Abstract
Identifying the proteins synthesized in defined cells at specific times in an animal will facilitate the study of cellular functions and dynamic processes. Here we introduce stochastic orthogonal recoding of translation with chemoselective modification (SORT-M) to address this challenge. SORT-M involves modifying cells to express an orthogonal aminoacyl-tRNA synthetase/tRNA pair to enable the incorporation of chemically modifiable analogs of amino acids at diverse sense codons in cells in rich media. We apply SORT-M to Drosophila melanogaster fed standard food to label and image proteins in specific tissues at precise developmental stages with diverse chemistries, including cyclopropene-tetrazine inverse electron demand Diels-Alder cycloaddition reactions. We also use SORT-M to identify proteins synthesized in germ cells of the fly ovary without dissection. SORT-M will facilitate the definition of proteins synthesized in specific sets of cells to study development, and learning and memory in flies, and may be extended to other animals.
Defining the proteins synthesized in specific cells, at specific times within animals will provide molecular insight into diverse biological processes including development,1 differentiation,1 circadian oscillations,2 morphogenesis3 and learning and memory.4 Despite the importance of this challenge, there are no general methods for identifying the proteins synthesized in specific cells at specific times within an animal.
One promising route to labelling proteins as they are synthesized in cells is the co-translational, statistical incorporation of analogs of natural amino acids at particular sense codons5. Alkyne- and azide-containing analogs of methionine (Met) have been incorporated at Met codons and labelled with probes via 3+2 cycloadditions with azide- or alkyne-bearing probes in approaches dubbed FUNCAT and BONCAT.5 It has been possible to label proteins in cell culture at Met codons using this approach, and to combine the approach with stable isotope labelling by amino acids in cell culture (SILAC) to obtain quantitative information in primary cells (QuanCAT).6 These approaches are based on selective pressure incorporation, which relies on the permissivity of Methionyl-tRNA synthetase (MetRS) for close analogs of methionine (Supplementary Fig. 1). These approaches do not require genetic manipulation and are useful for labelling all proteins synthesized in a population of cells. In Escherichia coli it has been possible to use variants of MetRS with expanded substrate tolerance to statistically label proteins in bacteria at methionine codons in the presence of mammalian cells, or in the presence of distinct bacteria.7,8 Recently this approach has been extended to the labelling on N-terminal methionines in mammalian cells using a variant of E. coli MetRS that recognizes azidonorleucine and aminoacylates the initiator tRNA with this unnatural amino acid.9 The incorporation of the amino acid analogs through selective pressure incorporation is commonly, though not exclusively,9 performed in minimal media under starvation conditions for the amino acid to be replaced.10 This limits the availability of the natural amino acid that otherwise efficiently outcompetes the analog within the synthetase active site (MetRS prefers methionine to a commonly used analog by a factor of approximately 400) and decreases labelling efficiency.11 Many of the limitations of these approaches for the stochastic incorporation of unnatural amino acids stem from co-opting endogenous aminoacyl-tRNA synthetase/tRNA pairs (or their derivatives) to statistically label proteins.
Genetic code expansion methods allow the quantitative, site-specific and genetically directed incorporation of unnatural amino acids with diverse chemical structures and bearing diverse functional groups. This is most commonly achieved by inserting the unnatural amino acid at an amber stop codon introduced into a gene of interest.12, 13 This approach does not allow proteome-wide labelling because it targets a single site in a single protein. Genetic code expansion is achieved through the introduction of an orthogonal aminoacyl-tRNA synthetase/tRNACUA pair into cells. The pyrrolysyl-tRNA synthetase (PylRS)/tRNACUA pair is amongst the most useful pairs for genetic code expansion,13 because it (i) can specifically recognize a range of useful unnatural amino acids, (ii) can be evolved to recognize an extended range of chemical structures, and (iii) can be used as an orthogonal pair for genetic code expansion in E. coli,14 yeast,15 mammalian cells,16-18 Caenorhabditis elegans19 and D. melanogaster.20
Here we describe stochastic orthogonal recoding of translation (SORT), which leverages advances in genetic code expansion to statistically label proteins (Fig. 1a) in ways that have been challenging to achieve with selective-pressure incorporation and its derivatives. Specifically we demonstrate that SORT can be used (i) to tag newly synthesized proteins with cyclopropene groups that can be labelled with tetrazine probes introduced via a chemoselective inverse electron demand Diels-Alder reaction (stochastic orthogonal recoding of translation with chemoselective modification, SORT-M), (ii) to label the proteome at a large set of sense codons, not just a single sense codon, (iii) to label the proteome of cells grown in rich media, (iv) as a modular and efficient approach for proteome labelling in the fly D. melanogaster, which allows selective labelling of proteins produced in a particular spatially or temporally defined set of cells in an intact animal fed its normal food supplemented with an unnatural amino acid, and (v) for the direct identification, from the total proteome, of proteins synthesized in a chosen tissue at a defined developmental stage in the fly.
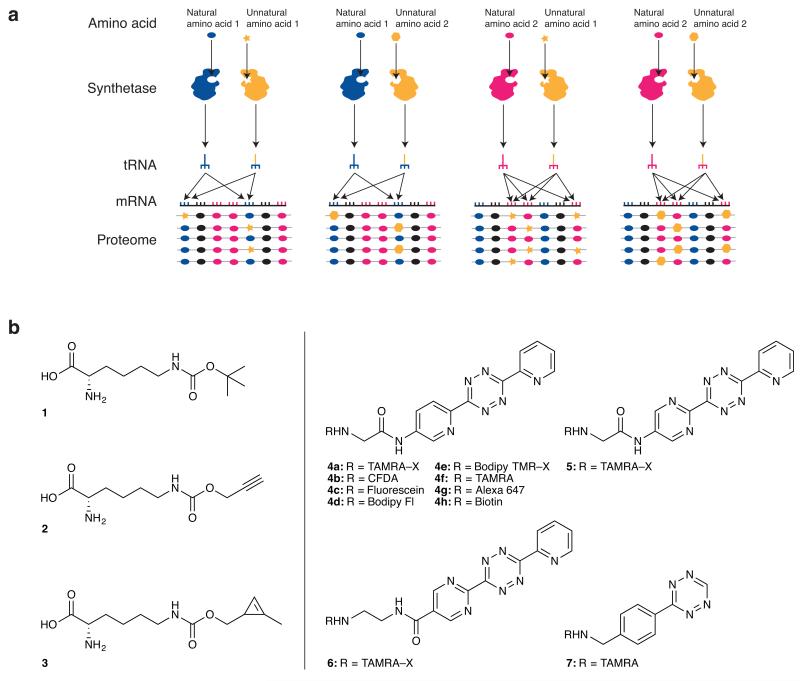
Figure 1. SORT-M enables proteome tagging and labelling at diverse codons, with diverse chemistries, and in genetically targeted cells and tissues.
(a) Proteome tagging via SORT (stochastic orthogonal recoding of translation) uses an orthogonal aminoacyl-tRNA synthetase/tRNA pair. The pyrrolysyl-tRNA synthetase/tRNA pair is used in this study. This synthetase (and its previously evolved active-site variants) recognizes a range of unnatural amino acids (yellow star, and yellow hexagon), does not aminoacylate endogenous tRNAs, but efficiently aminoacylates its cognate tRNA - without regard to anticodon identity; PyltRNA is not a substrate for endogenous aminoacyl-tRNA synthetases. Orthogonal pyrrolysyl-tRNA synthetase/tRNAXXX pairs (XXX indicates choice of anticodon, yellow) in which the anticodon has been altered compete for the decoding of sense codons (dark blue and pink) via a pathway that is orthogonal to that used by natural synthetases and tRNAs (dark blue and pink) to direct natural amino acids. SORT allows the incorporation of diverse chemical groups into the proteome, at diverse codons. Since there is no competition at the active site of the orthogonal synthetase, starvation and minimal media are not required. In addition the expression pattern of the orthogonal proteome tagging system can be genetically directed allowing tissue specific proteome labelling. Selective pressure incorporation approaches are shown in Supplementary Figure 1 for comparison to SORT. (b) The combination of encoding amino acids (1-3) across the proteome via SORT and chemoselective modification of 3 with tetrazine probes (4a-g, 5, 6 and 7) allows detection of labelled proteins via SORT-M (stochastic orthogonal recoding of translation and chemoselective modification). Amino acid structures: Nε-((tert-butoxy)carbonyl)-l-lysine 1, Nε-(1-propynlyoxy)carbonyl)-l-lysine 2 and Nε-(((2-methylcycloprop-2-en-1-yl)methoxy)carbonyl)-l-lysine.
Results
Encoding the site-specific incorporation of 3 in E. coli
A class of reaction with potential for proteome labelling is the very rapid and specific inverse electron demand Diels-Alder reaction between strained alkenes (or alkynes) and tetrazines.21-25 However, the incorporation of components for inverse electron demand Diels-Alder reactions into proteomes has not been possible using current approaches.
We, and others, have previously encoded unnatural amino acids bearing strained alkenes, alkynes and tetrazines using genetic code expansion, and we have demonstrated their use for site-specific protein labelling via inverse electron demand Diels-Alder reactions26-30. However, all the molecules used to date bear large groups (trans-cyclooctenes, bicyclononynes or norbornenes). These groups are not ideal for proteome labelling because they are much larger than natural amino acids and may therefore interfere with protein structure and function when introduced throughout the proteome. We have previously shown that a variety of carbamate derivatives of lysine are good substrates for PylRS,31 and it has been demonstrated that 1,3 disubstituted cyclopropenes, unlike 3,3 disubstituted cyclopropenes,32,24 react efficiently with tetrazines.22 We therefore designed and synthesized (Supplementary note 1) a carbamate derivative of lysine, bearing a 1,3 disubstituted cyclopropene (Nε-[((2-methylcycloprop-2-en-1-yl)methoxy)carbonyl]-l-lysine 3, Fig. 1b), for incorporation into proteins and labelling with tetrazines.
We demonstrated that 3 is efficiently and site-specifically incorporated into recombinant proteins at the amber stop codon using the PylRS/tRNACUA pair and an sfGFP gene bearing an amber codon at position 150 (Supplementary Fig. 2a). The yield of protein is 8 mg per litre of culture, which is greater than that obtained for a well-established efficient substrate for PylRS Nε-[(tert-butoxy)carbonyl]-l-lysine 1 (4 mg per litre of culture). 33 Electrospray ionisation mass spectrometry of sfGFP bearing 3 at position 150 (sfGFP-3) confirms the incorporation of the unnatural amino acid (Supplementary Fig. 2b). sfGFP-3 was specifically labelled with the fluorescent tetrazine probe 4a, whereas sfGFP-1 (sfGFP bearing 1 at position 150) was left unlabelled (Supplementary Fig. 2b). We found that 2 nmol of sfGFP-3 was quantitatively labelled with 10 equivalents of 4a in 30 minutes, as judged by both fluorescence imaging and mass spectrometry (Supplementary Fig. 2b). The second order rate constant for labelling SfGFP-3 with 4a was 27 ± 1.8 M−1s−1 (Supplementary Fig. 2c).26 Taken together, these results show that the PylRS/tRNACUA pair can be used to site specifically incorporate 3, and the encoded amino acid can be specifically and rapidly labelled with tetrazine probes.
