Abstract
Progressive liver fibrosis is characterized by the deposition of collagen by activated hepatic stellate cells (HSCs). Activation of HSCs is a multiple receptor-driven process in which profibrotic signals are enhanced, and anti-fibrotic pathways are suppressed. Here we report the discovery of a novel signaling platform comprised of G protein subunit, Gαi and GIV, its guanine exchange factor (GEF), which serves as a central hub within the fibrogenic signalling network initiated by diverse classes of receptors. GIV is expressed in the liver after fibrogenic injury and is required for HSC activation. Once expressed, GIV enhances the profibrotic (PI3K-Akt-FoxO1 and TGFβ-SMAD) and inhibits the anti-fibrotic (cAMP-PKA-pCREB) pathways to skew the signalling network in favor of fibrosis, all via activation of Gαi. We also provide evidence that GIV may serve as a biomarker for progression of fibrosis after liver injury and a therapeutic target for arresting and/or reversing HSC activation during liver fibrosis.
Introduction
Liver fibrosis is the consequence of an excessive deposition of collagen resulting from an imbalance between its synthesis and degradation. Upon liver injury, hepatic stellate cells (HSCs), the major collagen-synthesizing cells in the liver, become activated and transdifferentiate into myofibroblast-like cells which proliferate faster and display enhanced chemotaxis, survival and collagen production 1. Such transdifferentiation is triggered by the activation of a diverse classes of receptors, e.g., PDGFR 2, TGFβR 3, EGFR 4, VEGFR 5, IR/IGF1R 6, Gi-coupled chemokine/cytokine receptors (CCRs) 7 and Toll-like receptor 4 (TLR4) 8, and two of them (TGFβR and PDGFR) have been widely studied and established as major pro-fibrogenic receptors 2,3. Regardless of the type of receptor activated, two key profibrotic signals, the PI3K-Akt and the SMAD signaling cascades 9, must be enhanced to initiate and propagate HSC activation and collagen synthesis. Once activated, the PI3K-Akt pathway triggers fibrogenesis via 3 mechanisms-- 1) by inactivating Forkhead box protein O1 (FoxO1), an anti-fibrotic transcription factor that suppresses transdifferentiation and proliferation of HSCs 10, 2) by suppressing apoptosis 11, and 3) by further enhancing the TGFβ-SMAD-collagen pathway 11. Although widely recognized as a key profibrotic trigger, the precise multi-receptor mediated mechanism(s) that drives unrestricted enhancement of the PI3K-Akt pathway remains elusive.
Profibrotic pathways are antagonistically balanced by cyclic adenosine monophosphate (cAMP), a well known and conserved antifibrotic signaling second messenger of G protein signaling cascades 12. Accumulation of cAMP in HSCs is known to inhibit chemotaxis, proliferation, and collagen synthesis, while simultaneously increasing the removal of collagen by metalloproteases 13. Mechanistically, cAMP inhibits TGFβ-SMAD signaling at a transcriptional level via modulation of the protein kinase A (PKA)-cAMP response element binding protein (CREB)-dependent pathway 14. Although recognized as a key antifibrotic signal, the mechanism(s) by which cAMP levels are suppressed by multiple receptors during liver fibrosis remains unknown.
We recently established that GIV, a.k.a Gα-Interacting Vesicle-associated protein is a multimodular signal transducer and a guanine nucleotide exchange factor (GEF) for trimeric Gi that modulates key signaling events downstream of diverse classes of receptors, e.g., growth factor receptor tyrosine kinases (RTKs), e.g., EGFR, IGF1R, VEGFR and InsR 15-18 and chemotactic G protein-coupled receptors (GPCRs) like LPAR1 16,19,20. Working downstream of both RTKs and GPCRs, GIV serves as a common platform that enhances PI3K-Akt signals and reduces cAMP production, all via activation of Gi 16. GIV modulates signaling during diverse biological processes including epithelial wound healing, macrophage chemotaxis, development, autophagy, angiogenesis, vascular repair and tumor metastasis (reviewed in ref 21).
Because signaling pathways modulated by GIV are also major components of the fibrogenic network, here we investigated if GIV plays a role in the activation of HSCs during liver fibrosis. We found that GIV triggers HSC activation by coordinately reshaping the fibrogenic signaling network (enhances pro- and inhibits anti-fibrogenic signals) downstream of multiple receptors, all via activation of Gαi. Because the fundamental signaling pathways driven by GIV are common to fibrogenic diseases afflicting other organs, insights gained will impact the understanding, approach, and the paradigms of diverse fibrosis-related diseases.
