Abstract
Positive allosteric modulators of α-amino-3-hydroxy-5-methyl-isoxazole-propionic acid (AMPA) ionotropic glutamate receptors facilitate synaptic plasticity and contribute essentially to learning and memory, properties which make AMPA receptors targets for drug discovery and development. One region at which several different classes of positive allosteric modulators bind lies at the dimer interface between the ligand-binding core of the second, membrane-proximal, extracellular domain of AMPA receptors. This solvent-accessible binding pocket has been the target of drug discovery efforts, leading to the recent delineation of five “subsites” which differentially allow access to modulator moieties, and for which distinct modulator affinities and apparent efficacies are attributed. Here we use the voltage-clamp technique in conjunction with rapid drug application to study the effects of mutants lining subsites “A” and “B” of the allosteric modulator pocket to assess affinity and efficacy of allosteric modulation by cyclothiazide, CX614, CMPDA and CMPDB. A novel analysis of the decay of current produced by the onset of desensitization has allowed us to estimate both affinity and efficacy from single concentrations of modulator. Such an approach may be useful for effective high throughput screening of new target compounds.
Keywords: electrophysiology, mutagenesis, deactivation, desensitization, allosteric modulation, AMPAkine, ion channel gating, neuropharmacology
1. Introduction
Glutamate receptors are responsible for the majority of excitatory transmission throughout the CNS. The AMPA-subtype glutamate receptors (GluA) are ligand-gated ion channels composed of four transmembrane subunits (GluA 1–4); (Boulter et al., 1992; Mansour et al., 2001; Rosenmund et al., 1998). Each subunit has an alternatively-spliced “flip/flop” region between the M3 and M4 domains that has a major influence on receptor gating kinetics (Koike et al., 2000; Partin et al., 1996; Sommer et al., 1990; Sun et al., 2002). GluA receptors initiate rapid depolarization of excitatory post-synaptic potentials (EPSPs) and serve as a trigger for induction of long-term potentiation of synaptic strength (LTP), believed to be the cellular basis for learning and memory (Malinow et al., 2000). GluA receptors mediate changes in synaptic strength through changes in their copy number and subunit composition in the postsynaptic membrane (Malenka, 2003) and through post-translational modification (Anggono & Huganir, 2012). Subunit composition of GluA receptors varies by brain region throughout the CNS (Traynelis et al., 2010; Collingridge et al., 2009) and are implicated in and shown to be valuable therapeutic targets for a wide range of disease states, including Alzheimer's, Parkinson's, ADHD, autism, Schizophrenia, and stroke (Black, 2005; Menniti et al., 2013; Morrow et al., 2006; Ward et al., 2010). However, because GluA receptors are both ubiquitous and diverse, there is a pointed need for more selective drug design to match this diversity and better target these diseases (Schwenk et al., 2012).
Positive allosteric AMPA receptor modulators prolong the probability GluA receptors remain in the open state, thereby, prolonging EPSPs and increasing the likelihood of inducing LTP (Lynch et al., 2011; Staubli et al., 1994a; Staubli et al., 1994b). Like the GluA receptors they regulate, AMPA receptor modulators need to be diverse to selectively target specific GluA receptor subclasses. Known positive allosteric AMPA receptor modulators have been chemically classified as benzothiazides, benzamides, benzoxainones, and biarylsulfonamides, but have varying degrees of specificity, particularly splice-isoform selectivity for flip or flop GluA variants (e.g. CTZ and CX614) (Fernandez et al., 2006; Jin et al., 2005; Krintel et al., 2012; Krintel et al., 2013; Arai and Kessler, 2007; Harms et al., 2013; Mueller et al., 2011; Sun et al., 2002; Timm et al., 2011). Although this variation exists for modulators, finer selectivity is required for significant therapeutic value (Ward et al., 2010).
Experimentally, major differences in kinetics exist for the different GluA receptor isoforms. When exposed to brief pulses of agonist, channels will open and then close upon agonist removal (deactivation). However, when subjected to prolonged agonist exposure, channels will open briefly then close even with agonist bound (desensitization). Agonist dissociation is then required for recovery from desensitization. At present, AMPA receptor modulators have been shown to increase both deactivation and desensitization rates. A common concern about modulators that slow the onset of receptor desensitization is that substantially increased desensitization rates may result in excitotoxicity, making the drug, therapeutically, much less attractive (Moudy et al., 1994). Some discovery approaches have focused on the possibility of teasing apart the biophysical process of deactivation from that of desensitization. However, in no case has a positive allosteric modulator of deactivation been identified which does not also modulate desensitization (Ahmed et al., 2010; Arai et al., 2000; Mitchell and Fleck, 2007; Quirk and Nisenbaum, 2002; Sun et al., 2002; Timm et al., 2011).
The co-crystal structure of GluA2 ligand binding domain solved with several classes of modulator provides the opportunity to understand how GluA2 receptors interact with their modulators. These studies have revealed a hydrophilic modulator binding pocket which consists of 5 subsites that have been proposed to differentially affect receptor modulation and function (Ptak et al., 2009). Previously, we compared the structure and function of two novel positive allosteric modulators, CMPDA (phenyl-1,4-bis-alkylsulfonamide) and CMPDB (phenyl-1,4-bis-carboxythiophene) to cyclothiazide (CTZ) and the AMPAkine CX614 (2H,3H,6aH-pyrrolidino(2,1–3',2')1,3-oxazino(6',5'–5,4)benzo(e)1,4-dioxan-10-one) on WT GluA2 receptors (Timm et al., 2011). Here we extend that analysis using point mutations of subsites A and B of GluA2 receptors. Using patch-clamp electrophysiology, rapid-perfusion of modulators, and a novel method of data analysis, we examine the effects of several classes of positive allosteric modulator on GluA2 receptors with point mutations within the modulator-binding pocket (subsites A, B and B'). Computer simulations also provide a proof of concept study for our novel analysis which may serve as a beneficial tool for the future of characterizing and categorizing novel GluA receptor modulators.
2. Materials and Methods
2.1. Positive allosteric modulators
Four positive allosteric modulators were used for this study: cyclothiazide (CTZ; Tocris Bioscience; Bristol, UK); CX614 (kindly provided by Cortex Pharmaceuticals, Inc; Irvine, CA); and two compounds developed by Lilly Research Laboratories (Timm et al., 2011), bis-Alkylsulfonamide 506091 (referred to in this study as CMPDA) and bis-Carboxythiophene 2152080 (referred to in this study as CMPDB).
2.2 AMPA receptor cDNA plasmids
cDNAs were a gift of Dr. Peter Seeburg (University of Heidelberg, Germany). The flip and flop isoforms of rat GluA2 receptors were expressed in pRK5, a CMV expression vector, fused in-frame with yellow fluorescent protein (YFP). “WT” GluA2 cDNA was produced from the cDNA plasmids into which a pore mutation (R607Q) was made, using site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit, Stratagene; La Jolla, CA). Mutant receptors express easily-measured currents in HEK293 cells, while receptors with an arginine at position 607 express currents that are too small to measure with conventional patch-clamp electrophysiology (Hume et al., 1991; Verdoorn et al., 1991).
2.3. HEK293 cell culture and transient transfection
Human embryonic kidney 293 (HEK293) cells (CRL 1573; American Type Culture Collection; Manassas, VA) were cultured in 35 mm polystyrene dishes (Becton-Dickinson and Company; Lincoln Park, NJ) up through cell passage P45. Cells were transiently transfected using FuGene 6 reagent (Roche Diagnostic Corp., Indianapolis, IN) with GluA2 (i, flip or o, flop) cDNA and enhanced yellow fluorescent protein (EYFP) cDNA (1 and 0.2 μg/35 mm dish, respectively). Media was changed 4–5 hours post-transfection and NBQX was added to a final concentration of 10–20 μM in some experiments in order to prevent cyto-toxicity, but no NBQX was present when currents were measured.