Proteome labelling at diverse codons using SORT-M
As PylRS does not recognize the anticodon of its cognate tRNA34, we hypothesized that it may be possible to alter the anticodon of this tRNA to decode distinct codons. We created a new tRNA in which the anticodon of PyltRNACUA was converted from CUA to UUU (Supplementary Table 1), to decode a set of lysine codons. We added 0.1 mM 3 to cells containing PylRS, PyltRNAUUU and the gene for T4 lysozyme (to provide an overexpressed protein that we could subsequently use to assess sense codon incorporation efficiency). Following expression of T4 lysozyme, we detected proteins in the lysate bearing 3 with the tetrazine probe 4a (20 μM 1h, Supplementary Fig. 3). Control experiments show that the observed labelling requires the presence of the synthetase and tRNA, and electrospray ionization mass spectrometry demonstrates the incorporation of 3 in place of lysine in T4 lysozyme (Supplementary Fig. 4). The addition of 3 (0.1 or 0.5 mM) has little or no effect on cell growth (Supplementary Fig. 5) suggesting that the amino acid is not toxic at the concentration used for SORT, and there is substantial labelling within 1h of amino acid addition (Supplementary Fig. 6).
To demonstrate the generality of SORT-M at distinct codons, we repeated the proteome labelling using tRNAs with six additional Pyl tRNAXXX variants (where XXX represents the altered anticodons (Supplementary Fig. 3). For all anticodons, addition of 3 (0.1 mM) has little effect on cell growth (Supplementary Fig. 5), demonstrating that SORT-M is non-toxic with diverse codons in E. coli.
To estimate the frequency with which 3 is incorporated into proteins by SORT, we compared the fluorescence of T4 lysozyme labelled by SORT-M and T4 lysozyme quantitatively labelled with 4a (Supplementary Fig. 7). The incorporation frequencies ranged from 0.08% to 0.65% per codon. Labelling specificity is maintained with distinct tetrazine modules (Supplementary Fig. 8) and with distinct fluorophores (Supplementary Fig. 8), allowing imaging of proteomes labelled at orthogonal wavelengths (Supplementary Fig. 9). We also demonstrated proteome labelling with Nε-[(2-propynyloxy)carbonyl]-l-lysine (2), which we have previously shown to be a good substrate for the wild-type pyrrolysyl-tRNA synthetase31 (Supplementary Fig. 10). These results demonstrate that SORT provides a modular approach for introducing new functional groups into the proteome.
Genetic encoding of 3 and SORT-M in human cells
We expressed full-length mCherry-3-EGFP-HA (a protein fusion between a red fluorescent protein (mCherry) and an enhanced green fluorescent protein (EGFP) with a C-terminal human influenza hemagglutinin (HA) tag and amino acid 3 site specifically incorporated between mCherry and EGFP) was produced from HEK293 cells carrying the PylRS/tRNACUA pair and mCherry-TAG-EGFP-HA (a fusion between the mCherry gene and the EGFP gene with a C-terminal HA tag, separated by the amber stop codon (TAG)).18 Full-length protein was detected only in the presence of added 3 (Fig. 2a, full gels in Supplementary Fig. 11). mCherry-3-EGFP-HA was selectively labelled with 4a, whereas mCherry-1-EGFP-HA (with 1 incorporated in place of 3 between mCherry and EGFP) was not labelled (Fig. 2b)18 demonstrating the site-specific incorporation of 3 with the PylRS/tRNACUA pair in human cells.
Figure 2. Site-specific incorporation of 3 into proteins at diverse codons and specific proteome labelling using SORT-M in human cells.
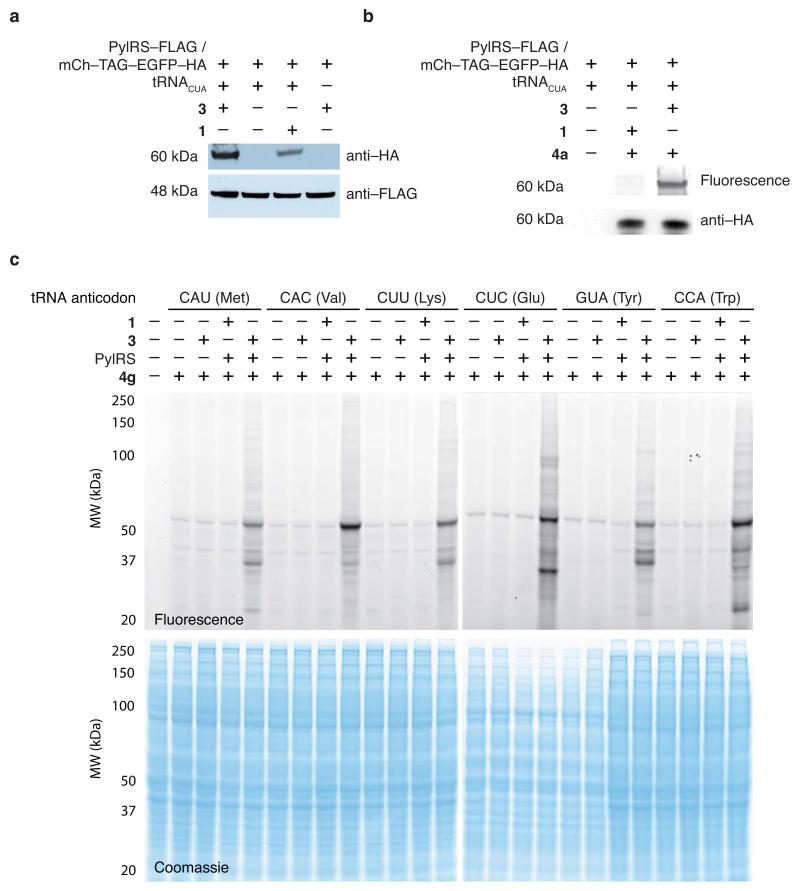
(a) Western blot analysis demonstrates the efficient amino acid dependant expression of an mCherry-EGFP fusion protein separated by an amber stop codon bearing a C-terminal HA-tag (mCh-TAG-EGFP-HA) in HEK293T cells. Anti-FLAG detected tagged PylRS (b) Specific labelling of mCh-TAG-EGFP-HA (immunoprecipitated from 106 cells) with 4a (20μM in 50μL PBS, 1h, RT) confirms the incorporation of 3 into protein in HEK293 cells. (c) SORT-M labelling of 3 that is statistically incorporated into newly synthesised proteins across the whole proteome of mammalian cells directed by six different PylRS/PyltRNAXXX mutants using 0.5 mM 3. Labeling with 4g (20μM in PBS, 1h, RT, as above). The amino acids in parentheses are the natural amino acids encoded by the endogenous tRNA bearing the corresponding anti-codon.
To demonstrate SORT-M for proteome labelling in mammalian cells, we transfected HEK293T cells with PylRS and a series of PyltRNAXXX constructs (Fig. 2c). The cells were incubated in growth media supplemented with 0.5 mM 3 for 48h. We labelled proteins in the lysate bearing 3 with the tetrazine probe 4g (4μM, 1h) (Fig. 2c). Labelling was amino acid-, tRNA- and synthetase-dependent. Additional experiments demonstrate clear labelling 12h after addition of 3, and demonstrate that higher concentrations of 3 do not substantially increase the amount of labelling (Supplementary Fig. 12).
We estimated the frequency of labelling as between 0.02% to 0.2% per codon (Supplementary Fig. 13). MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays35 demonstrated that SORT-M labelled cells are of comparable viability to cells that do not contain the synthetase/tRNA or amino acid (Supplementary Fig. 14). These experiments demonstrate SORT-M can be transferred from E.coli to mammalian cells, and that SORT is not toxic to mammalian cells at the concentrations used.
Genetic encoding of 3 in D. melanogaster
Next, we aimed to extend SORT-M to the fly to enable the labelling and identification of proteins expressed in particular tissues at particular times. The fly is particularly well suited to this because a large collection of tools exist,36-38 including a wealth of GAL4 lines, for specifically directing gene expression in specific cells at specific times.36 We envisioned that by directing PylRS expression into specific tissues we could direct tissue specific proteome labelling with 3 and use the highly specific labelling reaction of 3 with tetrazine probes to detect and identify proteins derived from a target tissue from the proteome of an entire fly.
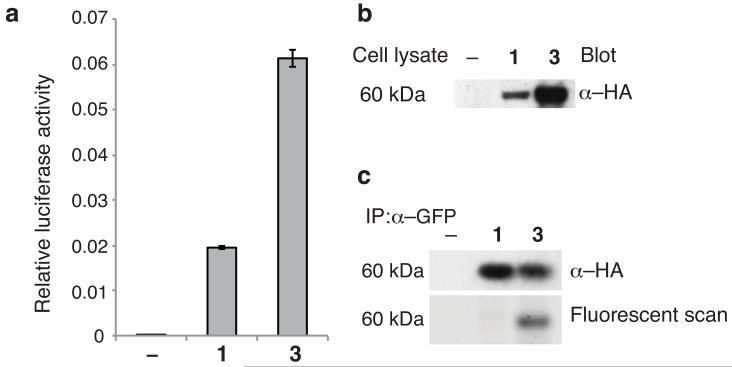
We first demonstrated that 3 can be site specifically incorporated into proteins in D. melanogaster. To achieve this, we used flies containing the PylRS/tRNACUA pair (with the tRNA expressed ubiquitously from a U6 promoter and UAS-PylRS expression directed to ovaries using a nos-vp16-GAL4 driver), and a dual luciferase reporter bearing an amber codon between firefly and renilla luciferase.20 We observe a strong luciferase signal that is dependent on the addition of 1 or 3, and the dual luciferase signal is larger with 3. These experiments demonstrate that 3 is taken up by flies and is more efficiently incorporated in vivo at an amber codon than 1 (Fig. 3a), a known excellent substrate for PylRS. In additional experiments, we demonstrated by western blot the efficient incorporation of 3 into a GFP-TAG-mCherry-HA construct (Supplementary Fig. 15) expressed in ovaries20 (Fig. 3b), and the specific fluorescent labelling of the incorporated amino acid with 4g (Fig. 3c).
Figure 3. Site-specific incorporation of amino acid 3 into protein produced in Drosophila melanogaster.
