Abstract
In obesity and type 2 diabetes, Glut4 glucose transporter expression is decreased selectively in adipocytes1. Adipose-specific knockout or overexpression of Glut4 alters systemic insulin sensitivity2. Here we show, using DNA array analyses, that nicotinamide N-methyltransferase (Nnmt) is the most strongly reciprocally regulated gene when comparing gene expression in white adipose tissue (WAT) from adipose-specific Glut4-knockout or adipose-specific Glut4-overexpressing mice with their respective controls. NNMT methylates nicotinamide (vitamin B3) using S-adenosylmethionine (SAM) as a methyl donor3,4. Nicotinamide is a precursor of NAD+, an important cofactor linking cellular redox states with energy metabolism5. SAM provides propylamine for polyamine biosynthesis and donates a methyl group for histone methylation6. Polyamine flux including synthesis, catabolism and excretion, is controlled by the rate-limiting enzymes ornithine decarboxylase (ODC) and spermidine–spermine N1-acetyltransferase (SSAT; encoded by Sat1) and by polyamine oxidase (PAO), and has a major role in energy metabolism7,8. We report that NNMT expression is increased in WAT and liver of obese and diabetic mice. Nnmt knockdown in WAT and liver protects against diet-induced obesity by augmenting cellular energy expenditure. NNMT inhibition increases adipose SAM and NAD+ levels and upregulates ODC and SSAT activity as well as expression, owing to the effects of NNMT on histone H3 lysine 4 methylation in adipose tissue. Direct evidence for increased polyamine flux resulting from NNMT inhibition includes elevated urinary excretion and adipocyte secretion of diacetylspermine, a product of polyamine metabolism. NNMT inhibition in adipocytes increases oxygen consumption in an ODC-, SSAT- and PAO-dependent manner. Thus, NNMT is a novel regulator of histone methylation, polyamine flux and NAD+-dependent SIRT1 signalling, and is a unique and attractive target for treating obesity and type 2 diabetes.
Nnmt is expressed at high levels in adipose tissue and liver and at lower levels in other organs3,4. Nnmt is increased in multiple cancers9,10, neuro-degenerative diseases11, and also in obesity and diabetes12–14. For example, Nnmt expression is increased in adipocytes of obese, compared to non-obese Pima Indians12. Metabolomic analyses reveal elevated levels of urinary N1-methylnicotinamide, the product of NNMT, in humans with type 2 diabetes, and in db/db mice and obese Zucker rats13, indicating increased NNMT activity in obesity and type 2 diabetes. Quantitative trait loci mapping in mice suggests a causative role of Nnmt in type 2 diabetes14. Therefore, we investigated the regulation and roles of NNMT in obesity and type 2 diabetes.
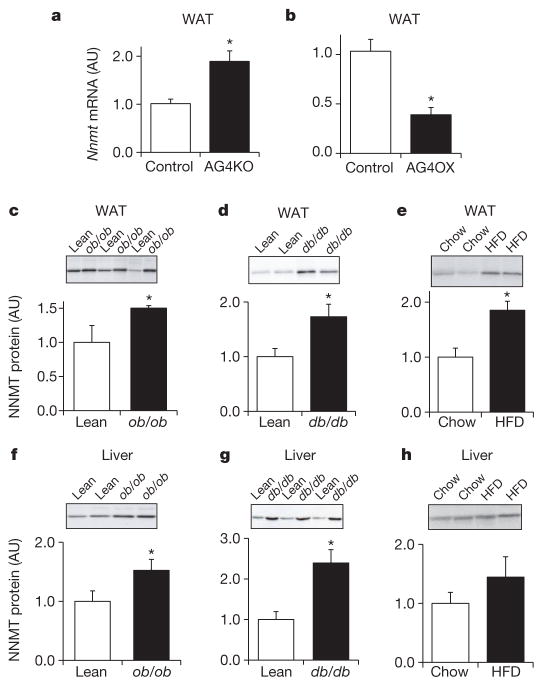
Adipose Nnmt messenger RNA levels were increased twofold in insulin-resistant adipose-specific Glut4-knockout mice (Fig. 1a), and reduced by 62% in insulin-sensitive adipose-specific Glut4-overexpressing mice (Fig. 1b). NNMT protein was increased 1.5- to 2-fold in WAT of ob/ob, db/db mice and high-fat diet (HFD)-fed mice (Fig. 1c–e), compared with lean, insulin-sensitive controls. Hepatic NNMT protein levels were increased in ob/ob and db/db mice (Fig. 1f, g) and tended to be higher in HFD-fed mice (Fig. 1h). Thus, NNMT is upregulated in adipose tissue and liver of mouse models of obesity and insulin resistance.
Figure 1. NNMT expression is increased in WAT and liver of obese and insulin-resistant mice.
a, b, Nnmt mRNA expression normalized by cyclophilin in WAT of adipose-specific Glut4 knockout (AG4KO) mice and aP2-Cre controls (n =4 per group) (a) and adipose-specific Glut4 overexpressing (AG4OX) and wild-type littermate controls (n =6 per group) (b). c–e, NNMT protein levels in WAT of ob/ob mice (n =8) and lean controls (n =4) (c); db/db mice and lean controls (n =7 per group) (d), and high-fat diet (HFD)-fed (n =6) and chow-fed mice (n =7) (e). f–h, NNMT protein levels in liver of ob/ob mice (n =9) and lean controls (n =6) (f); db/db mice and lean controls (n =7 per group) (g); and HFD-fed and chow-fed mice (n =6 per group) (h). Actin was used as a control for western blot analysis and the levels were not different between lean and obese mice. AU, arbitrary units. Error bars, ±s.e.m; *P <0.05.
Adipose and hepatic Nnmt expression varies highly among 25 different mouse strains15 (Supplementary Fig. 1). Adipose Nnmt expression is high in obesity-prone strains and low in obesity-resistant strains15–17 (Supplementary Fig. 1a). In contrast, liver Nnmt expression does not parallel the propensity for obesity (Supplementary Fig. 1b). Adipose Nnmt expression correlates highly with per cent fat mass in diet-induced obesity across 20 different mouse strains18, and with expression of retinol-binding protein 4 (Rbp4) (r =0.90, P <0.0001 by Pearson correlation coefficient, two-tailed test), an adipokine that contributes to insulin resistance2 (Supplementary Fig. 2a, b). Thus, we proposed that elevated NNMT levels in adipose tissue and/or liver may have a causative role in insulin resistance and obesity.
