenotype we used a hormone inducible construct in which Tfap2e was fused to the hormone-binding domain of human glucocorticoid receptor (Tfap2eGR). The NC expression of Snail2 was efficiently rescued in Tfap2e-depleted embryos by co-injection of 50-200 pg of Tfap2eGR mRNA, and dexamethasone treatment at the gastrula stage (Fig. 5J).
Tfap2e is sufficient to promote NC formation
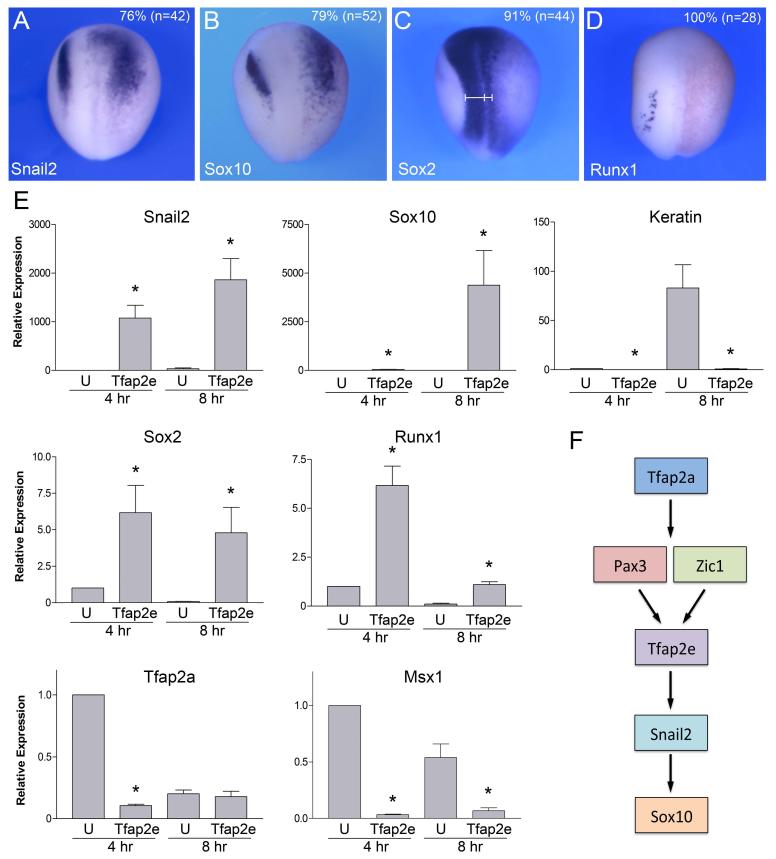
Using the hormone inducible construct we analyzed the consequences of Tfap2e expression on the embryos. Embryos injected with 0.5 ng of Tfap2eGR mRNA and treated with dexamethasone at the gastrula stage (stage 10.5-11) displayed an expansion of Snail2 and Sox10 expression domains (Fig 6A-B) in more than 70% of injected embryos. Sox2 and Runx1 were both down-regulated in these embryos (Fig 6C-D).
Figure 6. Tfap2e is sufficient to promote NC fate.
(A-D) Injection of 0.5 ng of Tfap2e-GR mRNA at the 2-cell stage expands Snail2 (A) and Sox10 (B) expression domains at the neural plate border, while reducing Sox2 (C) and Runx1 (D) in the neuroectoderm. (E) In animal cap explants Tfap2eGR is sufficient to strongly activate Snail2 and Sox10 expression after 4 hours (4 hr) and 8 hours (8 hr) in the presence of dexamethasone, respectively. Comparatively, Sox2 and Runx1 were only weakly activated by Tfap2eGR, while the epidermis marker, keratin, was reduced in these explants. Tfap2eGR was unable to activate the expression of Tfap2a and Msx1. The values were normalized to Ef1a and presented as mean ± s.e.m.; (*), p<0.05 versus uninjected (U), from four independent experiments. (F) Proposed model for the sequence of induction of Tpaf2e and its putative downstream targets in NC progenitors, based on this study and published work (O’Donnell et al., 2006; Hong and Saint-Jeannet, 2007; de Croze et al., 2011).