(a) Incorporation of 3 demonstrated by a dual luciferase reporter. Dual luciferase assay on ovary extract from 10 female flies expressing Triple-Rep-L in the presence or absence of 10 mM 1 or 10mM 3. The data show a representative example from 1 of 3 biological replicates. The error bars represent the standard deviation of 3 technical replicates from a single biological replicate. (b) Site-specific incorporation of 3 (or 1) into GFP_TAG_mCherry-HA in flies expressing PylRS/PyltRNACUA. The full-length protein resulting from unnatural amino acid incorporation is detected by anti-HA western blot. (c) Specific labelling of encoded 3 with tetrazine probes. Flies were fed with no amino acid, amino acid 1 (500 flies) or amino acid 3 (100 flies). 5 times more flies were fed with 1 in order to generate comparable amount of reporter protein. The full-length protein containing the unnatural amino acid was immunoprecipitated from lysed ovaries with anti-GFP beads. The beads were labelled (4g, 4μM, 200μL PBS, RT, 2h) washed. Full length protein was detected by anti-HA blot and the same gel imaged on a fluorescence scanner shows specific fluorescent labelling of the protein incorporating 3 but not 1, confirming the identity of the incorporated amino acid.
Tissue- and stage-specific proteome labelling in flies
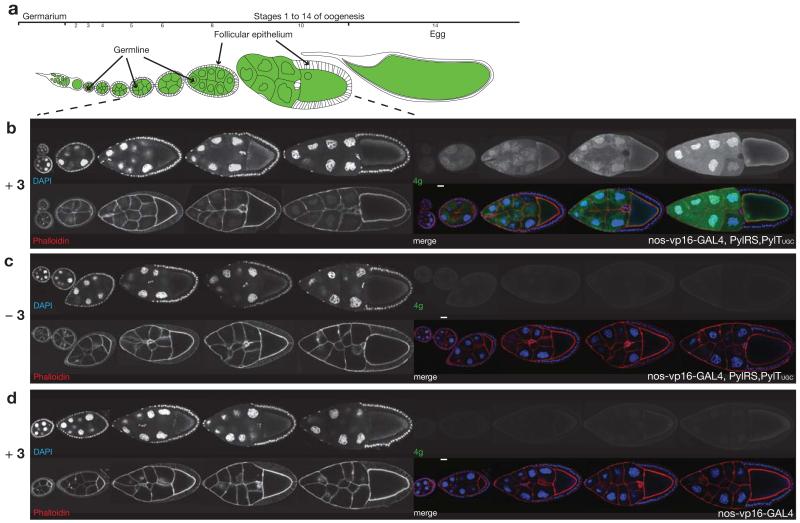
Next we demonstrated that SORT-M can be used to label newly synthesized proteins in the fly ovary with spatial (tissue) and temporal (developmental stage) specificity. To visualize cell-specific proteome labeling within the ovary, we generated transgenic D. melanogaster bearing PylRS on a GAL4 dependent promoter (upstream activating sequence, UAS) and four copies of PylTUGC (encoding PyltRNAUGC, which competes with Alanyl-tRNAs) on a ubiquitous U6 promoter. These transgenic flies were crossed with flies bearing the germline specific nos-vp16-GAL4 driver, leading to nos-vp16-GAL4, PylRS, PylTUGC flies. In parallel, we generated nos-vp16-GAL4/UAS-GFP flies and used these to define the spatial and temporal specificity of the nos-vp16-GAL4 driver within the ovary. GFP expression is strongest from stage five of oogenesis onwards, but is excluded from somatic cells in the egg chamber, as expected39 (Supplementary Fig. 16). These experiments define when and in which cells we expect to see labeling via SORT-M.
We fed nos-vp16-GAL4, PylRS, PylTUGC female flies food supplemented with amino acid 3 (10mM) for three days, extracted the ovaries and labeled them with 20μM 4g in PBS for 2h. We observe fluorescent labeling of proteomes in the germline cells of the egg chamber, which is strongest from stage 5 onwards, and no labeling of the proteome in the follicular epithelium that surrounds the germline cells or the border cells that migrate through the germline cells (Fig. 4b). Labelling is synthetase dependent and requires feeding the flies 3 (Fig. 4c, d), confirming that unnatural amino acid containing proteomes are specifically labelled. Labelling is brighter in the nuclei than the cytosol of nurse cells, consistent with nuclear endoreplication, which requires newly synthesized proteins in the nucleus40. We observe consistent temporally-specific and spatially-specific labeling of all the egg chambers derived from individuals within a population of nos-vp16-GAL4, PylRS,PylTUGC flies fed 3 (Supplementary Fig. 17). The egg chamber development in the presence of 3 and the synthetase and tRNA proceeds normally, and without growth defect demonstrating that the proteome labeling does not adversely affect oogenesis. This is particularly striking as oogenesis is an extremely complex process that requires several events: (i) all cells must perform consecutive mitoses and at least the beginning of a meiosis; (ii) the egg chamber must increase to more than five times its original size; (iii) differentiation of two different cell types, egg specification and the formation of the anterior-posterior and dorsoventral axis in the oocyte all must occur. If any of these processes are not performed correctly, the subsequent embryonic gastrulation would be impossible. The hatching rates for nos-vp16-GAL4, PylRS, PylTUGC flies and yellow/white “wild-type” embryos41 are similar (Supplementary Table 2), demonstrating that proteome labelling does not have a negative effect on hatching. As a newly hatched larva has, albeit in a smaller simplified form, already differentiated all the tissue of an adult animal, the study of the hatching rate shows that gastrulation and tissue differentiation are not affected by proteome labelling.
Figure 4. SORT-M enables selective imaging of proteins synthesized within the germ cells of the fly ovary from stage 5 onwards.
(a) Schematic representation of fly oogenesis, anterior to the left, green indicates the germline and white the somatic tissue (follicular epithelium). (b-d) Genetically directed, cell-type specific proteome labelling within an organ. Egg chambers from stage 4 to stage 10 of oogenesis of the indicated genotypes and feeding condition are shown. Anterior to the left, DAPI (blue), Phalloidin (red), 4g (green), (b) Egg chambers receiving 3 (10mM), expressing PylRS/PyltRNAUGC (nos-vp16-GAL4, PylRS,PylTUGC) were washed, fixed and labelled with 4g (4μM, PBS, RT, 2h). (c) Egg chambers not receiving 3, but expressing PylRS/PyltRNAUGC and labelled with 4g. (d) Egg chambers receiving 3, but not expressing PylRS/PyltRNAGCU, and labelled with 4g. scalebar shown on merge images is 20μm.
We created additional fly lines bearing PylRS/PylTGCU, PylRS/PylTCAG and PylRS/PylTCAU (in which PylRS is driven by a GAL4 UAS, and four copies of PylTXXX are transcribed by the U6 promoter) and crossed these with nos-vp16-GAL4 lines, creating nos-vp16-GAL4, PylRS, PylTXXX flies. Using these flies, we demonstrated that specific proteome labeling can also be achieved using tRNAs with additional anticodons GCU(Ser), CAG(Leu) and CAU(Met)), which decode additional distinct sense codons (Supplementary Fig. 18) and with distinct chemistry (Supplementary Fig. 19). Egg chamber development also proceeds normally for the additional anticodons tested and hatching rates are amino acid independent for all anticodons tested (Supplementary Table 2). Crossing PylRS/PylTCAU with a wing specific driver (MS1096-GAL4) enabled SORT-M labelling in the 3rd instar larval wing disc (Supplementary Fig. 20). These experiments demonstrate that the approach we have developed is modular: we can rapidly and straightforwardly change the sense codons labeled, the chemistry of labeling or the tissue and developmental stage targeted.
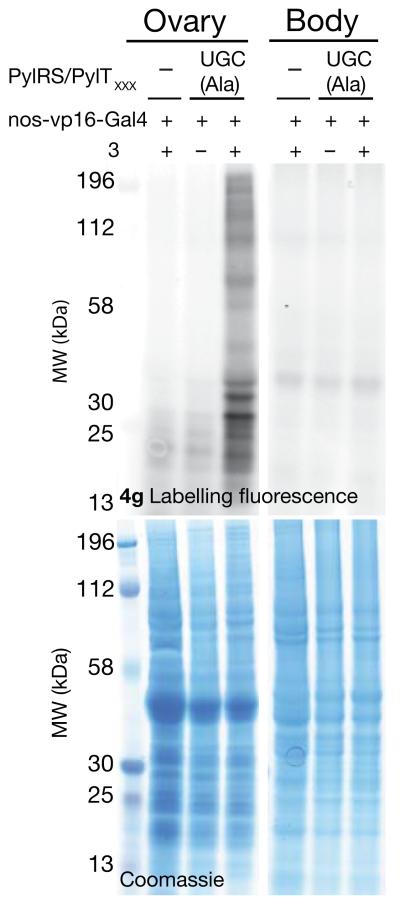
To examine the tissue specificity of the approach for labeling a target tissue versus proteins in the whole of the rest of the fly, we dissected ovaries from the rest of the fly body. We incubated proteins extracted from either the ovary or the rest of the body with 4g (20μM in PBS, 2h at RT) in parallel experiments. We observe clear strong, and specific labeling of the ovary proteome with minimal labeling of the rest of the body (Fig. 5a). The labeling is amino acid and PylRS dependent, further confirming the specificity of the approach (Fig. 5a and Supplementary Fig. 21a). Additional experiments demonstrated similar specific proteome labeling with a range of tetrazine-fluorophore conjugates (Supplementary Fig. 21b and Supplementary Fig. 22), and that the fly proteome is not substantially altered by the incorporation of 3 (Supplementary Fig. 23).
Figure 5. SORT-M facilitates tissue specific labelling of the fly proteome.
The ovary proteome, but not the rest of the body proteome from 20 female flies expressing PylRS/PyltRNAUGC (nos-vp16-GAL4, PylRS,PylTUGC) fed with 3 (10 mM) is labelled with 4g (20μM, PBS, RT, 2h, lysate 30 mg/ml)) . Labelling requires feeding 3 and the PylRS/PyltRNAUGC pair.
SORT-M defines tissue- and stage-specific proteins in flies
To demonstrate that SORT-M can be used to identify proteins from the proteome of the fly that are synthesized in a target tissue without dissection, we developed a two-dimensional difference gel electrophoresis (2D-DIGE)–based proteomic approach. We focused on demonstrating this approach for identifying proteins from the fly proteome that are synthesized in the germ cells of the ovary, because the ovary can be easily dissected from the rest of the body. This enables us to validate that proteins identified by SORT-M are indeed present in the dissected ovary, and define the utility of the approach for identifying proteins expressed in specific tissues at specific times without dissection.
We first dissected the ovary from the rest of the body and extracted proteins from each sample. The ovary proteome (follicular epithelium, germ cells and any ovary associated proteins carried over in dissection) was non-specifically labeled on lysine residues with a Cy3 dye, and in a parallel experiment the body proteome was non-specifically labeled on lysine residues with a Cy5 dye. Following the labeling step, the proteins were subject to 2D gel electrophoresis and Cy3 and Cy5 imaged independently to reveal the pattern of labeled proteins in the ovary and the pattern of labeled proteins in the rest of the body (Fig. 6a,b)42. This approach labels proteins present in either ovaries or bodies regardless of the timing or site of their synthesis, and allows us to define spots in the 2D gel labeled by this approach that correspond to proteins in the ovary and proteins in the body.