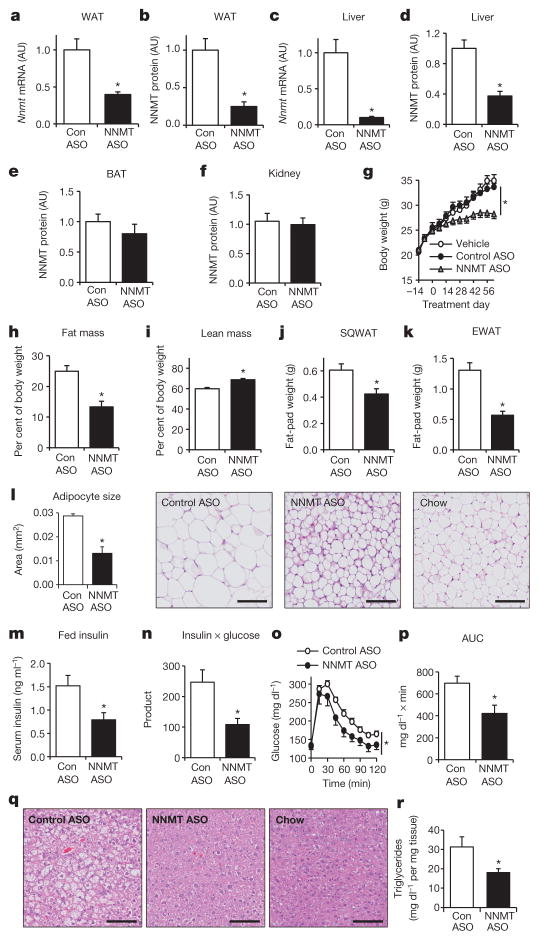
We knocked down Nnmt with antisense oligonucleotides (ASOs) in HFD-fed mice. ASOs regulate gene expression primarily in liver and fat19,20. Treating HFD-fed mice with Nnmt ASO reduced Nnmt mRNA and NNMT protein by 60 to 75% in WAT and 60% (protein) to 90% (mRNA) in liver (Fig. 2a–d), but not in brown adipose tissue and kidney (Fig. 2e, f). Serum transaminases and creatinine were normal, indicating no hepaticor renal toxicity with ASO treatment (Supplementary Fig. 3a–c).
Figure 2. Nnmt knockdown prevents diet-induced obesity and insulin resistance.
a–f, Knockdown efficiency of Nnmt-ASO. mRNA expression was normalized by cyclophilin and protein levels were corrected with actin levels: Nnmt mRNA (a) and NNMT protein in WAT (b); Nnmt mRNA (c) and NNMT protein in liver (d); NNMT protein in brown adipose tissue (BAT) (e) and kidney (f). g, Body weights of C57BL/6 mice fed a high-fat diet and treated with Nnmt ASO, control ASO, or vehicle (saline) for 8 weeks. h, Fat mass as a percentage of body weight. i, Lean mass as a percentage of body weight. j, Subcutaneous WAT (SQWAT) fat-pad weights. k, Epididymal WAT (EWAT) fat-pad weights. l, Epididymal adipocyte cross-sectional area and haematoxylin and eosin (H&E)-stained sections of SQWAT. m, Serum insulin levels. n, Glucose × insulin product (ng ml−1 × mg dl−1) in the fed state. o, Intraperitoneal glucose tolerance test. p, Area under the curve (AUC) of the glucose tolerance. q, H&E stain of liver sections of HFD-fed Nnmt-ASO-and control-ASO-treated mice, and of chow-fed mice. r, Hepatic triglyceride levels in Nnmt- and control-ASO-treated mice. The scale bars in l and q represent 100 μm; n =8 per group for a–p, n =13 per group for r. AU, arbitrary units. Error bars, ±s.e.m.; *P <0.05.
Nnmt knockdown in adipose tissue and liver protected mice from diet-induced obesity (Fig. 2g), causing a 47% reduction in relative fat mass and a 15% increase in relative lean mass (Fig. 2h, i). Free body water and hepatic glycogen content were not different (not shown) between control- and Nnmt-ASO-treated mice. Subcutaneous and epididymal fat-pad weights were lower in Nnmt-knockdown mice than in controls (Fig. 2j, k), largely owing to reduced adipocyte size (44% reduced area, Fig. 2l, and 70% reduced volume, Supplementary Fig. 4). Insulin sensitivity was enhanced, as evidenced by approximately 50 to 60% lower serum insulin levels and glucose-insulin product (Fig. 2m, n). Nnmt knockdown also improved glucose tolerance (Fig. 2o, p), prevented HFD-induced hepatic steatosis(Fig. 2q, r), and decreased serum triglycerides and free fatty acids (Supplementary Fig. 5a, b). Thus, Nnmt knockdown in WAT and liver protects against diet-induced obesity and its deleterious metabolic consequences.