2.4. Patch-clamp electrophysiology
Currents were recorded 1–2 days after transfection from cells expressing fluorescence arising from the YFP fusion protein. Outside-out membrane patches were held under voltage-clamp at −60 mV using an Axopatch 200B amplifier (Molecular Devices; Union City, CA). Synapse software (version 3.6d; Synergy Research, Inc., Silver Springs, MD) controlled data acquisition and movement of a two-barrel flowpipe perfusion system driven by a piezo-electric device (Burleigh Instruments; Fishers, NY). Micropipettes (TW150F; 2–5 MΩ; World Precision Instruments; Sarasota, FL) contained the following intracellular solution (in mM): 135 CsCl, 10 CsF, 10 HEPES, 5 Cs4BAPTA, 1 MgCl2, and 0.5 CaCl2, pH 7.2. Patches were perfused at 0.2 mL/min with solutions emitted from a two-barrel flow pipe made with theta tubing (BT150-10; Sutter Instruments; Novato, CA). One barrel contained control solution (in mM): 145 NaCl, 5.4 KCl, 5 HEPES, 1 MgCl2, 1.8 CaCl2, with 0.01 mg/mL phenol red, pH 7.3. The other barrel contained L-glutamate (10 mM) dissolved in the control solution. For drug studies, each barrel additionally contained either 100 μM cyclothiazide (CTZ; Tocris Bioscience; Bristol, UK), 100 μM CX614 (Cortex Pharmaceuticals, Inc; Irvine, CA), or 10 μM CMPDA or CMPDB (Eli Lilly & Company; Indianapolis, IN). CTZ and CX614 were dissolved in 1% DMSO. After attaining whole-cell voltage clamp, outside-out patches were pulled from cells, raised off the dish and positioned near the interface between the glutamate-free and glutamate-containing solutions of the flowpipe. Agonist applications were achieved by stepping into and out of the glutamate containing solution for either 1 or 500 ms to test for deactivation and desensitization, respectively. Because of extremely slow washout of modulator from cells and from tubing, only one modulator was used per cell, and control glutamate applications were applied first. Solution exchange was measured for each patch by measurement of junction potentials after obliterating the patch at the end of the experiment. Solution exchange times were approximately 500 μs. Responses were digitized at 20 kHz, and stored on a PowerPC Macintosh computer (Apple, Inc.; Cupertino, CA) using an ITC-16 interface (InstruTech, Port Washington, NY) connected through a USB-16 adapter (HEKA Instruments Inc.; Bellmore, NY).
2.5. Data analysis
Current traces and graphs were plotted using KaleidaGraph 3.6 (Synergy Software, Reading, PA). Traces were averaged (2–25 traces per sweep) and IPeak was calculated using an average of three points. ISS/IPeak was calculated using an averaged steady state current (SS). A single exponential was initially fit to the trace, but because most modulated responses were best fit with the sum of two exponentials, all data were fit with two exponentials and a weighted average was used for statistical comparisons of deactivation and desensitization kinetics.
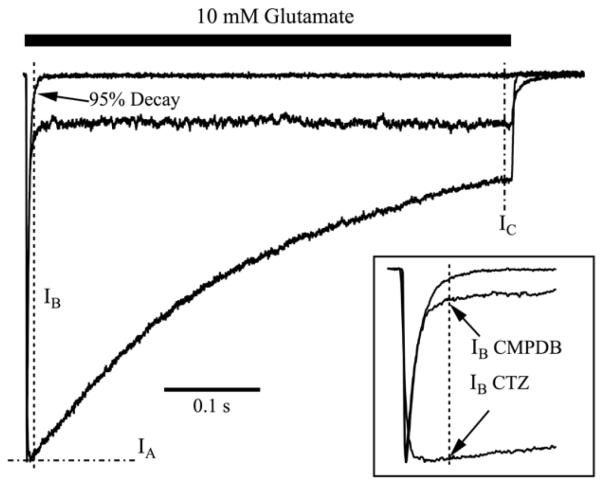
Additional analyses were run on a Macintosh MacBook Pro computer using Igor Pro 6.2 to determine the time to 95% decay of the peak current to the steady state current in glutamate alone. IA, IB and IC were then calculated, where IA is defined as the peak current response to glutamate and modulator, IB is the current amplitude for glutamate and modulator measured at the predetermined time of 95% decay in the absence of modulator, and IC is the averaged steady-state current in glutamate and modulator (see Figure 3).
Figure 3. Quantitative analysis of decay properties among modulators.
Shown are three different types of decay patterns that were observed for WT GluA2o. The control trace has a rapid decay of the current from the peak, and almost no sustained current during the pulse; the black bar represents application of 10 mM glutamate for 500 ms. The CTZ current trace shows a slow decay throughout the 500 ms pulse of glutamate. The CMPDB trace shows an immediate, rapid decay to a sustained current that is larger than baseline. These three traces illustrate the three representative patterns that were observed for various combinations of modulators and mutant receptors shown in Figure 2. Traces have been normalized to their peak amplitudes to emphasize the different kinetics of decay. To quantitatively analyze these differences, measurements were made of the peak amplitude (IA), the amplitude measured at the time of 95% decay of control responses in the absence of modulator (IB), and the amplitude just prior to termination of the pulse of glutamate (IC). For the control trace, the current at which there is 95% decay from the peak amplitude is marked with an arrow. The inset shows an expanded view of the peak amplitudes. Values for each condition are presented in Table 2. Traces with modulators are single sweeps. Trace of 10 mM glutamate alone is an average of 25 sweeps.
2.6. Kinetic simulations
Simulations of currents under voltage clamp to an AMPA receptor model were performed using code originally written by John Clements (Benveniste et al., 1990), revised and converted by M.B. to an Igor Pro XOP. Receptor state occupancies at each time point were determined numerically. The change in each state occupancy was calculated according to first order reaction rate kinetics for transitions into and out of each state, as detailed in Benveniste et al. (1990). This was done iteratively at least 20 times per time point. Simulations, fitting and related analyses were run on a Macintosh MacBook Pro computer utilizing Igor Pro 6.2.
2.7 Visualization of Protein Structures
Protein structures were visualized using The PyMol Molecular Graphics System (version 1.3, Schrödinger, LLC) in cartoon representation, using PDB files 2AL4 for CX614 (Jin et al., 2005); 3H6T for cyclothiazide (Hald et al., 2009); 3RN8 and 3RNN for CMPD A and B, respectively (Timm et al., 2011), obtained from the RCSB Protein Data Bank.
3. Results
3.1. Structure of subsite B of the positive allosteric binding pocket
The extracellular domain of the AMPA receptor subunit forms two globular regions – the N-terminal domain and the ligand binding domain (LBD). The LBD in each subunit forms a “clamshell” structure into which agonists permeate and bind. Upon association with agonist, the lower domain (D2) of the clamshell moves away from the lipid bilayer and toward the upper domain (D1) of the clamshell. New hydrogen bonds are formed between D1 and D2, and the energy of those interactions is thought to permit subsequent channel opening. However, the open conformation is strained, particularly at the interface between adjacent clamshells. The strain is relieved by disrupting the dimer interface such that, although glutamate remains bound to the clamshell, the receptor pore becomes nonconducting and the receptor enters a prolonged, desensitized state (Sun et al., 2002). If brief (1 ms) pulses of glutamate are applied, little receptor desensitization occurs. Rather, channels close, glutamate dissociates, and the receptor enters its “resting” conformation; this process is called receptor deactivation.
