Abstract
Of more than 150,000 published studies evaluating new biomarkers, fewer than 100 biomarkers have been implemented for patient care[1]. One reason for this is lack of rigorous testing by the medical community to validate claims for biomarker clinical relevance, and potential reluctance to publish negative results when confirmation is not obtained. Here we sought to determine the utility and reproducibility of immunohistochemical detection of hepatocyte growth factor (HGF) in melanoma tissue, an approach of potential assistance in defining patients with innate resistance to BRAF inhibitor therapy[2]. To this end, a published and a revised method that retained sensitivity but with greater specificity for HGF detection, were evaluated in cells known to endogenously express HGF, models where HGF is upregulated via cytokine induction, and via overexpression by gene transfection. Consequent patient evaluation in collaboration with the Melanoma Institute Australia of a cohort of 41 melanoma specimens with extensive clinical annotation failed to validate HGF immunohistochemistry as a predictor of response to BRAF inhibitors. Targeted therapies for advanced melanoma[3–5] and other cancers show great promise, and rigorous validation studies are thus indicated for approaches that seek to personalize such therapies in order to maximize therapeutic efficacy.
Keywords: HGF, melanoma, biomarker, RAF-inhibitor
Introduction
A critical determinant of effective deployment of recent promising treatments for advanced cancer is the use of tissue biomarkers that identify specific molecular and genomic targets or mediators of therapeutic resistance. Before a biomarker can be applied to personalize therapeutic approaches, rigorous validation is required[6]. An example of a potentially important mediator of cancer resistance to targeted therapy is the hepatocyte growth factor (HGF), which in vitro reactivates the mitogen-activated protein kinase (MAPK) pathway, a driver of BRAF-mutant melanoma progression, resulting in resistance to BRAF inhibitor therapy.[2, 7] It has been suggested[2] that the tumor microenvironment of metastatic melanoma elicits innate resistance to RAF inhibitors through the secretion of HGF. Early results indicate that RAF inhibitor therapy has the impressive ability to induce regression in BRAF-mutant metastatic melanoma,[4, 8–10] an otherwise fatal form of cancer, via inhibition of the MAPK pathway.[11–13] Thus, the ability of melanoma stromal cells, often few in number and located at the periphery of relatively large metastatic nodules, to counteract such treatment effects via HGF would have major implications for the potency of the peritumoral cancer niche in conferring resistance to current targeted therapies. Moreover, the prospect that immunohistochemical detection of any mediator in peritumoral stroma of melanoma metastases predicts patients that are either responsive or resistant to RAF inhibitors has major and pressing clinical implications for the use of such biomarkers in the field of personalized medicine. We thus sought to explore further the practical utility of HGF immunohistochemistry in defining candidates for RAF inhibitor therapy.
Materials and Methods
Cell Lines and Cell Culture
Human skin fibroblasts R2F1 (gift from Professor James G. Rheinwald, Brigham and Women’s Hospital and Harvard Medical School) were originally isolated from infant foreskin and cultured in a 1:1 mixture of M199 and M106 supplemented with 15% FBS, 10 ng/ml EGF, and 0.4 µg/ml hydrocortisone.
Viable cells were counted by Trypan blue exclusion assay under a hemocytometer.
HGF Overexpression
Stable overexpression of human HGF in fibroblasts was achieved using a retrovirus-based approach.[14] Retroviral particles were produced in HK293 cells by co-transfecting packaging vectors pCMV-VSV-G and pUMVC3 with HGF expressing vector (pBabe-puro HGF, plasmid 10901; Addgene, Cambridge, MA) or its control vector pBabe-puro (plasmid 1764, Addgene) as previous reported.[15] Viral supernatants were collected, filtered through 0.45µ m sterile filter, and added together with polybrene (8 mg/ml) to fibroblasts. Cells were selected with puromycin (1 mg/ml) starting at 48 hours post transfection. Manipulation of HGF levels was validated by quantitative RT-PCR and Western blot, and low-passage cell culture (passages≤12) were used for all experiments.
Quantitative RT-PCR
Total mRNA was extracted from subconfluent cell cultures using RNeasy Mini kit (Qiagen Valencia, CA), and first-strand cDNA was synthesized using High Capacity RNA-to-cDNA kit (Applied Biosystems; Life Technologies, Carlsbad, CA). HGF expression was quantified using HGF mRNA-specific primers (forward: TGATACCACACGAACACAGCTTTT; reverse: TCCATGAGACCTCGATAACTCTCC), with SYBR master mix (Qiagen) in 7300 Realtime PCR system (Applied Biosystems; Life Technologies) and calculated with ΔΔCt method.