To determine whether Tfap2e was sufficient to promote NC fate, embryos at the 2-cell stage were injected in the animal pole region with Tfap2eGR mRNA, animal explants were dissected at the blastula stage and cultured in vitro for 4 hours or 8 hours in the presence of dexamethasone and analyzed by Real-Time RT-PCR. Strong induction of Snail2 and Sox10 expression was observed in these explants 4 hours and 8 hours after addition of dexamethasone, respectively (Fig. 6E). Comparatively, Runx1 and Sox2 were only weakly activated in Tfap2eGR-injected explants, while the epidermis-specific gene, Keratin, was repressed (Fig 6E). Interestingly, Tfap2eGR was unable to activate the two early NPB specifiers, Tfap2a and Msx1, in these explants (Fig. 6E). Tfap2a and Msx1 are also expressed in the non-neural ectoderm (epidermis) in addition to the NPB, and as such their expression is repressed in these explants, as Tfap2eGR promote NC fate. In that respect Tfap2a and Msx1 behave like the epidermal marker Keratin. Altogether, these results suggest that Tfap2e is acting downstream of Tfap2a and Msx1 and upstream of Snail2 and Sox10 in the gene network controlling NC formation at the NPB (Fig. 6F).
Discussion
Here we report the expression and function of Tfap2e, the fourth member of the Tfap2 family in Xenopus laevis. At late gastrula/early neurula stage, Tfap2e is expressed in premigratory NC progenitors and Rohon-Beard sensory neurons. Later in development Tfap2e is primarily detected in migrating cranial NC cells and in the brain. All three Tfap2 family members described so far in Xenopus laevis, Tfap2a, Tfap2b and Tfap2c, are also expressed in premigratory NC cells at the neurula stage, as well as in the epidermis (Luo et al., 2002; 2003; Zhang et al., 2006). The fifth member of the family, Tfap2d, has not yet been identified in Xenopus laevis, however its ortholog exists in Xenopus tropicalis (Accession # XM_002933582).
The early expression of Tfap2e in NC progenitors is quite unique to frogs. In zebrafish, tfap2e expression starts around the time of NC cells migration and appears to be restricted to melanoblasts directing differentiation of melanophores (Van Otterloo et al., 2010). In mouse embryos, Tfap2e is not detected in the NC or its derivatives, rather Tfap2e is expressed in the mitral cell layer of the developing olfactory bulb (Feng and Williams, 2003), where it regulates lamination of the olfactory bulb (Feng et al., 2009). In chicken, Tfap2e is confined to the intermediate mesoderm and the pharyngeal arches and clefts (http://geisha.arizona.edu/geisha/search.jsp?gene=457711). The diversity of expression domains across species clearly indicates that Tfap2e function has not been conserved during evolution. Using the Genomicus Program (http://www.genomicus.biologie.ens.fr) we analyzed gene clustering in vertebrate genome around Tfap2e. We found that in human, TFAP2E is flanked by NCDN, oriented in the same direction, and PSMB2, oriented in the opposite direction. The same arrangement is conserved in the mouse, chicken and frog (Xenopus tropicalis) genomes, suggesting that the functional differences may not be the result of changes in gene synteny.
Tfap2e was isolated in a screen for targets of Pax3 and Zic1 in the NPB region (Bae et al., 2014). As such, Tfap2e expression in NC progenitors is regulated by signaling factors (canonical Wnt and Fgf) that activate these two NPB genes, and its expression depends on Pax3 and Zic1 function in this region of the ectoderm. Morpholino-mediated knockdown of Tfap2e resulted in a dramatic loss of Snail2 and Sox10, two NC specifier genes, and a subsequent loss of migratory NC cells in the head region. Tfap2e gain-of-function phenotype was equally dramatic causing an expansion of the NC progenitor pool, and a loss of neural plate tissue. Moreover, in animal cap explants, Tfap2e expression was sufficient to activate Snail2 and Sox10. Altogether, these observations establish Tfap2e as an important novel player in the gene regulatory network underlying NC specification in Xenopus downstream of Pax3 and Zic1 (Fig. 6F).