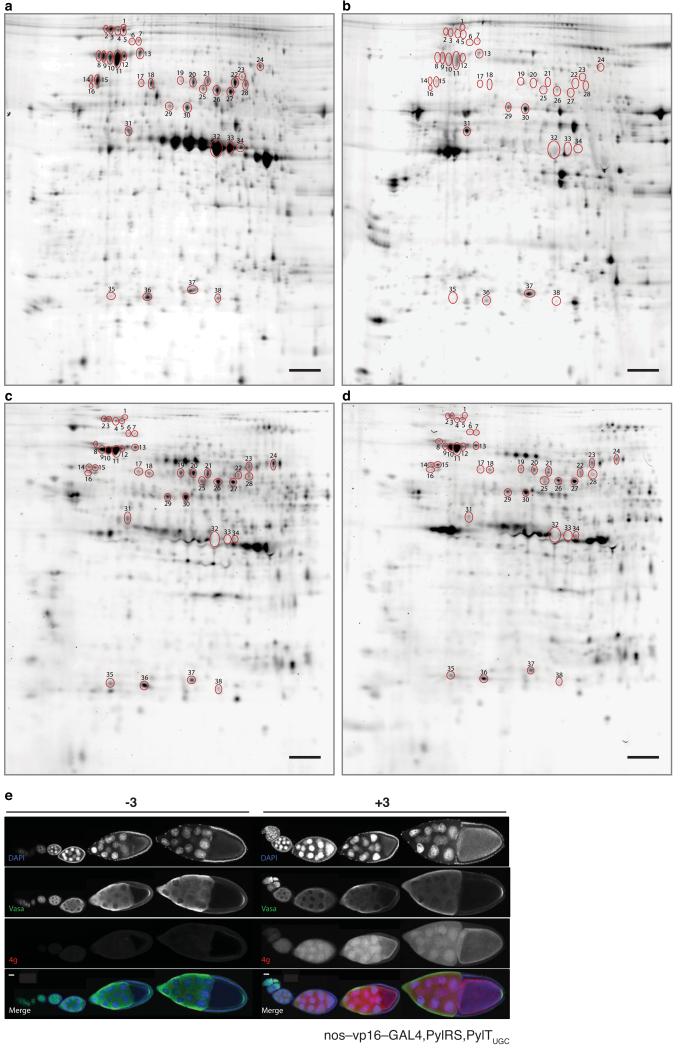
Figure 6. Tissue and developmental stage specific proteome labelling, protein identification and validation.
(a) In-gel fluorescence image of a 2D-gel of protein extracts from a dissected fly ovary that has been labelled non-specifically with an amine reactive NHS-Cy3 dye and then imaged. This gel acts as a reference for an ovary proteome. (b) In-gel fluorescence image of a 2D-gel of a protein extract from fly bodies that have had their ovaries removed. The extract has been labelled non-specifically with an amine reactive NHS-Cy5 dye. This gel acts as a reference for a fly body only proteome. (c) In-gel fluorescence image of protein extracts from ovaries dissected from flies fed with 3, then labelled with 4e and mixed with protein extracts from the remaining body parts. (d) In-gel fluorescence image of protein extracts from ovaries dissected from flies expressing PylRS/PyltRNAUGC fed with 3, mixed with protein extracts from the body and then labelled with 4e. The gut of body samples was removed to limit proteolysis in protein extracts. All experiments used nos-vp16-GAL4, PylRS/PylTUGC flies. Circled spots were excised for mass spectrometry. Scale bar on all 2D gels is 10 mm (e) The timing of Vasa protein expression and its pattern of immunostaining overlaps with that of SORT-M labelling, validating its identification by SORT-M. Ovaries from nos-vp16-GAL4, PylRS, PylTUGC flies fed with or without 3, were dissected and immunostained for Vasa (identified as Spot 13 from the 2D gels) and DAPI and labelled with 4g. Scale bar is 20 μm.
Next we dissected nos-vp16-GAL4, PylRS, PylTUGC flies fed 3 and labeled the proteome of the isolated ovary with 4e. We subjected this labeled sample, along with the unlabeled proteome from the rest of the body, to 2D gel electrophoresis. Many of the fluorescent spots on the cyclopropene incorporated sample labeled with 4e, (Fig. 6c) were matched to spots on the ovary sample non-specifically labeled with Cy3, as expected for specific ovary labeling with 4e. We also observe spots that are present in the Cy3 labeling but not in the tetrazine labeling and vice versa. We do not expect a perfect match between the Cy3 and 4e labeling because: (i) the labeling methods use distinct chemistry and will label different proteins with different efficiencies; (ii) 4e only labels protein synthesized in the germ cells of the ovary in a defined time period, while Cy dye-NHS labelling may label lysine containing proteins present in the whole ovary regardless of where or when they are synthesized; and (iii) Cy dye-NHS labelling of ovary samples will label protein contaminants that may be carried over from the body upon dissection.
We were particularly intrigued by a striking series of intense spots in the Cy3 labeling (Fig. 6a, spots 32, 33, 34) that were absent in the cyclopropene labeling of ovaries (Fig. 6c). To understand the basis of this observation, we picked these spots and identified the protein by mass spectrometry (Supplementary Table 3) as vitellogenins, proteins that make up the yolk in the ovary. The majority of vitellogenin in Drosophila is synthesized in the fat body, and accumulates in the ovary by pinocytosis,43 but vitellogenin is also synthesized within the follicular epithelium of the ovary leading to high levels of vitellogenin mRNA in the ovary44 (Supplementary Table 3). This result provides an example of how comparing our co-translational tagging and labeling approach to non-specifically labeled ovary proteins allows proteins that reside inside the ovary, but are synthesized outside the germ cells of the ovary, to be distinguished from proteins that are synthesized in the germ cells of the ovary. The abundance of vitellogenin, but its minimal labeling with tetrazine probes provides an additional measure of the specificity of the labeling process for the encoded cyclopropene.
To demonstrate that we can identify ovary proteins without dissection, we picked thirty-five additional protein spots that are labeled with 4e in the whole fly (Fig. 6, Supplementary Fig. 24 and Supplementary Table 3) from gels that contain both ovary and body proteins, and a gel on which only a dissected ovary sample had been run (Supplementary Fig. 24). Spots 1 to 28 were selectively labeled in the dissected ovary over the body (Compare Fig. 6a,b) and are synthesized in the germ cells of the ovary (Fig. 6c,d). We identified a range of proteins including a ubiquitin hydrolase (spot 1), TER94 (spot 2, an AAA+ ATPase implicated as a component of the fusome45) and hsp70 (spots 2–5), a dipetidyl-peptidase (spots 6 and 7), heat shock protein (hsp) 70 and hsp83 (spots 8–13), Vasa (spot 13, a dead box helicase that is essential for formation of the pole plasm in the posterior portion of the egg46, a Phosphotase 2A regulatory subunit A family protein (spots 14–16), proteins in the TCP-1 chaperone family complex (spots 14–27), inositol 3 phosphate synthetase (spot 17), Peroxiredoxin 1 (spots 35, 36 and 38) and a proteasome subunit (spot 36). Each spot contains proteins whose mRNA is enriched in the ovary relative to the carcass and other adult female tissue in modENCODE44 (Supplementary Table 3). Spots 29–31, 36 and 37 contain proteins with comparable or stronger labeling in the fly body than the ovary and proteins identified from these spots have comparable mRNA levels in the ovary and body.
To explicitly demonstrate that Vasa, one of the proteins we identified, is present in the egg chambers at the time, and in the cells where our protein labelling system is active, we immunostained for Vasa in the ovaries of nos-vp16-GAL4, PylRS,PylTUGC flies fed 3 and also labelled the ovaries with 4g (Fig. 6e). We observe Vasa protein in the germ cells, but not in the follicular epithelium, over all the stages where SORT-M is active in the ovary, consistent with the co-translational labelling of vasa by SORT-M. As expected, we observe Vasa staining in the cytoplasm of nurse cells throughout ovary development and its accumulation at the posterior of the oocyte for pole plasm assembly at later stages.46 These experiments clearly demonstrate that the power of the approach we have developed for identifying proteins present in a specific tissue of the whole animal without dissection.
DISCUSSION
We have characterized the genetically encoded, site-specific incorporation of a cyclopropene containing amino acid 3, and demonstrated the quantitative labelling of 3, with tetrazine probes, in proteins expressed in E. coli, mammalian cells and D. melanogaster.
We have defined a paradigm for tagging the proteome with the growing list of amino acids that can be incorporated using the orthogonal pyrrolysyl-tRNA synthetase/tRNA pair and its active site derivatives.13 The approach, SORT, requires the conversion of the anticodon of the pyrrolysyl tRNA from CUA to a sense decoding anticodon, and enables the stochastic tagging of newly synthesized proteins with an unnatural amino acid via an orthogonal translational pathway. SORT-M enables the proteomes of E. coli, mammalian cells and genetically defined sets of cells in the fly to be tagged with a cyclopropene group and selectively labelled with diverse tetrazine probes. Integration with powerful analytical methods allows the identification of proteins synthesized in defined cells.
By tagging the proteome at different codons in parallel experiments, and combining the information, it will be possible to provide a more complete description of the proteome than that obtained from single amino acid replacement approaches. This is likely to be crucial for obtaining good coverage in proteome tagging and modification, as although a single codon may be found at least once in a large fraction of open reading frames, not all proteins will incorporate the unnatural amino acid efficiently and not all the sites in the proteome that incorporate the unnatural amino acid will be accessible to labelling.6 Future extensions of SORT may include the proteome-wide incorporation of photocrosslinkers47 to map tissue-specific protein interactions and, the development of SORT based methods in other organisms.
Online Methods
Chemical syntheses - general methods
All chemicals and solvents were purchased from Sigma-Alrich, Alfa Aesar or Fisher Scientific and used without further purification unless otherwise stated. Qualitative analysis by thin layer chromatography (TLC) was performed on aluminium sheets coated with silica (Merck TLC 60F-254). The spots were visualized under short wavelength ultra-violet lamp (254nm) or stained with basic, aqueous potassium permanganate, ethanolic ninhydrin or vanillin. Flash column chromatography was performed with specified solvent systems on silica gel 60 (mesh 230-400).
