Abstract
Introduction
Pre-eclampsia (PE) has a familial association, with daughters of women who had PE during pregnancy having more than twice the risk of developing PE themselves. Through genome-wide linkage and genetic association studies in PE-affected families and large population samples, we previously identified the following as positional candidate maternal susceptibility genes for PE; ACVR1, INHA, INHBB, ERAP1, ERAP2, LNPEP,COL4A1 and COL4A2. The aims of this study were to determine mRNA expression levels of previously identified candidate maternal pre-eclampsia susceptibility genes from normotensive and severe PE (SPE) pregnancies and correlate mRNA expression levels with the clinical severity of SPE.
Methods
Third trimester decidual tissues were collected from both normotensive (n=21) and SPE pregnancies (n=24) and mRNA expression levels were determined by real-time PCR. Gene expression was then correlated with several parameters of clinical severity in SPE. Statistical significance was determined by Mann-Whitney U test and Spearman's Correlation.
Results
The data demonstrate significantly increased decidual mRNA expression levels of ACVR1, INHBB, ERAP1, ERAP2, LNPEP, COL4A1 and COL4A2 in SPE (p<0.05). Increased mRNA expression levels of several genes – INHA, INHBB, COL4A1 and COL4A2 were correlated with earlier onset of PE and earlier delivery of the fetus (p<0.05).
Conclusion
These results suggest altered expression of maternal susceptibility genes may play roles in PE development and the course of disease severity.
Keywords: clinical severity, decidua, gene expression, pre-eclampsia, susceptibility genes
Introduction
The key to pregnancy success is the establishment and growth of the placenta. When this is deficient, complications develop that compromise maternal and fetal health. The fundamental origin of pre-eclampsia (PE) lies in the placenta as PE only manifests during pregnancy and can occur in the absence of a fetus (i.e. in a molar pregnancy) [1]. While deficient placentation is characteristic of PE, the underlying causes are unknown [2]. PE has a familial association, with daughters of PE affected women having more than twice the risk of developing PE [3]. Delineating the key processes involved in the establishment of a healthy placenta and how they are altered by genetic variation is central to determining how PE develops.
The familial association of PE is well established. First-degree relatives of women who had a PE pregnancy have a two to five fold increased risk of also having a PE pregnancy [4]. A Norwegian study, examining 1.7 million births from 1967 to 1992, showed that the odds ratio of developing PE in a subsequent pregnancy was 2.2 for mothers with an affected sister [5]. A Swedish study estimated that maternal genetic factors alone accounted for almost half of PE familial aggregation while other factors such as environmental and paternal genetic factors accounted for the rest [6]. PE, like many common human diseases, is thus a complex trait that does not involve simple Mendelian monogenic inheritance [7].
Two approaches are commonly applied to identify causal genetic variation for heritable human diseases: candidate gene-based studies or positional cloning in affected families and/or large population-based cohorts of affected and unaffected individuals. In the candidate gene-based approach, genes are chosen based on the cumulative knowledge of disease pathways. Previous candidate gene studies for PE examined genes involved with thrombophilia, haemodynamics, endothelial function, inflammation and the immune response, oxidative stress, lipid metabolism, the endocrine system, angiogenesis and placentation [7-9]. Given the low probability of prioritising a functional gene with the incomplete and limited knowledge of the underlying causative mechanisms of PE, no single universally accepted susceptibility gene has been identified.
In the positional cloning approach, underlying disease susceptibility loci are first localised by a genome scan to a potentially large chromosomal region. This localisation is accomplished either by linkage mapping in affected families or by association analysis in large population cohorts of affected and unaffected individuals. The putative susceptibility loci identified are subsequently interrogated for plausible positional candidates that are then subjected to more rigorous genetic analyses. Several groups including our own adopted this positional cloning approach for studying PE. Genome-wide linkage scans were conducted in PE families from Iceland [10], Australia & New Zealand [11-13], the Netherlands [14] and Finland [15]. To date, our studies [12, 13, 16-19] have generated a range of plausible positional candidate genes identified through extensive genome-wide linkage and genome-wide association studies in multiple, well-defined, affected families and unrelated individuals from Australia and New Zealand.
Our genome-wide linkage analyses and fine mapping studies identified significant linkage for the severe PE (SPE) phenotype to chromosomes 2q, 5q and 13q [12, 13, 18]. Bioinformatic prioritisation of positional candidate genes in regions of linkage identified the following genes: on chromosome 2, the activin receptor gene ACVR1 and the inhibin subunit genes INHA and INHBB; on chromosome 5, the endoplasmic reticulum aminopeptidase genes ERAP1 and ERAP2 and the placental leucine aminopeptidase gene LNPEP; and on chromosome 13, the type IV collagen α chain genes COL4A1 and COL4A2. These genes can be functionally grouped as activin and inhibin signalling components (ACVR1, INHA,INHBB), M1 aminopeptidases (ERAP1, ERAP2, LNPEP) or connective tissue components (COL4A1, COL4A2).
By employing thorough genetic and bioinformatic analyses, we identified plausible positional candidate maternal susceptibility genes for PE. The aims of this current work were to determine the mRNA expression levels for these candidate genes in decidual tissue collected from third trimester normotensive and SPE pregnancies, and to correlate mRNA expression levels with the clinical severity of SPE.
Materials and Methods
Patient Samples
Third trimester decidua basalis samples were collected during Caesarean section from n=21 normotensive and n=24 SPE patients as described previously [18]. Normotensive patients underwent Caesarean section due to breech presentation, maternal request or previous history. Samples were classified according to the Australasian Society for the Study of Hypertension in Pregnancy criteria [20, 21], which were previously used to identify the susceptibility genes [12, 13, 18]. Clinical classifications of SPE used meet the current International Society for the Study of Hypertension in Pregnancy guidelines [22] (for summary of patient clinical characteristics see Table 1). Exclusion criteria consisted of factors known to predispose women to PE such as multiple pregnancies and diabetes. Blood pressures of normotensive patients were recorded as < 140/90mmHg. Patient records were independently verified by a non-treating obstetrician. All tissue samples were verified as decidual by the Royal Women's Hospital pathologists. Written informed consent was obtained from each patient. Research and ethics approval was obtained from the Royal Women's Hospital Research and Ethics Committees. Patient data were analysed for differences in maternal age, gestational age, infant birth weight, infant birth weight percentiles, placental weight, gravidity, parity, and infant sex. We included birth weight percentiles to account for the effect of gestation on infant birth weight [23].
