Abstract
Activation of caspase-mediated apoptosis is reported to be a hallmark of both granzyme B- and Fas- mediated pathways of killing by cytotoxic T lymphocytes (CTL); however, the kinetics of caspase activation remain undefined due to an inability to monitor target cell-specific apoptosis in real time. We have overcome this limitation by developing a novel biosensor assay that detects continuous, protease specific activity in target cells. Biosensors were engineered from a circularly-permuted luciferase, linked internally by either caspase 3/7 or granzyme B/caspase 8 cleavage sites, thus allowing activation upon proteolytic cleavage by the respective proteases. Co-incubation of murine CTL with target cells expressing either type of biosensor led to a robust luminescent signal within minutes of cell contact. The signal was modulated by the strength of TCR signaling, the ratio of CTL/target cells, and the type of biosensor utilized. Additionally, the luciferase signal at 30 minutes correlated with target cell death, as measured by 51Cr release assay. The rate of caspase 3/7 biosensor activation was unexpectedly rapid following granzyme B compared to Fas-mediated signal induction in murine CTL; the latter appeared gradually after a 90 minute delay in perforin or granzyme B deficient CTL. Remarkably, the Fas dependent, caspase 3/7 biosensor signal induced by perforin-deficient human CTL was also detectable after a 90 minute delay when measured by re-directed killing. Thus we have utilized a novel, real-time assay to demonstrate the distinct pattern of caspase activation induced by granzyme B versus Fas in human and murine CTL.
Introduction
Cytotoxic lymphocytes (CTL) are essential to eliminate virus-infected cells and malignant cells in the body. CTL kill target cells largely by one of two methods: 1) perforin-mediated granzyme delivery following exocytosis of cytotoxic granules and/or 2) Fas ligand (FasL) mediated coupling of target cell Fas death receptors. Immune synapse formation between CTL and target cells triggers the rapid orchestration of the granule exocytosis pathway. (1–3) At the immune synapse granzymes enter the target cells following membrane perturbation by perforin. The precise mechanism of how perforin delivers granzymes into the target cells is still under investigation. (4–6) Granzymes are serine proteases, capable of cleaving numerous substrates in the cytoplasm and nucleus required for induction of apoptosis. (7–9) Ten granzymes have been identified in the mouse and five in the human genome, suggesting redundancy. The targets of granzyme B proteolysis have been studied more extensively than the other granzymes. Direct cleavage and subsequent activation of caspase 3 by granzyme B is thought to be a critical first step in CTL-induced apoptosis.
Active caspase 3 is measurable in target cells by flow cytometry, representing one method by which CTL function may be assessed. (10–13) In addition, CTL function is routinely measured by loss of target cell membrane integrity or evidence of CTL degranulation by flow cytometry and/or by measurement of cytolysis by a gold standard, microplate assay of 51Cr release. (14–16) Each of these approaches utilizes an end point assay. None of the current methodologies permit kinetic, continuous evaluation of cytotoxic function. Although perforin mediated granzyme B delivery is described as an early event and Fas/FasL induced death is described as occurring late, the only available technologies available to evaluate this activity in real time require sophisticated microchip technology. (17) Additionally, monitoring of cytotoxicity by fluorescence based approaches leads to a relatively low signal to noise ratio, due to background fluorescence from live or fixed cells. We reasoned that a protease-inducible, luminescent biosensor introduced into target cells alone would allow us to measure CTL-induced caspase induction in a living cell continuously, in a convenient, non-radioactive, microplate format with minimal background signal.
Firefly luciferase (Photinus pyralis) has been successfully engineered using circular permutation to be a protease specific biosensor, in which residues near the native termini are joined by a protease cleavage site. The permuted luciferase is sterically constrained and exhibits low levels of activity until the joining sequence is cleaved by the corresponding protease. Previous work has shown successful engineering of a caspase 3/7 cleavable biosensor, designed to detect the protease activity in cell-free lysates. (18) We introduced a similar construct based on a thermal stable variant of Photuris pennsylvanica luciferase (GloSensor, GLS) with a DEVD proteolytic site, (GLS.DEVD) (19, 20) into EL4 target cells to detect intracellular caspase-3 activity following co-incubation with CTL in vitro. Within minutes of contact we observed a dramatic increase of luminescent signal. Importantly, the earliest activation of the biosensor was dependent on intact granzyme B and perforin and the activity of the biosensor correlated with the gold standard cytotoxic function assay, 51Cr release. Additional biosensors with granzyme B-cleavable luciferases yielded similar results, yet with distinctive kinetics in the early time period. In perforin-deficient murine and human CTL, caspase activity was detectable after a 90–120 minute delay and blocked by anti-FasL antibody, consistent with FasL/Fas induced activity. Thus we have created a non-radioactive assay capable of detecting the earliest changes in apoptosis, representing a major breakthrough to currently available CTL assays. We anticipate that this novel methodology will allow definition of genetic and pharmacologic modulators of CTL-mediated apoptosis in future studies.
Material and Methods
Mice, cell lines, and reagents
C57BL/6 wild type and PRF1 −/− mice were obtained from The Jackson Laboratory and bred in our animal facility. P14 transgenic mice were obtained from P. Marrack. (21) All murine studies were conducted in accordance with protocols approved by our institutional review committee. Granzyme B cluster knockout mice were obtained from T. Ley. (22) EL4 cells (ATCC) were maintained in RMPI-1640 10% Fetal Bovine Serum (FBS). Firefly luciferase antibody, Abcam (Cambridge, MA). GloSensor cAMP reagent (biosensor substrate), Promega (Madison, WI). Etoposide, APP Pharmaceuticals. Diluent for Etoposide: 0.2% citric acid; 3% Benzyl alcohol; 8% polysorbate 80; 65% polyethylene glycol 300; 30.5% (v/v) alcohol. Anti FasL antibody (clone MFL4- human, clone MFL3-murine) was obtained from BioLegend and used at 100μg/ml.
Constructs
The caspase 3/7 biosensor (GLS.DEVD) encoded by the pGloSensor-30F construct is a commercially available analog (GloSensor caspase 3/7, Promega) of the version previously described (18), based on a thermal stable variant of Photuris Pennsylvanica luciferase evolved for enhanced detection of caspase-3 activity in living cells. (19, 20) The plasmid containing the granzyme B/caspase 8 biosensor (GLS.IETD) was constructed by annealing the following oligos and ligating into the BamH1/HindIII sites of pGloSensor-30F plasmid: 5′-GATCCGGCATCGAGACCGACAGCGGA-3′; 5′-AGCTTCCGCTGTCGGTCTCGATGCCG-3′. The oligos utilized to generated GLS.IEAD were as follows: 5′-GATCCGGCCGGATCGAGGCCGACAGCGAA-3′; 5′-AGCTTTCGCTGTCGGCCTCGATCCGGCCG-3′; and GLS.VGPD: 5′-GATCCAAGAGCGTGGGCCCCGACTTCGGA3′; 5′-AGCTTCCGAAGTCGGGGCCCACGCTCTTG-3′. All four biosensors were also cloned into a retroviral vector pMIEG, allowing detection of transduced cells using green fluorescent protein (GFP) expressed in cis. (23) Oligo sequences utilized to generate control vectors may be obtained by contacting the authors.
In vitro testing of recombinant Glosensors
Recombinant Glosensor proteins were generated using the TNT T7 Coupled Wheat Germ Extract System (Promega) supplemented with the FluoroTect GreenLys in vitro Translation Labeling System (Promega). Following cell-free expression for two hours at 30 °C, aliquots of cell-free expression lysate were combined with an equal volume of murine granzyme B (Sigma) in assay buffer (100 mM HEPES, pH 7.4, 200 mM NaCl, 0.2% CHAPS, 2 mM EDTA, 20% glycerol). Following a one hour incubation at 37 °C, 10 μL of sample was combined with 90 μL of Luciferase Assay Reagent and luminescence was measured at room temperature on a GloMax Multi+ luminometer (0.5 second integration time).