Most members of the Tfap2 family in vertebrates have highly conserved functions in the development of the epidermis, NC and its derivatives (Hoffman et al., 2007). Among these factors, Tfap2a has been proposed as a “master regulator” of the NC regulatory cascade as it is required for the expression of NPB specifier genes such as Msx1, Pax3, Zic1 and Hairy2 (de Croze et al., 2011). Genome-wide analyses of chromatin marking patterns and transcription factors occupancy have shown that human NC enhancers are primarily occupied by TFAP2A (Rada-Iglesias et al., 2012), confirming that this transcription factor is a key regulator of NC fate. Both mouse and zebrafish Tfap2a mutants lack neural, skeletal, and pigment cells derived from cranial NC (Schorle et al., 1996; Knight et al., 2003, 2004; Zhang et al., 1996; Arduini et al., 2009; Van Otterloo et al., 2012), and in frogs Tfap2a knockdown prevents NC progenitors specification (Luo et al., 2003), by regulating multiple steps in the NC gene regulatory network (de Croze et al., 2011). Consistent with these observations, in our experiments Tfap2e knockdown did not affect Tfap2a expression levels in the embryo, and Tfap2e was unable to induce Tfap2a expression in animal cap explants, however, Tfap2a expression was sufficient to activate Tfap2e in these explants (not shown), thereby establishing Tfap2e as a genuine NC specifier downstream of Tfap2a and Pax3/Zic1 (Fig. 6F).
The extreme divergence of Tfap2e expression domains among vertebrates is very intriguing. It will be especially interesting to determine whether TFAP2e from other species have also NC-inducing properties in Xenopus embryos. TFAP2 proteins bind as dimer to a similar palindromic core “Tfap2 binding site” (GCCN3GGC) to activate transcription (Hilger-Eversheim et al., 2000), and in vitro studies have shown that all TFAP2 proteins, including TFAP2e, can equally bind to keratin-specific promoters for example (Tummala et al., 2003). Therefore we can predict that TFAP2e from various species may have the same ability to promote NC fate when placed in the proper context. Altogether our results point to species-specific differences in the relative importance of TFAP2 family members in the development of the NC; differences that are due to evolutionary divergences in the tissue-specific expression of individual Tfap2 factors rather than differences in the intrinsic activity of these factors.
Acknowledgements
We are grateful to Dr. Jane McCutcheon for comments on the manuscript, and to Dr. Tom Sargent for reagents. This work was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by Korean Government (MOE) to C-S H (2010-0025108), and a grant from the National Institutes of Health to J-P S-J (R01-DE014212).
References
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink EK, Lee Y-H, Credidio C, Saint-Jeannet J-P. Sox10 regulates the development of the neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Arduini BL, Bosse KM, Henion PD. Genetic ablation of neural crest cell diversification. Development. 2009;136:1987–1994. doi: 10.1242/dev.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C-J, Saint-Jeannet J-P. Induction and Specification of Neural Crest Cells: Extracellular Signals and Transcriptional Switches. In: Trainor P, editor. Neural Crest Cells: Evolution, Development and Disease. Elsevier; 2014. pp. 27–49. [Google Scholar]
- Bae C-J, Park B-Y, Lee Y-H, Tobias JW, Hong C-S, Saint-Jeannet J-P. Identification of targets of Pax3 and Zic1 in the developing neural crest. Dev Biol. 2014 doi: 10.1016/j.ydbio.2013.12.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Slack JMW. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev Biol. 1997;192:455–466. doi: 10.1006/dbio.1997.8732. [DOI] [PubMed] [Google Scholar]
- de Croze N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci U S A. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MPA, Tänzer M, Balluff B, Burgmeister E, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med. 2012;366:44–53. doi: 10.1056/NEJMoa1009473. [DOI] [PubMed] [Google Scholar]
- Feng W, Williams T. Cloning and characterization of the mouse AP-2 epsilon gene: a novel family member expressed in the developing olfactory bulb. Mol Cell Neurosci. 2003;24:460–475. doi: 10.1016/s1044-7431(03)00209-4. [DOI] [PubMed] [Google Scholar]
- Feng W, Simoes-de-Souza F, Finger TE, Restrepo D, Williams T. Disorganized olfactory bulb lamination in mice deficient for transcription factor AP-2epsilon. Mol Cell Neurosci. 2009;42:161–171. doi: 10.1016/j.mcn.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- Garnett AT, Square TA, Medeiros DM. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development. 2012;139:4220–4231. doi: 10.1242/dev.081497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Henry GL, Brivanlou IH, Kessler DS, Hemmati-Brivanlou A, Melton DA. TGF-beta signals and a pattern in Xenopus laevis endodermal development. Development. 1996;122:1007–1015. doi: 10.1242/dev.122.3.1007. [DOI] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. 2000;260:1–12. doi: 10.1016/s0378-1119(00)00454-6. [DOI] [PubMed] [Google Scholar]