Table 1.
Summary of patient characteristics: All patient samples
| Patient characteristicsa | Normotensive (n=20) | Pre-eclampsia (n=24) | P-valueb |
|---|---|---|---|
| Maternal age (years) | 31.810±0.89 | 30.29±0.98 | 0.26 |
| Gestational age (weeks) | 38.62±0.43 | 32.00±0.74 | <0.001 |
| Infant birth weight (g) | 3193.48±117.43 | 1662. 21±152.30 | <0.001 |
| Infant weight percentiles (%)c | 25-50 | 10-25 | <0.05 |
| Placental weight (g)d | 583.00±37.79 | 361.36±22.45 | <0.001 |
| Infant sex | 11F, 10M | 12F, 12M | 1.00 |
| Gravidity | 16 primi-, 5 multi- | 18 primi-, 6 multi- | 1.00 |
| Parity | 19 primi-, 2 multi- | 21 primi-, 3 multi- | 1.00 |
| Systolic blood pressure (mmHg)e | <140 | 168.45±2.56 | NA |
| Diastolic blood pressure (mmHg)e | <90 | 105.00±1.92 | NA |
| Antihypertensive treatment(s)f | Not given | 19 | NA |
| MgSO4 treatmentf | Not given | 17 | NA |
NA, not applicable.
Shown is the mean ± SEM unless stated otherwise.
Student's t test with Welch's Correction for parametric data and 2 X 2 contingency table with Fisher's Exact Test for categorical data were used.
Data shown as median.
Placental weights for n=3 normotensive and n=3 SPE patients were not recorded.
RNA Extraction, cDNA Synthesis and Real-time Polymerase Chain Reaction (PCR)
Decidual mRNA expression levels of maternal PE susceptibility genes were determined by real-time PCR on patient decidua basalis samples. Total RNA was extracted from decidual samples and cDNA was synthesised from the extracted RNA as described previously [18, 24]. Real-time PCR was performed using Applied Biosystems 7500 PCR System (Foster City, CA, USA). The reaction was carried out at 95°C for 10 minutes, followed by 95°C for 15 seconds and 60°C for 1 minute for 40 cycles. Pre-validated TaqMan® Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) with FAM-labelled probes for each gene of interest and VIC-labelled probe for the 18S rRNA housekeeping gene were used. The probes used were: Hs00153836 m1 ACVR1, Hs00171410 m1 INHA, Hs00173582 m1 INHBB, Hs00429970 m1 ERAP1, Hs01073631 m1 ERAP2, Hs00893646 m1 LNPEP, Hs00266237m1 COL4A1, Hs01098873m1 COL4A2 and Eukaryotic 18S rRNA Endogenous Control (VIC/MGB Probe, Primer Limited) for housekeeping. The average threshold cycle (Ct) value difference between FAM and VIC of the pooled normotensive duplicates was used as the calibrator for relative quantification. Relative mRNA expression levels were calculated using the 2-ΔΔCT method [25].
Clinical severity and characteristics analyses
Since the differentially expressed genes belonged to different functional groups, we next looked for relationships of maternal susceptibility gene expression with available patient data in our sample set. We analysed the correlation of mRNA expression levels of the susceptibility genes in the SPE sample set with different parameters of clinical severity and characteristics. The parameters studied were highest recorded systolic and diastolic blood pressures, highest recorded degree of proteinuria, lowest recorded platelet level, infant birth weight percentile, placental weight, gestation at onset of SPE, gestation at delivery of fetus, required treatment with magnesium sulphate, total number of antihypertensive medications prescribed, maternal age, gravidity and parity.
Immunohistochemistry
Serial 5 μm decidual sections were obtained from frozen decidual tissue of n=4 normotensive and n=4 SPE patients. All washes were performed three times in 1X Phosphate Buffered Saline. Sections were washed to remove embedding media, treated with 3% hydrogen peroxide in dH2O to block endogenous peroxidase and washed. Zymed Laboratories Histostain®-Plus Broad Spectrum Kit (Life Technologies, Carlsbad, CA, USA) was used thereafter following the manufacturer's protocol with the following modifications. Sections were covered with Solution A blocking solution in a humidified chamber for 1 hour at room temperature and then washed. Sections were then incubated in the humidified chamber overnight at 4°C with primary antibodies diluted in 1X Tris Buffered Saline (Supplementary Table 1). After overnight incubation, sections were washed, incubated with Solution B broad spectrum secondary antibody in the humidified chamber for 1 hour at room temperature and washed again. Sections were then incubated with Solution C containing horseradish peroxidase for 1 hour at room temperature and washed. Immunostaining was visualised using AEC substrate solution prepared from Zymed Laboratory AEC Red Substrate Kit (Life Technologies, Carlsbad, CA, USA) according to manufacturer's instructions. Sections were viewed under 200X magnification with an Axioskop 2 light microscope (Zeiss Gruppe, Oberkochen, Germany). Images were captured with a Nikon DXM1200C camera (Nikon Corp., Tokyo, Japan) attached to the microscope.
Statistical Analyses
Student's t test with Welch's Correction was used for comparing parametric data between two groups, Mann-Whitney U test was used for comparing non-parametric normalised data between two groups, 2 X 2 contingency table with Fisher's Exact Test was used for comparing categorical data between two groups and Spearman's correlation analysis was used to determine associations between data sets. All data are expressed as mean ± SEM unless stated otherwise. GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analyses. A value of p<0.05 was considered statistically significant.