Western Blotting
Cell culture medium (20 µl, equivalent to 2×105 viable cells/ml) was loaded to native, non-denaturing SDS-PAGE gel. Recombinant human HGF (rHGF), 0.1 µg, (PeProTech, Rocky Hill, NJ) was loaded as positive control. Proteins were separated on SDS-PAGE at constant 100V for 3.5 hours, and transferred to PVDF membrane at constant 340mA for 1.5 hours at 4°C. Membrane was blocked with 5% non-fat milk in TBS-Tween 20 at room temperature for 1 hour, incubated with 1µg/ml of goat anti-HGF polyclonal antibodies (R&D systems, Minneapolis, MN) over night at 4° C, and incubated with HRP-conjugated anti-goat antibodies (Vector Laboratories, Burlingame, CA) at room temperature for 1 hour. Membrane was washed with TBS-Tween 20 for 5 minutes, 3 times at room temperature between procedures. Signal was developed using chemiluminescent substrate (Thermo Scientific, Rockford, IL) at room temperature for 5 minutes and detected by ChemiDOC XRS+ imager (Bio-Rad Laboratories, Hercules, CA).
Normal Human Tissue and Tissue Culture
Normal human placenta was obtained from an electively terminated 9-week gestation, fixed overnight in 10% formalin and embedded in paraffin. Discarded normal human skin was acquired from a single abdominoplasty specimen, was trimmed to 1×0.5 cm sections and cultured at 37°C for 48 hours in cell culture media (DMEM [Lonza, Walkersville, MD]/10% FBS + 1% antibiotic-antimycotic [Gibco; Life Technologies]) alone and with 250UI/ml IL-1β (PeProTech). A portion of the specimen prior to culture, and explants after culture were fixed overnight in 10% formalin and embedded in paraffin.
All tissue was obtained according to IRB-approved protocols.
Patient Melanoma Samples
In conformity to IRB-approved protocol, clinically annotated patient BRAF-mutant formalin-fixed paraffin-embedded melanomas were obtained from the Melanoma Institute Australia (Sydney, AU). Twenty-three tumor samples corresponded to biopsies performed before (pre) BRAF inhibitor therapy (dabrafenib, 19 patients; vemurafenib, 4) and eighteen to biopsies obtained after initiation of treatment (dabrafenib, 16 patients; vemurafenib, 2) from tumors that were progressing on treatment and therefore resistant to BRAF inhibitor therapy (prog). Response to therapy was defined according to the Response Evaluation Criteria in Solid Tumors (RECIST).[16, 17] In order to maximize the likelihood of finding significant differences in responses to BRAF inhibitor when comparing HGF immunoreactivity, pre-treatment samples were selected to represent distant points in the spectrum of patient response and accordingly were grouped as good (at least 60% tumor size reduction) and poor (less than 20% tumor size reduction) responders to BRAF inhibitor.
Immunocytochemistry and Immunohistochemistry
Sensitivity and specificity of antibodies and staining protocols were established in three ways: 1) by evaluation of positive control human tissue (placenta) for stromal HGF reactivity;[18] 2) by assessment of HGF induction in dermal stromal cells after recombinant Il-1β exposure in vitro;[19, 20] and 3) by examination of cultured fibroblasts (R2F1 cells) transfected with HFG expressing vector. Two different immunohistochemical protocols were employed. The first was identical to that described by Straussman et al.[2], and the second consisted of a simplified protocol not involving a tertiary antibody in order to eliminate or diminish non-specific background staining present in control tissues when using the first staining protocol. Samples were incubated with 10% horse serum for 1 hour at room temperature (~25°C) followed by overnight incubation at room temperature with 5 µg/ml HGF specific antibody (R&D Systems) and subsequent incubation with alkaline phosphatase (AP)-coupled horse anti-goat antibody (Vector Laboratories) for 1 hour at room temperature. Signal detection was performed employing AP substrate Vector Red® (Vector Laboratories). Detection of E-selectin in formalin-fixed paraffin-embedded human skin explants was performed after deparaffinization and epitope retrieval achieved by heating tissue sections in 1mM EDTA (pH 8.0) incubating sections with 2.6µg/ml E-selectin-specific antibody (Neuromics, Edina, MN) overnight at room temperature and then for 2 hours with HRP-horse anti-mouse antibodies (Vector Laboratories). HRP substrate NovaRed® (Vector Laboratories) was employed for immunoreactivity detection.
Assessment of HGF Expression by Immunohistochemistry
The immunohistochemistry results were first reviewed by one observer (CL) using the four-tier system previously described[2] where a score of 0 indicates absence of staining; 1, weak; 2, moderate; and 3 strong staining, either in stromal or in tumor cells. When samples presented heterogeneity in staining intensity, score was given for the strongest reactivity present. The extent of staining was graded in four categories (1 to 10%, 11–24%, 25 to 50% and >50%) according to the proportion of stromal or melanoma cells that were positive within each cell population. The time spent analyzing each slide ranged from 15 to 30 minutes. Slides were also analyzed by a second observer (GFM) to exclude significant inter-observer variation and to establish consensus scoring.
Statistical Analysis
Two-sided t-tests were used for all comparisons. A p-value of < 0.05 was considered significant. Data are reported as sample means with error bars representing the standard error of the means.