EL4 target cell transfection and cloning
EL4 and P815 cells were transfected with GloSensor caspase 3/7 or derivatives using Fugene (Promega). Cells were selected and maintained with 600μg/ml of G418 starting three days after transfection. Clones were obtained by limiting dilution. For experiments using retroviral constructs, EL4 and YAC cells were infected with biosensor-expressing retrovirus and then sorted for GFP expressing cells 7 days later without antibiotic selection.
Generation of CTLs
Ex vivo CTL were generated from wild type and gene knockout C57BL/6 mice following LCMV infection (intraperitoneal, 200 PFU of LCMV-WE); mice were sacrificed 7 days after infection and total splenocytes were harvested and used directly for CTL assay. In vitro CTL were generated as previously described (24). Briefly, TCR transgenic P14 mice were sacrificed and splenocytes were harvested and stimulated with LCMV gp33–41 peptide (Anaspec) for 48 hours, followed by expansion in IL2-containing medium for 3–5 days before use. Murine NK cells were isolated from C57BL/6 mice using negative selection and cultured in murine IL2 at 1000 units/mL for 7 days before testing function. Human cytotoxic T cells were generated from peripheral blood mononuclear cells from healthy controls or a perforin deficient patient by transformation with Herpesvirus Saimiri as previously described (25) and maintained in 200 units/mL human IL-2. Permission to utilize human cells for research was obtained according to protocols approved by our institutional review board.
Biosensor CTL assay
CTL were washed once with PBS (pH7.4) and resuspended in growth medium without phenol red and supplemented with 20mM HEPES and 10% FBS. Cells were plated into 96-well, round-bottom, white plates (Corning) in replicate wells at 50 μl per well. Biosensor-expressing EL4 targets were harvested and washed with PBS, followed by incubation with 2% (v/v in phenol red free growth media) biosensor substrate at 37°C, 5% CO2 for 30 minutes and added to effector wells, 50μl per well containing 5,000–20,000 cells. The total volume per well for the assay was 100μl. Following centrifugation at 200 g for 5 minutes, the plate was incubated at 37°C in GloMax Multi Plus luminometer (Promega) and luminescence was measured kinetically every 3 minutes with integration time of 0.5 seconds. Each data point represents the mean of four replicate wells. A similar protocol was used for testing NK function against YAC target cells. For the re-directed killing assay, P815 cells were pre-incubated with OKT3 at 5 μg/ml for 30 minutes and then co-incubated with human T cells to trigger the biosensor.
51Cr release assay
51Cr release assay was performed as previously described. (26) EL4 target cells were pulsed with 100ng/mL of LCMV gp33–41 for one hour prior to 51Cr labeling. Effector and EL4 target cells (5,000 cells/well) were co-incubated for 4 hours at 37°C. 51Cr-release into supernatant was counted on a 96 well Lumaplate (PerkinElmer) by TopCount NXT. Cytotoxicity is reported as the % of 51Cr released to the supernatant according to the following calculation: (test − spontaneous)/(Max lysis − spontaneous) *100. Maximum lysis was determined by addition of 1% Triton X-100 (Sigma) to 5,000 EL4 target cells.
Results
Protease biosensors allow detection of perforin-mediated, granzyme B delivery by CTL in real time
Based on previous work that demonstrated caspase 3 activation was dependent upon perforin-mediated granzyme B delivery, we reasoned that a caspase 3/7 luciferase biosensor expressed in target cells would detect the earliest events of CTL-induced apoptosis. The caspase 3/7 biosensor, GLS.DEVD was constructed by insertion of a DEVD proteolytic site into a circularly-permuted, thermal stable variant of Photuris pennsylvanica luciferase (Figure 1A and Ref (20)). The GLS.DEVD has been validated in real-time, kinetic assays to monitor caspase 3/7 activity within living cells, both in vitro and in vivo. We also designed 3 novel biosensors, GLS.IETD, GLS.IEAD, and GLS.VGPD, to be directly cleavable by granzyme B. The IETD cleavage site is a known target for both granzyme B and caspase 8, (27) and IEAD is also a known target for caspase 10 (28). Using ExPasy Peptidecutter, we confirmed that the recognition sequence for GLS.VGPD is only predicted to be cleaved by granzyme B. (29, 30)
Figure 1. Design of caspase and granzyme B biosensors to measure CTL function.
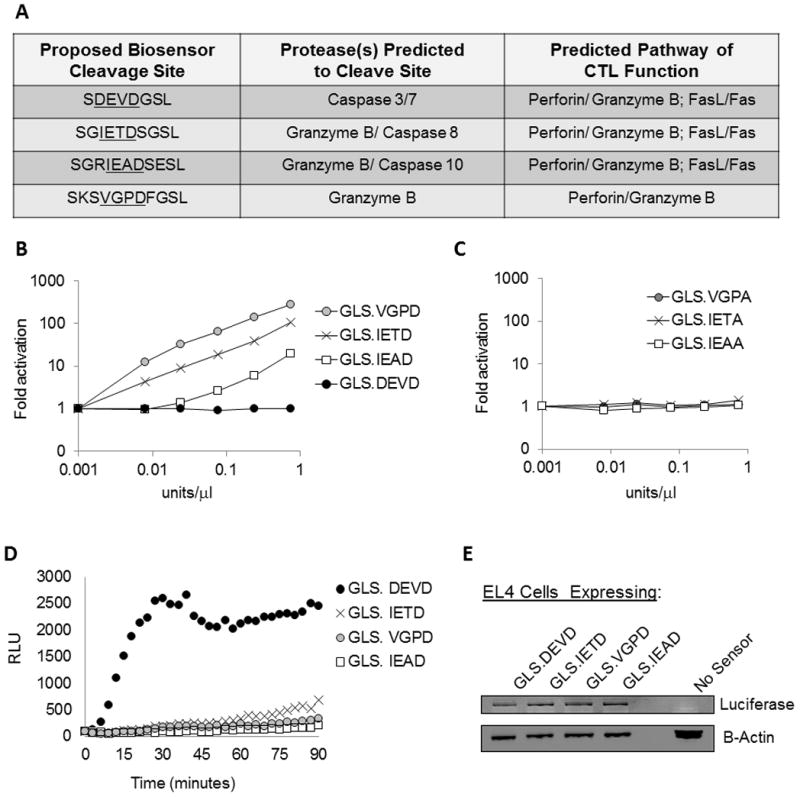
(A) We developed biosensors which directly measure granzyme B cleavage and/or caspase cleavage upon expression in target cells killed by CTL. The biosensors include a circularly-permuted luciferase containing a protease cleavable site that allows activation of the luciferase only upon cleavage. Shown is the engineered protease cleavage site, the proteases predicted to cleave the site, and the pathways predicted to activate the biosensor following CTL-target cell recognition. (B and C) In order to demonstrate granzyme B dependence upon the newly created biosensors, recombinant protein was generated in vitro and monitored for activation upon the addition of murine granzyme B. Granzyme B activated only the cleavage sites containing the required P1 aspartic acid residue (B); whereas the GLS.DEVD biosensor and control biosensors containing an alanine substitution for the P1 aspartic acid were inactive (B & C). (D) Biosensors were introduced into EL4 target cells by retroviral transduction and cells were sorted for GFP expression. In vitro CTLs were generated by antigen (LCMV gp33–41) stimulation of splenocytes from P14 TCR transgenic mice. CTLs were co-incubated with a biosensor-expressing EL4 target cells at effector/target ratios of 6:1, and luciferase function was measured as the amount of relative light units (RLU) emitted. Data shown is from one cell line, representative of 3 independent experiments. (E) Protein lysates analyzed by western blot using antibodies recognizing luciferase and actin demonstrated equivalent biosensor expression in each cell line.