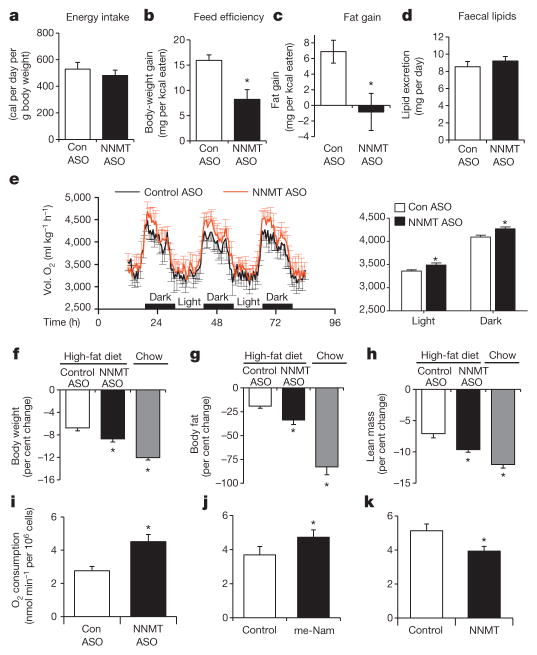
We sought to determine whether the leanness with Nnmt knockdown is due to reduced energy intake or increased energy expenditure. The NNMT substrate, nicotinamide, at pharmacological doses may suppress food intake and cause weight loss in rats21. However, Nnmt-ASO-treated mice consumed the same amount of calories as controls (Fig. 3a). Food intake was measured before body weights diverged, in order to eliminate confounding by body-weight differences. Feed efficiency (body weight change per kcal of food eaten) was reduced by approximately 50% in Nnmt-knockdown mice (Fig. 3b). Although control-ASO-treated mice gained about 7 mg of fat per kcal of food intake, Nnmt-ASO-treated mice gained no fat (Fig. 3c). Thus, body weight and fat gain were disproportionally low for the calorie intake in Nnmt-ASO-treated compared with control-ASO-treated mice. Faecal lipid excretion was not changed between control- and Nnmt-ASO-treated mice (Fig. 3d). Therefore, increased energy expenditure rather than decreased food intake or steatorrhoea explains the leanness in Nnmt-knockdown mice. This was further demonstrated by increased oxygen consumption (expressed per kg body weight or per mouse) during comprehensive laboratory animal monitoring system (CLAMS) analysis performed before body weights started to diverge (Fig. 3e and Supplementary Fig. 6a). Comparison of mice with similar body weight showed that Nnmt-ASO-treated mice had higher energy expenditure than control-ASO-treated mice (Supplementary Fig. 6b). Energy expenditure per gram of fat mass, but not per gram of lean mass was increased in Nnmt-ASO-treated mice compared with control-ASO-treated mice (Supplementary Fig. 6c, d). There was no measurable difference in food intake expressed per mouse or per g body weight, locomotor activity or respiratory exchange ratio (RER) (Supplementary Fig. 7a–d). As a third approach to assess the energy expenditure, we measured body weight and body composition before and after an overnight fast. In the absence of energy intake, increased loss of body fat indicates increased energy expenditure. After fasting, Nnmt-knockdown mice on HFD lost more body weight, body fat and lean mass than control-ASO-treated mice, and the response resembled that of chow-fed lean mice (Fig. 3f–h). Core body temperature, BAT weight, and uncoupling protein-1 levels in BAT (Supplementary Fig. 8a–c) were unchanged. Thus, Nnmt knockdown in WAT and liver increases energy expenditure, and this is likely to be independent of BAT-induced energy dissipation or heat production.
Figure 3. NNMT regulates energy expenditure.
a, Energy intake of HFD-fed control-ASO- and Nnmt-ASO-treated mice. b, Feed efficiency (body-weight gain per kilocalorie (kcal) consumed) of HFD-fed control-ASO- and Nnmt–ASO-treated mice. c, Fat mass gain per kcal consumed in HFD-fed mice treated with Nnmt ASO or control ASO. n =12 per group for a–c. d, Faecal lipid excretion (n =10 per group). e, Oxygen consumption (Vol. O2) measured by CLAMS (n =7 per group). f–h, Effects of a 16-h fast on body weight (f), fat mass (g), and lean body mass (n =8 per group) (h). i–k, Oxygen consumption in 3T3-L1 adipocytes transfected with control or Nnmt ASO (n =6 per group) (i), treated with 10 mM N1-methylnicotinamide (me-Nam) (n =6 per group) (j), or transfected with Nnmt cDNA (n =10 per group) (k). Error bars, ±s.e.m.; *P <0.05.
We asked whether NNMT regulates energy expenditure in a cell-autonomous manner. Nnmt knockdown in adipocytes led to a 60% increase in oxygen consumption (Fig. 3i). NNMT inhibition by N1-methylnicotinamide—the NNMT reaction product and a specific, potent inhibitor of NNMT at pharmacological doses3—also increased oxygen consumption (Fig. 3j). Conversely, Nnmt overexpression decreased oxygen consumption (Fig. 3k) in adipocytes. Similar results were obtained in cultured hepatoma cells (Supplementary Fig. 9). Therefore, NNMT regulates oxygen consumption in a cell-autonomous fashion in both adipocytes and hepatocytes.
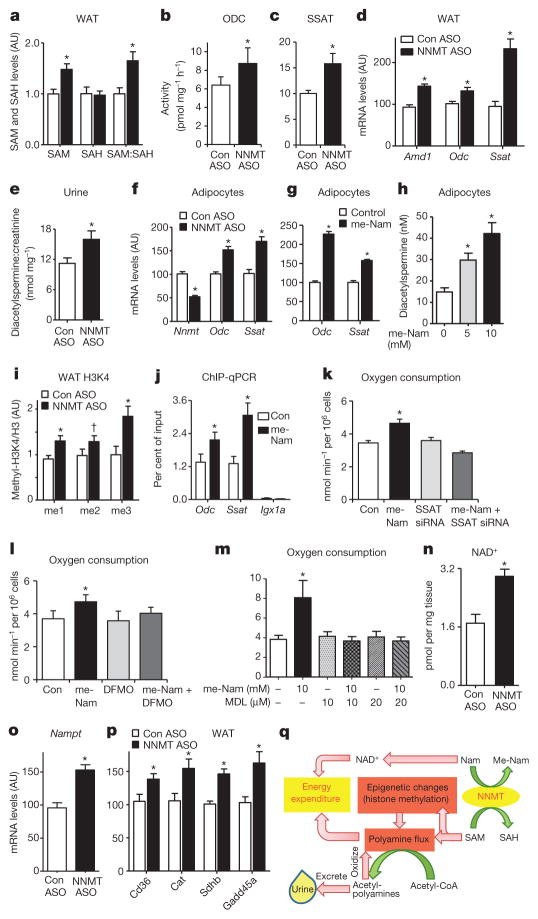
We sought to determine the molecular mechanisms for increased energy expenditure with NNMT inhibition. NNMT methylates nicotinamide using SAM as a methyl donor and generates S-adenosylhomocysteine (SAH). Adipose SAM, and the SAM:SAH ratio were increased by 50% in Nnmt-ASO-treated mice (Fig. 4a). In liver, SAM was not changed, but the SAM:SAH ratio was increased by 2.2-fold owing to a 48% reduction of SAH levels in Nnmt-ASO-treated mice (Supplementary Fig. 10). The unchanged hepatic SAM with Nnmt knockdown contrasts with knockdown of glycine N-methyltransferase (Gnmt), a dominant methyltransferase that accounts for >1% of total cytosolic protein in liver. Gnmt knockdown increases SAM by 40-fold and induces hepatic steatosis and fibrosis22. Hepatic Nnmt knockdown did not cause fibrosis as evidenced by normal expression of tissue inhibitor of metal-loproteinase 1 (Timp1) and collagen type 1 (Colla1), markers for hepatic fibrosis (Supplementary Fig. 11a, b).