Results
GIV expression is upregulated in liver after fibrotic injury
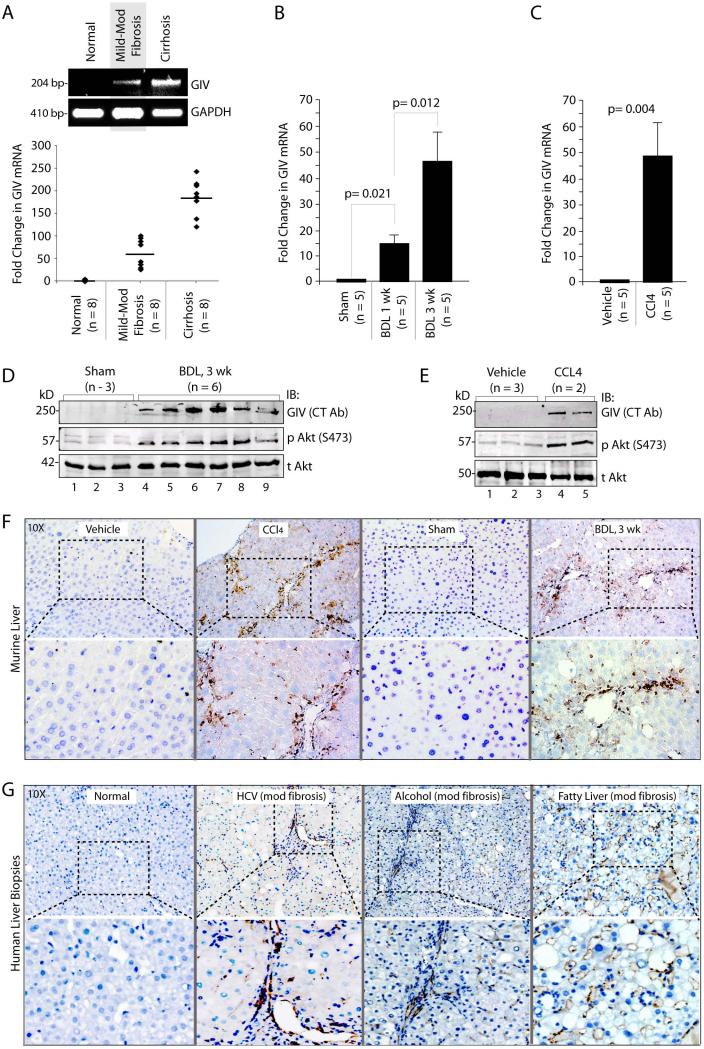
We previously showed that expression of GIV is upregulated during epithelial wound healing 22. Because liver fibrosis in response to chronic injury mimics the process of wound healing 23, we asked whether the level of GIV expression is altered during the course of chronic liver injury in patients with chronic HCV hepatitis, a leading cause of liver cirrhosis 24. We found that full-length GIV mRNA was virtually undetectable in normal livers, but elevated ~50 fold in livers with mild to moderate fibrosis (Ishak score 1-4), and ~175 fold in those with cirrhosis (Ishak scores 5, 6) (Figure 1A), indicating that GIV mRNA expression increases with increasing degree of liver fibrosis. When we analyzed liver samples from two well-accepted animal models for chronic liver injury 25, i.e., surgical bile duct ligation (BDL) and chronic injection of carbon tetrachloride (CCl4) we found that GIV mRNA was virtually undetectable in normal murine livers (sham and vehicle controls), but elevated ~15-45 fold in response to both types of injuries during progressive fibrosis (Figure 1B, 1C). Next we analyzed total liver lysates from BDL or CCl4-treated mice for GIV protein expression by immunoblotting with an antibody that recognizes the C-terminus of full-length GIV (CT-Ab) 26. GIV was detected exclusively after injury in both study models, increasing with progressive fibrosis (Figure 1D, 1E, Supplementary Fig. 1A), and such expression correlated with increased activation of the PI3K-Akt pathway, as determined by Akt phosphorylation (Figure 1D, 1E). GIV was also detected by immunohistochemistry (IHC) in mice livers exclusively after BDL or CCl4- injury (Figure 1F), and in liver biopsies from patients with liver fibrosis due to a variety of chronic injuries (Figure 1G). The staining pattern in injured livers was consistent with restricted expression of full-length GIV exclusively in non-parenchymal i.e., interstitial cells, whereas hepatocytes, which comprise ~85% of the total cell mass of the liver, do not express full-length GIV regardless of injury. These findings indicate: 1) that there is little or no expression of full-length GIV (neither mRNA nor protein) in normal livers; 2) that GIV expression is consistently induced after a variety of chronic injuries; 3) that the abundance of GIV mRNA increases with progressive fibrosis; 4) that the increase in mRNA is accompanied by translation of GIV protein; and 5) that increased GIV protein expression correlates with increased Akt activation in the injured livers.
Figure 1. Expression of GIV increases in liver after chronic fibrogenic injury.
(A) GIV mRNA levels were analyzed in livers from normal and HCV-infected patients with variable degrees of fibrosis by semi-quantitative (top) and quantitative PCR (bottom). Representative results for semi-quantitative PCR and a scattergram showing the fold induction of GIV mRNA are shown. Horizontal bar = mean. n = number of animals.
