Figure 1. Chemical structures of four positive allosteric modulators of AMPA receptors.
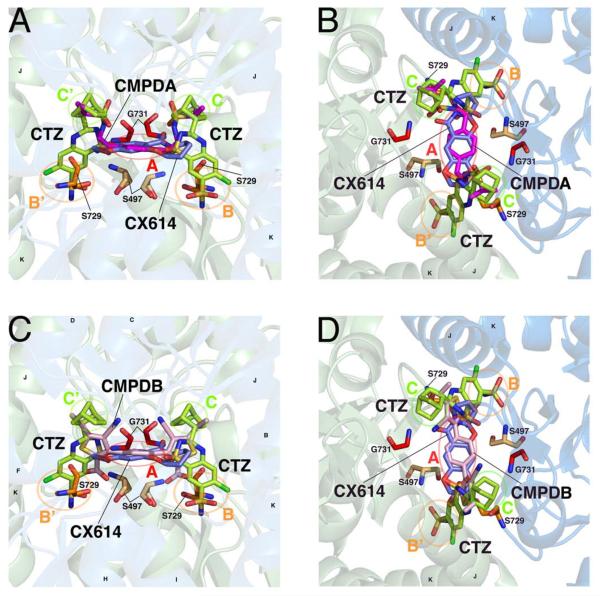
A solution-accessible binding site is formed between dimers of the ligand binding domain, allowing positive allosteric modulators to differentially bind and influence channel gating. A, B, Two views of CMPDA (magenta), overlaid with cyclothiazide (CTZ, green) and CX614 (blue). In each panel, the identity of subsites A, B, B', C, and C' are identified with circles and labels. Helices J and K, the flip/flop region, are identified. Residues hypothesized or known to be critical for modulator binding and/or function, and which are studied here, are shown. C, D, Two similar views of CMPDB (pink) in its binding site are shown, with similar labels as indicated above. The moieties of each modulator differentially occupy the five modulator subsites. Protein structures are visualized using PyMol with the PDB files 2AL4, 3H6T, 3RN8, 3RNN.