Figure 4. NNMT regulates SAM and NAD+ pathways in adipose tissue.
a, Adipose S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH) and SAM:SAH ratio measured by LC–MS/MS (n =8 for control-ASO-treated mice; n =12 for Nnmt-ASO-treated mice). b, c, ODC (b) and SSAT (c) activity (n =10 per group). d, Amd1, Odc and Ssat mRNA expression in adipose tissue of Nnmt-ASO- and control-ASO-treated mice (n =12 per group). e, Urinary diacetylspermine:creatinine ratio (n =22 for control-ASO-treated mice; n =29 for Nnmt-ASO-treated mice). f, g, Odc and Ssat mRNA levels in 3T3-L1 adipocytes with Nnmt knockdown (n =9 per group) (f) and N1-methylnicotinamide (me-Nam) (g) treatment (n =6 per group). h, Diacetylspermine secretion from 3T3-L1 adipocytes treated with me-Nam (n =10 per group). i, Expression of mono-, di- and tri-methylation of lysine histone 3 (H3K4) normalized to total H3 levels in adipose tissue (n =8 per group). j, H3K4me2 occupancy on Odc, Ssat genes and an open reading frame free region (Igx1a) as a negative control in adipocytes measured by ChIP-qPCR (n =12 per group). k–m, Oxygen consumption in adipocytes transfected with control or Ssat siRNA, and treated with or without 10 mM me-Nam (n =6 per group) (k); treated with 10 mM me-Nam with or without 5 mM difluoromethylornithine (DFMO), a specific ODC inhibitor (n =6 per group) (l); or treated with 10 mM me-Nam with or without MDL72527, a specific PAO inhibitor (n =6 per group) (m). n, NAD+ levels (n =7 Con-ASO; n =12 Nnmt ASO). o, mRNA levels of nicotinamide phosphoribosyltransferase (Nampt) (n =11 per group). p, mRNA levels of SIRT1 target genes: Cd36, catalase (Cat), succinate dehydrogenase B (SdhB) and growth arrest and DNA-damage-inducible protein (Gadd45a) in adipose tissue of NNMT-ASO- and control-ASO treated mice (n =11 per group). Error bars, ± s.e.m., *P <0.05, †P =0.06. q, Model of NNMT-regulated energy expenditure in adipocytes. NNMT methylates nicotinamide (Nam), a precursor of NAD+, using SAM as a methyl donor. SAM regulates polyamine flux by providing substrates and modulating histone methylation. Polyamine flux utilizes acetyl-CoA to generate acetyl-polyamines, which are oxidized or excreted in the urine. Taken together, this results in adipose metabolic substrate consumption and loss coupled with systemic alteration of energy expenditure.
SAM has two major functions: first, providing propylamine groups for polyamine biosynthesis; and second, donating methyl groups to substrates including histones6. Polyamines (putrescine, spermine and spermidine) are organic polycations that are essential for multiple cellular functions affecting cell growth, cancer and ageing23. Polyamine metabolism is tightly controlled (Supplementary Fig. 12)23. Synthesis is controlled by ODC producing putrescine, and by adenosylmethionine decarboxylase (AMD1) providing decarboxylated SAM for the synthesis of spermidine and spermine. Catabolism is controlled by spermidine–spermine N1-acetyltransferase (SSAT), which acetylates spermidine and spermine using acetyl-CoA as a substrate. The acetylated products including N1-acetylspermine, N1, N12-diacetylspermine, and N1-acetylspermidine, are either oxidized by PAO or excreted intact in urine. Polyamine flux has a major role in energy homeostasis. Genetic knockout of SSAT results in increased diet-induced obesity and transgenic SSAT overexpression causes leanness owing to altered energy expenditure7,8,24. As Nnmt knockdown increases adipose SAM levels (Fig. 4a), we reasoned that this may cause a ‘substrate shunt’ of SAM from the NNMT reaction to polyamine flux leading to increased energy expenditure in Nnmt-ASO-treated mice (Fig. 3). In support of this, Nnmt expression correlates negatively with Odc and Ssat expression in adipose tissue, but not in liver in 25 different mouse strains15 (Supplementary Fig. 13a–d).
Nnmt-ASO treatment in vivo augmented adipose ODC and SSAT activity (Fig. 4b, c) and mRNA expression (Fig. 4d). The expression of adenosylmethionine decarboxylase Amd1, which provides decarboxylated SAM for spermidine and spermine synthesis, was also increased (Fig. 4d). In liver, Nnmt ASO increased ODC, but not SSAT activity, or Odc or Ssat expression (Supplementary Fig. 14a–d). ODC and SSAT activation drives polyamine flux, which consumes the metabolic substrate, acetyl-CoA, for polyamine acetylation7. Consistent with reduced acetyl-CoA availability for lipogenesis, acetyl-CoA carboxylase 1 expression and fatty acid synthase expression and activity were decreased in adipose tissue of Nnmt-ASO-treated mice (Supplementary Fig. 15a–c). In addition, adipose Nnmt knockdown also decreased ATP, increased the AMP:ATP ratio and enhanced AMPK threonine 172 phosphorylation (Supplementary Fig. 16a–d). Increased urinary diacetylspermine provides direct evidence for enhanced adipose polyamine flux in Nnmt-ASO-treated mice (Fig. 4e). Furthermore, reduction in Nnmt increased Odc and Ssat expression in cultured adipocytes (Fig. 4f, g). N1-methylnicotinamide treatment promoted diacetylspermine secretion from adipocytes in a dose-dependent manner (Fig. 4h), complementing the increased urinary diacetylspermine excretion in Nnmt-ASO-treated mice (Fig. 4e). These data convincingly show that NNMT directly regulates polyamine flux in adipocytes.