To confirm that the newly created biosensors would be activated by granzyme B, we generated recombinant protein using in vitro translation and tested for murine granzyme B cleavage across a range of concentrations (0.001 to 0.75 units/microliter). Three additional control vectors were created, exchanging the critical aspartic acid within the proteolytic site with an alanine. As shown in Figures 1B and 1C, neither the control vectors nor the GLS.DEVD were cleaved by granzyme B. The three novel biosensors were activated by murine granzyme B as shown in Figure 1B. We predicted these novel biosensors would detect the earliest determinant of CTL function, granzyme B delivery into target cells, an event reported to precede activation of caspase 3. (7–9)
We introduced each biosensor into EL4 cells, a murine tumor cell line that is routinely used to present peptides in the context of H-2 Db. We generated stable lines by retroviral transduction followed by sorting of GFP expressing cells of equal intensity to yield cell lines with near equivalent levels of luciferase protein (Figure 1E). We first tested cytotoxic function of primary murine T cells cultured in IL-2 following antigen stimulation. These CTL were derived from transgenic mice expressing a single T cell receptor (P14) capable of recognizing LCMV gp33 peptide (referred hereafter as “in vitro P14 CTL”). The gp33 peptide was presented by EL4 cells expressing each of the four biosensors. Following co-incubation of CTL and EL4 at 37 degrees at an effector/target (E/T) ratio of 6:1, all four biosensors were activated within minutes by CTL. The data from a representative experiment is shown in Figure 1D and supplemental Figure 1. The GLS.DEVD biosensor yielded the most robust signal (nearly 30 fold induction over target cells alone), with the remaining biosensors yielding 10–15 fold (GLS.IETD), 5–7 fold (GLS.VGPD), and 3 fold (GLS.IEAD) signals above background (for background, see supplemental Figure 1). The maximum signal for GLS.DEVD peaked by 30 minutes, while the GLS.IETD, GLS.IEAD, and GLS.VGPD biosensor signals continued to rise during the 90 minute window of measurement. Each data point represents quadruplicate wells. See Supplemental Figure 1 for visualization of error bars from the biosensors and target cells alone, removed from Figure 1D and subsequent figures to decrease crowding.
In an effort to achieve a maximal signal-to-noise ratio, the two biosensors with highest signal, GLS.DEVD and GLS. IETD, were transfected into cells in a traditional expression vector, pF9A, and clonal cell lines obtained by limiting dilution in antibiotic selection. A maximally active clonal line was selected for further studies (Supplemental Figures 2A and B, for GLS.DEVD). Experiments were replicated using non-cloned cell cultures but the signal was consistently highest using clonal lines. We also expressed the biosensors in an NK target cell (YAC cells) and measured activity using IL2 activated NK cells from C57BL/6 mice which yielded a similar pattern of activation for GLS.DEVD, GLS.IEAD and GLS.IETD but the signal was relatively weak (Supplemental Figure 3). Therefore, we moved forward with studies restricted to CTL.
Biosensor(s) luminescent signals detect changes in CTL activation
We evaluated the capacity of the biosensor to measure ex vivo CTL function obtained from mice infected with LCMV. We isolated splenocytes one week after infection, at a time when LCMV-specific CTL have expanded to 0.5--2% of the total splenocytes. (31, 32) CTL function was measurable at day 7 and 8 post-infection at a total splenocyte: target cell ratio of 12:1, which equates to a CTL effector/target (E/T) ratio of approximately 0.1/1 with individual variation from mouse to mouse. The data from a representative experiment is shown in Figure 2. Both biosensors were activated within minutes by LCMV-specific CTL. LCMV-derived CTL initiated a peak response of the GLS.DEVD biosensor by 60 minutes; while the GLS.IETD biosensor signal continued to rise during the 90 minute window of measurement.
Figure 2. Strength of antigen recognition affects the induction of biosensors by ex vivo CTL.
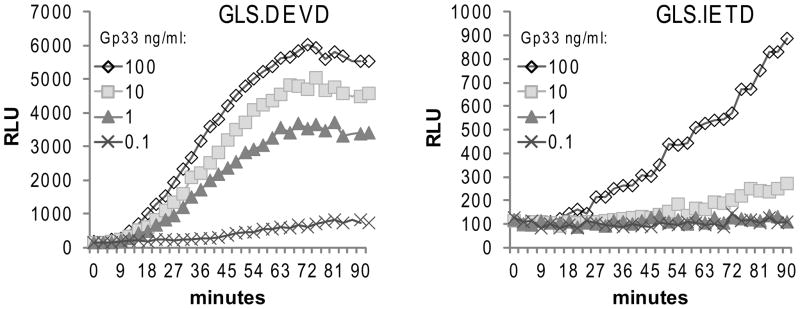
Ex vivo CTL from C57BL/6 mice infected with LCMV for 7 days were co-incubated with luciferin substrate-loaded EL4 cloned target cells expressing GLS.DEVD or GLS.IETD. EL4 cells were pulsed with gp33 peptide at titrating concentrations and co-incubated with ex vivo CTL at splenocyte/EL4 target ratio of 12:1. Luminescence (RLU) was measured every 3 minutes up to 90 minutes. Data is from one mouse, representative of 8 mice.
Epitope density on target cell MHC has been previously shown to influence the strength of the CTL response. (33, 34) In order to further test the capacity of the protease biosensors to detect changes in early CTL function, we titrated the concentration of the antigen (gp33) loaded onto EL4 target cells from 100ng/ml to 0.1ng/ml. We observed that the luminescent signal induced by ex vivo CTL decreased proportionally to the concentration of gp33 used to pulse target cells in EL4 expressing either the GLS.DEVD or GLS.IETD biosensor constructs. The GLS.DEVD signal was detectable down to peptide concentrations ≥1ng/ml; whereas, the GLS.IETD signal was detectable above the machine background when the gp33 concentration was ≥10ng/ml, likely due to the lower baseline signal generated by GLS.IETD. The limit of detection of the GLS.DEVD signal correlated well with 51Cr assays performed in parallel, with cytolysis detectable from 1–100ng/ml and absent at 0.1ng/ml (not shown). The GLS.DEVD biosensor signal was attenuated by a lower stimulating peptide concentration both in rate of induction (fold activation/time) and maximum signal obtained. It is apparent that T cell engagement/activation occurs over two log concentrations of gp33 (1–100ng/ml) with minimal attenuation of caspase 3/7 activation, accounting for the preservation of cytotoxicity in this peptide concentration range as well (data not shown).
CTL induction of biosensor luminescence correlates with 51Cr release assay of cytotoxic function
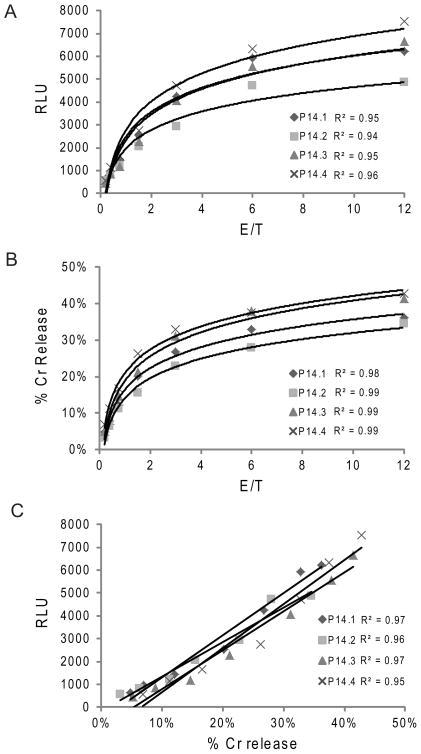
We predicted that the early proteolytic events detected by biosensor activation would correlate with CTL-induced cytolysis of target cells and thus be useful as a non-radioactive microplate assay to substitute for the gold standard, 4 hour 51Cr release assay. In order to control for cell number, we compared the luminescent signal to the 51Cr release assay for in vitro derived P14 CTL. As is typically done for 51Cr release assays, we evaluated function over a broad range of E/T ratios from CTL generated from four individual mice. The luminescence from the GLS.DEVD biosensor and the percentage of 51Cr release increased proportionally with increasing E/T ratio (Figure 3A and 3B). When we plotted percentage of total cell death in a 4 hour 51Cr release assay against the 30 minute luminescent signal in biosensor assay, we demonstrated remarkable correlation between the two assays with R2 ≥ 0.95 (Figure 3C) and comparable sensitivities when titrating to low E/T. Similar correlations were noted between the GLS.IETD signal and Cr51 assay (data not shown). Thus, when evaluating cytotoxic function in mice with intact perforin and granzyme B pathways, the 30 minute luminescent signal from the GLS.DEVD and GLS.IETD assays were equivalent to the gold standard cytolysis assay.