(B, C) GIV mRNA levels were analyzed in livers of mice subjected to BDL or sham surgery (B), or chronic injections of CCl4 or vehicle control (C) by qPCR as in A. Results are shown as fold change compared with sham- and vehicle-treated controls. . Error bars represent mean ± S.D. n = number of animals. Statistical significance was assessed with two-tailed Student's t test.
(D, E) Total liver lysates from mice subjected to BDL or sham surgery (D) chronic injections of CCl4 or vehicle control (E) were analyzed for full-length GIV, phospho-Akt (pAkt), and total Akt (tAkt) by immunoblotting (IB).
(F) FFPE liver sections from mice subjected to BDL or CCl4 injury and their respective controls were analyzed for expression of full-length GIV protein (CT-Ab) by IHC. Boxed area in top panel is magnified in bottom panels. Magnification = 10X.
(G) FFPE liver sections from normal subjects or from patients with a variety of chronic liver injuries were analyzed for GIV as in E. Images are representative of 4-11 livers analyzed in each category. Boxed area in top panel is magnified in bottom panels. Magnification = 10X.
Unlike chronic injuries, when microarrays performed on a variety of acute liver injuries were analyzed for the expression profile of GIV, we found that GIV expression was elevated in some, but not all cases (Supplementary Fig. 1B-E). GIV mRNA was minimally upregulated in patients with acute viral hepatitides (Supplementary Fig. 1B, C), which carries the potential of progressing to chronicity and liver fibrosis in some patients, whereas no such upregulation was observed in acute alcoholic hepatitis or in a murine model of acute injury by partial hepatectomy (Supplementary Fig. 1D, E), two conditions that heal without fibrosis 27. These findings indicate that GIV expression is induced during some, but not all injuries of the liver, and that persistent upregulation of GIV expression is primarily restricted only to chronic injuries of the liver.
GIV expression is upregulated in activated HSCs
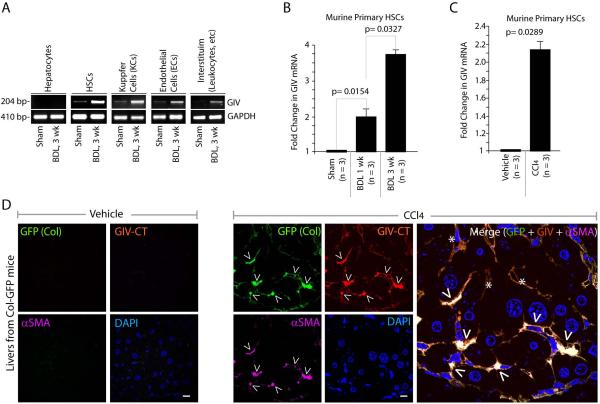
To determine which non-parenchymal cell type within the liver contributes to the observed increase in GIV we carried out liver cell fractionation from BDL-treated and control mice, and analyzed the isolated cell types for expression of GIV mRNA. Consistent with our IHC staining pattern, we found that hepatocytes have no detectable full-length GIV transcript regardless of injury (Figure 2A). Low levels of GIV mRNA were detected in all the other cells isolated from control livers, which significantly increased after BDL. Both HSCs and Kupffer cells (KCs; the liver resident macrophages), which drive liver fibrosis in response to chronic liver injury 28, showed significant increases after injury. We focused on the role of GIV expression in HSCs for two outstanding reasons: 1) they contribute to ~80% collagen produced during liver fibrosis, and 2) they are the final common target of cytokines and growth factors produced by other cells that contribute to liver fibrosis, e.g., KCs and sinusoidal endothelial cells (ECs) 28,29. When we quantitatively assessed GIV mRNA in isolated HSCs after BDL or CCl4 injuries, we found, similar to our results in whole liver, that expression of full-length GIV is upregulated ~2-4 fold during progressive fibrosis in both models of chronic injury (Figure 2B, 2C).
Figure 2. Expression of full-length GIV mRNA and protein increases in HSCs after injury.
(A) Isolated cell fractions from livers of mice subjected to BDL or sham surgery were analyzed for GIV and GAPDH mRNA levels by RT-PCR.
(B, C) Primary HSCs isolated from livers of mice subjected to BDL or sham surgery (B) or injections of CCl4 or vehicle control (C) were analyzed for GIV mRNA by qPCR. Results are shown as fold change compared with sham- and vehicle-treated controls, respectively. Error bars represent mean ± S.D. n = number of animals. Statistical significance was assessed with two-tailed Student's t test.
(D) Semi-thin liver sections from Col-GFP mice chronically treated with injections of CCl4 or vehicle control were stained for GIV (red), Collagen (GFP, green), α-SMA (purple) and DAPI/DNA (blue) and analyzed by confocal microscopy. Arrowheads = α-SMA pos, Col pos, GIV pos (i.e., HSCs); Stars = α-SMA neg, GIV pos, likely to be KCs and Sinusoidal ECs. Scale bar = 10 μm.