In addition to being a substrate for polyamine metabolism, SAM is a methyl donor for numerous methylation reactions including histone methylation, which is important for transcriptional regulation6. We measured eight types of histone methylation; the amounts of mono-, di- or tri-methylated lysine 4 of histone H3 (H3K4) were increased in adipose tissue of Nnmt-ASO-treated mice (Fig. 4i). Chromatin immunoprecipitation followed by quantitative PCR (ChIP–qPCR) revealed enrichment of methylated H3K4 on Odc and Ssat genes with NNMT inhibition in adipocytes (Fig. 4j). Thus, NNMT inhibition modifies histone methylation and increases Odc and Ssat expression, leading to activation of poly-amine flux.
To determine whether the increased energy expenditure with NNMT inhibition (Fig. 3) is polyamine-dependent, we used N1-methylnicotinamide to inhibit NNMT activity in adipocytes. This induced oxygen consumption (Fig. 4k–m). Knocking down Ssat (Supplementary Fig. 17), inhibiting ODC activity8 or blocking PAO activity abolished N1-methylnicotinamide-induced oxygen consumption (Fig. 4k–m). Thus, NNMT inhibition in adipocytes autonomously enhances oxygen consumption, and this depends on polyamine flux.
The other substrate of the NNMT reaction is nicotinamide, an NAD+ precursor. Nnmt ASO treatment did not alter adipose and hepatic nicotinamide levels (Supplementary Fig. 18a, b). This is likely to be because nicotinamide is metabolized to NAD+ in a salvage pathway, which is controlled by the rate-limiting enzymes nicotinamide phosphoribosyl-transferase (NAMPT) and three isoforms of nicotinamide mononucleotide adenylyltransferase (NMNAT1, NMNAT2 and NMNAT3)5. Nnmt knockdown increased NAD+ levels per mg of adipose tissue (Fig. 4n) and the expression of Nampt (Fig. 4o), cytosolic Nmnat2 and mitochondrial Nmnat3, but not nuclear Nmnat1 (Supplementary Fig. 19). NAD+ is a cofactor of SIRT1 deacetylase activity, and SIRT1 affects energy metabolism5. Expression of SIRT1 target genes, including Cd36, catalase (Cat), succinate dehydrogenase B (Sdhb) and growth-arrest and DNA-damage-inducible protein, were increased in adipose tissue of Nnmt-ASO-treated mice25–28 (Fig. 4p), consistent with SIRT1 activation. As Nnmt-ASO-treated mice are leaner, NAD+ levels per adipocyte may not be as elevated as per mg of adipose tissue. However, the increased gene expression upstream and downstream of NAD+ strongly supports enhanced NAD+ flux in Nnmt-ASO-treated adipocytes. In liver, Nnmt knockdown did not alter NAD+ levels or the expression of Nampt, Nmnat1, Nmnat2 or Nmnat3 (Supplementary Fig. 20a, b). In spite of this, Cd36, Cat and SdhB expression was decreased (Supplementary Fig. 20c), indicating reduced SIRT1 activity. Consistent with this, PGC-1α acetylation was enhanced in liver with Nnmt knockdown29 (Supplementary Fig. 20d). The decreased hepatic Sirt1 expression (Supplementary Fig. 20e) may contribute to the reduced SIRT1 activity with Nnmt knockdown. Despite the changes in SIRT1 and PGC-1α activity, hepatic Pck1 and G6pc expression were not altered by Nnmt ASO treatment (Supplementary Fig. 21).
We discovered that NNMT is a novel regulator of adiposity and energy expenditure. This involves modulating adipose SAM and NAD+, two fundamental metabolites for energy metabolism. NNMT also regulates hepatic energy metabolism (Supplementary Fig. 9), but most likely with different mechanisms, as Nnmt knockdown did not alter hepatic SAM and NAD+ levels. In adipocytes, SAM provides substrate for polyamine synthesis, and also modulates Odc and Ssat expression by modifying H3K4 methylation (Fig. 4q). SAM availability may alter histone methylation in a methylation-site-specific manner rather than affecting global histone methylation10. For example, threonine-regulated SAM specifically affects H3K4me3 in mouse embryonic stem cells30. NNMT-regulated H3K4 methylation may have broader effects on gene expression including effects associated with cancer10 and dementia, conditions in which NNMT activity is enhanced10,11.
Activation of adipose polyamine flux causes leanness by catalysing polyamine acetylation to generate acetylpolyamines using acetyl-coA as a metabolic substrate7. Acetylpolyamines are oxidized in a futile cycle or excreted in the urine, thereby reducing acetyl-CoA in cells (Fig. 4q and Supplementary Fig. 12). Although urinary excretion of acetyl-polyamines may not cause significant total-body calorie loss, the metabolic substrate consumption or loss in adipocytes seems to impact systemic energy expenditure. This is clearly demonstrated by leanness and increased energy expenditure in adipose Ssat transgenic mice24. Promoting consumption or loss of adipose acetyl-CoA as a metabolic substrate in the form of acetylpolyamines may be a novel strategy for obesity treatment (Fig. 4q).
NNMT regulates SAM and NAD+ primarily in adipose tissue, but not in liver, which has redundant regulatory pathways for SAM and NAD+ metabolism. Hepatic SAM is largely regulated by glycine N-methyltransferase22 and by methionine, an abundant SAM precursor in liver. When methionine is abundant, NNMT regulates only SAH, not SAM10. Liver NAD+ synthesis is controlled by a salvage pathway using nicotinamide as a precursor and a de novo pathway using tryptophan5. In contrast, adipose tissue barely expresses the key enzymes in the de novo NAD+ synthesis pathway (data not shown). Therefore, adipose NAD+ synthesis primarily relies on the salvage pathway using nicotinamide, for which NNMT is the only catabolic enzyme. The adipose-selective effects of NNMT inhibition on SAM and NAD+ levels are advantageous for drug development because increased SAM in liver causes hepatic steatosis and fibrosis22, and increased NAD+ may enhance hepatic gluconeogenesis29.
In summary, NNMT is a unique regulator of adiposity by directly altering NAD+ and SAM, which affect histone methylation, polyamine flux and SIRT1 signalling. NNMT inhibition leads to metabolic substrate consumption or loss from adipocytes coupled with increased energy expenditure in a cell-autonomous manner (Fig. 4q). These unique features render NNMT an attractive target for treating obesity and type 2 diabetes.