Figure 3. Biosensor signal correlates with gold standard 51Cr release assay.
In vitro CTLs were generated by antigen stimulation of splenocytes from 4 individual P14 TCR transgenic mice. CTLs were either co-incubated with biosensor expressing, cloned EL4 cells or 51Cr loaded EL4 target cells at effector/target ratios ranging from 12:1 to 0.19:1. 51Cr release was measured after 4 hours. (A) RLU at 30 minutes of the luminescence assay for all 4 mice with titration of E/T ratio; (B) 51Cr release percentage for all 4 mice with titration of E/T ratio; Best fit curves were logarithmic for A and B. (C) Correlation between biosensor assay and 51Cr assay. Data is representative of 3 independent experiments.
Target cell caspase 3/7 signal induced by CTL is abrupt and transient
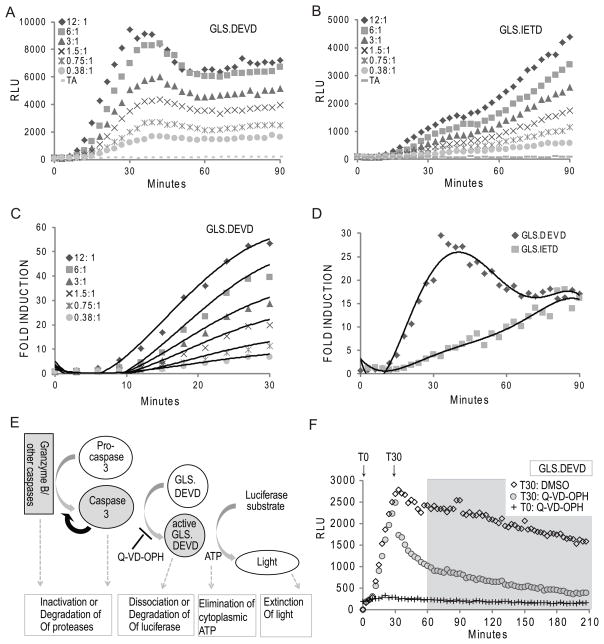
Whether using ex vivo CTL, or in vitro derived CTL, the area under the curve (luminescence versus time) was distinct when comparing the target cells expressing GLS.DEVD versus the granzyme B cleavable biosensors (Figures 1– 3). The luminescence from GLS.DEVD was detectable as early as 3 minutes following CTL/target co-incubation, rising sharply until a brief plateau and/or decline was reached at approximately 30–60 minutes; whereas the GLS.IETD signal rose more slowly and continued to rise during the first 90 minutes. After 60 minutes, the fold activation of both biosensors were similar (Figure 4D). We focused on the early activation signal of the GLS.DEVD biosensor, evaluating for the impact of effector number to determine if enhanced granzyme B delivery impacted the rate of rise. As shown in Figure 4A and B, the rate of luminescent activation was reduced at lower E/T, although the plateau still occurred at the same time. When we evaluated the earliest time period for induction (from 10–30 minutes, Figure 4C) we noted a range of GLS.DEVD activation from 7–70 fold induction (signal mediated by CTL/signal from target alone at the same time) over 20 minutes, dependent upon the E/T.
Figure 4. Caspase 3/7 activation of biosensor is rapid and transient from CTL induction.
CTLs derived in vitro from a representative P14 mouse were co-incubated with substrate-loaded target EL4 expressing GLS.DEVD (A) or GLS.IETD (B) as described in Figure 3. (C and D) Fold induction of GLS.DEVD and GLS.IETD. RLU were graphed as a fold induction (signal generated with CTL/signal generated by target alone) at each time point up to 90 minutes. C) The signal limited to the first 30 minutes of the GLS.DEVD assay is shown in higher detail to demonstrate the dependence of the fold induction on E/T. (D) Comparison of GLS.DEVD and GLS.IETD signals for the first 90 minutes illustrate the distinct kinetics of caspase 3/7 and granzyme B/caspase 8 activation. Data is representative of 3 independent experiments. (E) Model to show the potential mechanisms for acquisition and loss of GLS.DEVD signal. (F) Caspase dependency of GLS.DEVD activity. Using the same assay as described in A and B, CTL were co-incubated with peptide loaded EL4 cells and luminescence monitored from GLS.DEVD out to 4 hours. A pan caspase inhibitor, Q-VD-OPH, was added at the start of the assay, T0, or at 30 minutes, T30. There was complete inhibition of GLS.DEVD when Q-VD-OPH was added at T0. Following addition at T30, there was an immediate dramatic loss of signal to 60 minutes compared to cells exposed to DMSO, illustrating the dependence of the biosensor on caspase signaling from 0–60 minutes. There is a caspase independent loss of activity from 60–240 minutes (delineated by gray box) with nearly identical degradation in the presence or absence of caspase inhibition. (A–D); experiments utilized cloned EL4 cells expressing GLS.IETD or GLS.DEVD. (F) experiments utilized polyclonal, GFP sorted EL4 cells expressing GLS.DEVD.
The rapid increase of the GLS.DEVD signal within 3–10 minutes of co-incubation suggested that caspase 3/7 activation was more rapid than previously recognized following CTL-mediated granzyme B delivery. To compare the caspase 3/7 activation mediated by murine CTL to caspase activation induced by another method, we exposed the EL4 target cells to drugs known to induce apoptosis, such as etoposide (Supplemental Figure 4) or staurosporine (not shown). Etoposide-mediated caspase 3/7 activation was readily detectable in a kinetic assay, with induction of a dose-dependent signal at 200 minutes following etoposide exposure. The kinetic induction of apoptosis in EL4 cells initiated by etoposide was delayed and gradual by comparison to that induced by CTL (8 fold induction over 240 minutes for etoposide compared to 5–70 fold induction over 20 minutes for CTL). Additionally, the GLS.DEVD signal rose steadily from 200–440 minutes without decline, when apoptosis was induced by etoposide. During the staurosporine induction, no signal was detectable in the 4 hour window of measurement although the signal was induced after overnight incubation (not shown).
The termination of the GLS.DEVD induction at 30–60 minutes was unexpected. This was visualized at all E/T ratios and is associated with a steep decline in signal at 30 minutes at the highest E/T. A similar decline in signal was not observed for the GLS.IETD signal or other granzyme B activated biosensors; however, at higher effector concentrations, there is a subtle and transient plateau of GLS.IETD at 40–45 minutes followed by a continuous rise (Figure 4B, E/T 12:1 and 6:1) in luciferase. We do not know the precise mechanism for termination of the GLS.DEVD signal, but several factors may contribute such as inactivation or degradation of active caspases, loss of cytosolic ATP (luciferase is ATP-dependent), or degradation of the cleaved luciferase (modeled in Figure 4E). A similar phenomena was reported previously in two unrelated cell lines expressing GLS.DEVD where luminescence was rapidly triggered by TRAIL within 120 minutes, followed by a decline in RLU (20).
We tested the dependence of the luciferase signal on caspase function and noted that addition of a pan-caspase inhibitor led to immediate drop in signal only within the first 30 minutes (addition at time 0 and 30 minutes shown in Figure 4F). The remaining decay in the RLU signal was largely caspase-independent and therefore likely related to dissociation of pre-activated luciferase and/or loss of cellular ATP. The decline in signal was not due to a unique feature of cloned cell lines as the data in Figures 1d and 4F were obtained from polyclonal EL4 cells expressing GLS.DEVD. Thus, the kinetics for perforin-mediated, granzyme-activated caspase 3/7 is distinct, with rapid activation, followed by a plateau and/or decline of the early peak in caspase 3/7-induced biosensor signal.