- Hong C-S, Saint-Jeannet J-P. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C-S, Park B-Y, Saint-Jeannet J-P. Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development. 2008;135:3903–3910. doi: 10.1242/dev.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool part B: Mol Dev Evol. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Jonas EA, Sargent TD, Dawid IB. Epidermal keratin gene expressed in embryos of Xenopus laevis. Proc Natl Acad Sci USA. 1985;82:5413–5417. doi: 10.1073/pnas.82.16.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri C, Maeda R, Yamashika C, Mita K, Sargent TD, Yasumasu S. Molecular cloning of Xenopus hatching enzyme and its specific expression in hatching gland cells. Int J Dev Biol. 1997;41:19–25. [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. Lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Nelson S, Zhang T, Schilling T. Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Dev Dyn. 2004;229:87–98. doi: 10.1002/dvdy.10494. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171:267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- Lamborghini JE. Rohon-beard cells and other large neurons in Xenopus embryos originate during gastrulation. J Comp Neurol. 1980;189:323–333. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- Lamborghini JE. Disappearance of Rohon-Beard neurons from the spinal cord of larval Xenopus laevis. J Comp Neurol. 1987;264:47–55. doi: 10.1002/cne.902640105. [DOI] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Thomas ML, Weeks DL, Sargent TD. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev Biol. 2002;245:136–144. doi: 10.1006/dbio.2002.0621. [DOI] [PubMed] [Google Scholar]
- Luo T, Lee Y-H, Saint-Jeannet J-P, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc Natl Acad Sci USA. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Milet C, Maczkowiak F, Roche DD, Monsoro-Burq A-H. Pax3 and Zic1 drive induction and differentiation of multipotent, migratory, and functional neural crest in Xenopus embryos. Proc Natl Acad Sci USA. 2013;110:5528–5533. doi: 10.1073/pnas.1219124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A-H, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- O’Donnell M, Hong C-S, Huang X, Delnicki RJ, Saint-Jeannet J-P. Functional analysis of Sox8 during neural crest development in Xenopus. Development. 2006;133:3817–3826. doi: 10.1242/dev.02558. [DOI] [PubMed] [Google Scholar]
- Park B-Y, Saint-Jeannet J-P. Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev Biol. 2008;324:108–121. doi: 10.1016/j.ydbio.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-Y, Hong C-S, Weaver J, Rosocha E, Saint-Jeannet J-P. Xaml1/Runx1 is required for the specification of Rohon-Beard sensory neurons in Xenopus. Dev Biol. 2012;362:65–75. doi: 10.1016/j.ydbio.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev Biol. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–48. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;97:557–68. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Slack JM, Forman D. An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. J Embryol Exp Morphol. 1980;56:283–299. [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala R, Romano R-A, Fuchs E, Sinha S. Molecular cloning and characterization of AP-2e, a fifth member of the AP-2 family. Gene. 2003;321:93–102. doi: 10.1016/s0378-1119(03)00840-0. [DOI] [PubMed] [Google Scholar]
- Van Otterloo E, Li W, Bonde G, May KM, Hsu M-Y, Cornell RA. Differentiation of zebrafish melanophores depends on transcription Factors AP2 alpha and AP2 epsilon. PLoS Genetics. 2010;6:e1001122. doi: 10.1371/journal.pgen.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Otterloo E, Li W, Garnett A, Cattell M, Meulemans Medeiros D, Cornell RA. Novel Tfap2-mediated control of soxE expression facilitated the evolutionary emergence of the neural crest. Development. 2012;139:720–730. doi: 10.1242/dev.071308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winning RS, Shea LJ, Marcus SJ, Sargent TD. Developmental regulation of transcription factor AP-2 during Xenopus laevis embryogenesis. Nucleic Acids Res. 1991;19:3709–3714. doi: 10.1093/nar/19.13.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolda SL, Moody CJ, Moon RT. Overlapping expression of Xwnt-3a and Xwnt-1 in neural tissue of Xenopus laevis embryos. Dev Biol. 1993;155:46–57. doi: 10.1006/dbio.1993.1005. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Luo T, Sargent TD. Expression of TFAP2beta and TFAP2gamma genes in Xenopus laevis. Gene Expr. Patterns. 2006;6:589–595. doi: 10.1016/j.modgep.2005.11.011. [DOI] [PubMed] [Google Scholar]