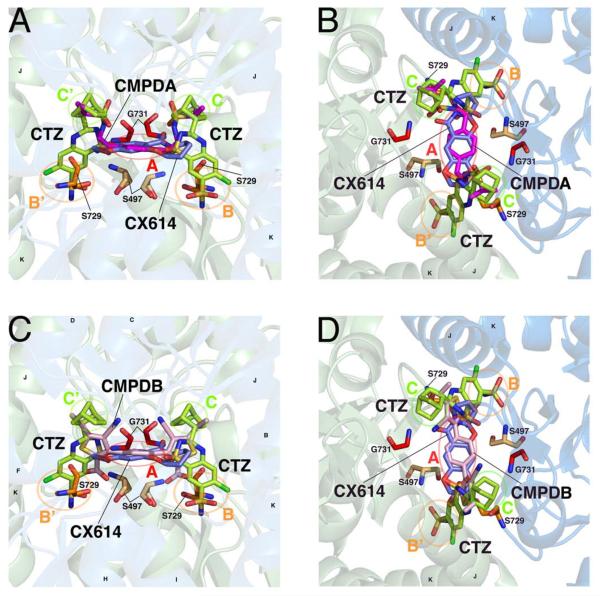
The allosteric binding pocket lies at the soluble interface that regulates receptor desensitization. Through the study of thiazide compounds, the structure of the interface has been delineated into five subsites: one central subsite, A, and two symmetrical copies of the smaller subsites (B, B', and C,C'; (Ptak et al., 2009)). A, the central, soluble subsite, lies at the axis of symmetry between subunits, and is formed by Pro494, Ser497, Gly731, and Ser729; B is a hydrophilic pocket formed by Tyr424, Phe495, Ser497, Ser729 and Lys763; and, C is a hydrophobic pocket formed by Ile481, Lys493, and Leu751, with residue Ser754 separating subsite B from subsite C. Figure 1 shows the five subsites at the dimer interface of two, adjacent subunits of GluA2 receptors, and how the four compounds studied here, CTZ, CX614, CMPDA and CMPDB, occupy these subsites. We and others have hypothesized that occupation of each subsite differentially impacts the efficacy for block of desensitization and/or the overall apparent affinity of modulator for binding GluA2 receptors (Timm et al., 2011). Based upon this structural information, a subset of important residues, most of which occupy subsite B, were chosen for further study with these four positive allosteric modulators.
Figure 1. Chemical structures of four positive allosteric modulators of AMPA receptors.
A solution-accessible binding site is formed between dimers of the ligand binding domain, allowing positive allosteric modulators to differentially bind and influence channel gating. A, B, Two views of CMPDA (magenta), overlaid with cyclothiazide (CTZ, green) and CX614 (blue). In each panel, the identity of subsites A, B, B', C, and C' are identified with circles and labels. Helices J and K, the flip/flop region, are identified. Residues hypothesized or known to be critical for modulator binding and/or function, and which are studied here, are shown. C, D, Two similar views of CMPDB (pink) in its binding site are shown, with similar labels as indicated above. The moieties of each modulator differentially occupy the five modulator subsites. Protein structures are visualized using PyMol with the PDB files 2AL4, 3H6T, 3RN8, 3RNN.
3.2. Functional analysis of the mutations
Initially, we recorded control responses elicited by a brief (1 ms) or prolonged (500 ms) pulse of 10 mM L-glutamate from outside-out membrane patches pulled from transiently transfected HEK293 cells expressing mutant receptors. In this way, we could ensure that the mutation did not kill receptor function. As shown in Table 1, all mutant receptors were functional, albeit some mutants had currents that were significantly reduced in amplitude (Gly731Ala in particular). In the absence of modulator, most mutant receptors had similar kinetics of receptor deactivation in response to 1 ms agonist pulses (Table 1). The only mutant receptor that had significantly altered deactivation kinetics was Gly731Ala, where currents decayed more rapidly (τ = 0.4 ms) than WT (τ = 0.8 ms). Similarly, the kinetics of the onset of desensitization were comparable to WT (τ = 6.7 ms), except Gly731Ala, which was extremely fast (τ = 0.5 ms) and Ser497Ala, which was slower (τ = 11.1 ms). From these data we conclude that the point mutations did not dramatically impede normal receptor gating in response to glutamate application.
Table 1.
Desensitization parameters for WT and mutant GluAZ flip and flop receptors with four modulators.
| GluA2i | Wild-type | S497A | S497T | S729A | G731A | K763Q |
|---|---|---|---|---|---|---|
| 10 mM Glu | ||||||
| Weighted τdes | 6.7 ± 0.4 (22) | 11.1 ± 2.2*** (5) | 6.7 ± 0.2 (4) | 6.2 ± 1.2 (4) | 0.54 ± 0.03*** (15) | 4.6 ± 0.3* (18) |
| % fast-component τdes | 51.8 ± 5.4 | 70.1 ± 9.5 | 68.9 ±10.2 | 57.4 ± 20.5 | 88.8 ± 2.7*** | 56.7 ± 5.8 |
| % Des | 96.3 ± 0.8 | 90.4 ± 4.8* | 97.9 ± 0.4 | 98.9 ± 0.5 | 96.8 ± 0.8 | 96.8 ± 0.6 |
| Weighted τdeact | 0.8 ± 0.1 (21) | 1.2 ± 0.4 (4) | 1.0 ± 0.1 (4) | 1.1 ± 0.3 (3) | 0.4 ± 0.04* (15) | 1.0 ± 0.1 (15) |
| % fast-component τdeact | 55.4 ± 6.3 | 79.2 ± 5.4 | 49.3 ± 17.3 | 54.6 ± 28.7 | 64.9 ± 6.5 | 78.9 ± 5.8 |
| 10 μM CMPDA | ||||||
| Weighted τdes | Non-Des (5) | Non-Des (1) | Non-Des (5) | Non-Des (2) | 8.8 ± 2.3 (7) | Non-Des (8) |
| % fast-component τdes | Non-Des | Non-Des | Non-Des | Non-Des | 92.6 ± 0.7* | Non-Des |
| % Des | 8.0 ± 2.9††† | 9.6 ± 0††† | 1.2 ± 0.6††† | −1.0 ± 0.4††† | 95.5 ± 0.6*** | 8.4 ± 2.8††† |
| Weighted τdeact | 1.8 ± 0.3 (5) | 7.8 ± 0†††* (1) | 3.2 ± 1.2 (5) | 6.1 ± 0.7†* (2) | 0.4 ± 0.07 (7) | 4.1 ± 0.7††† (7) |
| % fast-component τdeact | 53.1 ± 10.4 | 73.3 ± 0 | 35.3 ± 11.8 | 28.4 ± 8.8 | 81.8 ± 5.7 | 35.1 ± 7.8†† |
| 100 μM CX614 | ||||||
| Weighted τdes | 46.5 ± 12.0† (10) | Non-Des (4) | 65.0 ± 18.0† (3) | 114 ± 9.0†††* (4) | 0.9 ± 0.1 (6) | 62.9 ± 16.5† (9) |