Results
Patient Characteristics
Analysis for the n=21 normotensive and n=24 SPE patients showed significant differences in gestational age, birth weight, birth weight percentiles and placental weight, but no significant difference in maternal age, parity, gravidity or infant sex (Table 1).
To account for the significant difference in gestational age between SPE and normotensive patients, we selected a smaller subset of n=9 normotensive and n=7 SPE patients with similar gestational ages. Analysis of this smaller subset showed no significant difference in any patient characteristic analysed (Table 2). Analysis of the residual n=17 SPE patients have been included in Supplementary Table 2.
Table 2.
Summary of patient characteristics: Smaller subset
| Patient characteristicsa | Normotensive (n=9) | Pre-eclampsia (n=7) | P-valueb |
|---|---|---|---|
| Maternal age (years) | 33.00±1.40 | 30.57±2.34 | 0.39 |
| Gestational age (weeks) | 37.33±0.80 | 36.57±0.57 | 0.45 |
| Infant birth weight (g) | 3035.44±230.30 | 2495.57.±260.19 | 0.14 |
| Infant weight percentiles (%)c | 50-75 | 25-50 | 0.23 |
| Placental weight (g)d | 592.33±99.36 | 451.83±52.09 | 0.25 |
| Infant sex | 5F, 4M | 3F, 4M | 1 |
| Gravidity | 6 primi-, 3 multi- | 5 primi-, 2 multi- | 1 |
| Parity | 7 primi-, 2 multi- | 6 primi-, 1 multi- | 1 |
| Systolic blood pressure (mmHg) | <140 | 167.57±6.36 | NA |
| Diastolic blood pressure (mmHg) | <90 | 106.14±5.22 | NA |
| Antihypertensive treatment(s) | Not given | 5 | NA |
| MgSO4 treatment | Not given | 5 | NA |
NA, not applicable.
Shown is the mean ± SEM unless stated otherwise.
Student's t test with Welch's Correction for parametric data and 2 X 2 contingency table with Fisher's Exact Test for categorical data.
Data shown as median.
Placental weights for n=3 normotensive and n=1 SPE patients were not recorded.
Real-time PCR
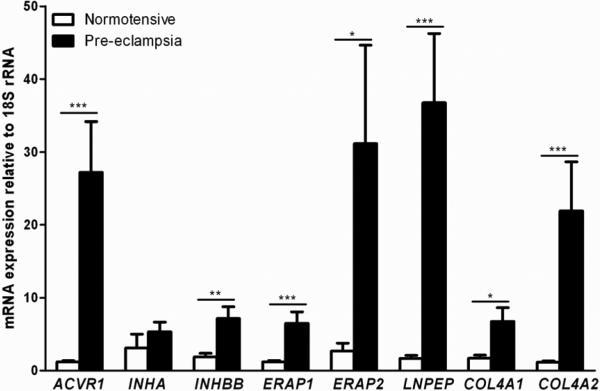
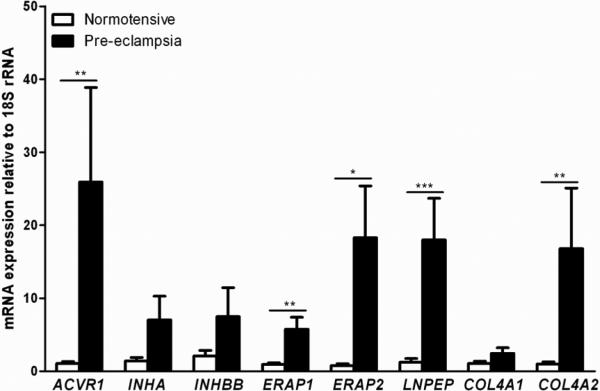
The results showed significantly increased mRNA expression levels of genes ACVR1, INHBB, ERAP1, ERAP2, LNPEP, COL4A1 and COL4A2 relative to housekeeping 18S rRNA in SPE compared with normotensive samples, while INHA showed no significant difference in the large sample set (Figure 1). Nevertheless, INHA showed a trend for increased expression with a p-value of 0.052. In the smaller subset, genes ACVR1, ERAP1, ERAP2, LNPEP and COL4A2 retained their significance (Figure 2). Analysis between the n=7 SPE samples used in the subset and the n=17 residual SPE samples showed no significant difference between the two groups (Supplementary Figure 1).
Figure 1. mRNA expression of maternal pre-eclampsia susceptibility genes: All patient samples.
Relative mRNA expression of maternal pre-eclampsia susceptibility genes in human third trimester decidua basalis samples from n=21 normotensive and n=24 severe pre-eclamptic pregnancies. Data presented as mean + SEM. * p<0.05, ** p<0.01, *** p<0.001, Mann Whitney U test.
Figure 2. mRNA expression of maternal pre-eclampsia susceptibility genes: Gestation matched subset.
Relative mRNA expression of maternal pre-eclampsia susceptibility genes in human third trimester decidua basalis samples from n=9 normotensive and n=7 severe pre-eclamptic pregnancies. Data presented as mean + SEM. * p<0.05, ** p<0.01, *** p<0.001, Mann Whitney U test.
In the large set, the activin receptor gene ACVR1 showed a 22.9 fold increase in relative mRNA expression levels. The inhibin gene INHBB showed a 3.8 fold increase in relative mRNA expression levels. The aminopeptidase genes, ERAP1, ERAP2 and LNPEP, showed 5.3, 11.5 and 21.7 fold increases in relative mRNA expression levels respectively. The collagen genes COL4A1 and COL4A2 showed 3.9 and 18.8 fold increases in relative mRNA expression levels respectively. Differential mRNA expression levels of ACVR1, ERAP1, ERAP2, LNPEP and COL4A2 were also detected in the smaller gestationally matched subset, with fold increases of 24.0, 6.1, 22.9, 14.5 and 16.5 in relative mRNA expression levels respectively.