To further corroborate whether GIV protein is expressed in HSCs after chronic liver injury we analyzed the expression of full-length GIV in situ in semi-thin sections of livers harvested from control and CCl4-treated Col-GFP mice by immunofluorescence. We used Col-GFP transgenic mice that express GFP driven by alpha1 (I) collagen promoter 30 because, in these mice, GFP serves as a surrogate marker for collagen expression in whole liver extracts and in isolated primary HSCs 30. As anticipated, GFP (collagen) and α-Smooth muscle actin (α-SMA), a bona fide marker for active HSCs 31 were detected exclusively after injury. GIV was undetectable in the control liver, but detected in activated HSCs, as determined by colocalization with α-SMA (Figure 2D). Expression of GIV correlated with collagen (GFP) expression in these α-SMA-positive HSCs (arrowheads in Figure 2D), indicating that upon injury, expression of GIV protein coincides with HSC activation and collagen synthesis. GIV was also detected at lower levels in α-SMA-negative cells that are likely to be ECs or KCs (stars in Figure 2D). Together these results demonstrate that GIV mRNA is upregulated in HSCs after injury and GIV protein is translated after HSC activation.
Expression of GIV precedes collagen synthesis during HSC activation
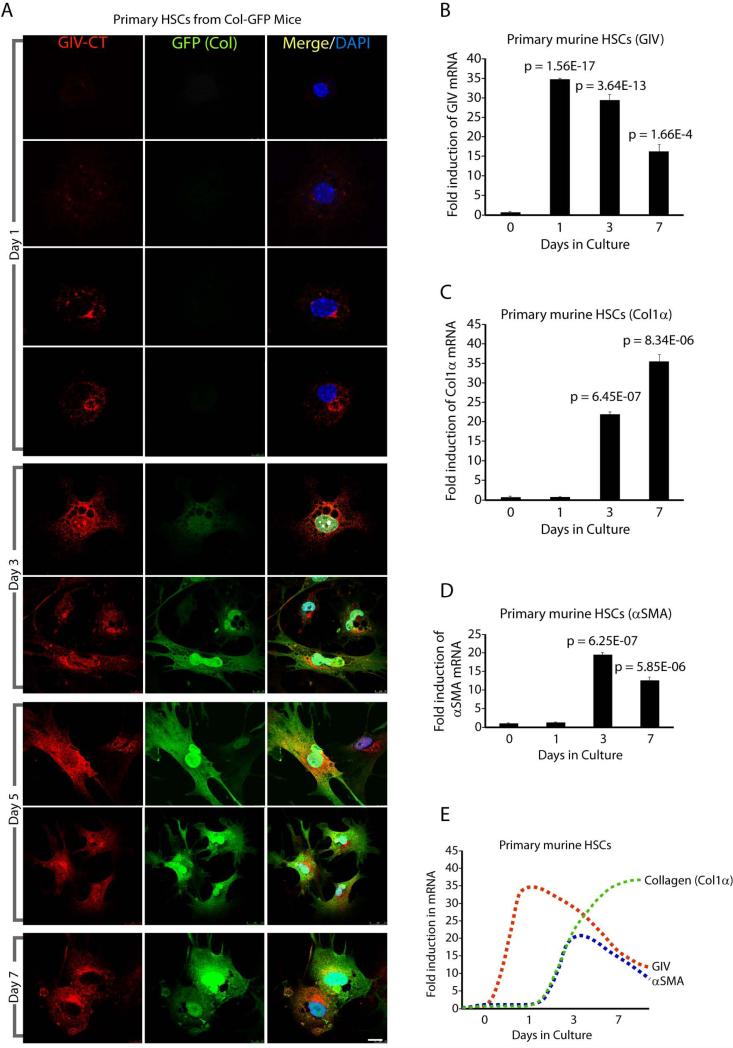
Next we investigated the temporal correlation between GIV expression, HSC activation (as determined by α-SMA expression) and collagen production in HSCs isolated from Col-GFP mice. HSCs were activated on plastic culture dishes and monitored for morphologic changes that are characteristic of transdifferentiation and by the abundance of collagen (GFP) synthesis (Supplementary Fig. 2). On day 1, as expected, inactive HSCs contained lipid droplets laden with retinyl esters and did not express collagen, however, ~50% of those collagen-negative cells already expressed GIV (Figure 3A). Both GIV and collagen expression increased robustly in all HSCs on days 3, 5 and 7. Noteworthy, there were no collagen positive HSCs that did not also express GIV, indicating that GIV expression invariably preceded expression of collagen. An analysis of mRNA levels confirmed that induction of GIV expression precedes collagen and α-SMA expression (Figure 3B-E) during HSCs activation; while GIV expression peaks on day 1 expression of collagen and α-SMA peak on day 3. These results demonstrate that in HSCs activated ex vivo, upregulation of GIV is likely to be an upstream event which precedes HSC activation and collagen synthesis.
Figure 3. Expression of GIV mRNA and protein precedes activation and collagen production in culture-activated HSCs.