| % fast-component τdes | 76.6 ± 3.4† | Non-Des | 66.9 ± 7.6 | 65.2 ± 3.7 | 76.3 ± 9.0 | 64.6 ± 4.4 |
| % Des | 34.4 ± 5.5††† | 10.7 ± 1.3†††* | 21.3 ± 6.0††† | 71.0 ± 2.7†††*** | 94.9 ± 2.3*** | 61.1 ± 2.8†††*** |
| Weighted τdeact | 1.9 ± 0.7 (10) | 4.2 ± 0.7†* (4) | 1.3 ± 0.02 (3) | 1.3 ± 0.1 (4) | 0.5 ± 0.06 (6) | 2.2 ± 0.3† (9) |
| % fast-component τdeact | 77.0 ± 6.8 | 41.9 ± 6.7†* | 74.4 ± 5.7 | 62.3 ± 4.6 | 49.5 ± 11.4* | 65.1 ± 5.9 |
| 10 μM CMPDB | ||||||
| Weighted τdes | 49.0 ± 20.8† (7) | 29.1 ± 2.8 (3) | Non-Des (6) | Non-Des (5) | 6.3 ± 1.6* (8) | 5.3 ± 0.2* (7) |
| % fast-component τdes | 57.6 ± 4.8 | 83.2 ± 1.3* | Non-Des | Non-Des | 89.6 ± 2.2*** | 73.6 ± 6.4 |
| % Des | 34.4 ± 10.9††† | 33.7 ± 1.0††† | 21.1 ± 1.4††† | 13.3 ± 0.8†††* | 93.7 ± 0.9*** | 94.4 ± 0.9*** |
| Weighted τdeact | 5.6 ± 1.2††† (7) | 3.7 ± 0.4† (3) | 3.6 ± 0.7 (6) | 5.3 ± 1.4† (5) | 0.5 ± 0.04*** (8) | 1.4 ± 0.3* (7) |
| % fast-component τdeact | 43.9 ± 10.2 | 29.3 ± 9.9† | 50.5 ± 10.9 | 38.4 ± 8.3 | 61.6 ± 6.5 | 68.3 ± 10.2 |
| 100 μM CTZ | ||||||
| Weighted τdes | Non-Des (8) | Non-Des (9) | Non-Des (3) | Non-Des (6) | 86.6 ± 35.6††† (6) | Non-Des (13) |
| % fast-component τdes | Non-Des | Non-Des | Non-Des | Non-Des | 39.7 ± 14.3††† | Non-Des |
| % Des | 4.3 ± 2.3††† | 8.3 ± 1.9††† | 0.9 ± 0.6††† | 8.9 ± 1.8††† | 27.7 ± 6.5†††*** | 20.3 ± 2.8†††* |
| Weighted τdeact | 1.3 ± 0.3 (8) | 1.4 ± 0.3 (6) | 1.9 ± 0.2 (3) | 2.6 ± 0.5* (6) | 0.8 ± 0.09†† (6) | 2.2 ± 0.2* (12) |
| % fast-component τdeact | 49.5 ± 12.1 | 75.5 ± 8.5 | 43.4 ± 4.2 | 36.5 ± 11.2 | 54.6 ± 6.7 | 51.6 ± 7.2† |
| GluA2o | Wild-type | S497A | S497T | S729A | G731A | K763Q |
|---|---|---|---|---|---|---|
| 10 mM Glu | ||||||
| Weighted τdes | 1.6 ± 0.1 (22) | 2.0 ± 0.1 (7) | 3.0 ± 0.7*** (3) | 1.0 ± 0.1* (5) | 0.8 ± 0.1*** (9) | 1.0 ± 0.1*** (25) |
| % fast-component τdes | 56.6 ± 4.5 | 58.9 ± 8.9 | 12.8 ± 5.8* | 37.4 ± 10.6 | 76.5 ± 4.5 | 71.7 ± 4.0 |
| % Des | 97.1 ± 0.7 | 95.8 ± 1.3 | 97.5 ± 1.9 | 99.2 ± 0.2 | 96.7 ± 1.1 | 98.0 ± 0.3 |
| Weighted τdeact | 0.7 ± 0.07 (18) | 1.0 ± 0.1 (7) | 1.0 ± 0.3 (3) | 0.6 ± 0.05 (5) | 0.5 ± 0.1 (9) | 0.7 ± 0.05 (23) |
| % fast-component τdeact | 62.0 ± 6.2 | 24.0 ± 5.4* | 24.2 ± 10.1* | 28.9 ± 3.2* | 75.6 ± 5.5 | 49.7 ± 5.4 |
| 10 μM CMPDA | ||||||
| Weighted τdes | Non-Des (12) | Non-Des (6) | 15.6 ± 5.7 (3) | Non-Des (4) | 1.1 ±0.2 (7) | Non-Des (6) |
| % fast-component τdes | Non-Des | Non-Des | 90.0 ± 3.7†† | Non-Des | 79.2 ± 2.5 | Non-Des |
| % Des | 6.1 ± 1.1††† | 11.2 ± 1.7††† | 40.4 ± 2.8†††*** | 8.9 ± 4.0††† | 97.5 ± 0.7*** | 4.8 ± 1.1††† |
| Weighted τdeact | 2.8 ± 0.4† (12) | 3.6 ± 0.5††† (6) | 3.5 ± 0.4†† (3) | 3.4 ± 0.8 (6) | 0.3 ± 0.02* (7) | 7.0 ± 1.2†††*** (6) |
| % fast-component τdeact | 45.6 ± 5.0 | 24.6 ± 6.7* | 22.5 ± 4.7 | 27.9 ± 7.2 | 71.2 ± 4.5* | 32.4 ± 6.8 |
| 100 μM CX614 | ||||||
| Weighted τdes | 121 ± 24.3††† (12) | Non-Des (3) | 2.1 ± 0.5* (3) | 108.5 ± 37.9 (5) | 1.0 ± 0.1* (7) | 120.9 ± 14.4††† (8) |
| % fast-component τdes | 31.1 ± 5.2†† | Non-Des | 55.0 ± 1.0† | 34.5 ± 16.7 | 74.7 ± 3.2*** | 26.1 ± 4.3††† |
| % Des | 33.4 ± 2.1††† | 10.5 ± 2.1†††*** | 94.8 ± 1.4*** | 86.3 ± 1.8†*** | 93.9 ± 1.8*** | 76.8 ± 2.4†††*** |
| Weighted τdeact | 2.4 ± 0.5† (14) | 6.1 ± 0.9†††* (3) | 0.8 ± 0.06 (3) | 2.7 ± 1.0 (5) | 0.5 ± 0.06* (8) | 4.3 ± 0.5†††* (8) |
| % fast-component τdeact | 37.6 ± 4.5† | 4.0 ± 2.0* | 97.5 ± 1.0††*** | 44.9 ± 15.8 | 59.0 ± 5.2* | 43.9 ± 1.2 |
| 10 μM CMPDB | ||||||
| Weighted τdes | 2.3 ± 0.2 (11) | 2.4 ± 0.4 (3) | 1.9 ± 0.2 (3) | 134.1 ± 45.4*** (3) | 1.5 ± 0.3 (8) | 0.9 ± 0.1 (9) |
| % fast-component τdes | 92.1 ± 2.6††† | 90.0 ± 2.5 | 58.9 ± 19.9†* | 32.9 ± 8.2*** | 71.3 ± 6.6 | 69.7 ± 6.8* |
| % Des | 81.5 ± 3.6††† | 75.1 ± 7.3†† | 99.2 ± 0.5* | 43.5 ± 3.2†††*** | 92.8 ± 2.8* | 97.4 ± 0.6* |
| Weighted τdeact | 3.7 ± 1.5†† (7) | 1.4 ± 0.2 (2) | 0.9 ± 0.06 (3) | 7.3 ± 1.8†† (3) | 0.3 ± 0.03* (8) | 0.6 ± 0.02* (8) |
| % fast-component τdeact | 72.0 ± 13.9 | 79.8 ± 6.1†† | 59.1 ± 10.5 | 13.6 ± 3.9* | 76.6 ± 6.1 | 38.0 ± 4.3* |
| 100 μM CTZ | ||||||
| Weighted τdes | 153 ± 12.3††† (8) | 148.6 ± 13.5††† (6) | 91.2 ± 14.0†††* (4) | 85.2 ± 9.1* (2) | 54.6 ± 4.7†††*** (8) | 52.4 ± 4.9†††*** (8) |
| % fast-component τdes | 11.9 ± 2.5††† | 13.9 ± 8.0† | 15.5 ± 7.9 | 21.6 ± 4.1 | 26.2 ± 2.2††† | 35.5 ± 2.9†††*** |
| % Des | 67.3 ± 5.1††† | 85.0 ± 2.8†* | 95.1 ± 1.6* | 93.3 ± 1.8* | 93.3 ± 2.9** | 87.6 ± 6.0†* |
| Weighted τdeact | 0.6 ± 0.07 (8) | 0.6 ± 0.1 (6) | 0.9 ± 0.4 (4) | 0.7 ± 0.06 (2) | 1.2 ± 0.3† (8) | 0.8 ± 0.07 (8) |
| % fast-component τdeact | 59.1 ± 9.6 | 42.3 ± 7.1 | 37.0 ± 11.2 | 77.1 ± 9.9† | 41.6 ± 11.71† | 55.3 ± 6.4 |
WT, but not mutant, data are reused from Timm et al. (2011) with permission from Molecular Pharmacology.