Results
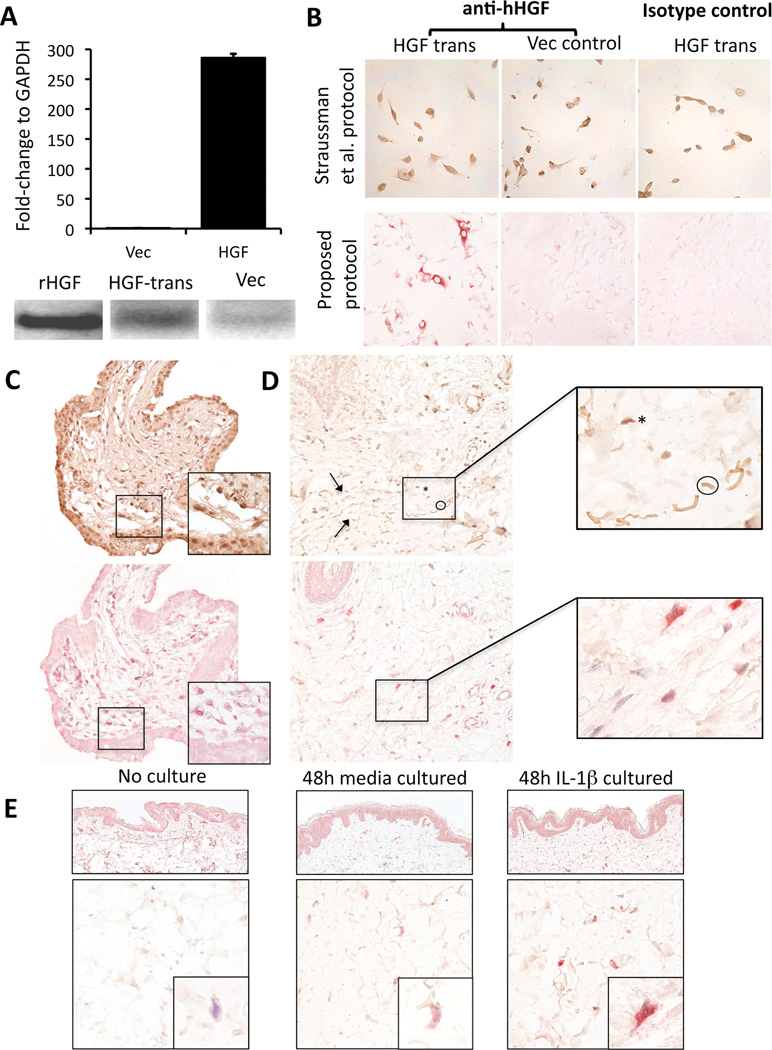
We initially examined a previously reported[2] immunohistochemistry protocol and evaluated its sensitivity and specificity in control cells and tissue with known HGF expression status. Accordingly, we examined human fibroblasts (R2F1) genetically modified to overexpress HGF, as confirmed by RT-PCR and Western blot analyses (Figure 1, A). Strong immunoreactivity in the HGF-expressing cells was documented. However, vector control cells and HGF-transfected cells incubated with isotype-matched irrelevant antibody both revealed positive reactivity with this staining protocol (Figure 1B, top row). We next examined human placenta employing this protocol and found HGF+ stromal cells in villous cores, as previously described.[18] However, there was high background staining as well as non-specific staining in isotype-matched irrelevant controls (Figure 1, C, top panel). Finally, normal human skin exposed to recombinant IL-1β, an established inducer of HGF expression in fibroblast cell lines,[19, 20] also showed high background staining as well as binding to elastic fibers (Figure 1, D, top panels) a finding further substantiated in medium only and normal skin controls (data not shown). As a result, the immunohistochemistry protocol was modified to eliminate the use of a tertiary antibody and a detection system (HRP-DAB) that could result in false positivity in melanoma samples due to presence of natural chromagen (e.g. melanin). By this second approach, HGF-transfected fibroblasts showed unequivocal positive immunoreactivity while vector and isotype-specific controls remained negative (Figure 1B, bottom row). HGF+ stromal cells were present in villous cores, and cytotrophoblasts remained appropriately negative, as previously described[18] (Figure 1, C, bottom panel). Human skin exposed to IL-1β demonstrated induction of HGF in stromal cells within the dermis, as previously documented[19, 20] (Figure 1, D, bottom panels). Normal human skin showed minimal HGF immunoreactivity, compared to skin explants exposed to media alone or IL-1β (Figure 1, E). E-selectin (E-sel) was positive in skin cultured in IL-1β consistent with culture viability and metabolic responsiveness, as previously described[21]. Intermediate HGF and E-selectin were also induced by media alone, consistent with previous observations of endogenous IL-1β release in this culture setting (data not shown).[21]
Figure 1. Validation of immunohistochemistry protocol for HGF.