(A) Primary HSCs isolated from livers of Col-GFP mice were culture-activated on uncoated plastic dishes for 7 days. Cells were fixed at various time points and stained for GIV (red) and DAPI/DNA (blue). Collagen production was detected by GFP signal as a surrogate marker. Representative images on different days are shown. Scale bar = 10 μm.
(B-E) Primary HSCs isolated from livers of Col-GFP mice were culture-activated on uncoated plastic dishes for 7 days. GIV (B), collagen (C) and α-SMA (D) mRNA levels were measured by qPCR at different time points. (E) Expression patterns of GIV, collagen and α-SMA in B-D are plotted as line graphs superimposed on each other. GIV expression precedes peak expression of α-SMA and collagen. Results are shown as fold change compared with day 0 for each target transcript.
GIV is required for activation of HSCs
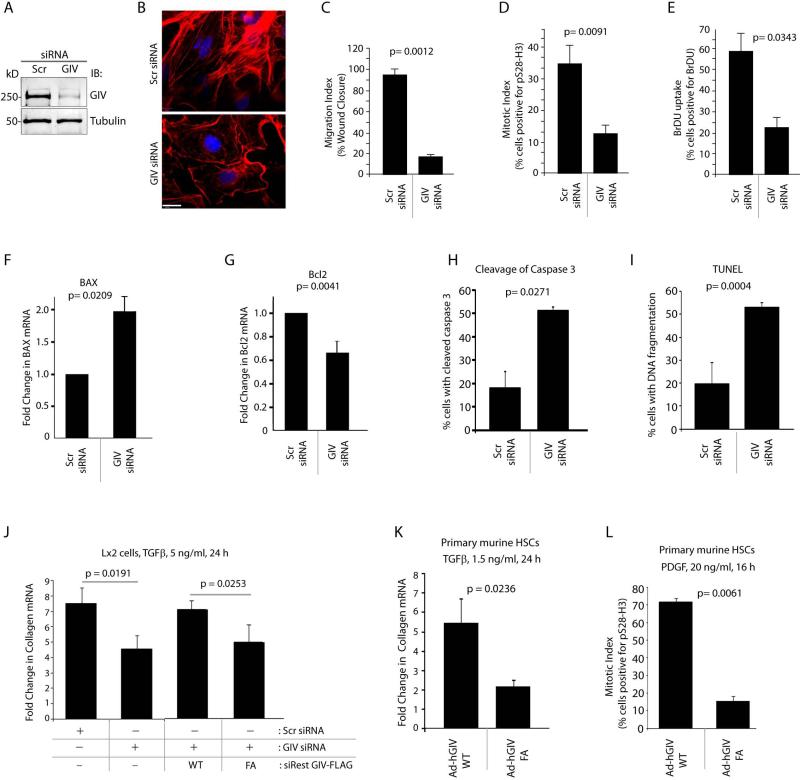
HSC activation is characterized by key phenotypes such as enhanced proliferation, cell migration, actin remodeling, resistance to apoptosis and collagen production 1. Next we asked if GIV is required for some or most of these phenotypes, and thereby, for HSC activation. First, we analyzed GIV expression, localization and properties in cultured human Lx2 HSC line, which has been extensively characterized and widely accepted as a model system to study fibrogenic pathways 31. We found that Lx2 cells express GIV as a ~250 kDa full-length protein (Figure 4A) which localizes to the Golgi and PM-associated actin bed (Supplementary Fig. 3A), as previously shown in other cell lines 19,32. Similar localization was also observed in primary murine HSCs (Supplementary Fig. 3A). Compared to control cells, GIV-depleted cells (~85-90% efficacy; Figure 4A) showed fewer actin stress fibers (Figure 4B), impaired cell migration (Figure 4C, Supplementary Fig. 3B), and suppressed mitosis, as determined by nuclear accumulation of phospho-histone H3 (Figure 4D, Supplementary Fig. 3C) and by BrDU uptake (Figure 4E, Supplementary Fig. 3D). These results demonstrate that GIV is required for cell migration, proliferation and cytoskeletal remodeling in HSCs.
Figure 4. GIV is required for actin remodeling, proliferation, migration, survival and collagen production in HSCs.
(A) Lx2 cells were treated either with control (Scr) or with G cvcIV siRNA, lysates were analyzed for GIV depletion by immunoblotting (IB).
(B) Control (Scr) and GIV siRNA-treated Lx2 cells were fixed and stained for Phalloidin (red) and DAPI/DNA (blue), and visualized by confocal microscopy. Scale bar = 20 μm.
(C) Control (Scr) or GIV siRNA-treated Lx2 cells were analyzed for their ability to migrate and close a scratch-wound by serial imaging of the wound area (see Supplementary Fig. 3B). Results are expressed as % wound closure.
(D, E) Lx2 cells as in C were analyzed for nuclear phospho-Histone H3 or BrDU uptake by immunofluorescence and analyzed by confocal microscopy (see Supplementary Fig. 3C, D). Results are expressed as % cells positive for staining.