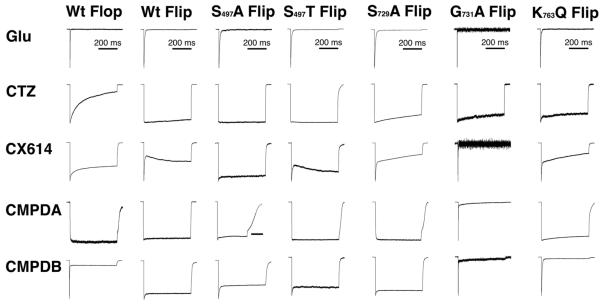
We next analyzed how the mutations influenced modulation of receptor desensitization (Table 1 and Figure 2). The mutation with the greatest impact was Gly731Ala, which forms part of subsite A, because the ability of any modulator that occupies subsite A (CX614, CMPDA, and CMPDB) to slow or block receptor desensitization was essentially lost with this mutation. Many of the other point mutations had a modest impact on allosteric modulation of receptor desensitization. For several mutations, including Ser497Ala or Thr, Ser729Ala, and Lys763Gln, the mutation actually enhanced the modulation.
Figure 2. Mutations in the modulator binding site differentially perturb receptor desensitization.
Outside-out membrane patches pulled from HEK293 cells transiently expressing WT GluA2o (flop) receptor isoforms and point mutations in the allosteric modulator binding site were screened for modulator efficacy, using 10 mM glutamate alone or in the presence of CTZ, CX614, CMPDA and CMPDB. Efficacious modulation of desensitization is observed as an absence of peak current decay during a prolonged (500 ms) pulse of 10 mM glutamate in the presence of 10 (CMPDA and B) or 100 (CTZ, CX614) μM. Traces shown are normalized to the largest peak amplitude, which was 500 pA, and ranged from10–500 pA. WT traces are re-published from Timm et al., 2011 with permission from the publisher.
Although we were seeking to understand residues that play a role in receptor-modulator affinity or efficacy, it was clear that performing experiments at a single concentration of modulator could not necessarily distinguish between altered efficacy or affinity. However, since many novel compounds in the drug discovery pipeline are synthesized only in very small amounts (including the novel compounds used in this study), we sought to understand if there are generalized rules for interpreting single-dose modulation data, based upon the kinetics of the response during the 500 ms pulse of glutamate (desensitization protocol).
3.3 Kinetics of desensitization can yield information about modulator efficacy, even at a single dose of modulator
To further analyze the relationship between the desensitization decay waveform and possible mechanisms of allosteric modulation, we sought to establish objective measures that might reflect changes in modulator affinity versus efficacy. We noticed that in the presence of modulator, there were three basic responses to a sustained application of glutamate (Figure 2): 1) little or no decay of current, representing nearly full block of desensitization (e.g. CMPDA on WT receptors); 2) a relatively slow decay of current representing the onset of desensitization (e.g. CTZ on WT Flop receptors); or 3) a very rapid decay of current followed by a slower decay (e.g. CMPDB on WT Flop receptors). Because the current decay of non-modulated AMPA receptors to a sustained pulse of glutamate has a relatively fast time-course (Control WT receptors), we reasoned that in cases where modulator was used, a rapidly decaying component could represent a population of receptors for which modulator was not bound, as one might expect modulator binding to slow these kinetics.
As shown in Figure 3, this initial rapid decay could be independently assessed by comparing the change in the peak amplitude (IA) versus the amplitude after a very brief time (IB), which we chose as the time for control glutamate responses in the absence of modulator to decay 95% of their peak amplitude. We hypothesized that the greater the difference between IA and IB, the fewer receptors are bound with modulator, suggesting that the modulator being tested has a lower affinity for the receptor. The trace for CMPDB in Figure 3 shows a clear example in which 80% of receptors are not bound by modulator. In contrast, the difference between IB and IC on the CTZ trace might reflect the low efficacy of the modulator to block desensitization, as the change in current amplitude suggests that receptors are still desensitizing, over time. The absence of an extremely rapid decay component suggests that only a minority of receptors remain unbound by CTZ.
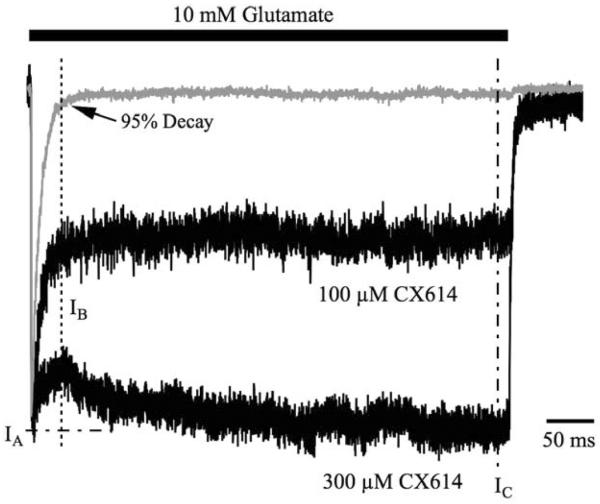
Support for this hypothesis was provided by the data shown in Figure 4, where an increase in concentration of CX614 from 100 to 300 μM, (a concentration at which solubility becomes an issue) increased the steady state current (IC) but did not change the ratio of IC to IB (mean ratio of IC/IB = 114.7 ± 4.4% at 100 μM CX614). The lack of change in IC/IB ratio, with the change in IA/IB ratio, suggests that the former represents a measure of efficacy whereas the latter represents a measure of affinity. In this instance, adding more CX614 increased the receptor occupancy by modulator but did not change its inherent efficacy. To analyze this fully, we calculated IA, IB, and IC for all conditions, and then turned to computational modeling to seek mechanistic insight into the mutant data.
Figure 4. Rapid decay to a sustained current likely represents a sub-saturating concentration of allosteric modulator.
Shown are three averaged traces representing representing a 500 ms pulse of glutamate in the absence of modulator, or in the presence of 100 or 300 μM CX614. These traces are normalized to their peak responses to easily compare the kinetics of decay from peak. IA, IB and IC are shown to demarcate the peak amplitude, amplitude of 95% decay of control and the steady state current, respectively. IB Rapid decay to IB signifies desensitization of receptors not bound by CX614. Increasing the concentration of CX614 reduces the amplitude of this rapid decay without significantly affecting its kinetics. Traces for control, 100 and 300 μM CX614 are averages of 2, 3 and 8 sweeps, respectively.
3.4 Simulations indicate that onset of desensitization can yield information on drug affinity and efficacy
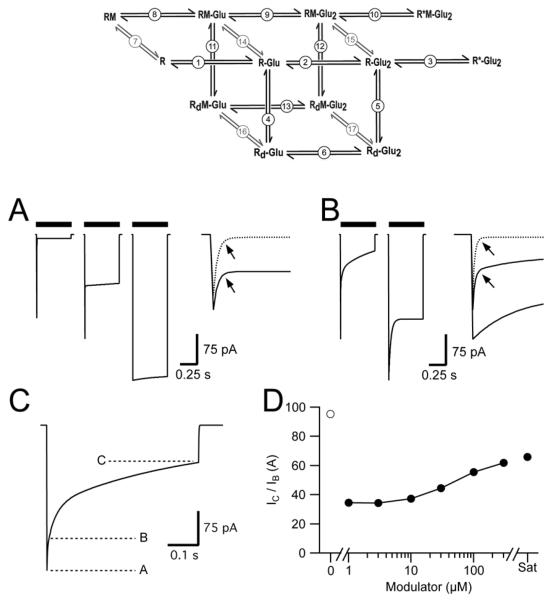
We used a simplified gating scheme to simulate (Figure 5) the impact of changes in modulator affinity and efficacy due to the point mutations. The model was fully described and characterized (Harms et al., 2013), with control kinetics of 0.9 ms for receptor deactivation and 1.4 ms for receptor desensitization. The model entails six unbound (control) states (receptor with no glutamate, a receptor with one or two glutamate molecules bound, a receptor with one and two glutamate molecules bound but in the desensitized state, and a single open state); six modulator-bound states that parallel the control states, and five transitions between bound and unbound states that represent relative modulator affinity (Figure 5 and Supplemental Table 1).
Figure 5. Simulations of High and Low Efficacy Modulators at Different Receptor Occupancies.