Distinguishing two independent mechanisms for CTL killing
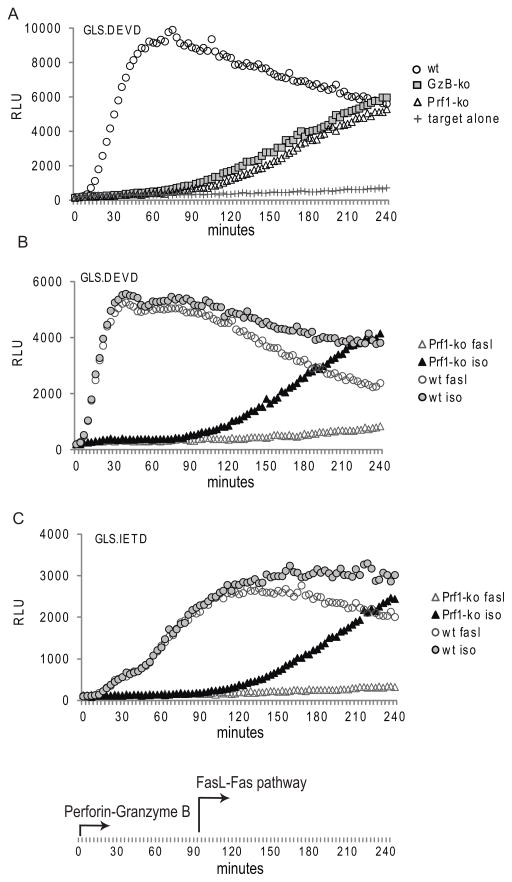
CTL killing is mediated largely by two different pathways, the secretory granule (perforin/granzyme B) and the death receptor (Fas/FasL) pathways. Both mechanisms of cytotoxicity lead to caspase 3 activation, although the timing of the death receptor pathway is reported to be delayed and secondary to caspase 8 activation rather than granzyme B cleavage. (35, 36) (37) Due to the rapid kinetics of the luciferase induction noted from either biosensor, we predicted that the perforin-mediated pathway would be the sole determinate of GLS.DEVD and GLS.IETD biosensor activation in the first 90 minutes; while FasL mediated killing would be detectable if the assay were extended beyond this early time point. We tested this by generating ex vivo CTL from perforin deficient and granzyme B deficient mice. Co-incubation of ex vivo CTL from either immunodeficient strain with gp33-pulsed EL4 expressing GLS.DEVD or GLS.IETD led to luciferase induction only after the first 90–120 minutes (Figure 5A (GLS.DEVD); GLS.IETD not shown).
Figure 5. Caspase 3/7 activation mediated by perforin deficient murine CTL is detectable following late phase Fas-mediated induction.
(A) Ex vivo CTL were generated in wild type (WT) C57BL/6, perforin deficient (prf1 ko), and granzyme B deficient knockout (GzB ko) mice infected with LCMV for 7 days. Splenocyte/EL4 target ratio of 12:1 is shown for a representative mouse (4 mice/group tested in 3 independent assays). Evaluation of RLU out to 240 minutes reveals a late activation of the GLS.DEVD (A) and GLS.IETD biosensors (not shown) in mice lacking perforin-mediated granzyme B delivery. (B and C) P14 in vitro CTL from WT and perforin deficient mice were also tested as effectors. Addition of anti-FasL antibody to the reaction prevented the delayed rise in GLS.DEVD (B) and GLS.IETD (C) signal in perforin deficient mice, illustrating the delayed onset of the death receptor pathway (≥ 90 minutes). Target cells were polyclonal EL4, sorted by GFP following retroviral transduction of luciferase biosensors.
CTL from perforin deficient mice triggered a delayed GLS.DEVD and GLS.IETD signal that continued to rise out to 4 hours. To determine if the delayed biosensor signal in GLS.DEVD and GLS.IETD was due to Fas/FasL mediated activation, we tested CTL function in the presence of an anti-FasL antibody in perforin sufficient and deficient CTL (P14 crossed to perforin deficient mice). This was tested for both ex vivo CTL (not shown) and in vitro-derived CTL as shown in Figure 5B and 5C. In vitro derived CTL were obtained from antigen-stimulated, IL-2 augmented splenocyte cultures from transgenic P14 mice. For both biosensors, at an E/T of 1.5:1, the delayed biosensor signal from perforin deficient CTL was completely blocked in the presence of anti-FasL antibodies. A portion of the decaying signal (starting at 90– 120 minutes) from wild type CTL was also blocked. The biosensor activation from P14 CTL was clearly perforin-dependent in the first 90 minutes, with a delayed signal occurring from Fas/FasL signaling. Thus the biosensors detect both the rapid, perforin-dependent granzyme B induced caspase activation as well as the delayed, Fas/FasL mediated caspase 3/7 (GLS.DEVD) or caspase 8 (GLS.IETD) activation. We also tested for delayed FasL/Fas activation in EL4 cells expressing GLS.IEAD and GLS.VGPD but did not observe a late signal. This suggests that these two biosensors are granzyme B specific and therefore unable to detect other caspase activity in apoptosis cells, although we cannot exclude the possibility of a low detection sensitivity of the biosensors.
The rate of activation of the GLS.DEVD signal by the FasL/Fas pathway in murine CTL was slow in comparison to the induction mediated by the perforin/granzyme B mediated pathway; whereas the rate of induction of GLS.IETD was similar for both pathways and the maximum signal was the same. The maximum signal obtained by the GLS.DEVD induced by either pathway was the same but took two hours when mediated by Fas versus thirty minutes induced by Granzyme B. These studies define for the first time the exact kinetics of FasL/Fas induced- caspase activation, confirm the absolute dependence of early caspase 3/7 activation on granzyme B delivery, and demonstrate the accelerated rate of caspase 3/7 induction by the secretory granule pathway. Finally the lack of early signal in the GLS.IETD (not shown) and GLS.DEVD (Fig 5A) sensors following induction of apoptosis by granzyme B deficient CTL demonstrates the absolute dependence of these biosensors on granzyme B delivery as predicted. We conclude that the GLS.DEVD and GLS.IETD biosensors may be utilized to study both perforin/granzyme mediated apoptosis as well as late phase, death receptor-induced apoptosis.
In order to generalize our findings obtained using murine CTL, we tested the capacity of the GLS.DEVD biosensor to detect caspase activation from human CTL. We tested both primary T lymphoblast cultures obtained following mitogen stimulation (not shown) and transformed, stable human T cell cultures obtained following herpesvirus saimiri (HVS) transformation. The advantage of the latter is the stability of these cultures over time and tolerance for freeze/thaw. They have previously been shown to exhibit strong cytolysis following re-directed killing, a method which relies on activation of T cells via antibody stimulation of CD3, rather than an antigen- specific T cell. (38) We derived HVS CTL from both a healthy control and a perforin deficient patient who presented with hemophagocytic lymphohistiocytosis due to biallelic, truncating perforin mutations (50delt) and absent NK function.
