Abstract
Introduction
The role of interleukin (IL)-1β –31T/C promoter polymorphism in the pathogenesis of chronic obstructive pulmonary disease (COPD) has been studied with inconsistent results. This meta-analysis was performed to assess the association of IL-1β –31T/C promoter polymorphism with COPD susceptibility.
Material and methods
Published case-control studies from PubMed and China National Knowledge Infrastructure (CNKI) databases were retrieved. Data were extracted and pooled odds ratios (OR) with 95% confidence intervals (CI) were calculated.
Results
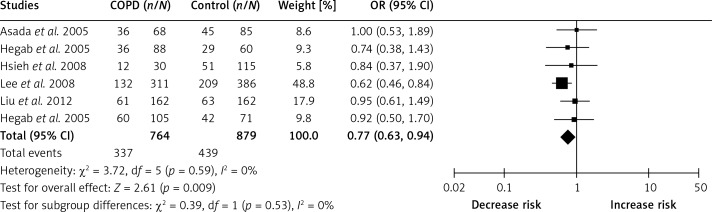
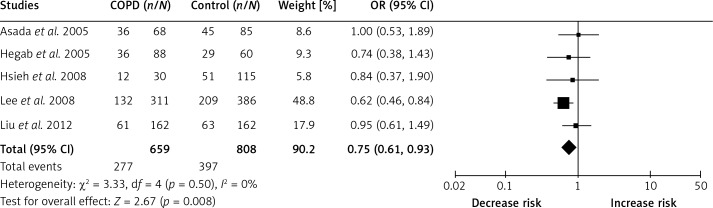
Six case-control studies were included in this meta-analysis. The pooled effect size showed that IL-1β -31T/C was significantly associated with COPD susceptibility in an overdominant genetic model (CC+TT vs. TC, OR: 0.77, 95% CI: 0.63–0.94), indicating that homozygotes (CC and TT) had a decreased risk for COPD compared with heterozygotes (TC). In the subgroup analysis by ethnicity, the results indicated that IL-1β –31T/C was significantly correlated with COPD susceptibility in Asians (overdominant model, OR: 0.75, 95% CI: 0.61–0.93), further suggesting a protective role of IL-1β –31T/C in COPD pathogenesis in Asians. Moreover, after excluding the study without Hardy-Weinberg equilibrium, the pooled results were robust and no publication bias was found in this study.
Conclusions
This meta-analysis suggests that IL-1β –31T/C promoter polymorphism confers protection against COPD in Asians.
Keywords: chronic obstructive pulmonary disease, interleukin-1β, polymorphism
Introduction
Chronic obstructive pulmonary disease (COPD) is a worldwide disease, characterized by persistent airflow limitation that is progressive and associated with an enhanced inflammatory response in airways and lungs [1]. To date, although the underlying mechanisms of COPD remain not fully elucidated, a genetic predisposition of COPD has been strongly evidenced [2–4].
Interleukin (IL)-1β, a proinflammatory cytokine, is encoded by the IL1B gene on chromosome 2q14, which is identified as a candidate gene of COPD [5]. In the past decade, the relationship of polymorphisms in the IL1B gene with COPD risk has been well studied, including the –31T/C promoter polymorphism in the IL1B gene with the potential to alter the transcriptional activity [6]. However, inconsistent results were reported [7–11], which may be owing to differences in ethnicity and sample size in individual studies, resulting in lower statistical power. It is suggested that meta-analysis could be a useful means to pool the independent statistical powers and thus achieve a quantitative understanding of the associations. Accordingly, in the present study, a meta-analysis was performed to determine IL-1β –31T/C promoter polymorphism and the risk of COPD.
Material and methods
Search strategy
The literature search was conducted using PubMed and China National Knowledge Infrastructure (CNKI) (http://www.cnki.net/) databases. CNKI database was founded by Tsinghua University of China in 1996 and includes over 8000 Chinese journals covering natural and social sciences. The languages were limited to English and Chinese. The following search terms were utilized: interleukin 1beta or interleukin 1b or IL1beta or IL1b, and polymorphism or variant or SNP or genotype, and chronic obstructive pulmonary disease or COPD.
Data extraction
Two independent reviewers collected the data according to the inclusion and exclusion criteria. For inclusion in the meta-analysis, retrieved articles had to inform about the number of cases and controls, and the number of individual genotypes in cases and controls. Exclusion criteria in the meta-analysis were 1) not case-control genetic study, 2) duplicated report, 3) no useful data reported, 4) other IL-1β polymorphisms than –31T/C. Unpublished data were not considered. Disagreement was resolved by discussion before reaching a consensus. If more than one article was published by the same group using the same cases, the study with a higher sample size was selected.