Clinical severity and characteristics analyses
Correlation analyses of SPE samples in both the large set and smaller subset mostly showed positive associations of increased gene expression with greater disease severity (Table 3). In the large set, decreased ERAP2 mRNA expression levels were associated with increased systolic blood pressure and a greater number of antihypertensive medications prescribed. Increased COL4A1 mRNA expression levels were associated with an earlier onset of SPE. Increased LNPEP mRNA expression levels were associated with an earlier delivery of the fetus. Increased COL4A2 mRNA expression levels were associated with increasing parity of the mother. In the subset, increased COL4A2 mRNA expression levels were associated with an earlier onset of SPE. Increased mRNA expression levels of INHA, INHBB and COL4A2 were associated with an earlier delivery of the fetus. Analysis for the residual n=17 SPE samples is presented in Supplementary Table 3.
Table 3.
Associations of decidual mRNA expression levels to different parameters of clinical severity and characteristics
| Clinical severity parameters and characteristics analysed a | Associated genes (all SPE samples) | Associated genes (smaller subset SPE samples) |
|---|---|---|
| Increasing systolic blood pressure (mmHg) | ERAP2↓ | NIL |
| Increasing diastolic blood pressure (mmHg) | NIL | NIL |
| Increasing proteinuria (dipstick +) | NIL | NIL |
| Decreasing level of platelets | NIL | NIL |
| Greater number of antihypertensive medications | ERAP2↓ | NIL |
| Required MgSO4 treatment | NIL | NIL |
| Earlier onset of SPE (weeks) | COL4A1↑ | COL4A2↑ |
| Earlier delivery of fetus (weeks) | LNPEP↑ | INHA↑, INHBB↑, COL4A2↑ |
| Decreasing infant weight percentile (%) | NIL | NIL |
| Decreasing placental weight (g) | NIL | NIL |
| Increasing maternal age (years) | NIL | NIL |
| Increasing gravidity | NIL | NIL |
| Increasing parity | COL4A2↑ | NIL |
Spearman's correlation for correlation of non-parametric data. ↑ denotes increasing expression of gene. ↓ denotes decreasing expression of gene.
Immunohistochemistry
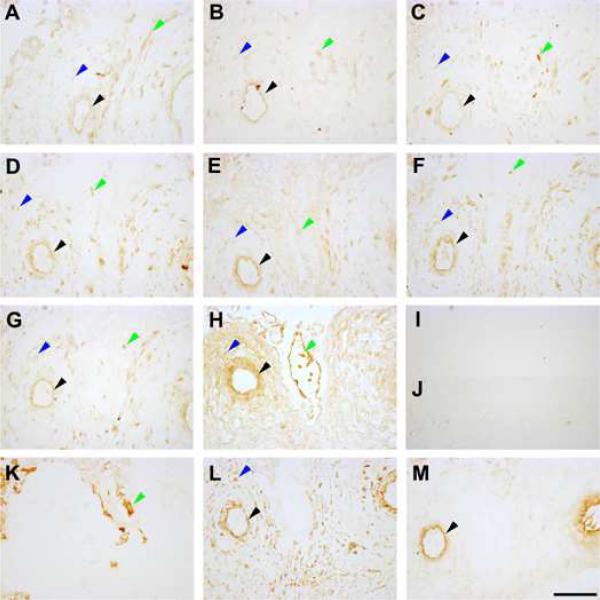
Immunohistochemistry performed on serial decidual sections from normotensive and SPE pregnancies showed decidual localisation of all genes in the decidual stromal cells, the maternal endothelium and the cytotrophoblast cells. Representative sections from a normotensive patient are presented in Figure 3. There were no obvious staining pattern differences between the normotensive and SPE patients (data not shown).
Figure 3. Protein localisation of maternal pre-eclampsia susceptibility genes in the decidua.
Decidual sections shown are from a representative patient sample. A: Activin type I receptor (ACVR1) expression. B: Inhibin α subunit (INHA) expression. C: Inhibin βB subunit (INHBB) expression. D: Endoplasmic reticulum aminopeptidase 1 (ERAP1) expression. E: Endoplasmic reticulum aminopeptidase 2 (ERAP2) expression. F: Placental leucine aminopeptidase (LNPEP) expression. G: Collagen type IV α1 chain (COL4A1) expression. H: Collagen type IV α2 chain (COL4A2) expression. I: Non-immune mouse IgG control. J: Non-immune rabbit IgG control. K: Cytokeratin 7 staining. L: Vimentin staining. M: von Willebrand factor staining. Black arrowheads denote endothelium staining, blue arrowheads denote decidual stromal cell staining and green arrowheads denote cytotrophoblast cell staining. Images were taken under 200X magnification. Scale bar denotes 100 μm.
Discussion
This study examined the mRNA expression levels and protein localisation of candidate maternal PE susceptibility genes in the maternal decidua. We showed differential expression of 7 out of our 8 candidate genes, ACVR1, INHBB, ERAP1, ERAP2, LNPEP, COL4A1 and COL4A2, in SPE compared with normotensive maternal decidua. The differentially expressed genes belong to several functional groups as described below. We also showed correlations of mRNA expression levels of susceptibility genes with various parameters of clinical severity and characteristics. No difference in protein localisation was qualitatively observed between SPE and normotensive decidua.