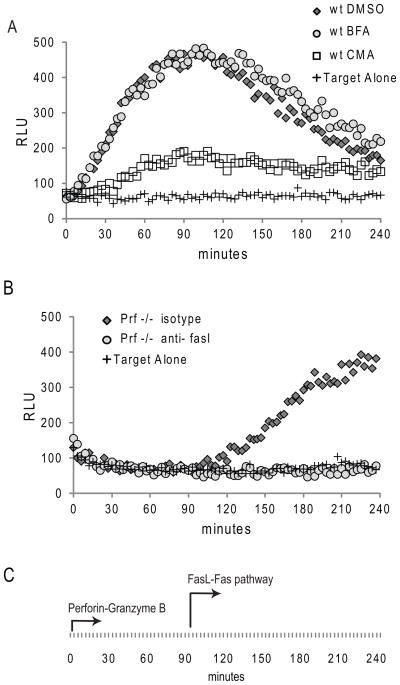
For assays of human CTL function by re-directed killing, the GLS. DEVD biosensor was cloned by limiting dilution into P815, a murine tumor line that binds murine immunoglobulin via Fc receptor. Upon co-incubation of P815 cells with anti-CD3 antibody and HVS T cells from a healthy donor at an E/T of 1, we saw activation of the biosensor within 10 minutes, plateauing at 90 minutes, followed by a gradual, persistent decline in luciferase signal (Figure 6A). The luciferase signal was inhibited by concanamycin A, a compound previously shown to ameliorate perforin mediated cytotoxic function; (39) whereas brefeldin A, which inhibits FasL upregulation on T cells, had no impact. We then tested the function of perforin deficient HVS T cells and noted that these cells activated the GLS.DEVD biosensor only after a 90 minute delay (Figure 6B). This rise in signal was blocked using anti-FasL antibody (Figure 6B) or brefeldin A (not shown). The rate of induction of the Fas mediated signal from the perforin deficient T cells was slower than the induction of the granzyme mediated signal from the perforin sufficient cell line, although this difference was not as pronounced as the differences seen in murine CTL. Remarkably the timing of induction following Fas activation was the same as that seen using perforin deficient murine CTL, with a gradual rise in signal occurring only after a 90–120 minute delay.
Figure 6. Redirected killing: human CTL utilize secretory granule and Fas-mediated pathways to activate Caspase 3/7 in target cells conjugated by anti-CD3.
(A) HVS transformed human CTL were co-incubated at 1:1 with GLS.DEVD expressing, cloned P815 cells, pre-incubated with anti-CD3 antibody. P815 cells without added effectors are shown as a negative control. Evaluation of RLU out to 240 minutes reveals early activation of GLS.DEVD in P815 incubated with wild type (wt) CTL contrasting with (B) late activation following co-incubation with perforin deficient CTL (Prf −/−). The early activation in the wt cell line was prevented by the addition of concanamycin A (CMA) but not brefeldin (BFA), demonstrating the dependence upon the secretory granule pathway. Addition of anti-FasL antibody to the PRF −/− CTL prevented the delayed rise in GLS.DEVD (B) confirming the role of the delayed onset death receptor pathway. Shown is a representative figure from 3 independent experiments utilizing these two HVS lines.
This technology allows for the first time, the capacity to precisely time the induction of delayed Fas/FasL mediated caspase activation following the immediate induction achieved by perforin-mediated granzyme B delivery. Additionally, the method allows comparison of the rate of induction of caspase induction by the two different pathways of cytotoxicity and signaling via two different immune synapses (MHC-peptide versus anti-CD3 antibody).
Discussion
We have shown for the first time that circularly-permuted luciferase proteins may be engineered as protease biosensors to measure CTL-mediated granzyme B delivery and subsequent caspase activation in live target cells. This novel luminescence-based CTL assay will be an invaluable tool for mechanistic assessment of CTL cytotoxicity. Utilizing a combination of caspase 3/7 and granzyme B cleavable biosensors, we have shown for the first time that the caspase 3/7 signal induced by CTL is extremely rapid and transient, contrasting to the signal obtained utilizing granzyme B cleavable biosensors.
Previous studies revealed the absolute dependence of murine granzyme B to induce caspase 3 activation following recognition of target cells (12, 40, 41) by CTL, and demonstrated caspase 3 activation in target cells by flow cytometry following CTL contact. (10–12) Caspase 3 activation is also noted in activated T cells, (42–46) limiting the utility of caspase 3 activation as a measure of CTL function using freely diffusible fluorescent substrates in a 96 well format. As such, this luciferase biosensor assay is the first cell-based method that detects CTL activation of caspase 3/7 in target cells in a microplate format.
We engineered a novel biosensor with a cleavage site for both granzyme B and caspase 8 (IETD). In the absence of granzyme B, there was no activation GLS.DEVD or GLS.IETD within 90 minutes, confirming the absolute dependence on granzyme B for caspase 3/7 and caspase 8 activation induced by secretory granule-mediated killing. Additional protease recognition sites specific for alternative granzymes may also be introduced in future studies to investigate their role in apoptosis in the presence or absence of granzyme B. Two such sites, IEAD and VGPD, were screened in the current study, but not pursued due to their suboptimal signal-to-noise ratio in EL4 target cells.
The luciferase biosensor approach provides several advantages over conventional radioactive and flow based CTL cytotoxic assays: (1) homogenous, microplate format without radioactivity; (2) real-time assessment of early events of target cell apoptosis in live cells, and (3) is amenable to a high throughput format. The main limitation of the assay relates to standardization of the assay results. Unlike a 51Cr release or flow based assay, there is no “maximum” signal. One approach is to perform the assay using a range of E/T to find the maximum signal per assay. Indeed, we have found that beyond CTL/target ratios of 6–12:1 there is no increase in luminescent signal (not shown). An ideal approach would induce apoptosis in target cells by a cell-free method (such as shown with etoposide) to provide a CTL independent, maximum signal in parallel with the CTL cytotoxicity assay. We are currently investigating a compound that may induce apoptosis within minutes, analogous to the CTL-induced pathway. Alternative standardization may utilize measures of fold induction compared to the basal signal in the negative control as shown for Figure 4 and Supplemental Figure 4.
We have measured caspase 3 activation in EL4 cells using other methodologies to evaluate the phenomenon of rapid caspase 3 activation mediated by murine CTL. Unfortunately, measuring active caspase 3 by fluorogenic substrate or antibody recognition of the active form was not sensitive at early time points to show induction of caspase 3 in EL4 cells within the first 30 minutes of CTL co-incubation (data not shown). This is likely because the fluorogenic substrates have substantial background in EL4 cells. (12) Evaluating caspase 3 cleavage in EL4 target cells by western blot following co-incubation with CTL was unreliable for two reasons: 1) CTL have significant amounts of caspase 3 and, 2) cleavage of caspase 3 by granzyme B occurs during the lysate preparation. The latter could be overcome with the use of a granzyme-specific inhibitor, but none are currently available for murine granzyme B.
The luminescent assay permits serial measurements of caspase activation over the entire time course of the assay. Real time evaluation of kinetics is straightforward, using a heated platform and buffered media to preserve CTL function outside of the tissue culture incubator. Our data demonstrate that delivery of granzyme B to the target cell by the effector CTL is extremely rapid, consistent with previous studies using 51Cr release assays or direct visualization. (5, 6, 47) Therefore both biosensors detect the earliest events of cytotoxicity. Indeed, the signal at 30, 60, and 90 minutes correlates well with the eventual cytolysis of target cells at 4 hours (Figure 3 and data not shown).
The kinetics for GLS.DEVD induction in target cells was slightly different when comparing in vitro derived and ex vivo murine CTL, highlighting the capacity of the biosensors to distinguish CTL function. The in vitro-derived CTL induced a caspase-mediated signal that peaked at 30–40 minutes while the ex vivo CTL induced a peak signal at 60 minutes. The difference in the kinetics was not explained by the lower E/T for the ex vivo CTL, as the cultured CTL behaved similarly at low E/T (although the peak was less distinct). However, we did not try to dilute the target cells with an irrelevant cell to test whether the delayed timing was related to a delay in cell contact for the ex vivo CTL, which were diluted in the assay by non-responding splenocytes. Future studies may be directed to understand if the differences are related to differences in cellular adhesion or perforin/granzyme B levels.