Statistical analysis
Categorical variables are presented as odds ratio (OR) with 95% confidence interval (CI). CC, TC and TT are the genotypes of IL-1β –31T/C polymorphism. OR1, OR2 and OR3 were calculated as follows: OR1: CC vs. TT; OR2: TC vs. TT; OR3: CC vs. TC. These pairwise differences (OR1, OR2 and OR3) were used to indicate the most appropriate genetic model as follows: if OR1 = OR2 ≠ 1 and OR3 = 1, then a dominant model was suggested; if OR1 = OR3 ≠ 1 and OR2 = 1, then a recessive model was suggested; if OR2 = 1/OR3 ≠ 1 and OR1 = 1, then a complete overdominant model was suggested; if OR1 > OR2 > 1 and OR1 > OR3 > 1 (or OR1 < OR2 < 1 and OR1 < OR3 < 1), then a codominant model was suggested. Once the best genetic model was identified, this model was used to collapse the three genotypes into two groups (except a codominant model) and to pool the results again. Pooled ORs with 95% CI were calculated and p < 0.05 was accepted as statistical significance. Heterogeneity was checked by the Q test. Meta-analysis was done with the fixed-effects model when there was no heterogeneity (p ≥ 0.1). Otherwise, the random-effects model was used. Subgroup analysis was performed by ethnicity to assess the effect of possible clinical heterogeneity on the summary ORs. Pearson's χ2 test was used to determine whether the observed frequencies of genotypes in controls conformed to the Hardy-Weinberg equilibrium (HWE). Studies with controls that depart from HWE (p < 0.05) were subjected to a sensitivity analysis in order to check the consistency of the overall effect size. Funnel plots, as well as Begg's rank correlation test and Egger's linear regression test, were used to assess the potential publication bias, and p < 0.05 was considered significant publication bias. All analyses were conducted using Revman 5.0 (Oxford, UK, The Cochrane Collaboration) and Stata 11.0 (StataCorp LP, College Station, TX, USA) [12].
Results
Studies included in the meta-analysis
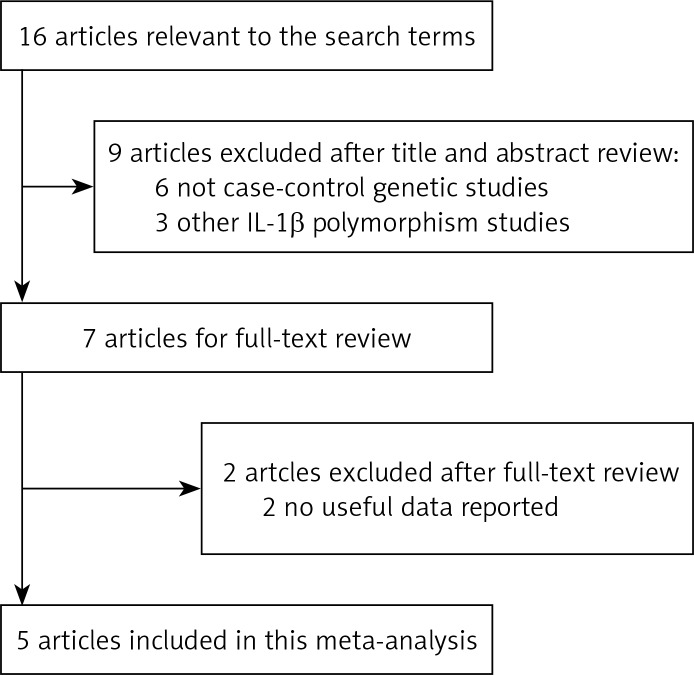
Sixteen studies were relevant to the search terms. After reviewing the titles, abstracts and articles, 11 studies were excluded; thus 5 articles with 6 case-control studies for –31T/C matched the inclusion criteria (Figure 1). Of the 5 included articles, 4 were published in English, and 1 was published in Chinese. These studies were carried out in China, Japan, Korea, Egypt, and Taiwan. Notably, the study by Hegab et al. [8] was performed in two ethnic populations (Japan and Egypt). The main features of the studies included in this meta-analysis are presented in Table I.
Figure 1.
Flow diagram of search process
Table I.