All genes significantly differentially expressed in the large set were also differentially expressed in the smaller gestation matched subset with the exception of two genes. The slight variation in the number of differentially expressed genes between the large set and the smaller subset is probably due to sample size. In the large set, gestational age, placental weight, infant weight and infant weight percentiles were all significantly different between SPE and normotensive patients, but not observed in the smaller subset. As SPE often results in preterm birth, fewer SPE patients go to term and this is thus reflected in the small sample size of the subset. Many published studies have similar differences when using term normotensives as the control comparison group for SPE studies [26-29]. Other studies used a preterm labour group to negate the potential effect of gestational age on gene expression [30-32]. Nevertheless, what triggers preterm labour is a confounder in itself and thus it is debatable as to whether these patients can truly be considered comparable normotensive controls. Additionally, none of the genes in our study were reported to change expression throughout gestation based on a comparative study conducted by Winn et al. [33], which compared gene expression between mid-gestation decidual tissues and term decidual tissues. A recent study, which analysed transcription profiles by merging data from different genome-wide platforms, showed that differential decidual gene expression observed in PE can be influenced by gestational age, highlighting the importance of gestational age matching between sample groups [34]. Their analyses did not show the genes examined in this study to be influenced by gestation [34]. All genes ACVR1, ERAP1, ERAP2, LNPEP and COL4A2 differentially expressed in the large set with the exception of INHBB and COL4A1, were also differentially expressed in the smaller subset, which was gestationally matched, suggesting gestation had no effect on our gene expression and that it is PE state that changes the gene expression.
ACVR1 and INHBB are genes encoding an activin type I receptor and an inhibin β subunit respectively involved in the activin and inhibin signalling pathways [35]. The increased decidual ACVR1 mRNA expression level in SPE is in contrast to two other activin receptor genes, ACVR1C and ACVR2A, which were previously found to be decreased in SPE [18]. The increased ACVR1 expression observed in our study may be a compensatory mechanism to counter-act the decreased expression of ACVR1C and ACVR2A. The increased INHBB expression in SPE decidua is novel. Studies have reported lower or no difference in maternal serum levels of inhibin B, of which INHBB is a subunit, in PE as compared normotensive pregnancies [36, 37]. The increased expression observed in our data may be tissue specific and thus not reflected in the circulating maternal blood.
The genes ERAP1, ERAP2 and LNPEP that showed significantly increased expression in both sample sets, belong to the M1 family of aminopeptidases, which are involved in regulating blood pressure and parturition through cleaving peptide hormones such as the angiotensins, vasopressin and oxytocin, as well as processing MHC I antigens [38]. ERAP1 is associated with chronic hypertension and a genetic variant shows reduced enzymatic activity, thus highlighting its role in blood pressure regulation [39, 40]. ERAP1 is also differentially expressed in essential hypertension [41]. ERAP2 shares similar functions with ERAP1 in blood pressure regulation and MHC I antigen presentation [42]. To date, fetal ERAP2 is associated with PE in an African American population and is altered in first trimester placentae that later develop PE [43, 44]. LNPEP activity is lower in PE pregnancies than in normotensive pregnancies [45]. Hence, altered enzymatic activity and expression may lead to compromised blood pressure regulation and contribute to the hypertension observed in PE. These studies and our work provide strong evidence for a role of the aminopeptidases in PE development.
The expression of collagen type IV α chain genes COL4A1 and COL4A2 are significantly increased in SPE decidua. Increased atypical expression of collagen may lead to poor basement membrane remodelling and result in shallow trophoblast invasion, a key feature of PE pregnancies [12]. The decidua and myometrium that trophoblast cells invade during early pregnancy are rich in collagen type IV, so increased collagen IV expression could alter the extracellular matrix properties [46]. Our data suggest improper collagen expression exists in PE that may contribute to the shallow trophoblast invasion. Additionally, fragments of the non-collagenous domain of COL4A1 and COL4A2, known as arresten and canstatin respectively, have anti-angiogenic effects such as increased apoptosis and decreased proliferation of endothelial cells [47]. In PE, serum levels of these fragments are elevated [48]. Increased collagen expression in the basement membrane would consequently lead to more degradation fragments and the anti-angiogenic effect of these fragments may thus contribute to the endothelial dysfunction observed in PE.
Our data show novel associations of susceptibility gene mRNA expression levels with various clinical parameters of severity and characteristics. Increased expression of the susceptibility genes suggests a compensatory mechanism occurs in PE. Compensation may or may not be sufficient to account for an inherent gene defect, which results in altered gene function such as receptor signalling or enzymatic activity. The associations of decreased expression of blood pressure regulator ERAP2 with increased systolic blood pressure and number of antihypertensive medications may be due to a genetic defect in ERAP2 resulting in decreased enzymatic activity. Hence as a compensatory mechanism, there is an increase in the enzyme levels to account for decreased activity. Women with an insufficient increase in enzyme levels are unable to successfully modulate blood pressure, resulting in a higher blood pressure and a greater number of antihypertensive medications required to regulate blood pressure. Nevertheless, the association with systolic blood pressure must take into consideration that most patients were treated with medications, particularly antihypertensives. However, the parameters of the onset of SPE and the delivery of the fetus are unmodifiable by treatments and do reflect the severity of the maternal condition. The negative correlations observed for onset and the delivery of fetus suggest that the increased expression of susceptibility genes may compensate insufficiently, resulting in a correlation with a more severe phenotype with earlier onset and delivery. Increased parity is protective of PE [2], and our data show increased expression of COL4A2 associates with increased parity, suggesting that increased expression of COL4A2 is protective and the higher expression observed in our SPE patients may be due to this compensatory mechanism. The exact mechanism that these genes work through in PE remains to be elucidated.
Most PE expression studies have looked at gene expression in the fetal placenta rather than in the maternal decidua [26, 29-31, 49, 50]. Nevertheless, several groups worldwide have conducted several gene expression studies in normotensive and PE decidual samples, mainly employing microarray analyses to identify differentially expressed genes [18, 27, 28, 32]. None of our investigated genes except INHA was significantly differentially expressed in these studies [32]. This is due to two main reasons. Firstly, PE covers a whole spectrum of clinical characteristics from mild to severe PE. The cohorts used in these studies and ours are different. We used a SPE cohort in contrast to a PE cohort used by Løset et al.[28] and while Winn et al.[32] used a SPE cohort, they used a different set of criteria to define SPE. Additionally, Winn et al.'s study [32] uses a preterm labour control group resulting in a different baseline for comparison. Hence, with cohort variability, it is unsurprising that little overlap exists between our study and those in the literature. There is also emerging evidence that PE may be several diseases appearing as one, thus different genes may contribute to the underlying aetiology [51, 52].