METHODS
Mice
Adipose-specific Glut4-knockout and Glut4-overexpressing mice were described previously31,32. All other mice were purchased from Jackson Laboratories or Charles River Laboratories. For measurements of Nnmt expression in adipose tissue and liver, the ob/ob and db/db mice were killed at 10 to 12 weeks of age. High-fat diet (HFD)-fed male mice in C57BL/6 background were treated with a HFD for 12 weeks starting at 6 to 8 weeks of age. For Nnmt ASO experiments, mice were fed standard chow providing 17% calories from fat (LabDiet Formulab 5008) or a HFD providing 55% calories from fat (Harlan Teklad, TD93075) starting at 6 to 7 weeks of age. Nnmt and control ASOs were injected at a dose of 37.5 mg kg–1 intraperitoneally twice per week starting 2 weeks after the initiation of HFD feeding. Mouse studies were conducted in accordance with federal guidelines and were approved by the Beth Israel Deaconess Medical Centre (BIDMC) Institutional Animal Care and Use Committee.
Selection of mouse Nnmt ASO
Rapid-throughput screens were performed in primary hepatocytes to identify mouse Nnmt antisense oligonucleotides. In brief, 80 ASOs were designed to the Nnmt mRNA sequence and tested in primary hepatocytes to identify Nnmt antisense oligonucleotides. Nnmt gene knockdown was screened by quantitative PCR. Eight ASOs were selected and further characterized in a dose–response screen in vivo. Two Nnmt ASOs with greatest effects on Nnmt knockdown in liver and adipose tissue with no toxicity to liver and kidney were used to treat mice on a HFD. Treatment with either of the Nnmt ASOs resulted in leanness compared with a control-ASO-treated group. The most potent Nnmt ASO (GAAATGAACCAGCAGGCCTT) was chosen for subsequent studies. A control ASO (ISIS 425851), which has no complementarity to any known gene sequence, was used for the control group. Nnmt and control ASOs have a uniform phosphorothioate backbone and a 20-base chimaeric design with a 2′-O-(methoxy)-ethyl (2′-MOE) modification on the first five and the last five bases. This modification enhances their binding affinity to complementary sequences and their resistance to the action of nucleases. ASOs may down regulate their target genes through an RNase H-dependent cleavage mechanism or a non-RNA-degrading mechanism by blocking translation of mRNA. Two pathways may be involved ASO entry into cells: first, a productive pathway using a vesicular transport system; and second, a non-productive pathway that results in accumulation of ASOs in lysosomal structures19.
CLAMS
Energy expenditure was evaluated using a Comprehensive Lab Animal Monitoring System (Columbia Instruments)33. HFD-fed mice were treated with Nnmt- or control-ASO for 3 weeks. Body weights were not significantly different when the mice were subjected to CLAMS study. The mice were acclimated in the metabolic chambers for 2 days before the experiments. CO2 and O2 levels were collected every 32 min for each mouse during a period of 3 days. Food intake and activity were measured at regular intervals.
RNA extraction and quantitative PCR
RNA extraction and Taqman quantitative PCR were performed as described previously2. All Taqman primers were purchased from Applied Biosytems, except for the probe for adenosylmethionine decarboxylase (Amd1), which was designed using IDT software: forward 5′-GAG AGTGGAATTCGTGACCTG-3′; probe 5′-ACTGTTCAATCCTTGTGGCTAC TCGATG-3′; reverse 5′-TTCTGGTTCTGGAGTGATGTG-3′.
Western blot analysis and immunoprecipitation
NNMT was detected using a chicken anti-NNMT antibody from GenWay Biotech or the rabbit anti-NNMT antibody kindly provided by R. Weinshilboum34. Goat anti-UCP1 (M-17) and rabbit anti-PGC1α (H-300) were from Santa Cruz Biotechnology. Rabbit anti-acetylated lysine was from Cell Signaling Technology. For detecting PGC-1α acetylation, PGC-1α was immunoprecipitated from liver or cultured-cell lysates with anti-PGC-1α. The precipitated protein was subjected to western blot analysis using anti-acetylated lysine.
Body composition, body temperature, and faecal lipid excretion
Body composition was analysed in mice treated with Nnmt and control ASO for 5 weeks using an EchoMRI 3-in-1 instrument (Echo Medical Systems). To measure body composition after fasting, food was removed from Nnmt-ASO- or control-ASO-treated mice for 16h. Body temperature was measured intrarectally using a Thermalert TH-5 thermometer (Physitemp). To analyse faecal lipid excretion, lipid content of faeces was extracted using chloroform:methanol (2:1) and air-dried under a fume hood.
Histology and determination of cell size
Tissues were fixed in 10% buffered formalin and subjected to H&E staining. To measure adipocyte cross-sectional area, four fields of vision of H&E-stained adipose tissue were digitally photographed under a microscope. Images were analysed using the software ImageJ (National Institutes of Health) using a previously published technique35. Images were converted to 16-bit grayscale and inverted; a threshold was applied to identify cell boundaries, and the cells were analysed with the ‘Analyze Particles’ command to obtain adipocyte area. Adipocyte volume was calculated from the radius using the formula v =4/3πr3.
Intraperitoneal glucose tolerance test
The food was removed from mice for 5 h. Intraperitoneal glucose tolerance test (IPGTT) was performed by intraperitoneal injection of 1 mg glucose per g of body weight. Glucose levels were measured at the indicated times.
Feed efficiency
Total calorie intake was computed from the measured food intake during the second and third weeks of ASO treatment. Feed efficiency was expressed as the body weight gain divided by the total calorie intake between the start and the end of the food intake measurement36.
Urinary diacetylspermine measurement
Spot urine was collected from Nnmt-ASO-and control-ASO-treated mice. Urinary diacetylspermine levels were measured using an ELISA kit from Abnova. An enzymatic mouse creatinine assay (Crystal Chem) was used for measuring creatinine levels. Because the mice drank water ad libitum, the urinary diacetylspermine levels were corrected by creatinine levels. Urinary creatinine levels were not different between Nnmt-ASO- and control-ASO-treated mice.