Main characteristics of included studies
| Ref. | Country | Race | Genotyping | Source of control | COPD | Control | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | CC | TC | TT | Total | CC | TC | TT | HWE (p) | |||||
| [4] | Japan | Asian | PCR+RFLP | Healthy population | 68 | 20 | 32 | 16 | 85 | 20 | 40 | 25 | 0.8771 |
| [5] | Japan | Asian | PCR+RFLP | Healthy population | 88 | 16 | 52 | 20 | 60 | 8 | 31 | 21 | 0.8082 |
| [5] | Egypt | Caucasian | PCR+RFLP | Healthy population | 105 | 11 | 45 | 49 | 71 | 16 | 29 | 26 | 0.3734 |
| [6] | Korea | Asian | ABI sequencer | Healthy men | 311 | 74 | 179 | 58 | 386 | 109 | 177 | 100 | 0.2696 |
| [7] | China | Asian | PCR+RFLP | Healthy population | 162 | 29 | 101 | 32 | 162 | 27 | 99 | 36 | 0.0159 |
| [8] | Taiwan | Asian | PCR+RFLP | Healthy population | 30 | 6 | 18 | 6 | 115 | 23 | 64 | 28 | 0.4665 |
HWE – Hardy-Weinberg equilibrium, PCR – polymerase chain reaction, Ref – reference, RFLP – restriction fragment length polymorphism
Quantitative synthesis
In order to indicate the most appropriate genetic model, OR1, OR2 and OR3 were calculated. Results showed OR1 = 1.12, OR2 = 1.38 and OR3 = 0.80 for –31T/C, suggesting an overdominant genetic model (CC+TT vs. TC). The pooled effect size showed that there was a significant association of IL-1β –31T/C with the risk of COPD (OR: 0.77, 95% CI: 0.63–0.94, p = 0.009, fixed model, Figure 2). In the subgroup analysis by ethnicity, the results indicated that IL-1β –31T/C was significantly correlated with COPD susceptibility in Asians (OR: 0.75, 95% CI: 0.61–0.93, p = 0.008, fixed model, Figure 3).
Figure 2.
Forest plots of OR with 95% CI for the association of IL-1β –31T/C and COPD risk (CC+TT vs. TC)
Figure 3.
Forest plots of OR with 95% CI for the association of IL-1β –31T/C and COPD risk in Asians (CC+TT vs. TC)
Test of heterogeneity
Significant heterogeneity was not revealed between all studies in CC+TT vs. TC comparison (overdominant model) for –31T/C (I2 = 0%, p = 0.59). When stratified by ethnicity, no heterogeneity was observed between the studies in Asians.
Sensitivity analyses
In the present meta-analysis, in only one study [10] was there lack of HWE, which had a potential to influence the robustness of the present meta-analysis. After exclusion of this study, the pattern of the pooled effect size persisted, while the overall heterogeneity was not significantly altered for –31T/C (I2 = 0%, p = 0.62), indicating that this study might not contribute to the overall heterogeneity.
Publication bias
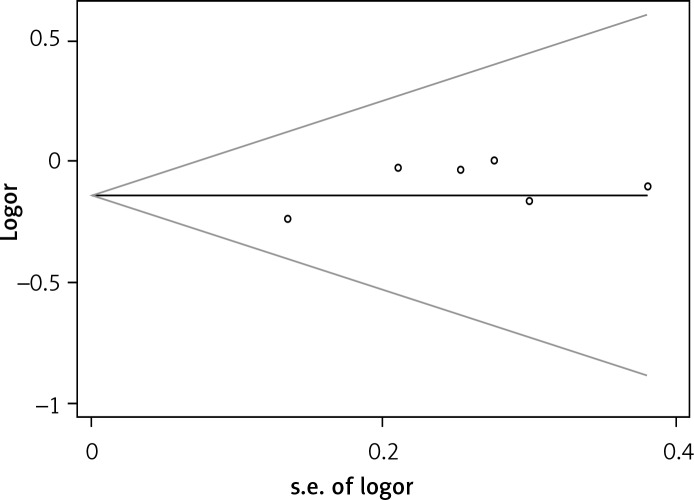
The funnel plots showed no significant asymmetry in this meta-analysis (Figure 4). Moreover, publication bias was not suggested by Begg's rank correlation test (p = 1.000) or Egger's linear regression test (p = 0.138).
Figure 4.
Begg's funnel plot for evaluation of publication bias in the included studies on the associations of IL-1β –31T/C with COPD risk
Discussion
IL-1β, a proinflammatory cytokine, contributes to the pathogenesis of COPD. Recently, association of IL-1β –31T/C promoter polymorphism with COPD risk caught more attention, owing to its potential to alter the transcriptional activity.
The pooled effect size showed that IL-1β –31T/C was significantly associated with COPD susceptibility in an overdominant genetic model, indicating that homozygotes (CC and TT) had a decreased risk for COPD compared with heterozygotes (TC). Significant heterogeneity was not revealed between all studies in CC+TT vs. TC comparison for –31T/C. In the subgroup analysis by ethnicity, the results indicated that IL-1β –31T/C was significantly correlated with COPD susceptibility in Asians, further suggesting a protective role of IL-1β –31T/C in COPD pathogenesis in Asians. After exclusion of the study without HWE by Liu et al. [10], the pattern of the pooled effect size persisted, while the overall heterogeneity was not significantly altered for –31T/C, indicating that this study might not contribute to the overall heterogeneity. Publication bias was not suggested in the present study, possibly owing to the deliberate search strategy and data extraction.