(F, G) Control (Scr) or GIV siRNA-treated Lx2 cells were analyzed for BAX (F) and Bcl-2 (G) mRNA by qPCR. Results are displayed as fold change compared to their respective Scr-treated controls.
(H, I) Control (Scr) or GIV siRNA-treated Lx2 cells were analyzed for cleaved Caspase 3 (I) by immunofluorescence and fragmented DNA by TUNEL staining (H). Bar graphs display % cells with cleaved Caspase 3 (I) or fragmented DNA (H; TUNEL) on Y axis.
(J) Lx2 cells were treated with control (Scr) or GIV siRNA or transfected with siRNA-resistant GIV-WT or GIV-FA, as indicated, starved and subsequently treated with TGF-β1. Collagen α1 mRNA was determined by qPCR and results are displayed as fold change compared to starved controls (Y axis).
(K, L) Primary murine HSCs were infected with adenoviruses expressing HA-tagged GIV-WT or GIV-FA, starved, and subsequently treated with TGF-β1 (I) or PDGF-BB (J). Collagen α1 mRNA level was determined by qPCR. Results are displayed as fold change compared to starved controls (Y axis) (I) and mitotic index was analyzed using phospho-Histone H3 (J) as in D. Results are displayed as % of cells positive for nuclear phospho-Histone H3 (Y axis). Error bars represent mean ± S.D. n = 3. Statistical significance was assessed with two-tailed Student's t test.
Because activated HSCs acquire resistance to apoptosis 1 we analyzed the expression of two bona fide pro- (Bax) and anti- (Bcl-2) apoptotic signaling intermediates 33. In GIV-depleted HSCs, proapoptotic Bax was upregulated ~2 fold (Figure 4F) and anti-apoptotic Bcl-2 was decreased by ~40% (Figure 4G) compared to controls, indicating that in the absence of GIV cells are overall pro-apoptotic. Moreover, in HSCs treated with gliotoxin 34, an inducer of apoptosis, GIV expression was reduced by ~50%, alongside the expected increase of Bax and decrease in Bcl-2 (Supplementary Fig. 3E), indicating that changes in GIV expression profile after a pro-apoptotic injury mirrors that of an anti-apoptotic gene, i.e., Bcl-2, and not Bax. To confirm GIV's anti-apoptotic role in HSCs we carried out TUNEL assays and analyzed cleavage of Caspase 3, two widely used approaches for measuring cellular apoptosis35. We found that compared to control cells, GIV-depleted cells had increased Caspase 3 activity, as determined by the presence of the cleaved enzyme (Figure 4H; ~ 2.5 fold) and increased, fragmented DNA, as determined by TUNEL (Figure 4I; ~ 2.5 fold). These results demonstrate that depletion of GIV triggers apoptosis, and that GIV is required for survival of HSCs after injury.
Because GIV modulates signal transduction primarily via its GEF property 16 we asked if two key HSC phenotypes, i.e., proliferation and collagen production requires GIV's GEF function. To investigate this, we took advantage of previously validated GIV constructs: wild-type GIV (GIV-WT), and GEF-deficient F1685A (GIV-FA) mutant 16. Depletion of GIV from Lx2 HSCs suppressed collagen production in response to transforming growth factor-β1 (TGF-β1) by ~40-50% compared to controls (Figure 4J), an effect that was reversed by exogenous expression of siRNA-resistant GIV-WT, but not GIV-FA mutant. Similar results were obtained also in TGF-β1-stimulated primary HSCs, i.e., expression of GIV-FA suppressed collagen synthesis by ~60% compared to GIV-WT (Figure 4K). Finally, with regard to mitogenic response to platelet-derived growth factor (PDGF), expression of GIV-FA suppressed mitosis by ~7 fold compared to GIV-WT (Figure 4L), indicating that GIV's GEF function is required for growth factor-stimulated collagen synthesis and proliferation in HSCs. Together these results demonstrate that GIV and its GEF function is required for several key phenotypes that are critical for transdifferentiation and sustained activation of HSCs during progressive fibrosis.