ODC and SSAT activity
ODC and SSAT activities were assayed from tissue supernatant fractions under initial rate conditions with non-limiting substrate concentrations as described previously37,38. In brief, ODC was assayed by measuring the release of 14CO2 in a reaction with DL-[1-14C]ornithine as the substrate. SSAT was assayed by measuring the formation of acetylated spermidine in a reaction with 14C-CoA and spermidine as co-substrates.
NAD+ measurement
NAD+ levels were determined according to instructions provided with the NAD/NADH assay kit purchased from Abcam.
Histone methylation and ChIP-qPCR
Histone protein was extracted from adipose tissue treated with Nnmt ASO and control ASO using the EpiQuik Total Histone Extraction Kit from Epigentek. Eight types of histone methylation were measured by western blot analysis using the following antibodies: H3K4me2, H3K9me2, H3K27me2, H3K36me2 and H3K79me2 from Cell Signaling; and H3K4me1, H3K4me3 and H3K9me3 from Abcam. The levels of histone methylation were normalized to total H3 expression. For ChIP, chromatin was first extracted from 3T3-L1 adipocytes treated with or without 10 mM N-methylnicotinamide for 48 h. Immunoprecipitation was performed using EpiQuik Methyl-Histone H3-K4 ChIP kit from Epigentek. Enrichment of methylated H3K4 on Odc and Ssat genes was measured by real-time PCR using EpiTect ChIP qPCR primers from Qiagen (Odc, GPM1030188(+)01A; Ssat, GPM1055920(+)01A). An open reading frame free region (Igx1a) was used as a negative control for ChIP-qPCR. Control IgG showed minimum background among all the regions analysed.
Targeted mass spectrometry
Metabolites nicotinamide, S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), AMP and ATP were measured using tandem mass spectrometry30,39. Metabolite extracts using 80% methanol (−80 °C) were dried by nitrogen. Samples were re-suspended using 20 μl LC/MS grade water, of which 10 μl were injected and analysed using a 5500 QTRAP triple quadrupole mass spectrometer (AB/Sciex) coupled to a Prominence HPLC system (Shimadzu) via selected reaction monitoring (SRM). The dwell time was 4 ms per SRM transition and the total cycle time was 1.89 s. Approximately 8 to 11 data points were acquired per detected metabolite. Samples were delivered to the MS using a 4.6 mm internal diameter × 10 cm Amide XBridge HILIC column (Waters) at 300 μl min−1. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 35% B from 0 to 3.5 min; 35% B to 2% B from 3.5 to 11.5 min; 2% B was held from 11.5 to 16.5 min; 2% B to 85% B from 16.5 to 17.5 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A consisted of 20 mM ammonium hydroxide and 20 mM ammonium acetate (pH 9.0) in 95:5 water:acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/Sciex).
Cell culture
3T3-L1 cells were differentiated to adipocytes using a standardized protocol40. Five to six days after differentiation, cells were transfected using Amaxa electroporation41. Hepatoma H2.35 cells were transduced with NNMT or β-galacto-sidase adenovirus for 24 h, and oxygen consumption was measured as described below.
Oxygen consumption
Oxygen consumption was measured with a Clark-type oxygen electrode (Rank Brothers). Cultured cells were trypsinized, counted in a hemocytometer, spun down, and resuspended in respiration buffer (PBS, 2% BSA, 4.5 g l-1 glucose, 120 mg l−1 sodium pyruvate) to 1 × 106 ml−1. Partial oxygen pressure was recorded using a Rank Brothers digital model 10 controller and a PowerLab 4/30 data acquisition system (ADInstruments)42.
Statistical test
All data are expressed as mean mean ±s.e.m. Two-tailed Student’s t-tests were used for single comparisons. Analyses of variance were performed followed by Bonferroni-Holm post-hoc tests for multiple comparisons. Statistical significance is assumed at P <0.05. Sample size was chosen based on results from pilot studies and our extensive experience in investigating metabolic physiology in mice. The sample exclusion criteria were determined before experiments for technical failures such as mis-injection of glucose in glucose tolerance test. For randomization, experimental groups of mice were stratified according to body weight using an algorithm based on Kullback–Leibler divergences43. The CLAMS study, metabolic analyses and oxygen consumption in adipocytes were carried out in a blinded fashion, others were not.
Supplementary Material
Acknowledgments
We thank R. Weinshilboum for NNMT antibody; P. Woster for DFMO; M. Yuanfor tandem mass spectrometry; A. Karppinen, A. Korhonen, T.Reponen, A. Uimari, S. Pirnes-Karhuand T. Koponen for measurements of ODC and SSAT activity; C. Semenkovich and S. Fried for protocols for FAS activity measurements; and P. Aryal for assistance with real-time qPCR. D.Kr. is supported by the Deutsche Forschungsgemeinschaft (KR 3475/1-1) and American Heart Association (AHA) (09POST2250499); Q.Y. is a Klarman Scholar at the Beth Israel Deaconess Medical Center. This work is supported by grants from the NIH (R37 DK43051, P30 DK57521) and a grant from the JPB foundation to B.B.K.; grants from the NIH (KO8 DK090149, R01 DK100385, BNORC P30 DK046200 and NORCH P30 DK040561) to Q.Y.; grant RO1 DK69966 to P.P.; P01CA120964 and P30CA006516-46 to J.M.A.; AHA 13SDG14620005 and P&F P30 DK0460200 to D.K.; the Ellison Medical Foundation New Scholar in Aging Award to A.A.S.; and academy of Finland grant 118590 to L.A.
Footnotes
Online Content
Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions
Q.Y. discovered NNMT from the initial microarray analysis. D.Kr., Q.Y. and B.B.K designed the experiments, interpreted the data and wrote the paper. D.Ko. performed oxygen consumption experiments in adipocytes. A.S.B. performed CLAMS studies. L.Z., T.C.P., F.G., YC.W. and O.D.P. provided assistance with cell culture and animal experiments. O.D.P. also performed the microarray studies. J.T.R. and P.P. performed PGC-1α acetylation experiments. E.P. and L.A. provided expertise on polyamines and measured ODC and SSAT activity. Y.C. and A.A.S. measured nicotinamide and metabolites. J.M.A. performed metabolomics studies. B.P.M and S.B. provided Nnmt and control ASOs.