LC-MS analysis was performed on Agilent 1200 machine. The solvents used consisted of 0.2 % formic acid in water (buffer A) and 0.2 % formic acid in acetonitrile (buffer B). LC was performed using Phenomenex Jupiter C18 column (150 × 2 mm, 5μm) and monitored using variable wavelengths. Retention times (Rt) are recorded to a nearest 0.1 min and m/z ratio to nearest 0.01 mass units. The following programme was used for small molecule LC gradient: 0-1 min (A:B 10:90-10:90, 0.3 mL/min), 1-8 min (A:B 10:90-90:10, 0.3 mL/min), 8-10 min (A:B 90:10-90:10, 0.3 mL/min), 10-12 (A:B 90:10-10:90, 0.3 mL/min).
Mass spectrometry analysis following LC was carried out in ESI mode on a 6130 Quadrupole spectrometer and recorded in both positive and negative ion modes. NMR analysis was carried out on a Bruker 400MHz instrument. All reported chemical shifts (δ) relative to TMS were referenced to the residual protons in deuterated solvents used: d1 – chloroform (1H δ = 7.26 ppm, 13C δ = 77.16 ppm), d6 – dimethylsulfoxide (1H δ = 2.49 ppm, 13C δ = 39.52 ppm), D2O (1H δ = 4.70). APT or two-dimensional experiments (COSY, HSQC) were always performed to provide additional information used for analysis where needed. Coupling constants are given in Hz and described as: singlet – s, doublet – d, triplet – t, quartet – q, broad singlet – br, multiplet – m, doublet of doublets – dd, etc. and combinations thereof.
Protein expression, purification and labelling of site-specifically incorporated 3 in E. coli
Expression and purification of sfGFP-3 from E. coli
Electrocompetent E. coli DH10B cells were co-transformed with pBK-MbPylRS and psfGFP150TAG PylT14, 26. Transformed cells were recovered in S.O.B. (1 mL, supplemented with 0.2% glucose) for 1 h at 37 °C and used to inoculate LB containing 50 μg/mL kanamycin and 25 μg/mL tetracycline (LB-KT). The cells were incubated with shaking overnight at 37 °C, 250 r.p.m. 1 mL of overnight culture was used to inoculate 100 mL of LB-KT½, the day culture was then incubated (37 °C, 250 r.p.m). At O.D.600 ~0.3, the culture was divided equally and supplemented with either 3 (1 mM) or H2O (500 μL) and incubated further (37 °C, 250 r.p.m). At O.D.600 ~0.6 protein expression was induced by the addition of arabinose (0.2%), after 4 h, the cells were harvested by centrifugation (4000 r.p.m, 20 min) and the pellet frozen until further use.
The frozen bacterial pellet was thawed on ice and resuspended in 2.5 mL lysis buffer (Bugbuster®, Novagen®, 50 μg/mL DNAse 1, Roche inhibitor cocktail and 20 mM imidazole). Cells were incubated (4 °C, 30 minutes) then clarified by centrifugation (16000 g, 4 °C, 30 minutes). The clarified lysates were transferred to fresh tubes and 100 μL Ni-NTA slurry added. The mixtures was incubated with agitation (4 °C, 1 h) and then collected by centrifugation (1000 g, 4 °C, 5 min). The beads were resuspended three times in 500 μL wash buffer (10 mM Tris-HCL, 40 mM imidazole, 200 mM NaCl, pH 8) and collected by centrifugation (1000 g, 4 °C, 5 min). Finally, the beads were resuspended in 100 μL elution buffer (10 mM Tris-HCL, 300 mM imidazole, 200 mM NaCl, pH 8), pelleted by centrifugation (1000 g, 4 °C, 5 min) and the supernatant collected into fresh tubes. The elution was repeated three times with 100 μL of elution buffer. The purified proteins were analysed by 4-12% SDS-PAGE and LC-MS.
Protein Mass Spectrometry
Using an Agilent 1200 LC-MS system, ESI-MS was additionally carried out with a 6130 Quadrupole spectrometer. The solvent system consisted of 0.1 % formic acid in H2O as buffer A, and 0.1 % formic acid in acetonitrile (MeCN) as buffer B. Protein UV absorbance was monitored at 214 and 280 nm. Protein MS acquisition was carried out in positive ion mode and total protein masses were calculated by deconvolution within the MS Chemstation software (Agilent Technologies).
In vitro labeling of purified sfGFP150-3
To Purified sfGFP150-1 or sfGFP150-3 protein (~30 μM, in elution buffer) was added 4a (10 molar equivalents, from a 2 mM stock solution in DMSO). The reactants were mixed by aspirating several times and the mixture then incubated at room temperature for 2 hours, a sample was analysed by ESI-MS. Following incubation the proteins were separated by 4-12% SDS-PAGE and analysed by using Typhoon Trio phosphoimager (GE Life Sciences).
Time course of sfGFP150-3 and sfGFP150-NorK labelling and rate constant determination
2 nmol sfGFP-3 (10.6 μM) was labeled at room temperature by the addition of 20 nmol of tetrazine-dye conjugate 4a (10μl of a 2 mM solution in DMSO) the samples were mixed by aspirating several times. At different time points, 8 μL aliquots were taken from the solution and quenched with a 700-fold excess of bicyclo[6.1.0]non-4-yn-9-ylmethanol (BCN) and plunged into liquid nitrogen. Samples were mixed with NuPAGE LDS sample buffer supplemented with 5 % β-mercaptoethanol, heated for 10 min to 90°C and analyzed by 4-12% SDS page. The amounts of labelled proteins were quantified by scanning the fluorescent bands with a Typhoon Trio phosphoimager (GE Life Sciences). Bands were quantified with the ImageQuant™ TL software (GE Life Sciences) using rubber band background subtraction. The rate constant was determined by fitting the data to a single-exponential equation. The calculated observed rate k’ was divided by the concentration of 4a to obtain rate constant k for the reaction. Measurements were done in triplicate. All data processing was performed using Kaleidagraph software (Synergy Software, Reading, UK). For comparison the rate of labelling sfGFP bearing Ne-5-norbornene-2-yloxycarbonyl-L-lysine (NorK), a known substrate for PylRS, was determined in a similar way using 11.25 mM sfGFP bearing NorK at position 150 (SfGFP-NorK) and 20 equivalents of 4a.
E. coli SORT-M
Plasmid construction for pBAD_wtT4L_MbPylTXXX
pBAD_T4L83TAG_MbPylTCUA was digested with NcoI and KpnI restriction enzymes. The same restriction enzymes were also used to digest the wild-type T4 lysozyme from (D67) pBAD_wtT4L. The insert and backbone were ligated in 3:1 ratio using T4 DNA ligase (RT, 2 hours), transformed into chemically competent DH10B cells and grown on Tetracycline agar plates (37°C, 18 hours). Single colonies were picked and the correct sequence was confirmed by DNA sequencing (GATC Gmbh.), this step created intermediate pBAD_wtT4L_MbPylTCUA. This vector was subjected to site directed mutagenesis using primers targeting the CUA anticodon sequence of the tDNA in order to replace the amber anticodon with anticodons that decode sense codons creating a set of pBAD_wtT4L_MbPylTXXX, all final constructs were confirmed by DNA sequencing. A list of primers used for mutagenesis can be found in Supplementary Table 1.
Proteomic incorporation of 3 via SORT in E. coli expressing T4 lysozyme
Electrocompetent E. coli DH10B cells (50 μL) were either doubly transformed with pBAD_wtT4L_MbPylTXXX plasmid (2 μL, necessary for expression of PyltRNAXXX and expresses T4 lysozyme under arabinose control) and pBKwtPylS plasmid (2 μL necessary for expression of PylRS) or singly transformed with pBAD_wtT4L_MbPylTXXX alone. Transformed cells were recovered in 1 mL S.O.B. (supplemented with 0.2% glucose) for 1 h at 37 °C. 100 μL of the recovery was used to inoculate 5 mL LB-KT (50 μg/mL kanamycin and 25 μg/mL tetracycline) or LB-T (25 μg/mL tetracycline). Cultures were incubated overnight (37 °C, 250 r.p.m.). 1 mL of each overnight culture was used to inoculate 15 mL ½ strength antibiotic containing media LB-T or LB-KT. Cultures were incubated at 37 °C until O.D.600 ~0.3 was reached, at this time each culture was divided into 5 mL aliquots and supplemented with either 3 (0.1 mM final conc.) or H2O (50 μL). Cultures were then incubated (37 °C, 250 r.p.m.). At O.D.600 0.6. T4 lysozyme expression was initiated by the addition of arabinose (0.2% final conc.) and cultures incubated for a further 4 hours. Cells were harvested by centrifugation (4000 rpm, 4 °C, 20 minutes) and then resuspended three times in 1 mL of ice cold PBS and collected by centrifugation (4000 rpm, 4 °C, 20 minutes). The final bacterial pellets were immediately frozen for storage.
E.coli SORT-M: Chemoselective labelling proteomes tagged with 3 by SORT with tetrazine-dye conjugates
Frozen bacterial pellets were resuspended in 500 μL PBS and lysed using a bath sonicator (energy output 7.0, 90 s total sonication time. 10 s blasts and 20 s breaks, Misonix Sonicator 3000). The lysate was cleared by centrifugation (4 °C, 14000 r.p.m., 30 min) and the supernatant aspirated to a fresh tube. To 50 μL of cleared cell lysate was added 4a (2 mM, stock in DMSO, final concentration – 20 μM). The reactions were mixed by aspirating several times and the samples then incubated in the dark (room temperature, 1 h). After this time 17 μL of 4× LDS sample buffer supplemented (6 mM BCN and 5% BME) was added and mixed by vortexing gently. Samples were incubated for 10 min before boiling at 90 °C for 10 min. Samples were analysed by 4-12% SDS-PAGE and fluorescent images were acquired using Typhoon Trio phosphoimager (GE Life Sciences).
The same protocol for fluorescent labelling of the E. Coli proteome was applied for all tetrazine-dye conjugates.
E.coli SORT growth curve analysis for toxicity assessment of SORT
Chemically competent DH10B cells (50 μL) were transformed with two plasmids: pBKwtPylRS (2 μL, necessary for expression of PylRS) and pBAD_wtT4L_MbPylTXXX (2 μL, necessary for expression of PyltRNAXXX and expresses T4 lysozyme under arabinose control). The cells were recovered in 1 ml SOB medium for one hour at 37°C prior to aliquoting to 10 ml LB-KT (LB media with 50 μg ml−1 kanamycin, and 25 μg ml−1 tetracycline) and incubated overnight (37°C, 250 rpm, 12 h). The overnight culture (OD600~3) was diluted to a OD600~0.3 in 10 mL LB-KT1/2 (LB media with 25 μg ml−1 kanamycin, and 12.5 μg ml−1 tetracycline) supplemented with 3 at different concentrations, 0, 0.1, 0.5 mM. 200 μL aliquots of these cultures were transferred into a 96-well plate and OD600 measured using a Microplate reader, Infinite 200 Pro (TECAN). OD600 was measured for each sample every 10 min with linear 1 mm shaking between the measurements.