Secondly, the candidate genes were identified in the Australian/New Zealand families and individuals through bioinformatics prioritisation of positional candidate genes, and may be specific to these populations. Although possible, this is unlikely as at least one of these genes is also associated with PE in the Norwegian population [17]. Since these genes are susceptibility genes, not every patient would have all genes differentially expressed, and some variation is expected. Interestingly, all but one of our genes showed significantly increased expression in SPE and the one that did not also showed a similar trend to be increased, validating the methods used to identify these genes. Moreover, this expression study was undertaken in patients unrelated to those involved in the previous genetic studies and thus further supports the potential roles of these genes in the underlying aetiology of PE.
Conclusion
In summary, this study demonstrates differential decidual mRNA expression levels of candidate maternal PE susceptibility genes, identified using a strategy combining quantitative bioinformatics, transcriptional profiling in pregnancy-specific tissue and gene-centric SNP associations with PE. Thus, the gene expression data from our study adds more candidates to the many already identified [53]. The possible consequence of altered expression of these genes on successful placentation and the clinical course of severity remains to be further investigated.
Supplementary Material
Acknowledgements
The authors would like to thank the patients and clinical research midwives who were involved with sample donation and collection. This work was supported by funding from the Royal Women's Hospital, Parkville, Australia, the National Institutes of Health Grant HD049847 (to E.K. Moses and S.P. Brennecke). H.E.J. Yong was supported by the University of Melbourne's Melbourne International Fee Remission Scholarship and the Felix Meyer Scholarship in Obstetrics and Gynaecology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no conflict of interest.
Author Contributions
HEJ Yong: I declare that I participated in the substantial contribution to acquisition, analysis and interpretation of data, drafting and revision of the manuscript and that I have seen and approved the final version. I have no conflicts of interest.
P Murthi: I declare that I participated in the substantial contribution to conception and design, data analysis and interpretation, drafting the manuscript and critical revision for intellectual content and that I have seen and approved the final version. I have no conflicts of interest.
A Borg: I declare that I participated in the substantial contribution to acquisition and analysis of data, revision of the manuscript and that I have seen and approved the final version. I have no conflicts of interest.
B Kalionis: I declare that I participated in the substantial contribution to conception and design, data analysis and interpretation, drafting the manuscript and critical revision for intellectual content and that I have seen and approved the final version. I have no conflicts of interest.
EK Moses: I declare that I participated in the substantial contribution to conception and design, data analysis and interpretation, critical revision of the manuscript for intellectual content and that I have seen and approved the final version. I have no conflicts of interest.
SP Brennecke: I declare that I participated in the substantial contribution to conception and design, data analysis and interpretation, critical revision of the manuscript for intellectual content and that I have seen and approved the final version. I have no conflicts of interest.
RJ Keogh: I declare that I participated in the substantial contribution to conception and design, data analysis and interpretation, drafting the manuscript and critical revision for intellectual content and that I have seen and approved the final version. I have no conflicts of interest.
References
- 1.Roberts JM, Redman CW. Pre-eclampsia: more than pregnancy-induced hypertension. Lancet. 1993;341(8858):1447–51. doi: 10.1016/0140-6736(93)90889-o. [DOI] [PubMed] [Google Scholar]
- 2.Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-eclampsia. Lancet. 2010;376(9741):631–44. doi: 10.1016/S0140-6736(10)60279-6. [DOI] [PubMed] [Google Scholar]
- 3.Skjaerven R, Vatten LJ, Wilcox AJ, Ronning T, Irgens LM, Lie RT. Recurrence of pre-eclampsia across generations: exploring fetal and maternal genetic components in a population based cohort. BMJ. 2005;331(7521):877. doi: 10.1136/bmj.38555.462685.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mutze S, Rudnik-Schoneborn S, Zerres K, Rath W. Genes and the preeclampsia syndrome. J Perinat Med. 2008;36(1):38–58. doi: 10.1515/JPM.2008.004. [DOI] [PubMed] [Google Scholar]
- 5.Lie RT, Rasmussen S, Brunborg H, Gjessing HK, Lie-Nielsen E, Irgens LM. Fetal and maternal contributions to risk of pre-eclampsia: population based study. BMJ. 1998;316(7141):1343–7. doi: 10.1136/bmj.316.7141.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cnattingius S, Reilly M, Pawitan Y, Lichtenstein P. Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: a population-based Swedish cohort study. Am J Med Genet A. 2004;130A(4):365–71. doi: 10.1002/ajmg.a.30257. [DOI] [PubMed] [Google Scholar]
- 7.Chappell S, Morgan L. Searching for genetic clues to the causes of pre-eclampsia. Clin Sci (Lond) 2006;110(4):443–58. doi: 10.1042/CS20050323. [DOI] [PubMed] [Google Scholar]