Author Information
The microarray data from adipose tissue of adipose-specific knockout and adipose-specific overexpression have been published by our laboratory and are available in NCBI Gene Expression Omnibus under accession number GSE35378. Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to B.B.K. (bkahn@bidmc.harvard.edu) or Q.Y. (qin.yang@uci.edu).
References
- 1.Shepherd PR, Kahn BB. Glucose transporters and insulin action—implications for insulin resistance and diabetes mellitus. N Engl J Med. 1999;341:248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 2.Yang Q, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 3.Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem. 1994;269:14835–14840. [PubMed] [Google Scholar]
- 4.Riederer M, Erwa W, Zimmermann R, Frank S, Zechner R. Adipose tissue as a source of nicotinamide N-methyltransferase and homocysteine. Atherosclerosis. 2009;204:412–417. doi: 10.1016/j.atherosclerosis.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jell J, et al. Genetically altered expression of spermidine/spermine N1-acetyltransferase affects fat metabolism in mice via acetyl-CoA. J Biol Chem. 2007;282:8404–8413. doi: 10.1074/jbc.M610265200. [DOI] [PubMed] [Google Scholar]
- 8.Pirinen E, et al. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Mol Cell Biol. 2007;27:4953–4967. doi: 10.1128/MCB.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartini D, et al. Nicotinamide N-methyltransferase in non-small cell lung cancer: promising results for targeted anti-cancer therapy. Cell Biochem Biophys. 2013;67:865–873. doi: 10.1007/s12013-013-9574-z. [DOI] [PubMed] [Google Scholar]
- 10.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nature Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams AC, Cartwright LS, Ramsden DB. Parkinson’s disease: the first common neurological disease due to auto-intoxication? QJM. 2005;98:215–226. doi: 10.1093/qjmed/hci027. [DOI] [PubMed] [Google Scholar]
- 12.Lee YH, et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salek RM, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics. 2007;29:99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- 14.Yaguchi H, Togawa K, Moritani M, Itakura M. Identification of candidate genes in the type 2 diabetes modifier locus using expression QTL. Genomics. 2005;85:591–599. doi: 10.1016/j.ygeno.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander J, Chang GQ, Dourmashkin JT, Leibowitz SF. Distinct phenotypes of obesity-prone AKR/J, DBA2J and C57BL/6J mice compared to control strains. Int J Obes (Lond) 2006;30:50–59. doi: 10.1038/sj.ijo.0803110. [DOI] [PubMed] [Google Scholar]
- 17.Svenson KL, et al. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- 18.Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2009;37:D720–D730. doi: 10.1093/nar/gkn778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 20.Erion DM, et al. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci USA. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang-Lee YA, et al. Metabolic effects of nicotinamide administration in rats. J Nutr. 1983;113:215–221. doi: 10.1093/jn/113.2.215. [DOI] [PubMed] [Google Scholar]
- 22.Varela-Rey M, et al. Fatty liver and fibrosis in glycine N-methyltransferase knockout mice is prevented by nicotinamide. Hepatology. 2010;52:105–114. doi: 10.1002/hep.23639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegg AE, Casero RA., Jr Current status of the polyamine research field. Methods Mol Biol. 2011;720:3–35. doi: 10.1007/978-1-61779-034-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koponen T, et al. The activation of hepatic and muscle polyamine catabolism improves glucose homeostasis. Amino Acids. 2011;42:427–440. doi: 10.1007/s00726-011-1013-0. [DOI] [PubMed] [Google Scholar]
- 25.Alcendor RR, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 26.Finley LW, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS ONE. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi Y, et al. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16:237–243. [PubMed] [Google Scholar]
- 28.Stein S, et al. SIRT1 decreases Lox-1-mediated foam cell formation in atherogenesis. Eur Heart J. 2010;31:2301–2309. doi: 10.1093/eurheartj/ehq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 30.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 32.Shepherd PR, et al. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- 33.Bubolz AH, et al. Activation of endothelial TRPV4 channels mediates flow-induced dilation in human coronary arterioles: role of Ca2+ entry and mitochondrial ROS signaling. Am J Physiol Heart Circ Physiol. 2012;302:H634–H642. doi: 10.1152/ajpheart.00717.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan L, Otterness DM, Craddock TL, Weinshilboum RM. Mouse liver nicotinamide N-methyltransferase: cDNA cloning, expression, and nucleotide sequence polymorphisms. Biochem Pharmacol. 1997;54:1139–1149. doi: 10.1016/s0006-2952(97)00325-0. [DOI] [PubMed] [Google Scholar]
- 35.Chen HC, Farese RV., Jr Determination of adipocyte size by computer image analysis. J Lipid Res. 2002;43:986–989. [PubMed] [Google Scholar]
- 36.Bence KK, et al. Neuronal PTP1B regulates body weight, adiposity and leptin action. Nature Med. 2006;12:917–924. doi: 10.1038/nm1435. [DOI] [PubMed] [Google Scholar]
- 37.Bernacki RJ, et al. Preclinical antitumor efficacy of the polyamine analogue N1, N11-diethylnorspermine administered by multiple injection or continuous infusion. Clin Cancer Res. 1995;1:847–857. [PubMed] [Google Scholar]
- 38.Jänne J, Williams-Ashman HG. On the purification of L-ornithine decarboxylase from rat prostate and effects of thiol compounds on the enzyme. J Biol Chem. 1971;246:1725–1732. [PubMed] [Google Scholar]
- 39.Yang X, et al. Using tandem mass spectrometry in targeted mode to identify activators of class IA PI3K in cancer. Cancer Res. 2011;71:5965–5975. doi: 10.1158/0008-5472.CAN-11-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan QW, et al. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes. 2007;56:2533–2540. doi: 10.2337/db07-0007. [DOI] [PubMed] [Google Scholar]
- 41.Eguchi J, et al. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7:86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulinilkunnil T, et al. Adrenergic regulation of AMP-activated protein kinase in brown adipose tissue in vivo. J Biol Chem. 2011;286:8798–8809. doi: 10.1074/jbc.M111.218719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endo A, Nagatani F, Hamada C, Yoshimura I. Minimization method for balancing continuous prognostic variables between treatment and control groups using Kullback–Leibler divergence. Contemp Clin Trials. 2006;27:420–431. doi: 10.1016/j.cct.2006.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.