E.coli SORT-M time dependence and concentration dependence of 3 at the AAA codon
Chemically competent DH10B cells (50 μL) were transformed with two plasmids: pBKwtPylRS (2 μL, necessary for expression of PylRS) and pBAD_wtT4L_MbPylTUUU (2 μL, necessary for expression of PyltRNAUUU and expresses T4 lysozyme under arabinose control). After transformation, cells were recovered in 1 ml SOB medium for one hour at 37°C prior to inoculation in 10 ml LB-KT (LB media with 50 μg ml−1 kanamycin, and 25 μg ml−1 tetracycline). The culture was incubated overnight (37°C, 250 rpm, 12 h) and subsequently diluted to an OD600~0.3 in 30 mL LB-KT1/2 (LB media with 25 μg ml−1 kanamycin, and 12.5 μg ml−1 tetracycline) then supplemented with 3 at different concentrations, 0, 0.1, 0.5 mM. The cultures were incubated (37°C, 250 rpm) for 1 h, when OD600~0.6 was reached, 2 ml culture aliquots were collected in a separate tube for each of three cultures. Subsequently arabinose was added at a final concentration of 0.2% (v/v) to induce expression of T4 lysozyme and culture aliquots of 2 mL were collected every hour. For each of the collected cultures, bacterial cells were pelleted by centrifugation at 4°C, washed with ice cold PBS (3 × 1 mL) and subsequently the pellets were frozen and stored at −20°C. The pellets were then thawed in 200 μL of ice cold PBS and lysed by sonication (9 × 10 s ON / 20 s OFF, 70% power). The lysates were clarified by centrifugation at 15,000 RPM, 4 °C for 30 minutes. The supernatants were transferred to fresh 1.5 mL tubes. 50 μL of supernatant was transferred to a new tube for the labeling reactions, and the rest was frozen in liquid nitrogen and stored at −80 °C. To the 50 μL of supernatant, 0.5 μL of 2 mM 4a (20 μM) was added and the lysates were incubated at 25 °C for 1 hour. After 1h, 17 μL of 4× LDS sample buffer supplemented (6mM BCN and 5% BME) was added and mixed by vortexing gently. Samples were incubated for 10min before boiling at 90 °C for 10 min. Samples were analysed by 4-12% SDS-PAGE and fluorescent images were acquired using Typhoon Trio phosphoimager (GE Life Sciences).
Proteomic incorporation of 2 via SORT in E. coli
Chemically competent E. coli DH10B cells were transformed with the plasmids pCDF_Duet_MbPylTXXX and pBKwtPylS. The transformation was recovered in 600 μl SOC media for 1 hr at 37°C. 200 μl of the recovered cells were used to inoculate 100 ml of (1× LB-KS). The culture was grown overnight before inoculation of 15 ml of LB-KS½. The culture was grown to O.D.600 ~0.4 before it was divided in two 5 ml subcultures and 2 was added to appropriate tubes to final concentration of 10 mM. The cultures were grown for 4 hours (37°C, 250 r.p.m.) before the cells were spun down for 15 mins, (4000 r.p.m., 4°C). The cell pellet was washed twice with 5 ml of PBS before it was frozen at −20°C or further processed.
E. coli SORT-M: Chemoselective labelling proteomes tagged with 2 by SORT in a copper catalysed 3+2 biotinylation
Cells were re-suspended in ice cold 500 μl PBS (pH 8) and lysed using a bath sonicator (energy output 7.0, 90 s total sonication time. 10 s blasts and 20 s breaks, Misonix Sonicator 3000). The cell lysate was spun at 4°C for 15 minutes at 14000 rpm and the cleared cell lysate was aspirated to a fresh tube. Lysate was then incubated with 10 uL of neutravidin agarose beads suspension (Thermo Scientific cat.n. 29200) for 2h at room temperature on a rotating shaker. After 2h samples were centrifuged for 30s on a table top centrifuge at room temperature to pellet endogenously biotinylated proteins, and the supernatant was collected. Protein concentration of all samples was then determined using a BCA assay (Thermo Scientific) according to manufacturer’s protocol for microplate settings.
Click reaction pre-mix was prepared as follows: 1 aliquot of 10 mM CuSO4 was mixed with 1 aliquot 10 mM TCEP and left to react with occasional vortexing for approximately 5 min, 1/10 of the aliquot of the TBTA ligand (100 mM stock solution) is then added and the solution vortexed for another 5 min until off-white solution is formed. 21 μl of the pre-mix was then mixed with 78.5 μl of each sample and 0.5 μl of 10mM DMSO stock Biotin-azide-PEG4 (from Life Technologies Ltd.) before the samples were allowed to react at 24°C for 2 hours with vigorous shaking. The reaction was stopped by addition of NuPAGE LDS Sample buffer (4×) supplemented with β-mercaptoethanol, the sample was heated to 90°C for 10 min and centrifuged for another 10 min at 13200rpm. The samples were separated on 4 – 12% Bis-Tris 15-well gel using NuPAGE MES SDS 1× Running buffer. Freshly run gels were carefully transferred onto a nitrocellulose membrane supplied as part of the iBlot device for semi-dry Western blotting. The membrane was then washed for 1 hr with TBS-T (20mM Tris.HCl pH = 7.4, 137mM NaCl, 0.1% Tween 20) before blocking with 5% BSA in TBS-T solution. Primary rabbit-anti-biotin antibody (1:1,000) was used for detection of tagged proteins and allowed to bind overnight at 4°C with shaking, washed 3 times with TBS-T before the secondary antibody (anti-rabbit-HRP 1:10,000) was added for 1hr and washed extensively with TBS-T. The membrane was developed using ECL western blotting detection system from GE Healthcare in PeqLab Fusion high sensitivity CCD device.
Site-specific incorporation of 3 in HEK293 cells and chemoselective labelling with tetrazine probes
Site specific incorporation of 3 in HEK cells
HEK293 Cells (ATCC CRL-1573) were plated on 24 well plates and grown to near confluence. The cells were transfected using Lipofectamine 2000 (Invitrogen) with the pMmPylS-mCherry-TAG-EGFP-HA construct and the p4CMVE-U6-PylT construct.18 After 16hrs growth with or without 1mM 3 or with 1mM 1 the cells were lysed on ice using RIPA buffer (Sigma). The lysates were spun down and the supernatant was added to 4× LDS sample buffer (Life technologies). The samples were run out by SDS-PAGE, transferred to a nitrocellulose membrane and blotted using primary rat anti-HA(clone 3F10, Roche, No. 1 867 423) and mouse anti-FLAG (clone G191, Abnova, cat. MAB8183), the secondary antibodies were anti-rat (Invitrogen, A11077) and anti-mouse (Cell Signaling Technologies, No. 7076S).
Labelling site-specifically incorporated 3 from HEK 293 cells
Adherent HEK293T cells (ATCC CRL-11268; 4×106 per immunoprecipitation) were transfected with 7.5 μg p4CMVE-U6-PylT and 7.5ug pPylRS-mCherry-TAG-EFGP-HA 18 using TransIT-293 transfection reagent according to the manufacturer’s protocol and cultured for 48 hours in DMEM/10%FBS, supplemented with 0.5 mM 1 or 2 mM 3 where indicated. Cells were washed twice with PBS and lysed on ice for 30 minutes in 1mL Lysis Buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris HCl (pH 8.0). After clarifying the lysate by centrifugation (10 min at 16000g), HA-tagged proteins were captured using 50 μL μMACS HA-tag MicroBeads (Miltenyl Biotec) per transfection, washed with 0.5 mL RIPA (150 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris HCl (pH 8.0) and 0.5 mL PBS (pH 7.4). The suspension of MicroBeads was incubated with 50 μL PBS (pH 7.4), 20 μM 4a for 1 hour and subsequently washed with 0.5 mL RIPA to remove excess dye. HA-tagged proteins were eluted from beads using SDS sample buffer and separated on a 4-12% Bis-Tris PAGE gel (Invitrogen), imaged using a Typhoon imager (GE Healthcare) and subsequently stained with DirectBlue or transferred for western blotting with Anti-HA-tag pAb-HRP-DirecT (MBL).
SORT-M in mammalian cells
Cells were plated on 24 well plates and grown to near confluence. The cells were transfected using Lipofectamine 2000 or TurboFect (Thermo Scientific) with the pCMV-MmPylS18 construct and either the pU6-PyltRNACAU, pU6-PyltRNACAC, pU6-PyltRNACUU, pU6-PyltRNACUC, pU6-PyltRNAGUA, or pU6-PyltRNACCA constructs. After 48 hours growth with or without 0.5 mM 3 or with 0.5 mM 1 the cells were lysed by sonication in ice cold PBS. The lysates were clarified by centrifugation at 4°C and 21,000 × g, and the tetrazine-dye conjugate 4g was added to the supernatants to a final concentration of 4 μM. The labelling reaction was incubated at 25°C for 60 minutes. The reactions were then quenched by heating to 95°C in 4× LDS sample buffer (Life technologies) supplemented with BCN to give a final concentration of 1 mM, and analyzed by SDS-PAGE. After destaining in ultrapure water, the gels were imaged on a Typhoon Trio imaging system (GE Healthcare).
Expression and purification of SfGFP from mammalian cells
HEK293T were transfected in a 10cm tissue culture dish with 15ug DNA using PEI and incubated for 72 hours with 3 (0.5 μM). Cells were washed twice with PBS and lysed in 1mL RIPA buffer. Cleared lysate was added to 50μL GFP-Trap® M (ChromoTek) and incubated for 4h. Beads were washed with 1mL RIPA, 1mL PBS, 1mL PBS+500mM NaCl, 1mL ddH2O and eluted in 1%Acetic Acid/ddH2O. Purified protein was labeled with 2μM 4a for 4h and loaded on a 4-12% Bis-Tris PAGE gel. Fluorescence of 4a-labeled sfGFP was detected on a Typhoon imager and gel was stained subsequently with DirectBlue.
Fly plasmids, transgenic flies and culture
For all fly experiments no randomisation or blinding was used within this study.