A: Quantitative RT-PCR analysis (top) of R2F1 fibroblasts showing mRNA for control vector (Vec) and HGF-expressing vector (HGF); Western blot (bottom) reveals band from recombinant HGF (rHGF), transfected cell supernatant (HGF-trans), and vector control (Vec). B: R2F1 transfected fibroblasts were stained for HGF applying the immunostaining protocol proposed by Straussman et al. (top row). Note strong staining in HGF transfected cells (HGF trans), strong staining in fibroblasts transfected with control vector (Vec control) and also high background staining in the isotype-matched irrelevant control. A simplified staining protocol (bottom row) showed immunoreactivity only in HGF transfected fibroblasts and not in vector control transfected cells; the isotype control was negative.
C: Human normal placenta stained with Straussman et al. method (upper panel) and with a modified protocol that eliminates the tertiary antibody and uses an alkaline phosphatase detection system with a red chromagen (lower panel); insets show cytotrophoblasts (lower third) and mesenchymal cells D: Human normal skin exposed to IL-1β for 48 hours stained for HGF following Straussman et al. (upper panels). Note the similarity at 400× between an HGF+dermal cell (asterisk) and an elastic fiber (encircled, arrows point other fibers). Oil immersion (1000×) microphotograph (inset) allows distinction between the otherwise equivocal tissue components. Same sample of human skin exposed to IL-1β for 48 hours stained for HGF using the modified protocol does not present elastic fiber staining that could be misinterpreted as HGF+dermal cells (lower panels). E: Normal skin showed lightly counterstained nuclei with minimal immunoreactivity (left column); skin cultured in media alone and with IL-1β for 48 hours showed progressive increase in expression of HGF (middle and right columns), with IL-1β–treated skin containing cells that showed maximal (score 3) HGF immunoreactivity.
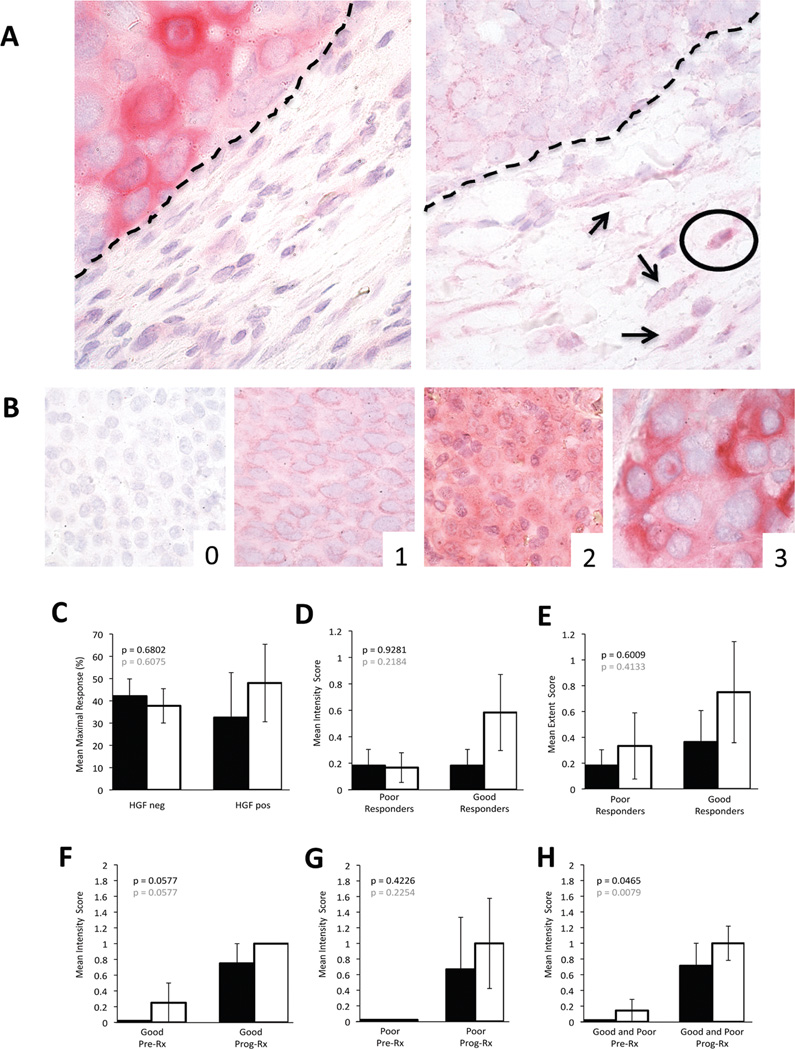
Employing the modified staining protocol that enhanced specificity and diminished false-positivity, without sacrificing sensitivity, we next assessed HGF biomarker expression by peri-tumoral stromal cells and melanoma cells from clinically-annotated patient samples before BRAF inhibitor therapy (n=23) and after disease progression while on BRAF inhibitor therapy (n=18). A four-tier scale previously described[2] was employed to assess staining intensity as “negative”, “weak”, “moderate” or “strong”. The proportion of positive cells was stratified in 1 to 10%, 11 to 24%, 25 to 50% and more than 50%. Four of 23 pre-BRAF inhibitor samples had weak HGF positivity in peri-tumoral stromal cells, three of them with <10% staining extent (Figure 2, A and Table 1). In 6 of the 23 pre-BRAF inhibitor samples, HGF+melanoma cells were observed. Three of these cases showed both HGF+ stromal and melanoma cells. Staining intensity in melanoma cells ranged from weak (n=4), to moderate (n=1) to strong (n=1) (Figure 2, B and Table 1). The proportion of HGF+melanoma cells represented up to 10% (n=2), between 11 and 24% (n=2), between 25 and 50% (n=1) and more than 50 % (n=1) of all tumor cells. Employing the RECIST system[16, 17] for evaluation of BRAF inhibitor response, we found no significant differences in maximal tumor size reduction when comparing presence or absence of HGF immunoreactivity in peri-tumoral stromal or melanoma cells themselves (Figure 2, C black and white bars, respectively). Furthermore, no significant differences were found between patients that had good (at least 60% tumor size reduction) and poor (less than 20% tumor reduction) responses when compared with intensity or extent of HGF immunoreactivity in peri-tumoral stromal cells or melanoma cells. (Figure 2, D and E, respectively; black bars=stromal cells, white bars=melanoma cells).