Comparison of GLS.DEVD versus GLS.IETD signals in the earliest time points following in vitro derived CTL induction also revealed distinct kinetics, including the rate of luminescence activation and the rate of luminescence decline. Although both the GLS.DEVD and GLS.IETD sensors were activated upon contact of effector CTL and target cells, the overall signal induction was faster and 2–5 times higher for GLS.DEVD, which likely reflects the auto-amplification property of caspase 3 following granzyme B cleavage. (48, 49) Following contact with cultured, P14 CTL, the GLS.DEVD signal declined rapidly beyond 30 minutes; whereas, the GLS.IETD signal plateaued briefly (30–45 minutes) and then continued to rise during the entire 90 minutes. Previous studies have predicted CTL: target cycles of contact lasting approximately 20–30 minutes. (50–52) The brief plateau in the IETD signal at 30 minutes may thus reflect the beginning of the second cycle or a pause between cell contact. The subsequent decline noted in the EL4 cells expressing the GLS.DEVD suggests that the initial caspase 3/7 activation signal may be turned off in a fraction of the earliest responding cells at 30 minutes. This may be related to inactivation of caspase 3/7 by endogenous inhibitors (53, 54), dissociation of the luciferase, or related to rapidly declining ATP levels in these early responding cells. Although we would expect the GLS.IETD signal to similarly decline if related to cell death/ATP loss, the loss of signal may be too low to detect in this biosensor where there is no amplification of the signal from caspases other than granzyme B. Further studies are needed to investigate this phenomenon.
The kinetics of caspase 3/7 activation following Fas/FasL mediated apoptosis pathway was also distinct from that induced by the secretory granule pathway. Our data in perforin deficient murine and human CTL suggests that in the first 90 minutes, there is no detection of Fas/FasL mediated activation of caspase 3/7 or caspase 8 (Figure 5 and 6). As CTL from perforin deficient animals are unable to kill target cells as measured by 51Cr release assay (data not shown), our data confirms that perforin deficiency is not compensated by Fas/FasL activation in the first 90 minutes of CTL: target contact. Late activation of the Fas/FasL path was clearly visualized in perforin and/or granzyme deficient CTL. The delay in initiation of the death receptor pathway and the gradual induction of caspase activation observed may be interrogated further with this novel system of measuring caspase activation.
In summary, using a novel, luminescence-based assay for measuring CTL function we have detected granzyme B mediated apoptosis within live target cells in real time. This approach is an important technological advance, since measurement of target cell protease activity in a typical CTL assay (with two independent cell populations present) has only been feasible by flow cytometry. The rate of activation was modulated by the number of CTL interacting with each target cell and the strength of TCR signaling. We predict that the luciferase biosensor assay will rapidly become a widely utilized method for measuring cytotoxic function in CTL. Our initial studies with murine NK were somewhat disappointing, but may require use of a different target cell line. Indeed, we have already adapted the assay to human NK targets (manuscript in preparation). We also predict that the microplate assay will be readily adaptable to a high throughput screening format, allowing transformative studies to address genetic and pharmacologic modifiers of cytotoxic function.
Supplementary Material
Acknowledgments
IL-2 was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Financial support: KAR was supported by University of Cincinnati/Cincinnati Children’s Hospital Medical Center for Clinical and Translational Research “T1” Award. JL was supported by a NIH training award, T32 AI-060515. Promega Corporation generated the granzyme B cleavable biosensor vectors and tested in vitro translated products.
Footnotes
Author roles: JL, SKF, MBJ, AV, BLB, BFB and KAR designed and executed experiments and interpreted data. AF and YT generated the human cell lines from healthy adults and patients with perforin mutations. KAR and BFB designed the granzyme B cleavable biosensors. All authors contributed to the writing and review of the manuscript.
References
- 1.Jenkins MR, Griffiths GM. The synapse and cytolytic machinery of cytotoxic T cells. Curr Opin Immunol. 2010;22:308–313. doi: 10.1016/j.coi.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dustin ML, Long EO. Cytotoxic immunological synapses. Immunol Rev. 2010;235:24–34. doi: 10.1111/j.0105-2896.2010.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Saint Basile G, Menasche G, Fischer A. Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat Rev Immunol. 2010;10:568–579. doi: 10.1038/nri2803. [DOI] [PubMed] [Google Scholar]
- 4.Praper T, Sonnen AF, Kladnik A, Andrighetti AO, Viero G, Morris KJ, Volpi E, Lunelli L, Dalla Serra M, Froelich CJ, Gilbert RJ, Anderluh G. Perforin activity at membranes leads to invaginations and vesicle formation. Proc Natl Acad Sci U S A. 2011;108:21016–21021. doi: 10.1073/pnas.1107473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery J, Keefe D, Boulant S, Boucrot E, Walch M, Martinvalet D, Goping IS, Bleackley RC, Kirchhausen T, Lieberman J. Perforin pores in the endosomal membrane trigger the release of endocytosed granzyme B into the cytosol of target cells. Nat Immunol. 2011;12:770–777. doi: 10.1038/ni.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez JA, Susanto O, Jenkins MR, Lukoyanova N, Sutton VR, Law RH, Johnston A, Bird CH, Bird PI, Whisstock JC, Trapani JA, Saibil HR, Voskoboinik I. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood. 2013;121:2659–2668. doi: 10.1182/blood-2012-07-446146. [DOI] [PubMed] [Google Scholar]
- 7.Ewen CL, Kane KP, Bleackley RC. A quarter century of granzymes. Cell Death Differ. 2012;19:28–35. doi: 10.1038/cdd.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Susanto O, Trapani JA, Brasacchio D. Controversies in granzyme biology. Tissue Antigens. 2012;80:477–487. doi: 10.1111/tan.12014. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Chahroudi A, Silvestri G, Wernett ME, Kaiser WJ, Safrit JT, Komoriya A, Altman JD, Packard BZ, Feinberg MB. Visualization and quantification of T cell-mediated cytotoxicity using cell-permeable fluorogenic caspase substrates. Nature medicine. 2002;8:185–189. doi: 10.1038/nm0202-185. [DOI] [PubMed] [Google Scholar]
- 11.Jerome KR, Sloan DD, Aubert M. Measurement of CTL-induced cytotoxicity: the caspase 3 assay. Apoptosis: an international journal on programmed cell death. 2003;8:563–571. doi: 10.1023/A:1026123223387. [DOI] [PubMed] [Google Scholar]
- 12.Pardo J, Bosque A, Brehm R, Wallich R, Naval J, Mullbacher A, Anel A, Simon MM. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J Cell Biol. 2004;167:457–468. doi: 10.1083/jcb.200406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L, Hakimi J, Salha D, Miron I, Dunn P, Radvanyi L. A sensitive flow cytometry-based cytotoxic T-lymphocyte assay through detection of cleaved caspase 3 in target cells. J Immunol Methods. 2005;304:43–59. doi: 10.1016/j.jim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Betts MR, Koup RA. Detection of T-cell degranulation: CD107a and b. Methods Cell Biol. 2004;75:497–512. doi: 10.1016/s0091-679x(04)75020-7. [DOI] [PubMed] [Google Scholar]
- 15.Brunner KT, Mauel J, Cerottini JC, Chapuis B. Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology. 1968;14:181–196. [PMC free article] [PubMed] [Google Scholar]
- 16.Sheehy ME, McDermott AB, Furlan SN, Klenerman P, Nixon DF. A novel technique for the fluorometric assessment of T lymphocyte antigen specific lysis. J Immunol Methods. 2001;249:99–110. doi: 10.1016/s0022-1759(00)00329-x. [DOI] [PubMed] [Google Scholar]