PDGFRβ directly binds and phosphorylates GIV in activated HSCs
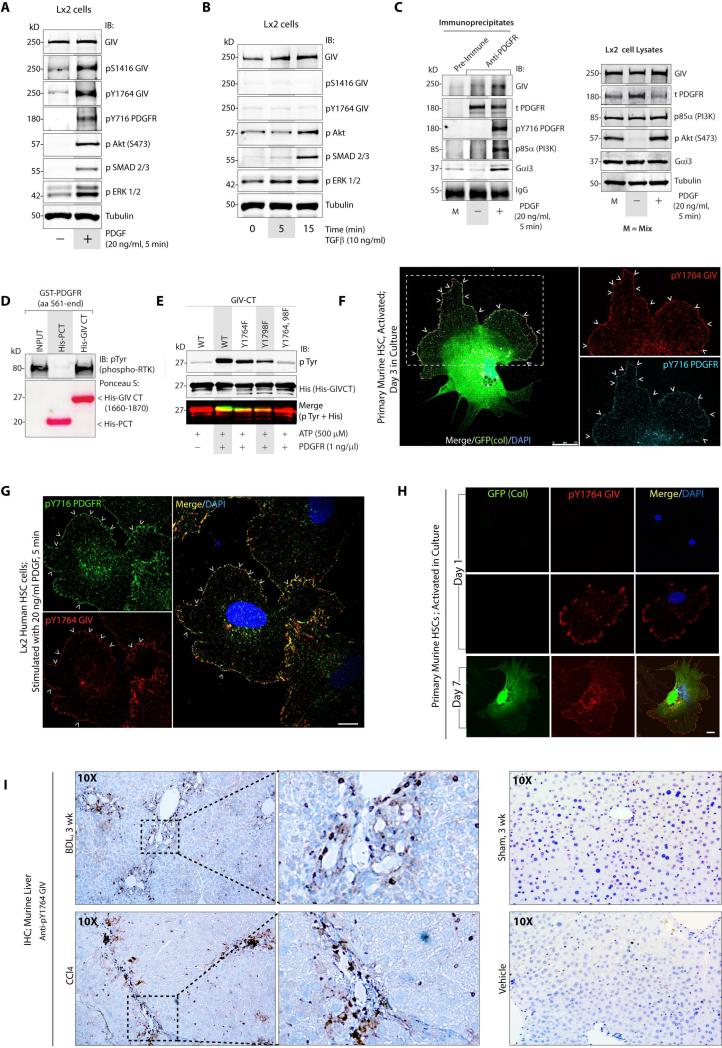
Next we asked which receptor(s) and ligands activate GIV-dependent signaling to potentiate HSC activation. First we analyzed signaling events triggered in Lx2 cells by PDGF and TGF-β1, the two major pro-fibrotic stimuli 2,3. We found that PDGF, but not TGFβ-1 triggered phosphorylation of GIV at Tyr(Y)1764, a substrate site for RTKs 17 and Ser(S)1416, a substrate site for Akt 32, that are established surrogate markers of activation of the RTK-GIV-PI3K-Akt signaling pathway 17 (Figure 5A-B). However, both growth factors triggered phosphorylation of Akt, SMAD and ERK proteins (Figure 5A-B). Consistent with such activation, GIV co-immunoprecipitated with ligand-activated PDGFRβ (Figure 5C), along with Gαi and PI3K. Using recombinant proteins in pulldown assays we confirmed that the interaction between autophosphorylated PDGFRβ and GIV's C-terminus is direct (Figure 5D), and found that recombinant PDGFRβ phosphorylated GIV on tyrosines 1764 and 1798 in vitro (Figure 5E); the same residues that are targeted by other RTKs 17. We found that tyrosine-phosphorylated GIV partially colocalized with activated PDGFRβ on patches at the PM in culture-activated primary HSCs (arrowheads, Figure 5F), as well as in PDGF-stimulated Lx2 cells (arrowheads, Figure 5G). These results demonstrate that upon PDGF stimulation, GIV and Gαi interact with activated PDGFRβ, presumably at the PM. Subsequent phosphorylation of GIV by PDGFR triggers the previously defined RTK(active)-pTyrGIV-Gi-PI3K pathway 17. By contrast, neither GIV, nor Gαi could be detected in complex with TGFβR under the same conditions, suggesting that the Ser/Thr TGFβR kinase may not bind or directly transduce signal via GIV.
Figure 5. PDGFRβ directly binds and phosphorylates GIV in HSCs during liver injury.
(A, B) Whole cell lysates of Lx2 cells stimulated with either PDGF-BB (A) or with TGF-β1 (B) were analyzed for various signaling pathways by immunoblotting (IB).
(C) Immunoprecipitation was carried out on lysates (right panel) of serum-starved or PDGF-BB treated Lx2 cells with anti-PDGFR or control IgG. Bound immune complexes (left) were analyzed for GIV, total (t) and activated (pY716) PDGFR, p85α(PI3K) and Gαi3 by immunoblotting (IB). M = Mix of starved and stimulated Lx2 lysates.
(D) GST-PDGFR was autophosphorylated in vitro and used in pull-down assays with purified His-GIV CT or His-PCT (Podocalyxin Tail; a negative control). Bound receptor was visualized by immunoblotting (IB) using phospho-Tyr (pTyr) antibody. Equal amounts of His-tagged ligands were confirmed by Ponceau S stain.
(E) In vitro kinase assays were carried out with recombinant PDGFR on wild-type (WT) and phosphomutants of His-GIV CT. Reactions were analyzed for pTyr and His-GIV-CT by immunoblotting (IB). Yellow pixels in the merged panel (pTyr and His) shows phosphoGIV-CT.