Figure 2. HGF expression in BRAF mutant patient melanomas.
A: Example of tumor HGF expression juxtaposed with negative stroma (left panel), scattered stromal cells with weak (arrows) to moderate (encircled) stromal HGF reactivity (right panel) [dashed line= tumor/stroma interface]. B: Example of tumor HGF grading (0–3).
C: Correlation of mean maximal response to BRAF inhibitor with peri-tumoral stromal (black bars) and melanoma cell (white bars) HGF expression in pre-treament samples. D: Mean intensity of HGF immunoreactivity for peri-tumoral stromal (black bars) and melanoma cells (white bars) in pre-treatment biopsies of poor versus good responders. E: Mean extent of HGF immunoreactivity for peri-tumoral stromal (black bars) and melanoma cells (white bars) in pre-treatment biopsies of poor versus good responders. F–H: Mean intensity of stromal cell HGF expression in good responders (F), in poor responders (G), and in good and poor responders combined (H) before BRAF inhibitor therapy (Pre) and upon progressive disease while on BRAF inhibitor (Prog). Note trend to an increase in HGF expression in progressing lesions in both stromal (black bars) and tumor cells (white bars) that reaches statistical significance when good and poor response groups are combined (H). Data for extent of HGF immunoreactivity in peri-tumoral stromal and melanoma cells before BRAF inhibitor therapy and upon progressive disease while on BRAF inhibitor treatment showed similar trends for good and poor responders when analyzed separately; the increase in HGF expression extent in progressing tumor samples reached statistical significance when good and poor responder groups were combined (data not shown).
Table 1.
Pre-BRAF inhibitor patient samples and HGF immunoreactivity assessment.
| HGF in stromal cells | HGF in tumor cells | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Genotype | BRAF inhibitor |
% tumor size1 |
Site | Intensity | Extent | Intensity | Extent | |
| Good responders | 1 | V600E | Dab | −60 | LN | 0 | 0 | 0 | 0 |
| 2 | V600E | Dab | −62 | SQ | 0 | 0 | 1 | 1 | |
| 3 | V600E | Dab | −63 | SQ | 0 | 0 | 0 | 0 | |
| 4 | V600E | Dab | −63 | Adrenal | 0 | 0 | 0 | 0 | |
| 5 | V600E | Dab | −71 | Brain | 0 | 0 | 0 | 0 | |
| 6 | V600K | Dab | −67 | Lung | 1 | 1 | 1 | 1 | |
| 7 | V600E | Dab | −71 | LN | 0 | 0 | 0 | 0 | |
| 8 | V600E | Dab | −67 | SQ | 1 | 3 | 2 | 4 | |
| 9 | V600E | Dab | −76 | SQ | 0 | 0 | 0 | 0 | |
| 10 | V600E | Vem | −76 | LN | 0 | 0 | 0 | 0 | |
| 11 | V600E | Dab | −78 | Rectum | 0 | 0 | 0 | 0 | |
| 12 | V600E | Vem | −100 | Muscle | 0 | 0 | 3 | 3 | |
| Poor responders | 13 | V600K | Dab | 10 | Adrenal | 1 | 1 | 1 | 2 |
| 14 | V600E | Dab | 4 | LN | 0 | 0 | 0 | 0 | |
| 15 | V600E | Vem | −2 | Brain | 0 | 0 | 1 | 2 | |
| 16 | V600E | Dab | −5 | SQ | 0 | 0 | 0 | 0 | |
| 17 | V600E | Dab | −6 | LN | 1 | 1 | 0 | 0 | |
| 18 | V600E/W604C | Dab | −6 | SQ | 0 | 0 | 0 | 0 | |
| 19 | V600E | Dab | −10 | SQ | 0 | 0 | 0 | 0 | |
| 20 | V600K | Dab | −14 | SQ | 0 | 0 | 0 | 0 | |
| 21 | V600E | Dab | −15 | LN | 0 | 0 | 0 | 0 | |
| 22 | V600E | Dab | −15 | SQ | 0 | 0 | 0 | 0 | |
| 23 | V600E | Vem | −17 | SQ | 0 | 0 | 0 | 0 | |
Measured according to the RECIST criteria.