- 17.Hassin D, Garber OG, Meiraz A, Schiffenbauer YS, Berke G. Cytotoxic T lymphocyte perforin and Fas ligand working in concert even when Fas ligand lytic action is still not detectable. Immunology. 2011;133:190–196. doi: 10.1111/j.1365-2567.2011.03426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV. Novel genetically encoded biosensors using firefly luciferase. ACS chemical biology. 2008;3:346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- 19.Hoff BA, Chughtai K, Jeon YH, Kozloff K, Galban S, Rehemtulla A, Ross BD, Galban CJ. Multimodality imaging of tumor and bone response in a mouse model of bony metastasis. Translational oncology. 2012;5:415–421. doi: 10.1593/tlo.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galban S, Jeon YH, Bowman BM, Stevenson J, Sebolt KA, Sharkey LM, Lafferty M, Hoff BA, Butler BL, Wigdal SS, Binkowski BF, Otto P, Zimmerman K, Vidugiris G, Encell LP, Fan F, Wood KV, Galban CJ, Ross BD, Rehemtulla A. Imaging proteolytic activity in live cells and animal models. PLoS One. 2013;8:e66248. doi: 10.1371/journal.pone.0066248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pircher H, Michalopoulos EE, Iwamoto A, Ohashi PS, Baenziger J, Hengartner H, Zinkernagel RM, Mak TW. Molecular analysis of the antigen receptor of virus-specific cytotoxic T cells and identification of a new V alpha family. European journal of immunology. 1987;17:1843–1846. doi: 10.1002/eji.1830171226. [DOI] [PubMed] [Google Scholar]
- 22.Shresta S, MacIvor DM, Heusel JW, Russell JH, Ley TJ. Natural killer and lymphokine-activated killer cells require granzyme B for the rapid induction of apoptosis in susceptible target cells. Proceedings of the National Academy of Sciences. 1995;92:5679–5683. doi: 10.1073/pnas.92.12.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, Jia B, Zheng Y, Ambruso DR, Lowe JB, Atkinson SJ, Dinauer MC, Boxer L. Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood. 2000;96:1646–1654. [PubMed] [Google Scholar]
- 24.Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J Clin Invest. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biesinger B, Muller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers RC, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci U S A. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urrea Moreno R, Gil J, Rodriguez-Sainz C, Cela E, LaFay V, Oloizia B, Herr AB, Sumegi J, Jordan MB, Risma KA. Functional assessment of perforin C2 domain mutations illustrates the critical role for calcium-dependent lipid binding in perforin cytotoxic function. Blood. 2009;113:338–346. doi: 10.1182/blood-2008-08-172924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. The Journal of biological chemistry. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 28.Fischer U, Stroh C, Schulze-Osthoff K. Unique and overlapping substrate specificities of caspase-8 and caspase-10. Oncogene. 2006;25:152–159. doi: 10.1038/sj.onc.1209015. [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Packard BZ, Telford WG, Komoriya A, Henkart PA. Granzyme B activity in target cells detects attack by cytotoxic lymphocytes. J Immunol. 2007;179:3812–3820. doi: 10.4049/jimmunol.179.6.3812. [DOI] [PubMed] [Google Scholar]
- 31.Lykens JE, Terrell CE, Zoller EE, Risma K, Jordan MB. Perforin is a critical physiologic regulator of T-cell activation. Blood. 2011;118:618–626. doi: 10.1182/blood-2010-12-324533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallimore A, Glithero A, Godkin A, Tissot AC, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med. 1998;187:1383–1393. doi: 10.1084/jem.187.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gairin JE, Mazarguil H, Hudrisier D, Oldstone MB. Optimal lymphocytic choriomeningitis virus sequences restricted by H-2Db major histocompatibility complex class I molecules and presented to cytotoxic T lymphocytes. J Virol. 1995;69:2297–2305. doi: 10.1128/jvi.69.4.2297-2305.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valitutti S, Muller S, Dessing M, Lanzavecchia A. Different responses are elicited in cytotoxic T lymphocytes by different levels of T cell receptor occupancy. J Exp Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowin B, Hahne M, Mattmann C, Tschopp J. Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature. 1994;370:650–652. doi: 10.1038/370650a0. [DOI] [PubMed] [Google Scholar]
- 36.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 37.Shresta S, Russell JH, Ley TJ. Mechanisms responsible for granzyme B-independent cytotoxicity. Blood. 1997;89:4085–4091. [PubMed] [Google Scholar]
- 38.Mittrucker HW, Muller-Fleckenstein I, Fleckenstein B, Fleischer B. Herpes virus saimiri-transformed human T lymphocytes: normal functional phenotype and preserved T cell receptor signalling. Int Immunol. 1993;5:985–990. doi: 10.1093/intimm/5.8.985. [DOI] [PubMed] [Google Scholar]
- 39.Kataoka T, Shinohara N, Takayama H, Takaku K, Kondo S, Yonehara S, Nagai K. Concanamycin A, a powerful tool for characterization and estimation of contribution of perforin- and Fas-based lytic pathways in cell-mediated cytotoxicity. J Immunol. 1996;156:3678–3686. [PubMed] [Google Scholar]
- 40.Adrain C, Murphy BM, Martin SJ. Molecular ordering of the caspase activation cascade initiated by the cytotoxic T lymphocyte/natural killer (CTL/NK) protease granzyme B. The Journal of biological chemistry. 2005;280:4663–4673. doi: 10.1074/jbc.M410915200. [DOI] [PubMed] [Google Scholar]
- 41.Darmon AJ, Nicholson DW, Bleackley RC. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature. 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 42.Sabbagh L, Kaech SM, Bourbonniere M, Woo M, Cohen LY, Haddad EK, Labrecque N, Ahmed R, Sekaly RP. The selective increase in caspase-3 expression in effector but not memory T cells allows susceptibility to apoptosis. Journal of immunology. 2004;173:5425–5433. doi: 10.4049/jimmunol.173.9.5425. [DOI] [PubMed] [Google Scholar]
- 43.Miossec C, Dutilleul V, Fassy F, Diu-Hercend A. Evidence for CPP32 activation in the absence of apoptosis during T lymphocyte stimulation. J Biol Chem. 1997;272:13459–13462. doi: 10.1074/jbc.272.21.13459. [DOI] [PubMed] [Google Scholar]
- 44.Wilhelm S, Wagner H, Hacker G. Activation of caspase-3-like enzymes in non-apoptotic T cells. European journal of immunology. 1998;28:891–900. doi: 10.1002/(SICI)1521-4141(199803)28:03<891::AID-IMMU891>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 45.Alam A, Cohen LY, Aouad S, Sékaly RP. Early Activation of Caspases during T Lymphocyte Stimulation Results in Selective Substrate Cleavage in Nonapoptotic Cells. The Journal of experimental medicine. 1999;190:1879–1890. doi: 10.1084/jem.190.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy NJ, Kataoka T, Tschopp J, Budd RC. Caspase Activation Is Required for T Cell Proliferation. The Journal of experimental medicine. 1999;190:1891–1896. doi: 10.1084/jem.190.12.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lees RK, MacDonald HR, Sinclair NR. Inhibition of clone formation as an assay for T cell-mediated cytotoxicity: short-term kinetics and comparison with 51Cr release. Journal of immunological methods. 1977;16:233–244. doi: 10.1016/0022-1759(77)90201-0. [DOI] [PubMed] [Google Scholar]
- 48.Metkar SS, Wang B, Ebbs ML, Kim JH, Lee YJ, Raja SM, Froelich CJ. Granzyme B activates procaspase-3 which signals a mitochondrial amplification loop for maximal apoptosis. The Journal of cell biology. 2003;160:875–885. doi: 10.1083/jcb.200210158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, Green DR, Martin SJ. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. The Journal of cell biology. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macken CA, Perelson AS. A multistage model for the action of cytotoxic T lymphocytes in multicellular conjugates. Journal of immunology. 1984;132:1614–1624. [PubMed] [Google Scholar]
- 51.Deguine J, Breart B, Lemaitre F, Di Santo JP, Bousso P. Intravital imaging reveals distinct dynamics for natural killer and CD8(+) T cells during tumor regression. Immunity. 2010;33:632–644. doi: 10.1016/j.immuni.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 52.Mrass P, Takano H, Ng LG, Daxini S, Lasaro MO, Iparraguirre A, Cavanagh LL, von Andrian UH, Ertl HC, Haydon PG, Weninger W. Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J Exp Med. 2006;203:2749–2761. doi: 10.1084/jem.20060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.