Upper Panel, 12-state model used to simulate effect of low and high efficacy modulators on AMPA receptor desensitization. Circled numbers indicate transitions whose rate constants can be found in Supplementary Table 1. R = receptor; M = modulator; Glu = glutamate; Rd = desensitized receptor. R*-Glu2 and R*M-Glu2 represent the open states for receptors without and with modulator bound. A, Simulations with a high efficiency modulator in which a saturating concentration of modulator yields full block of desensitization. Simulated AMPA receptor currents in the absence of modulator are depicted in the left panel. Middle and right panels represent AMPA receptor currents in the presence of modulator where approximately half the receptors or all the receptors are occupied by modulator. Inset. Expanded scale of the rapid component from the left (dotted line) and middle panels (solid line). B, Simulations utilizing a low-efficacy modulator. Left and right panels represent < 50% occupancy and full occupancy by modulator, respectively. Inset: Expanded scale of initial decay of currents. Dotted line represents a simulation in the absence of modulator (A, left panel). Arrows in A and B indicate the time to 95% decay where IB is measured and represents the fast component in the absence of modulator. C, Quantification of unoccupied receptors and the degree of desensitization. Unoccupied receptors can be quantified as (IA - IB) / IA while desensitization of receptors occupied by modulator (modulator efficacy) is characterized as IC / IB. D, The efficacy of the modulator measured as % desensitization of occupied receptors (IC / IB) does not change substantially with modulator concentration. Open circle indicates % Desensitization in the absence of modulator. Sat = saturating concentration of modulator.
Both modulator occupancy of AMPA receptors and their efficacy to block desensitization would affect the onset of desensitization. Thus, the concentration of modulator was varied to vary occupancy, and the rate constants for onset and recovery from desensitization (transitions 11 and 12, Figure 5) were varied to reproduce a low and a high efficacy block of desensitization. “Flat” current responses during the pulse of agonist likely represent compounds that have full efficacy, independent of receptor occupancy (left, middle and right traces in Figure 5A). The rapid component of decay in the two left traces in Figure 5B likely represent receptors unoccupied by modulator at the beginning of the pulse of glutamate. In contrast, lowering the efficacy of the modulator by increasing the rate of recovery from desensitization (Figure 5B and Supplemental Table 1) results in a slower decaying component from the peak response than is observed for responses in the absence of modulator (Figure 5A, left trace). Responses with a lack of a fast component at the onset of the glutamate pulse are likely indicative of a compound with lower efficacy, but for which there is near full receptor occupancy (higher affinity). On the other hand, responses with a moderate to large rapid decay, as well as an additional slower decay waveform, are likely indicative of low occupancy, but may or may not be indicative of low efficacy. These conclusions held when the simulations were run with a range of on and off-rates for modulator (0.5–30 for the on-rates), with a modulator pre-incubation time of 10 s. Thus, we can estimate the receptor occupancy (affinity) by measuring (IA−IB)/IA, whereas we can measure the efficacy of the modulator by looking at the desensitization of the occupied receptors, IC/IB.
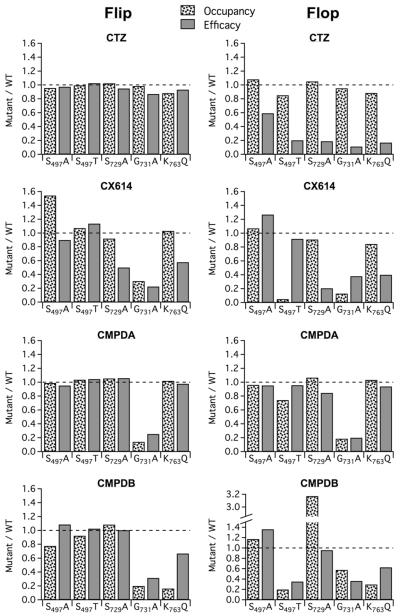
We reexamined the experimental data from the mutations from our measures of modulator occupancy and efficacy (Table 2). The impact of a mutation on the effectiveness of each modulator could then be assessed by determining the ratio of these measures with respect to wildtype (Figure 6). These data suggest that subsite B mutations alter efficacy and affinity of modulators more for flop than flip isoforms. An unanticipated finding was that CMPDB and CTZ, which have the most extensive pharmacophore (number of residues that are within 5 angstroms of the compound (Timm et al., 2011)), could have their efficacy and affinity severely impacted by a single point mutation. In contrast, modulators with smaller pharmacophores appeared to have more resiliency with regard to perturbation of the modulator binding site. More classes of modulators need to be tested to determine if this notion is generalizable. We note that binding is necessary but not sufficient for high efficacy, and therefore our estimates of impact of a mutation on efficacy may be an underestimate. Nevertheless, the results in Figure 6 support the idea that a “snapshot” of the promise of a new compound can be taken, at a single concentration of drug, by converting the waveforms of the decay during a long pulse of glutamate to simple numbers that represent receptor occupancy and drug efficacy.
Table 2.
Efficacy and Occupancy Analysis
| Receptor + Drug | Decay to 95% (ms) | IC/IB (%) (efficacy) | 1 – (IA – IB)/IA (%) (occupancy) | n |
|---|---|---|---|---|
| GluA2i WT | 29 | 22 | ||
| + CTZ | 101.4 ± 2.0 | 91.9 ± 2.1 | 8 | |
| + CX614 | 114.7 ± 4.4 | 55.1 ± 4.3 | 10 | |
| + CMPDA | 95.7 ± 2.4 | 93.6 ± 1.8 | 5 | |
| + CMPDB | 94.8 ± 1.7 | 83.8 ± 1.4 | 7 | |
| GluA2i S497A | 31 | 5 | ||
| + CTZ | 98.9 ± 2.1 | 87.9 ± 1.9 | 9 | |
| + CX614 | 103.1 ± 2.8 | 85.0 ± 1.5 | 4 | |
| + CMPDA | 91.1 | 92.3 | 1 | |
| + CMPDB | 102.9 ± 4.5 | 65.1 ± 2.0 | 3 | |
| GluA2i S497T | 26 | 4 | ||
| + CTZ | 103.9 ± 1.9 | 91.3 ± 4.2 | 3 | |
| + CX614 | 130.2 ± 1.5 | 59.0 ± 4.8 | 3 | |
| + CMPDA | 100.0 ± 0.8 | 96.9 ± 0.8 | 5 | |
| + CMPDB | 97.3 ± 1.6 | 77.5 ± 2.4 | 6 | |
| GluA2i S729A | 19 | 4 | ||
| + CTZ | 96.3 ± 3.4 | 94.2 ± 3.5 | 6 | |
| + CX614 | 57.6 ± 4.9 | 50.7 ± 1.5 | 4 | |
| + CMPDA | 101.3 | 98.7 | 2 | |
| + CMPDB | 95.0 ± 2.0 | 90.8 ± 1.7 | 5 | |
| GluA2i G731A | 2 | 15 | ||
| + CTZ | 88.2 ± 2.5 | 90.5 ± 3.5 | 5 | |
| + CX614 | 25.8 ± 5.6 | 16.8 ± 3.3 | 5 | |
| + CM PDA | 24.3 ± 10.6 | 13.1 ± 2.8 | 5 | |
| + CMPDB | 29.8 ± 3.5 | 16.7 ± 1.6 | 8 | |
| GluA2i K763Q | 15 | 18 | ||
| + CTZ | 94.6 ± 1.4 | 81.0 ± 1.6 | 13 | |
| + CX614 | 66.3 ± 4.5 | 56.8 ± 1.2 | 9 | |
| + CMPDA | 93.6 ± 2.4 | 95.4 ± 1.2 | 8 | |
| + CMPDB | 63.2 ± 16.7 | 13.6 ± 2.0 | 7 | |
| GluA2o WT | 5 | 22 | ||
| + CTZ | 31.0 ± 5.4 | 88.1 ± 2.8 | 7 | |
| + CX614 | 72.9 ± 2.6 | 88.9 ± 1.5 | 12 | |
| + CMPDA | 103.6 ± 4.7 | 90.6 ± 2.3 | 12 | |
| + CMPDB | 68.3 ± 2.6 | 26.1 ± 4.2 | 11 | |
| GluA2o S497A | 7 | 7 | ||
| + CTZ | 18.4 ± 1.9 | 95.3 ± 1.9 | 6 | |
| + CX614 | 92.6 ±2.2 | 94.9 ± 1.4 | 3 | |
| + CMPDA | 98.8 ± 0.8 | 87.2 ± 3.6 | 4 | |
| + CMPDB | 93.1 ± 11.6 | 30.6 ± 4.3 | 3 | |
| GluA2o S497T | 10 | 3 | ||
| + CTZ | 6.3 ± 1.4 | 75.0 ± 5.5 | 4 | |
| + CX614 | 66.9 ± 7.2 | 4.4 ± 0.5 | 3 | |
| + CMPDA | 99.2 ± 3.2 | 67.2 ± 3.8 | 3 | |
| + CMPDB | 24.0 ± 3.6 | 5.2 ± 3.5 | 3 | |
| GluA2o S729A | 3 | 5 | ||
| + CTZ | 5.9 | 92.7 | 2 | |
| + CX614 | 14.9 ± 1.3 | 80.8 ± 8.5 | 4 | |
| + CMPDA | 87.6 ± 2.4 | 96.6 ± 0.9 | 5 | |
| + CMPDB | 65.5 ± 2.6 | 82.8 ± 2.4 | 3 | |
| GluA2o G731A | 3 | 9 | ||
| + CTZ | 3.5 ± 1.4 | 83.9 ± 2.6 | 7 | |
| + CX614 | 27.7 ± 3.6 | 11.4 ± 1.4 | 5 | |
| + CMPDA | 20.6 ± 5.1 | 16.8 ± 1.3 | 6 | |
| + CMPDB | 24.8 ± 3.2 | 15.0 ± 2.0 | 6 | |
| GluA2o K763Q | 3 | 25 | ||
| + CTZ | 5.2 ± 1.4 | 78.0 ± 3.6 | 8 | |
| + CX614 | 29.2 ± 2.7 | 75.1 ± 1.3 | 8 | |
| + CMPDA | 97.3 ± 1.7 | 93.6 ±2.0 | 6 | |
| + CMPDB | 42.8 ± 8.6 | 7.7 ± 0.9 | 9 |
Figure 6. Comparison of modulator affinity and efficacy for different AMPA receptor mutations.