Dab= Dabrafenib. Vem= Vemurafenib. LN= Lymph node. SQ= subcutis.
Samples obtained from tumors that were progressing on BRAF inhibitor treatment (n=18) were also evaluated. Nine of these cases had at least some HGF+ peri-tumoral stromal cells, 6 with weak staining and 3 with moderate staining. The proportion of positive peri-tumoral stromal cells varied from less than 10% (n=6) to between 11 and 24% (n=3). Fourteen of 18 samples of progressing tumors had at least some HGF+ melanoma cells. The intensity of the staining varied from weak (n=12) to moderate (n=2). The proportion of positive melanoma cells varied from less than 10% (n=6), to 11 to 24% (n=4), to 25 to 50% (n=1) to more than 50% (n=3) (data not shown).
Seven patients had paired samples of pre-treatment and progressing on BRAF i therapy tumor lesions. Four of them had at least 60% maximal response by RECIST; and three patients, less than 20% tumor size reduction. When HGF expression in either stromal cells or tumor cells was compared between pre and progression samples, there was a consistent trend toward increased HGF expression in progressing lesions that was independent of degree of therapeutic response and consistent within both HGF expression parameters analyzed, intensity and percent area of reactivity (Figure 2, F and G, for good and poor responders respectively; black bars=stromal cells, and white bars=melanoma cells; data for percent area of reactivity not shown). This trend was non-statistically significant when good and poor responders were analyzed separately, and reached statistical significance when these groups were combined (Figure 2, H; black bars=stromal cells, and white bars=melanoma cells).
Discussion
Here we show that biomarker detection of stromal or tumor HGF in pre-therapy specimens of metastatic melanoma fails to predict response to RAF inhibitor therapy. Negative results traditionally have been published and cited less than those that support a stated hypothesis, and this trend has been averred potentially to produce skewing that may contribute to scientific bias.[22] Indeed negative findings are becoming increasingly recognized as crucial to scientific progress that is made possible only through rigorous self-correction.[23] It also has been posited that the so-called 'decline effect', where discoveries may gradually diminish over time as they are eventually repeated, may be fueled by limited incentive for more immediate testing and validation that carries with it the potential to produce negative outcomes.[24] With respect to biomarkers, Marchio[25] has emphasized that although great emphasis has been given to discovery, technical validation assays have not been embraced with equal enthusiasm, although the process of assay validation is critical for the clinical introduction of any new biomarker. We therefore believe that findings such as those reported herein, while negative, ultimately may serve to accelerate progress toward refining pathology-based approaches that seek to personalize novel therapies for metastatic melanoma.
Lim et al.[26] have emphasized the critical need for fully appreciating technical and experimental design complexities inherent to biomarker validation in human tissues. In our study, reasons that might explain the disparate conclusions regarding the predictive value for HGF melanoma stromal biomarker expression[2] include differences in 1) biospecimen preservation and archiving, 2) detection systems employed, 3) treatment protocols, and finally 4) the nature of the target antigen itself. Firstly, the Melanoma BioSpecimen Bank of the Melanoma Institute Australia has been a source for collaborative research worldwide, supplying a wide range of clinically-annotated biomaterials. Specimen procurement, preparation protocols, and archiving methods are rigidly controlled, and the bioarchive successfully has supported a number of recent melanoma findings, including those involving sensitive detection of the SOX2 transcription factor and the epigenetic mark 5-hydroxymethylcytosine (5-hmC).[15, 27] Second, the detection system that we employed was developed to eliminate false positivity while retaining the ability to sensitively detect HGF-expressing stromal and tumor cells, an approach fundamental to clinical biomarker validation and application The use of multiple complementary positive control tissues and cells is regarded as critical to the validation process of tissue biomarkers such as HGF, and this component of our study may indeed account for results that differed from previous reports. Third, our patient cohort was treated mainly with the BRAF inhibitor, dabrafenib, rather than vemurafenib that was employed in previous studies[2] However, both have similar potency for BRAFV600E inhibition, and treatment outcomes are similar.[8, 10, 28–30] Moreover, the previous study found no differences in correlation of the impact of HGF stromal expression in relationship to the use of different MAPK inhibitors.[2] Finally, HGF is a secreted protein synthesized by tumor and stromal cells, as well as other cell types, including hematopoietic cell lines and bone marrow stroma.[31, 32] Because protein secretion is a dynamic process, detection of intracellular stores from a single cellular microenvironment may not always be as reliable as quantification of levels in serum[7] that likely reflect aggregate secretion from multiple sources.