Plasmid construction for transgenic fly line generation
The PyltRNACUA anticodon was mutated using the QuikChange mutagenesis kit and pSG108 (pJet 1.2-U6-PylT, gift from S. Greiss) as a template. This contains the PylT gene without its 3′ terminal CCA fused to the Drosophila U6-b promoter. Primers FMT19 and FMT20 were used to generate PyltRNATGC to decode alanine codons (creating pFT18); primers FMT23 and FMT24 were used to generate PyltRNAGCT to decode serine codons (creating pFT20); primers FMT27 and FMT28 were used to generate PyltRNACAG to decode leucine codons (creating pFT22) and primers FMT29 and FMT30 were used to generate PyltRNACAT to decode methionine codons (creating pFT23). The mutated tRNA expression cassettes were subcloned from pFT18, pFT20, pFT22 and pFT23 into pUC18 using EcoRI and HinDIII then multimerised using AsiSI, BamHI and BglII to create 2, then 4 copies of the tRNA. The 4 copy versions of the tRNA cassette were subcloned into pSG118 using AsiSI and MluI to create pFT58 (Ala), pFT60 (Ser), pFT62 (Leu) and pFT63 (Met). pSG118 contains the M.mazei PylRS gene.20
Fly lines and culture conditions
Transgenic lines were created by P element insertion using a Drosophila embryo injection service (BestGene Inc.). Lines were generated using the following plasmids: pFT58 (Ala), pFT60 (Ser), pFT62 (Leu) and pFT63 (Met). nos-Gal4-VP16 (Bloomington 4937) and MS1096-Gal4 (Bloomington 8860) were used as Gal4 drivers. All flies were grown at 25°C on standard Iberian medium. Flies were fed unnatural amino acids by mixing dried yeast with the appropriate concentration of amino acid (usually 10mM) diluted in dH2O to make a paste. Ovaries were prepared from females that were grown on Iberian fly food supplemented with a yeast paste with or without the amino acid for a minimum of 48 hours. For proteome labelling experiments transgenic male flies of constructs FT58, FT60, FT62 and FT63 were crossed with nos-vp16-GAL4 virgins to generate FT58/nos-vp16-GAL4, FT60/nos-vp16-GAL4, FT62/nos-vp16-GAL4 and FT63/nos-vp16-GAL4 respectively.
Site specific incorporation of 3 in D. melanogaster
Luciferase assays
Ovaries from 10 females of Triple Rep-L flies recombined with nos-Gal4-VP16 fed 3, 1 or no amino acid were dissected in 100μl 1× Passive lysis buffer and processed for luciferase assays as previously described 20.
Immunoprecipitation and labelling of site specifically incorporated 3
Ovaries from 100 (for control and 3) or 500 (for 1) females were dissected in PBS then lysed in 300 or 1500 μl RIPA buffer containing 1× complete protease inhibitor cocktail (Roche). A sample was taken into 4 × LDS buffer as a total lysate control then the remainder was used for immunoprecipitation with GFP-TRAP agarose beads (Chromotek) following the manufacturer’s instructions. The total volume of the IP was 3ml. After overnight incubation, the beads were washed 2 × with RIPA buffer then 2 × with PBS. For tetrazine labeling, the beads were resuspended in 200μl PBS + 4μM 4g and incubated for 2 hours on a roller at RT. The beads were washed 3 times with 500 μL of wash buffer then resuspended in 4× LDS sample buffer.
Tissue and cell-type specific SORT-M in D. melanogaster
Tissue specific SORT-M in fly ovary extract
For SORT-M transgenic male flies of constructs FT58, FT60, FT62 and FT63 were crossed with nos-vp16-GAL4 virgins to generate FT58/nos-vp16-GAL4, FT60/nos-vp16-GAL4, FT62/nos-vp16-GAL4 and FT63/nos-vp16-GAL4 respectively. In feeding experiments adult females were fed the indicated amino acid in a yeast paste at a concentration of 10 mM.
20 female flies of the indicated genotype and with the indicated feeding regime were dissected and their ovaries harvested in PBS. The remnant of the body excluding the gut was also harvested in PBS. Ovaries and body remnants were smashed using a pestle in PBS and the sample was centrifuged at 4°C for 10 min at 13,000 rpm. The Supernatant was filtered through a DNA extraction column (Qiagen) then the concentration of the samples was measured using a Nanodrop. The samples were adjusted to the same concentration (typically 30mg/ml) using PBS then incubated for 30 min at 37°C with DNase I. The samples were then incubated on a shaker with either 4d or 7 or 4g at a concentration of 20 μM for 2h at room temperature in the dark. Samples were boiled in LDS and loaded on a SDS PAGE gel. Equal amounts of each sample were loaded onto two gels, one of which was stained using Instant Blue (Expedeon cat. N. ISB1L), the other was imaged using a Typhoon Trio scanner (GE Healthcare) detecting absorption at 488, 594 and 633 nm for 4d, 7 and 4g respectively.
Imaging cell-type specific SORT-M in ovaries
15 female flies of the indicated genotype and with the indicated feeding regime were dissected and their ovaries harvested in PBS. Ovaries were fixed for 20 min in 4% paraformaldehyde in PBS then permeabilized in PBS + 0.1% Triton-X-100 (PBT) for 2 × 20 min at room temperature. Ovaries were washed for 20 min at room temperature in PBS then incubated overnight at 4°C in fresh PBS. In the case of flies fed with 2, ovaries were washed 3 × 15 min in PBS pH 8 and then incubated in PBS pH 8, 1mM CuSO4, 1mM TCEP, 100μM TBTA, 1mM Alexa 488-azide (Life Technologies, A10266) for 2h at room temperature in the dark. In the case of flies fed with 3, ovaries were incubated on a shaker in PBS + 20 μM 4g for 2h at room temperature in the dark. Ovaries were washed 3 × PBT and incubated for 4 hours in PBT with DAPI (Invitrogen, D3571) and Alexa-549 conjugated Phalloidin (Life Technologies, A12381). The ovaries were then washed 3 × 20 min with PBT, then mounted in Vectashield Mounting medium (Vector Laboratories). Images were collected using a Zeiss 780 confocal microscope with a 20× or 40× magnification. For the anti-Vasa staining the ovaries were incubated with primary antibody prior to the labelling reaction. Following the 2 × PBT washes the ovaries were blocked for 1 hour in PBT + 10% BSA, then incubated overnight at 4°C with rat anti-Vasa (Developmental Studies Hybridoma Bank) diluted 1:10 in PBT + 1% BSA. The ovaries were washed 2 × PBT then 2 × PBS and incubated with 20μM 4g as described above. Secondary antibody Alexa 568 anti-rat (Invitrogen A11077) was then included in the incubation with DAPI in place of the Alexa-549 conjugated Phalloidin.
Imaging cell-type specific SORT-M in wing imaginal discs
Wing imaginal discs were dissected from 3rd instar larvae resulting from a cross between MS1096-Gal4 virgins and FT63 transgenic males. The discs were fixed 2 × 20 min in 4% paraformaldehyde in PBS, then washed 3 × 20 min in PBS + 0.3% Triton-X-100 (PBT3) then 2 × 20 min in PBS. They were then incubated for 2 hours on a shaker at 25°C with 4μM 4g diluted in PBS, then washed 3 × 20 min in PBT3 and mounted in Vectashield Mounting medium (Vector Laboratories). Images were collected on a Zeiss 780 confocal microscope with a 40× or 20× magnification.
Analysis of biological variation between nos-vp16-GAL4, PylRS, PylTUGC flies fed 3 and the parent strain
To quantitatively compare the proteome of the nos-vp16-GAL4, PylRS, PylTUGC flies fed normal food supplemented with 3 and nos-vp16-GAL4 flies fed normal food we performed multi-replicate 2D-DIGE experiments with internal standards and analyzed the data in the DeCyder software package, using the differential in-gel analysis- and biological variation analysis-modules.
Flies were grown and proteomes prepared as described in the section on non-specific labelling for 2D gel electrophoresis (below). The proteomes were extracted from 3 biological replicates of nos-vp16-GAL4, PylRS, PylTUGC flies and 3 biological replicates of the nos-vp16-GAL4 flies (50 flies per replicate). An internal standard was created containing equal amounts of protein (50 μg) from each of the six samples (300 μg total fly protein) and labelled with Cy5-NHS ester. In 6 seperate 50 μg (10 μL) reactions 1 μL (0.4 mM) Cy5-NHS ester (Minimal Labelling Kit, Cyanagen) was added and the reactions incubated in the dark for 30 minutes before being quenched by the addition of 1 μL lysine (1 mM) the 6 reactions were then pooled to create an internal standard stock. In a parallel labelling reaction, 50 μg (10 μL) of each biological replicate was labelled with 1 μL (0.4 mM) Cy3-NHS ester (Minimal Labelling Kit, Cyanagen) for 30 minutes in the dark before quenching with 1 μL lysine (1 mM). The Cy3 labelling of each biological replicate was mixed with 50 μg of the internal standard and subject to 2D-DIGE with isoelectric focusing in the first dimension using 13 cm IPG strips (pH 3-10, non-linear), followed by a second dimension separation by SDS-PAGE.
The gels were imaged using a Typhoon scanner to visualize and quantify the Cy3 and Cy5 fluorescence. Using the DeCyder software package (GE) we identified 900-1000 spots per gel. We used the following exclude filter criteria to define spots and remove dust spot data: slope >1.5, peak height<130, peak area<240, peak volume<50,000. For each gel, peak volume ratios (Cy3/Cy5) were extracted by DeCyder. The maximum volume was plotted against the log of volume ratio and the distribution of log volume ratios determined and compared to a standard normal distribution. We used the differential in gel analysis module within DeCyder to analyze the data. “Nanos” on plots refers to nos-vp16-GAL4 flies, while “Ala” on plots refers to nos-vp16-GAL4, PylRS, PylTUGC flies fed 3. The number of spots falling within two-fold deviations of a log volume ratio of zero on each gel were as follows: nanos gel 1(915 spots): 97.3%, (>1.9%, <0.9%); nanos 2 (889 spots): 97.1% %, (>2.6%, <0.3%); nanos gel 3 (723 spots) 95%, (>3.6%, <1.4%); Ala gel 1 (925 spots): 91.7%, (>0.5%, <7.8%); Ala gel 2 (981 spots), 98.3%, (>0.9%, <0.8%); Ala gel 3 (785 spots) 98.2 % (>1.1%, <0.6%). These data indicate that the nanos and Ala proteome spots are both very similar to the internal standard and are normally distributed around the mean.
To further investigate proteome similarity on a per spot basis and define the number of spots that are statistically different in the two fly genotypes we used biological variation analysis to compare variation between the biological replicates for each genotype and between genotypes. The biological variation analysis module within DeCyder used to match the spots between each of the six gels using the internal standard as a reference. This generated a list of approximately 600 matched spots across the 12 images. For all gels an average volume ratio for each matched spot was generated along with a variance. These data were used to perform a paired Student’s t test, testing the null hypothesis that the matched spot volume ratios of the nos-vp16-GAL4 flies and nos-vp16-GAL4, PylRS, PylTUGC flies fed 3 are the same. The data were filtered to indicate spots showing a greater than 2.3 fold difference in the mean ratio (p < 0.05, power >0.8). Power was calculated using online tools (http://homepage.stat.uiowa.edu/~rlenth/Power/). Storey’s q-value analysis was used to account for multisampling false discovery rates, as previously described for 2D-DIGE data sets48.