Efficacy and occupancy data from Table 2 for the effect of a given modulator on a given mutation in either flip (left) or flop (right) isoforms of GluA2 receptors has been normalized to the same modulator treatment for the respective wild type receptor isoform. Receptor occupancy is indicative of relative changes in modulator apparent affinity.
4. Discussion
The availability of structural information, based upon co-crystals of the ligand-binding core of AMPA receptors with positive allosteric modulators, brings a new level of understanding about the molecular interactions between receptor and compound. However, understanding the molecular mechanism of modulation requires a further, functional assessment of the role of each residue in the binding pocket or within the pharmacophore. Initially, we sought to do straight-forward structure-function analysis of residues interacting with well-known (CTZ and CX614) and newer (CMPDA and CMPDB) positive allosteric modulators. Given the relatively large size of some of the drugs (such as CMPDB) and the overlapping nature of the subsites, the data demanded greater depth of analysis than we anticipated.
Many positive allosteric modulators, except for the thiazides, slow both deactivation and desensitization. Kinetic transitions may be dependent on concentration of agonist or modulator. For electrophysiology experiments, however, near saturating concentrations of modulator are generally used so that the waveform of the decay represents the highest modulatory occupancy possible, and the data therefore are well-fit by a single decay function that accurately represents the transition being studied. In this study we focused primarily on receptor desensitization, which is technically a less challenging measurement to make.
We noticed that two different modulators could have vastly differing desensitization kinetics (eg. Figure 2, CMPDB versus CTZ on WT Flop receptors) but still yield similar steady state to peak ratios for currents elicited by a 500 ms pulse of glutamate in the presence of modulator. This suggested that we were losing mechanistic information by virtue of oversimplification of our measurements. Refining the analysis to include three points of measurement, carefully recorded, represents a novel approach for interpreting data for new drugs. In fact, the approach is only generalizable if there are distinct kinetic components that are measurable, as is the case for AMPA receptors.
Screening new drugs using a functional assay can be time consuming. If full dose response curves are required, dose ranges can be limited by compound solubility. For electrophysiology experiments, which can consume vast quantities of a drug that may have been synthesized in small quantities, performing a full dose response profile with a significant power to make conclusions remains a challenge in the field. In particular, saturating concentrations of the test compound are needed to get consistent measures of the kinetics of deactivation and desensitization. We believe that the approach outlined in this paper allows one to create a meaningful “profile” of a new compound, a characteristic which can readily discern between drugs that have altered affinity or efficacy versus the parent compound. We suggest that this approach might be well-suited for high-throughput screening of novel compounds, and would help to identify compounds with functionally relevant properties. This experimental approach is complementary to the overall “structure based drug design” methods that have been recently described (Jamieson et al., 2011).
One goal of this study was to use a mutational approach to gain an understanding of the relative contributions of specific residues in permitting high affinity binding, as well as efficacy, for specific allosteric modulators. That goal was hampered by the fact that certain amino acid residues within the pharmacophore had putative interactions with overlapping “subsites”, as they are defined (Ptak et al., 2009; Timm et al., 2011). For example, atomic moieties of Ser497 and Ser729 have interactions with modulator within subsites A and B. However, Gly731 and Lys763 reside distinctly within subsite A and B, respectively. It is interesting to note for these two residues that the results of the mutations are parsimonious with a simple interpretation: mutation of Gly731 significantly disrupts modulator affinity and efficacy for CX614, the compound that resides only in subsite A, whereas mutation of Lys763, which lies outside of subsite A, is modulated by CX614 similarly to WT receptors. However, the mutation to Lys763 impacts efficacy of CMPDB, which fully occupies subsite B.
Inspection of Figure 6 reveals how different the snapshot of modulation of flop isoforms is compared with flip isoforms. Three of the modulators, CTZ, CMPDB and CX614 are less efficacious and have lower apparent affinities for the flop isoform. The functional differences are striking, in light of the structural similarity shared by crystal structures of the flip and flop isoforms (Ahmed et al., 2009). The molecular basis of these differences is unclear and is believed to be largely governed by the S/N754 residue (Ahmed et al., 2010; Kessler et al., 2000; Partin et al., 1996); however, why there are differences in the thermodynamics of the flip and flop receptors that might impact compound efficacy or affinity remains poorly understood.
Supplementary Material
Highlights.
Mutational analysis of a positive allosteric binding site of AMPAR was done.
Cyclothiazide, CX614, CMPDA and CMPDB were compared.
Computational modeling of results characterized drug efficacy and affinity.
Results may suggest a new approach to high throughput drug screening.
Acknowledgements
NIHR01 MH064700 (KMP); NINDS S11NS055883 and U54NS083932, (MB), NIA SC1AG046907, (MB). The authors thank John Gieser for his contributions to the artwork. We thank Cortex Pharmaceuticals, Inc. for providing CX614 and Lilly Research Laboratories for providing CMPDA and CMPDB.