Current therapeutic strategies focused on metastatic melanoma are seeking to target immunosuppressive co-stimulatory molecules,[3, 33, 34] melanoma-initiating cells,[35] and oncogenic pathways.[8, 28, 36] Despite early success, cancers appear to be able to thwart therapy through new virulence mechanisms that evolve during treatment; recent examples being increased expression of melanoma stem cells displaying the biomarker ABCB5 during chemotherapy,[37] and upregulation of CD274 (PD-L1) in melanomas during BRAF inhibitor treatment.[38] Our finding in the present study that HGF expression by tumor and stromal cells is significantly enhanced at disease progression is potentially relevant to such therapy-associated virulence acquisition by melanoma cells and requires further inquiry and validation. The literature clearly supports a role for HGF and other tyrosine-kinase ligands in conferring resistance to targeted therapies (such as inhibitors of the MAPK pathway), and the in vitro findings of Straussman et al.[2] represent an important contribution to this dataset.[7, 36, 39–47] Although our analysis clarifies the limited of applicability of the HGF biomarker to melanoma stromal cells, it remains of key importance to continue to consider how HGF in the living patient affects the MAPK pathway, and how this may be most reliably measured. In this regard, a recent study by Jubb and coworkers[48] examining the potential value of immunohistochemical detection of the HGF receptor tyrosine kinase, MET, in defining melanoma resistance to BRAF therapy failed to show prognostic significance in terms of response rate, progression-free survival, or overall survival, and thus is consistent with our data indicating limited applicability of markers of the HGF-MET pathway in determining such therapeutic responses.
Integration of biomarkers into drug development and clinical trials requires quality assurance and assay validation to establish standardized guidelines for broad spectrum application.[1, 6, 49] Studies that seek to correlate compelling in vitro data with the possibility of practical application to predict responses in patients represent novel and exciting directions in deployment of personalized pathology-based techniques. Indeed, while our study did not confirm the utility of immunohistochemical detection of stromal HGF as a means of defining melanoma patients who are resistant to BRAF therapy, it in no way diminishes the potentially key role of HGF in conferring such resistance, as suggested in the important study by Straussman and colleagues.[2] In aggregate, in this report focusing on HGF we more broadly seek to emphasize that with continued rigor in biomarker validation, appreciation of the complementary value of both positive and negative findings, and aggressive translation of basic mechanistic insights into practical therapeutic applications, there will exist significant and imminent promise for ultimate control of melanoma once it spreads beyond the primary site.
Acknowledgments
NIH-P50 CA93683 supported the present work. We thank Julie Howle, Jessica Hyman, Tracy Liaw, Katherine Carson, Raghwa Sharma and the staff at Melanoma Institute Australia and Westmead Medical Oncology Trials Centre.
Dr. Murphy has since received a grant from Bristol-Myers Squibb focused on detection of immune cell targeting in melanomas from patients undergoing specific immunomodulatory therapies.
Footnotes
Disclosures
The authors have no conflicts of interest during the course of the study
References
- 1.Poste G. Bring on the biomarkers. Nature. 2011;469:156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 2.Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sondak VK, Flaherty LE. Targeted therapies: Improved outcomes for patients with metastatic melanoma. Nat Rev Clin Oncol. 2011;8:513–515. doi: 10.1038/nrclinonc.2011.119. [DOI] [PubMed] [Google Scholar]
- 6.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–S32. doi: 10.6004/jnccn.2011.0137. quiz S3. [DOI] [PubMed] [Google Scholar]
- 7.Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falchook GS, Long GV, Kurzrock R, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan RJ, Flaherty K. MAP kinase signaling and inhibition in melanoma. Oncogene. 2013;32:2373–2379. doi: 10.1038/onc.2012.345. [DOI] [PubMed] [Google Scholar]
- 12.Trunzer K, Pavlick AC, Schuchter L, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013;31:1767–1774. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 13.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta PB, Kuperwasser C, Brunet JP, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girouard SD, Laga AC, Mihm MC, et al. SOX2 contributes to melanoma cell invasion. Lab Invest. 2012;92:362–370. doi: 10.1038/labinvest.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 17.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Kauma S, Hayes N, Weatherford S. The differential expression of hepatocyte growth factor and met in human placenta. J Clin Endocrinol Metab. 1997;82:949–954. doi: 10.1210/jcem.82.3.3806. [DOI] [PubMed] [Google Scholar]
- 19.Tamura M, Arakaki N, Tsubouchi H, Takada H, Daikuhara Y. Enhancement of human hepatocyte growth factor production by interleukin-1 alpha and-1 beta and tumor necrosis factor-alpha by fibroblasts in culture. J Biol Chem. 1993;268:8140–8145. [PubMed] [Google Scholar]