However, some limitations should be considered in this meta-analysis. First, large sample size studies were lacking and the pooled statistical power might be insufficient. Second, the pooled estimates were not based on adjustment by confounding factors, such as sex, age, and smoking history. Third, the lack of original data in the studies limited our further analysis of the potential interactions between gene and gene, or gene and environment, which might modulate COPD risk.
In conclusion, although the pooled estimates should be interpreted with caution, our meta-analysis suggests that IL-1β –31 T/C promoter polymorphism confers protection against COPD in Asians. However, large sample size studies with unbiased genotyping methods, standardized defined COPD cases and matched controls in different populations, as well as more detailed data about individual and environment, are warranted.
Acknowledgments
Min Xiao and Lingli Guo equally contributed.
This study was supported in part by an Application Infrastructure Project (2013JY0032) from the Science and Technology Department of Sichuan Province and grant 81230001 from the National Natural Science Foundation of China.
References
- 1.Global Strategy for Diagnosis, Management, and Prevention of COPD (Updated 2013); [Google Scholar]
- 2.Ezzeldin N, Shalaby A, Saad-Hussein A, et al. Association of TNF-alpha –308 G/A, SP-B 1580 C/T, IL-13 –1055 C/T gene polymorphisms and latent adenoviral infection with chronic obstructive pulmonary disease in an Egyptian population. Arch Med Sci. 2012;8:286–95. doi: 10.5114/aoms.2012.28556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurowski M, Majkowska-Wojciechowska B, Wardzynska A, Kowalski ML. Associations of allergic sensitization and clinical phenotypes with innate immune response genes polymorphisms are modified by house dust mite allergen exposure. Arch Med Sci. 2011;7:1029–36. doi: 10.5114/aoms.2011.26616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antczak A, Ciebiada M, Pietras T, Piotrowski WJ, Kurmanowska Z, Górski P. Exhaled eicosanoids and biomarkers of oxidative stress in exacerbation of chronic obstructive pulmonary disease. Arch Med Sci. 2012;8:277–85. doi: 10.5114/aoms.2012.28555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung KF, Adcock IM. Multifaceted mechanisms in COPD: inflammation, immunity, and tissue repair and destruction. Eur Respir J. 2008;31:1334–56. doi: 10.1183/09031936.00018908. [DOI] [PubMed] [Google Scholar]
- 6.Smolonska J, Wijmenga C, Postma DS, Boezen HM. Meta-analyses on suspected chronic obstructive pulmonary disease genes: a summary of 20 years’ research. Am J Respir Crit Care Med. 2009;180:618–31. doi: 10.1164/rccm.200905-0722OC. [DOI] [PubMed] [Google Scholar]
- 7.Asada M, Yamaya M, Ebihara S, et al. Interleukin-1beta gene polymorphisms associated with COPD. Chest. 2005;128:1072–3. doi: 10.1378/chest.128.2.1072. [DOI] [PubMed] [Google Scholar]
- 8.Hegab AE, Sakamoto T, Saitoh W, et al. Polymorphisms of TNFalpha, IL1beta, and IL1RN genes in chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 2005;329:1246–52. doi: 10.1016/j.bbrc.2005.02.099. [DOI] [PubMed] [Google Scholar]
- 9.Lee JM, Kang YR, Park SH, et al. Polymorphisms in interleukin-1B and its receptor antagonist genes and the risk of chronic obstructive pulmonary disease in a Korean population: a case-control study. Respir Med. 2008;102:1311–20. doi: 10.1016/j.rmed.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Liu X, Li H, Wang L. Relationship between polymorphism of IL-1beta gene and chronic obstructive pulmonary disease in Han population. Zhong Guo Man Xing Bing Yu Fang Yu Kong Zhi. 2012;20:128–31. [Google Scholar]
- 11.Hsieh MH, Chong IW, Hwang JJ, et al. Lack of associations between several polymorphisms in cytokine genes and the risk of chronic obstructivepulmonary diseases in Taiwan. Kaohsiung J Med Sci. 2008;24:126–37. doi: 10.1016/S1607-551X(08)70140-2. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Shen Y, Liu L, Li X, Wang T, Wen F. Interleukin-13 –1112 C/T promoter polymorphism confers risk for COPD: a meta-analysis. PloS One. 2013;8:e68222. doi: 10.1371/journal.pone.0068222. [DOI] [PMC free article] [PubMed] [Google Scholar]