Tissue and cell-type specific SORT-M in D. melanogaster with protein identification by 2D-gel electrophoresis and tandem mass spectrometry
Non-specific labelling and 2D-gel electrophoresis of dissected fly ovaries and fly body parts
To generate reference 2D-gels for a fly ovary proteome and a fly body proteome, FT58/nos-vp16-GAL4 adult female flies were fed with 3 in a yeast paste at a concentration of 10 mM, for 3 days. Their ovaries were harvested in PBS and the remnant of the body (excluding the gut) was also harvested in PBS. Ovaries and body remnants were smashed using a pestle in 6M Urea, 2M thiourea, 4% CHAPS, 15mM Tris, pH 8.5 and the sample was centrifuged in a table top centrifuge at top speed at 4°C. The supernatants were submitted for analysis to The Cambridge Centre for Proteomics. Non-specific labelling of the ovary and body extracts was done with Cy3-NHS and Cy5-NHS respectively along with 2D-gel electrophoresis and imaging, using their standard protocols.49 Typically 100 μg of protein was loaded onto 13 cm pH 3-10 gradient IPG strips for first dimension isoelectric focusing. For the second dimension, these strips are transferred to a polyacrylamide gel for SDS-PAGE. Gels were imaged on a Typhoon scanner using standard settings for each dye.
Tissue specific SORT-M in ovary and whole fly extracts with protein identification by 2D-gel electrophoresis and tandem mass spectrometry
FT58/nos-vp16-GAL4 adult female flies were fed with 3 in a yeast paste at a concentration of 10 mM, for 3 days. Their ovaries were harvested in PBS and the remnant of the body excluding the gut was also harvested in PBS. Ovaries and body remnants were smashed using a pestle in 6M Urea, 2M thiourea, 4% CHAPS, 15mM tris, pH 8.5 and the sample was centrifuged in a table top centrifuge at top speed at 4°C. For gel 1 (Supplementary Fig. 24) 100 μg ovary extracts (20 μL) were labelled with 1 μL (400 μM in DMSO) 4e at a final concentration of 20 μM for 2h at room temperature in the dark then quenched by the addition of 1 μL BCN (2 mM, in DMSO). These extracts were then mixed in equal volume with the corresponding body extracts (~ 5 μg/μL). For gel 2 (Supplementary Fig. 24) ovary extracts were first mixed in equal volume with body extracts and then labelled with 4e using our protocol above. For gel 3 (Supplementary Fig. 24) ovary extracts were labelled with 4e and not mixed with any body extracts. All samples were submitted to The Cambridge Centre for Proteomics for 2D-gel electrophoresis, in-gel fluorescence imaging and silver staining. These were performed using their standard protocols;49 dye 4e was imaged using settings equivalent to those for a Cy3 dye.
Tandem mass spectrometry
To identify specific proteins, spots of interest were excised from the silver stained gels. Polyacrylamide gel slices (1-2 mm) containing the purified proteins were prepared for mass spectrometric analysis by manual in situ enzymatic digestion. Briefly, the excised protein gel pieces were placed in a well of a 96-well microtitre plate and destained with 50% v/v acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, proteins were digested with 6 ng/μL Trypsin (Promega, UK) overnight at 37 °C. The resulting peptides were extracted in 2% v/v formic acid, 2% v/v acetonitrile. The digest was analysed by nano-scale capillary LC-MS/MS using a Ultimate U3000 HPLC (ThermoScientific Dionex, San Jose, USA) to deliver a flow of approximately 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm × 20 mm nanoViper (ThermoScientific Dionex, San Jose, USA), trapped the peptides prior to separation on a C18 Acclaim PepMap100 3 μm, 75 μm × 250 mm nanoViper (ThermoScientific Dionex, San Jose, USA). Peptides were eluted with a gradient of acetonitrile. The analytical column outlet was directly interfaced via a modified nano-flow electrospray ionisation source, with a hybrid dual pressure linear ion trap mass spectrometer (Orbitrap Velos, ThermoScientific, San Jose, USA). Data dependent analysis was carried out, using a resolution of 30,000 for the full MS spectrum, followed by ten MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 300–2000. MS/MS scans were collected using a threshold energy of 35 for collision induced dissociation. LC-MS/MS data were then searched against a protein database (UniProt KB) using the Mascot search engine programme (Matrix Science, UK). Database search parameters were set with a precursor tolerance of 5 ppm and a fragment ion mass tolerance of 0.8 Da. Two missed enzyme cleavages were allowed and variable modifications for oxidized methionine, carbamidomethyl cysteine, pyroglutamic acid, phosphorylated serine, threonine and tyrosine were included. MS/MS data were validated using the Scaffold programme (Proteome Software Inc., USA. All data were additionally interrogated manually. D. melanogaster proteins identified with greater than 95% certainty, a spectral count greater than 5 and a unique peptide count greater than 3, as identified within Scaffold, were considered positively identified in a gel spot.
Supplementary Material
Genetic code expansion meets proteomics.
By incorporating unnatural amino acids into proteins, Elliott et al. label, image and identify proteins in specific cells in a developing fly under physiological conditions.
Acknowledgements
We are exceptionally grateful to Dr Mark Skehl and the LMB Mass Spectrometry service for mass spectrometry. We are grateful to the Medical Research Council (U105181009, UD99999908) and the European Research Council for funding.
Footnotes
The authors declare no competing financial interests
References
- 1.Sopko R, Perrimon N. Receptor tyrosine kinases in Drosophila development. Cold Spring Harbor perspectives in biology. 2013;5:a009050. doi: 10.1101/cshperspect.a009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keene AC, Sprecher SG. Seeing the light: photobehavior in fruit fly larvae. Trends in neurosciences. 2012;35:104–110. doi: 10.1016/j.tins.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Tepass U. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annual review of cell and developmental biology. 2012;28:655–685. doi: 10.1146/annurev-cellbio-092910-154033. [DOI] [PubMed] [Google Scholar]
- 4.Dubnau J. Neuroscience. Ode to the mushroom bodies. Science. 2012;335:664–665. doi: 10.1126/science.1218171. [DOI] [PubMed] [Google Scholar]
- 5.Ngo JT, Tirrell DA. Noncanonical Amino Acids in the Interrogation of Cellular Protein Synthesis. Accounts of Chemical Research. 2011;44:677–685. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howden AJ, et al. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Truong F, Yoo TH, Lampo TJ, Tirrell DA. Two-Strain, Cell-Selective Protein Labeling in Mixed Bacterial Cultures. Journal of the American Chemical Society. 2012;134:8551–8556. doi: 10.1021/ja3004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngo JT, Schuman EM, Tirrell DA. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:4992–4997. doi: 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinz FI, D D, Tirrell DA, Schuman EM. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci. 2012;18:40–49. doi: 10.1021/cn2000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Link AJ, et al. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci U S A. 2006;103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin JW. Reprogramming the genetic code. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]
- 13.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 14.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 15.Hancock SM, Uprety R, Deiters A, Chin JW. Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair. Journal of the American Chemical Society. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukai T, et al. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochemical and Biophysical Research Communications. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 17.Chen PR, et al. A Facile System for Encoding Unnatural Amino Acids in Mammalian Cells. Angewandte Chemie International Edition. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier A, et al. Genetically Encoded Photocontrol of Protein Localization in Mammalian Cells. Journal of the American Chemical Society. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 19.Greiss S, Chin JW. Expanding the Genetic Code of an Animal. Journal of the American Chemical Society. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]
- 21.Blackman ML, Royzen M, Fox JM. Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels–Alder Reactivity. Journal of the American Chemical Society. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Šečkutė J, Cole CM, Devaraj NK. Live-Cell Imaging of Cyclopropene Tags with Fluorogenic Tetrazine Cycloadditions. Angewandte Chemie International Edition. 2012;51:7476–7479. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole CM, Yang J, Šečkutė J, Devaraj NK. Fluorescent Live-Cell Imaging of Metabolically Incorporated Unnatural Cyclopropene-Mannosamine Derivatives. ChemBioChem. 2013;14:205–208. doi: 10.1002/cbic.201200719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA. Functionalized Cyclopropenes As Bioorthogonal Chemical Reporters. Journal of the American Chemical Society. 2012;134:18638–18643. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- 25.Devaraj NK, Weissleder R. Biomedical Applications of Tetrazine Cycloadditions. Accounts of Chemical Research. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang K, et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang K, et al. Genetic Encoding of Bicyclononynes and trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels–Alder Reactions. Journal of the American Chemical Society. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borrmann A, et al. Genetic Encoding of a Bicyclo[6.1.0]nonyne-Charged Amino Acid Enables Fast Cellular Protein Imaging by Metal-Free Ligation. ChemBioChem. 2012;13:2094–2099. doi: 10.1002/cbic.201200407. [DOI] [PubMed] [Google Scholar]
- 29.Plass T, et al. Amino Acids for Diels–Alder Reactions in Living Cells. Angewandte Chemie International Edition. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 30.Seitchik JL, et al. Genetically Encoded Tetrazine Amino Acid Directs Rapid Site-Specific in Vivo Bioorthogonal Ligation with trans-Cyclooctenes. Journal of the American Chemical Society. 2012;134:2898–2901. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen DP, et al. Genetic Encoding and Labeling of Aliphatic Azides and Alkynes in Recombinant Proteins via a Pyrrolysyl-tRNA Synthetase/tRNACUA Pair and Click Chemistry. Journal of the American Chemical Society. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]
- 32.Yu Z, Pan Y, Wang Z, Wang J, Lin Q. Genetically Encoded Cyclopropene Directs Rapid, Photoclick-Chemistry-Mediated Protein Labeling in Mammalian Cells. Angewandte Chemie International Edition. 2012;51:10600–10604. doi: 10.1002/anie.201205352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 34.Ambrogelly A, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci U S A. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 36.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 37.Hadjieconomou D, et al. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- 38.Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, D L, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- 40.Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Von Stetina JR, Lafever KS, Rubin M, Drummond-Barbosa D. A Genetic Screen for Dominant Enhancers of the Cell-Cycle Regulator alpha-Endosulfine Identifies Matrimony as a Strong Functional Interactor in Drosophila. G3. 2011;1:607–613. doi: 10.1534/g3.111.001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lilley KS. Current Protocols in Protein Science. John Wiley & Sons, Inc.; 2001. [Google Scholar]
- 43.Bownes M. Expression of the genes coding for vitellogenin (yolk protein) Annual Review of Entomology. 1986;31:507–531. [Google Scholar]
- 44.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leon A, McKearin D. Identification of TER94, an AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Mol Biol Cell. 1999;10:3825–3834. doi: 10.1091/mbc.10.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 47.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 48.Karp NA, Kreil DP, Lilley KS. Determining a significant change in protein expression with DeCyder during a pair-wise comparison using two-dimensional difference gel electrophoresis. Proteomics. 2004;4:1421–1432. doi: 10.1002/pmic.200300681. [DOI] [PubMed] [Google Scholar]
- 49.Harper S, Mozdzanowski J, Speicher D. Current Protocols in Protein Science. John Wiley & Sons, Inc.; 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.