- 20.Kikuchi T, Abe T, Yaekashiwa M, et al. Secretory leukoprotease inhibitor augments hepatocyte growth factor production in human lung fibroblasts. Am J Respir Cell Mol Biol. 2000;23:364–370. doi: 10.1165/ajrcmb.23.3.3942. [DOI] [PubMed] [Google Scholar]
- 21.Messadi DV, Pober JS, Fiers W, Gimbrone MA, Jr, Murphy GF. Induction of an activation antigen on postcapillary venular endothelium in human skin organ culture. J Immunol. 1987;139:1557–1562. [PubMed] [Google Scholar]
- 22.Fanelli D. Do pressures to publish increase scientists' bias? An empirical support from US States Data. PLoS One. 2010;5:e10271. doi: 10.1371/journal.pone.0010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanelli D. Negative results are disappearing from most disciplines and countries. Scientometrics. 2012;90:891–904. [Google Scholar]
- 24.Schooler J. Unpublished results hide the decline effect. Nature. 2011;470:437. doi: 10.1038/470437a. [DOI] [PubMed] [Google Scholar]
- 25.Marchio C, Dowsett M, Reis-Filho JS. Revisiting the technical validation of tumour biomarker assays: how to open a Pandora's box. BMC Med. 2011;9:41. doi: 10.1186/1741-7015-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim MD, Dickherber A, Compton CC. Before you analyze a human specimen, think quality, variability, and bias. Anal Chem. 2011;83:8–13. doi: 10.1021/ac1018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lian CG, Xu Y, Ceol C, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menzies AM, Long GV, Murali R. Dabrafenib and its potential for the treatment of metastatic melanoma. Drug Des Devel Ther. 2012;6:391–405. doi: 10.2147/DDDT.S38998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauschild A, Grob JJ, Demidov LV, et al. An update on BREAK-3, a phase III,randomized trial: Dabrafenib (DAB) versus dacarbazine (DTIC) in patients with BRAF V600E-positive mutation metastatic melanoma (MM) J Clin Oncol. 2013;31 abstract 9013. [Google Scholar]
- 30.Chapman PB, Hauschild A, Robert C, et al. Updated overall survival (OS) results for BRIM-3, a phase III randomized, open-label, multicenter trial comparing BRAF inhibitor vemurafenib (vem) with dacarbazine (DTIC) in previously untreated patients with BRAFV600E-mutated melanoma. J Clin Oncol. 2012;30 abstract 8502. [Google Scholar]
- 31.Matsuda-Hashii Y, Takai K, Ohta H, et al. Hepatocyte growth factor plays roles in the induction and autocrine maintenance of bone marrow stromal cell IL-11, SDF-1 alpha, and stem cell factor. Exp Hematol. 2004;32:955–961. doi: 10.1016/j.exphem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Pons E, Uphoff CC, Drexler HG. Expression of hepatocyte growth factor and its receptor c-met in human leukemia-lymphoma cell lines. Leuk Res. 1998;22:797–804. doi: 10.1016/s0145-2126(98)00071-x. [DOI] [PubMed] [Google Scholar]
- 33.Razzak M. From ASCO-targeted therapies: Anti-PD-1 approaches-important steps forward in metastatic melanoma. Nat Rev Clin Oncol. 2013 doi: 10.1038/nrclinonc.2013.98. [DOI] [PubMed] [Google Scholar]
- 34.Hamid O, Robert C, Daud A, et al. Safety and Tumor Responses with Lambrolizumab (Anti-PD-1) in Melanoma. N Engl J Med. 2013 doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chartrain M, Riond J, Stennevin A, et al. Melanoma chemotherapy leads to the selection of ABCB5-expressing cells. PLoS One. 2012;7:e36762. doi: 10.1371/journal.pone.0036762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frederick DT, Piris A, Cogdill AP, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19:1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClaine RJ, Marshall AM, Wagh PK, Waltz SE. Ron receptor tyrosine kinase activation confers resistance to tamoxifen in breast cancer cell lines. Neoplasia. 2010;12:650–658. doi: 10.1593/neo.10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 42.Yano S, Yamada T, Takeuchi S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol. 2011;6:2011–2017. doi: 10.1097/JTO.0b013e31823ab0dd. [DOI] [PubMed] [Google Scholar]
- 43.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 44.Sierra JR, Cepero V, Giordano S. Molecular mechanisms of acquired resistance to tyrosine kinase targeted therapy. Mol Cancer. 2010;9:75. doi: 10.1186/1476-4598-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones HE, Goddard L, Gee JM, et al. Insulin-like growth factor-I receptor signalling and acquired resistance to gefitinib (ZD1839;Iressa) in human breast and prostate cancer cells. Endocr Relat Cancer. 2004;11:793–814. doi: 10.1677/erc.1.00799. [DOI] [PubMed] [Google Scholar]
- 46.Mahadevan D, Cooke L, Riley C, et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene. 2007;26:3909–3919. doi: 10.1038/sj.onc.1210173. [DOI] [PubMed] [Google Scholar]
- 47.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jubb AM, Ribas A, Sosman JA, et al. Impact of MET expression on outcome in BRAF advanced melanoma. Histopathology. 2013 doi: 10.1111/his.12169. [DOI] [PubMed] [Google Scholar]
- 49.Chau CH, Rixe O, McLeod H, Figg WD. Validation of analytic methods for biomarkers used in drug development. Clin Cancer Res. 2008;14:5967–5976. doi: 10.1158/1078-0432.CCR-07-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]