- 8.Goddard KA, Tromp G, Romero R, Olson JM, Lu Q, Xu Z, Parimi N, Nien JK, Gomez R, Behnke E, Solari M, Espinoza J, Santolaya J, Chaiworapongsa T, Lenk GM, Volkenant K, Anant MK, Salisbury BA, Carr J, Lee MS. Vovis GF and Kuivaniemi H. Candidate-gene association study of mothers with pre-eclampsia, and their infants, analyzing 775 SNPs in 190 genes. Hum Hered. 2007;63(1):1–16. doi: 10.1159/000097926. [DOI] [PubMed] [Google Scholar]
- 9.Wilson ML, Goodwin TM, Pan VL, Ingles SA. Molecular epidemiology of preeclampsia. Obstet Gynecol Surv. 2003;58(1):39–66. doi: 10.1097/00006254-200301000-00022. [DOI] [PubMed] [Google Scholar]
- 10.Arngrimsson R, Siguroardottir S, Frigge ML, Bjarnadottir RI, Jonsson T, Stefansson H, Baldursdottir A, Einarsdottir AS, Palsson B, Snorradottir S, Lachmeijer AM, Nicolae D, Kong A, Bragason BT, Gulcher JR, Geirsson RT, Stefansson K. A genome-wide scan reveals a maternal susceptibility locus for pre-eclampsia on chromosome 2p13. Hum Mol Genet. 1999;8(9):1799–805. doi: 10.1093/hmg/8.9.1799. [DOI] [PubMed] [Google Scholar]
- 11.Harrison GA, Humphrey KE, Jones N, Badenhop R, Guo G, Elakis G, Kaye JA, Turner RJ, Grehan M, Wilton AN, Brennecke SP, Cooper DW. A genomewide linkage study of preeclampsia/eclampsia reveals evidence for a candidate region on 4q. Am J Hum Genet. 1997;60(5):1158–67. [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson MP, Fitzpatrick E, Dyer TD, Jowett JB, Brennecke SP, Blangero J, Moses EK. Identification of two novel quantitative trait loci for pre-eclampsia susceptibility on chromosomes 5q and 13q using a variance components-based linkage approach. Mol Hum Reprod. 2007;13(1):61–7. doi: 10.1093/molehr/gal095. [DOI] [PubMed] [Google Scholar]
- 13.Moses EK, Lade JA, Guo G, Wilton AN, Grehan M, Freed K, Borg A, Terwilliger JD, North R, Cooper DW, Brennecke SP. A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia, on chromosome 2. Am. J Hum Genet. 2000;67(6):1581–5. doi: 10.1086/316888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachmeijer AM, Arngrimsson R, Bastiaans EJ, Frigge ML, Pals G, Sigurdardottir S, Stefansson H, Palsson B, Nicolae D, Kong A, Aarnoudse JG, Gulcher JR, Dekker GA, ten Kate LP, Stefansson K. A genome-wide scan for preeclampsia in the Netherlands. Eur J Hum Genet. 2001;9(10):758–64. doi: 10.1038/sj.ejhg.5200706. [DOI] [PubMed] [Google Scholar]
- 15.Laivuori H, Lahermo P, Ollikainen V, Widen E, Haiva-Mallinen L, Sundstrom H, Laitinen T, Kaaja R, Ylikorkala O, Kere J. Susceptibility loci for preeclampsia on chromosomes 2p25 and 9p13 in Finnish families. Am J Hum Genet. 2003;72(1):168–77. doi: 10.1086/345311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenstad MH, Johnson MP, Roten LT, Aas PA, Forsmo S, Klepper K, East CE, Abraham LJ, Blangero J, Brennecke SP, Austgulen R, Moses EK. Genetic and molecular functional characterization of variants within TNFSF13B, a positional candidate preeclampsia susceptibility gene on 13q. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson MP, Roten LT, Dyer TD, East CE, Forsmo S, Blangero J, Brennecke SP, Austgulen R, Moses EK. The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum Genet. 2009;126(5):655–66. doi: 10.1007/s00439-009-0714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moses EK, Fitzpatrick E, Freed KA, Dyer TD, Forrest S, Elliott K, Johnson MP, Blangero J, Brennecke SP. Objective prioritization of positional candidate genes at a quantitative trait locus for pre-eclampsia on 2q22. Mol Hum Reprod. 2006;12(8):505–12. doi: 10.1093/molehr/gal056. [DOI] [PubMed] [Google Scholar]
- 19.Johnson MP, Brennecke SP, East CE, Goring HH, Kent JW, Jr., Dyer TD, Said JM, Roten LT, Iversen AC, Abraham LJ, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Laivuori H, Austgulen R, Blangero J, Moses EK. Genome-Wide Association Scan Identifies a Risk Locus for Preeclampsia on 2q14, Near the Inhibin, Beta B Gene. PLoS One. 2012;7(3):e33666. doi: 10.1371/journal.pone.0033666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown MA, Gallery EDM, Gatt SP, Leslie G, Robinson J. Management of Hypertension in Pregnancy - Executive Summary. Med J Australia. 1993;158(10):700–2. [PubMed] [Google Scholar]
- 21.Brown MA, Hague WM, Higgins J, Lowe S, McCowan L, Oats J, Peek MJ, Rowan JA, Walters BNJ. The detection, investigation and management of hypertension in pregnancy: executive summary. Australian and New Zealand Journal of Obstetrics and Gynaecology. 2000;40(2):133–8. doi: 10.1111/j.1479-828x.2000.tb01136.x. [DOI] [PubMed] [Google Scholar]
- 22.Tranquilli AL, Brown MA, Zeeman GG, Dekker G, Sibai BM. The definition of severe and early-onset preeclampsia. Statements from the International Society for the Study of Hypertension in Pregnancy (ISSHP). Pregnancy Hypertension: An International Journal of Women's Cardiovascular Health. 2013;3(1):44–7. doi: 10.1016/j.preghy.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Guaran RL, Wein P, Sheedy M, Walstab J, Beischer NA. Update of growth percentiles for infants born in an Australian population. Aust N Z J Obstet Gynaecol. 1994;34(1):39–50. doi: 10.1111/j.1479-828x.1994.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 24.Murthi P, Fitzpatrick E, Borg AJ, Donath S, Brennecke SP, Kalionis B. GAPDH, 18S rRNA and YWHAZ are suitable endogenous reference genes for relative gene expression studies in placental tissues from human idiopathic fetal growth restriction. Placenta. 2008;29(9):798–801. doi: 10.1016/j.placenta.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199(5):566, e1–11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herse F, Dechend R, Harsem NK, Wallukat G, Janke J, Qadri F, Hering L, Muller DN, Luft FC, Staff AC. Dysregulation of the circulating and tissue-based renin-angiotensin system in preeclampsia. Hypertension. 2007;49(3):604–11. doi: 10.1161/01.HYP.0000257797.49289.71. [DOI] [PubMed] [Google Scholar]