Abbreviations
- AMPA
(alpha-amino-3-hydroxy-5-methyl-isoxazole-propionic acid)
- CTZ
(cyclothiazide; 2,4-benzothiadiazine-3-bicyclo[2.2.1]hept-5-en-2-yl-6-chloro-4-dihydro-2h-1)
- CX614
(2H,3H,6aH-pyrrolidino(2,1–3',2')1,3-oxazino(6',5'-5,4)benzo(e)1,4-dioxan-10-one)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed AH, Ptak CP, Oswald RE. Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA. Biochemistry. 2010;49:2843–2850. doi: 10.1021/bi1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Wang Q, Sondermann H, Oswald RE. Structure of the S1S2 glutamate binding domain of GluR3. Proteins. 2009;75:628–637. doi: 10.1002/prot.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggano V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Current Opinions in Neurobiology. 2012;22:461–9. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai A, Kessler M, Rogers G, Lynch G. Effects of the potent ampakine CX614 on hippocampal and recombinant AMPA receptors: interactions with cyclothiazide and GYKI 52466. Molecular Pharmacology. 2000;58:802–813. doi: 10.1124/mol.58.4.802. [DOI] [PubMed] [Google Scholar]
- Arai AC, Kessler M. Pharmacology of ampakine modulators: from AMPA receptors to synapses and behavior. Curr Drug Targets. 2007;8:583–602. doi: 10.2174/138945007780618490. [DOI] [PubMed] [Google Scholar]
- Benveniste M, Clements J, Vyklicky L, Mayer ML. A kinetic analysis of the modulation of N-methyl-D-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. Journal of Physiology (London) 1990;428:333–357. doi: 10.1113/jphysiol.1990.sp018215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MD. Therapeutic potential of positive AMPA modulators and their relationship to AMPA receptor subunits. A review of preclinical data. Psychopharmacology (Berl) 2005;179:154–163. doi: 10.1007/s00213-004-2065-6. [DOI] [PubMed] [Google Scholar]
- Boulter J, Bettler B, Dingledine R, Edgebjerg J, Hartley M, Hermans-Borgmeyer I, Hollmann M, Hume RI, Rogers S, Heinemann S. Molecular biology of the glutamate receptors. Clinical Neuropharmacology. 1992;15(Suppl 1 Pt A):60A–61A. doi: 10.1097/00002826-199201001-00033. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Olsen R, Peters JA, Spedding M. Ligand gated ion channels. Neuropharmacology. 2009;56:1. doi: 10.1016/j.neuropharm.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Fernandez MC, Castano A, Dominguez E, Escribano A, Jiang D, Jimenez A, Hong E, Hornback WJ, Nisenbaum ES, Rankl N, et al. A novel class of AMPA receptor allosteric modulators. Part 1: design, synthesis, and SAR of 3-aryl-4-cyano-5-substituted-heteroaryl-2-carboxylic acid derivatives. Bioorg Med Chem Lett. 2006;16:5057–5061. doi: 10.1016/j.bmcl.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Hald H, Ahring PK, Timmermann DB, Liljefors T, Gajhede M, Kastrup JS. Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2. J Mol Biol. 2009;391:906–917. doi: 10.1016/j.jmb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Harms JE, Benveniste M, Maclean JK, Partin KM, Jamieson C. Functional analysis of a novel positive allosteric modulator of AMPA receptors derived from a structure-based drug design strategy. Neuropharmacology. 2013;64:45–52. doi: 10.1016/j.neuropharm.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- Jamieson C, Maclean JK, Brown CI, Campbell RA, Gillen KJ, Gillespie J, Kazemier B, Kiczun M, Lamont Y, Lyons AJ, et al. Structure based evolution of a novel series of positive modulators of the AMPA receptor. Bioorg Med Chem Lett. 2011 doi: 10.1016/j.bmcl.2010.11.098. [DOI] [PubMed] [Google Scholar]
- Jin R, Clark S, Weeks AM, Judman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Rogers G, Arai A. The norbornenyl moiety of cyclothiazide determines the preference for flip-flop variants of AMPA receptor subunits. Neuroscience Letters. 2000;287:161–165. doi: 10.1016/s0304-3940(00)01180-0. [DOI] [PubMed] [Google Scholar]
- Koike M, Tsukada S, Tsuzuki K, Kijima H, Ozawa S. Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J Neurosci. 2000;20:2166–2174. doi: 10.1523/JNEUROSCI.20-06-02166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krintel C, Frydenvang K, Olsen L, Kristensen MT, de Barrios O, Naur P, Francotte P, Pirotte B, Gajhede M, Kastrup JS. Thermodynamics and structural analysis of positive allosteric modulation of the ionotropic glutamate receptor GluA2. Biochem J. 2012;441:173–178. doi: 10.1042/BJ20111221. [DOI] [PubMed] [Google Scholar]
- Krintel C, Harpsoe K, Zachariassen LG, Peters D, Frydenvang K, Pickering DS, Gajhede M, Kastrup JS. Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain. Acta Crystallogr D Biol Crystallogr. 2013;69:1645–1652. doi: 10.1107/S0907444913011839. [DOI] [PubMed] [Google Scholar]
- Lynch G, Palmer LC, Gall CM. The likelihood of cognitive enhancement. Pharmacol Biochem Behav. 2011;99:116–129. doi: 10.1016/j.pbb.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC. Synaptic plasticity and AMPA receptor trafficking. Ann NY Acad Sci. 2003;1003:1–11. doi: 10.1196/annals.1300.001. [DOI] [PubMed] [Google Scholar]
- Malinow R, Mainen ZF, Hayashi Y. LTP mechanisms: from silence to four-lane traffic. Curr Opin Neurobiol. 2000;10:352–357. doi: 10.1016/s0959-4388(00)00099-4. [DOI] [PubMed] [Google Scholar]
- Mansour M, Nagarajan N, R.B. N, Clements J, Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32:841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Lindsley CW, Conn PJ, Pandit J, Zagouras P, Volkmann RA. Allosteric modulators for the treatment of schizophrenia: targeting glutamatergic networks. Current Topics in Medicinal Chemistry. 2013;13:26–54. doi: 10.2174/1568026611313010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell NA, Fleck MW. Targeting AMPA receptor gating processes with allosteric modulators and mutations. Biophys J. 2007;92:2392–2402. doi: 10.1529/biophysj.106.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JA, Maclean JK, Jamieson C. Recent advances in positive allosteric modulators of the AMPA receptor. Curr Opin Drug Discov Devel. 2006;9:571–579. [PubMed] [Google Scholar]
- Moudy AM, Yamada KA, Rothman SM. Rapid desensitization determines the pharmacology of glutamate neurotoxicity. Neuropharmacology. 1994;33:953–962. doi: 10.1016/0028-3908(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Mueller R, Rachwal S, Lee S, Zhong S, Li YX, Haroldsen P, Herbst T, Tanimura S, Varney M, Johnson S, et al. Benzotriazinone and benzopyrimidinone derivatives as potent positive allosteric AMPA receptor modulators. Bioorg Med Chem Lett. 2011;21:6170–6175. doi: 10.1016/j.bmcl.2011.07.098. [DOI] [PubMed] [Google Scholar]
- Partin KM, Fleck MF, Mayer ML. AMPA receptor flip/flop mutants affecting deactivation, desensitization and modulation by cyclothiazide, aniracetam and thiocyanate. Journal of Neuroscience. 1996;16:6634–6647. doi: 10.1523/JNEUROSCI.16-21-06634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak CP, Ahmed AH, Oswald RE. Probing the allosteric modulator binding site of GluR2 with thiazide derivatives. Biochemistry. 2009;48:8594–8602. doi: 10.1021/bi901127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk JC, Nisenbaum ES. LY404187: A novel positive allosteric modulator of AMPA receptors. CNS Drug Reviews. 2002;8:255–282. doi: 10.1111/j.1527-3458.2002.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Staubli U, Perez Y, Xu FB, Rogers G, Ingvar M, Stone-Elander S, Lynch G. Centrally active modulators of glutamate receptors facilitate the induction of long-term potentiation in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1994a;91:11158–11162. doi: 10.1073/pnas.91.23.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubli U, Rogers G, Lynch G. Facilitation of glutamate receptors enhances memory. Proceedings of the National Academy of Sciences of the United States of America. 1994b;91:777–781. doi: 10.1073/pnas.91.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer ML, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Timm DE, Benveniste M, Weeks AM, Nisenbaum ES, Partin KM. Structural and functional analysis of two new positive allosteric modulators of GluA2 desensitization and deactivation. Mol Pharmacol. 2011;80:267–280. doi: 10.1124/mol.110.070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991;252:1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- Ward SE, Bax BD, Harries M. Challenges for and current status of research into positive modulators of AMPA receptors. Br J Pharmacol. 2010;160:181–190. doi: 10.1111/j.1476-5381.2010.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.