- 28.Loset M, Mundal SB, Johnson MP, Fenstad MH, Freed KA, Lian IA, Eide IP, Bjorge L, Blangero J, Moses EK, Austgulen R. A transcriptional profile of the decidua in preeclampsia. Am J Obstet Gynecol. 2011;204(1):84, e1–27. doi: 10.1016/j.ajog.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitras V, Paulssen RH, Gronaas H, Leirvik J, Hanssen TA, Vartun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30(5):424–33. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18(1):46–56. doi: 10.1177/1933719110374115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta.;2007;28(5-6):487–97. doi: 10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K, Kramer A, Rumer KK, Haimov-Kochman R, Yeh RF, Overgaard MT, Varki A, Oxvig C, Fisher SJ. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150(1):452–62. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, Feng KT, Bernlohr DA, McDonagh S, Pereira L, Sali A, Fisher SJ. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148(3):1059–79. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- 34.Lian IA, Langaas M, Moses E, Johansson Å. Differential Gene Expression at the Maternal-Fetal Interface in Preeclampsia Is Influenced by Gestational Age. PLoS One. 2013;8(7):e69848. doi: 10.1371/journal.pone.0069848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pangas SA, Woodruff TK. Activin signal transduction pathways. Trends Endocrinol Metab. 2000;11(8):309–14. doi: 10.1016/s1043-2760(00)00294-0. [DOI] [PubMed] [Google Scholar]
- 36.Petraglia F, Luisi S, Benedetto C, Zonca M, Florio P, Casarosa E, Volpe A, Bernasconi S, Genazzani AR. Changes of dimeric inhibin B levels in maternal serum throughout healthy gestation and in women with gestational diseases. J Clin Endocrinol Metab. 1997;82(9):2991–5. doi: 10.1210/jcem.82.9.4241. [DOI] [PubMed] [Google Scholar]
- 37.Yair D, Eshed-Englender T, Kupferminc MJ, Geva E, Frenkel J, Sherman D. Serum levels of inhibin B, unlike inhibin A and activin A, are not altered in women with preeclampsia. Am J Reprod Immunol. 2001;45(3):180–7. doi: 10.1111/j.8755-8920.2001.450310.x. [DOI] [PubMed] [Google Scholar]
- 38.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751(1):9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto N, Nakayama J, Yamakawa-Kobayashi K, Hamaguchi H, Miyazaki R, Arinami T. Identification of 33 polymorphisms in the adipocyte-derived leucine aminopeptidase (ALAP) gene and possible association with hypertension. Hum Mutat. 2002;19(3):251–7. doi: 10.1002/humu.10047. [DOI] [PubMed] [Google Scholar]
- 40.Goto Y, Hattori A, Ishii Y, Tsujimoto M. Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (ALAP)/ER-aminopeptidase-1. FEBS Lett. 2006;580(7):1833–8. doi: 10.1016/j.febslet.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Korkor MT, Meng FB, Xing SY, Zhang MC, Guo JR, Zhu XX, Yang P. Microarray analysis of differential gene expression profile in peripheral blood cells of patients with human essential hypertension. Int J Med Sci. 2011;8(2):168–79. doi: 10.7150/ijms.8.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M. Human leukocyte-derived arginine aminopeptidase - The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem. 2003;278(34):32275–83. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 43.Hill LD, Hilliard DD, York TP, Srinivas S, Kusanovic JP, Gomez R, Elovitz MA, Romero R, Strauss JF., 3rd. Fetal ERAP2 variation is associated with preeclampsia in African Americans in a case-control study. BMC Med Genet. 2011;12:64. doi: 10.1186/1471-2350-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Founds SA, Conley YP, Lyons-Weiler JF, Jeyabalan A, Hogge WA, Conrad KP. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30(1):15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landau R, Laverriere A, Bischof P, Irion O, Morales M, Cohen M. Alteration of circulating Placental Leucine Aminopeptidase (P-LAP) activity in preeclampsia. Neuro Endocrinol Lett. 2010;31(1):63–6. [PubMed] [Google Scholar]
- 46.Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266(5190):1508–18. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 47.Mundel TM, Kalluri R. Type IV collagen-derived angiogenesis inhibitors. Microvasc Res. 2007;74(2-3):85–9. doi: 10.1016/j.mvr.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bieglmayer C, Hofer G. Radioimmunoassay for immunoreactive non-collagenous domain of type IV collagen (NC1) in serum: normal pregnancy and preeclampsia. J Clin Chem Clin Biochem. 1989;27(3):163–7. doi: 10.1515/cclm.1989.27.3.163. [DOI] [PubMed] [Google Scholar]
- 49.Hoegh AM, Borup R, Nielsen FC, Sorensen S, Hviid TV. Gene expression profiling of placentas affected by pre-eclampsia. J Biomed Biotechnol. 2010;2010:787545. doi: 10.1155/2010/787545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pang ZJ, Xing FQ. Expression profile of trophoblast invasion-associated genes in the pre-eclamptic placenta. Br J Biomed Sci. 2003;60(2):97–101. doi: 10.1080/09674845.2003.11783682. [DOI] [PubMed] [Google Scholar]
- 51.Ness RB, Roberts JM. Heterogeneous causes constituting the single syndrome of preeclampsia: a hypothesis and its implications. Am J Obstet Gynecol. 1996;175(5):1365–70. doi: 10.1016/s0002-9378(96)70056-x. [DOI] [PubMed] [Google Scholar]
- 52.Vatten LJ, Skjaerven R. Is pre-eclampsia more than one disease? Bjog-Int J Obstet Gy. 2004;111(4):298–302. doi: 10.1111/j.1471-0528.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 53.Founds SA. Bridging global gene expression candidates in first trimester placentas with susceptibility loci from linkage studies of preeclampsia. J Perinat Med. 2011;39(4):361–8. doi: 10.1515/jpm.2011.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.