(F) Culture-activated primary HSCs from Col-GFP mice were fixed and stained for active pY1764GIV (red) and active pY716PDGFR (cyan). Merge panel shows colocalization of red and cyan pixels at the PM (arrowheads). Arrowheads = Plasma membrane. Scale bar = 25 μm.
(G) Lx2 cells stimulated with PDGF-BB were fixed and stained for pY1764GIV (red), active pY716PDGFR (green) and DAPI/DNA (blue). Merge panel shows yellow pixels indicative of colocalization at the PM (arrowheads). Arrowheads = Plasma membrane. Scale bar = 10 μm.
(H) Culture-activated primary HSCs were fixed and stained for active pY1764GIV (red) and DAPI/DNA (blue). Collagen production was detected by GFP signal. Scale bar = 10 μm. F, G and H were analyzed by confocal microscopy.
(I) Left: Liver sections from mice subjected to BDL or CCl4 injury were analyzed for active pY1764GIV by IHC. Boxed area on left panels are magnified on the right. Right: No staining was observed in the respective controls for both experimental models. Magnification = 10X.
Next we investigated the temporal correlation between activation of the RTK(active)-pTyrGIV-PI3K pathway (using phosphoTyr 1764 GIV as a surrogate marker) and collagen synthesis in culture-activated primary HSCs. On day 1, when collagen is still undetectable, pY1764-GIV is detected in ~50% of HSCs (Figure 5H, Supplementary Fig. 4). By day 3, ~15-20% HSCs express collagen, whereas 100% express pY1764-GIV (Supplementary Fig. 4). Thus, early expression of GIV during HSC activation we observed before (Figure 3A) is also accompanied by activation of the PDGFR-GIV-PI3K pathway, and both are upstream events that precede collagen synthesis during HSC activation. We confirmed that this pathway is activated in vivo during liver fibrosis because pY1764-GIV was detected exclusively in livers from BDL or CCl4-treated mice and not in control livers by IHC (Figure 5I). These results demonstrate that upon ligand stimulation, GIV directly binds autophosphorylated PDGFRβ, which allows: a) GIV to link G protein activation to RTK signaling, and b) phosphorylation of GIV by PDGFRβ; two events that synergistically enhance Akt signals via GIV 17.
GIV enhances the pro-fibrotic PI3K-Akt and SMAD signals downstream of multiple receptors
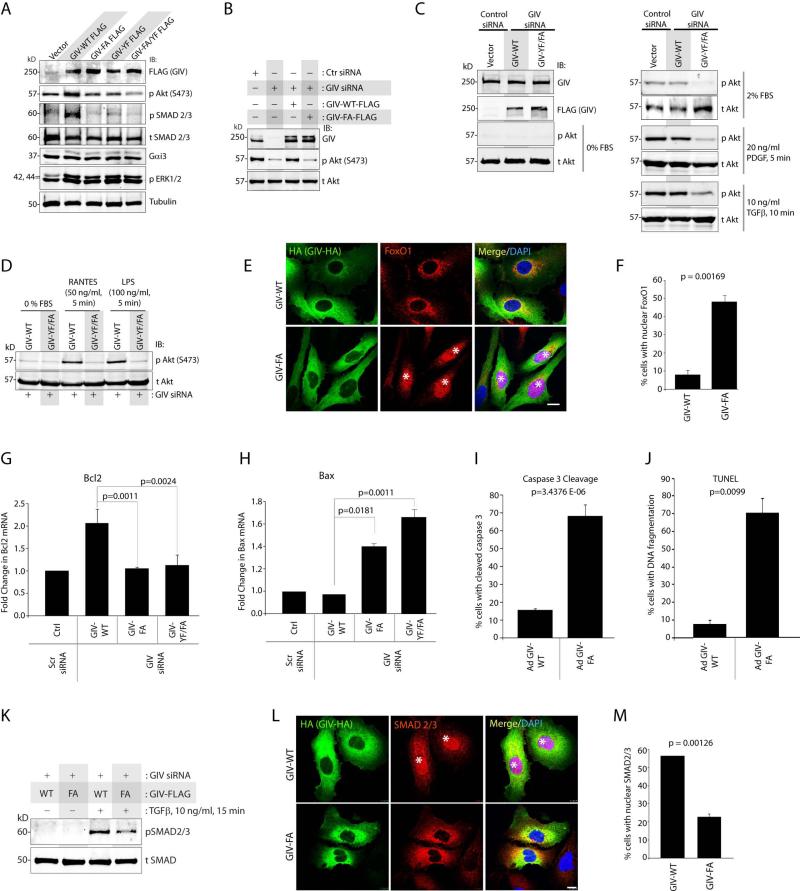
Because the PI3K-Akt and TGFβ-SMAD pathways are major profibrotic signals that drive liver fibrosis 36 we asked if GIV and, more specifically, its GEF function or tyrosine phosphorylation modulates these signals in HSCs. To investigate this, we took advantage of two more previously validated GIV constructs: non-phosphorylatable Y1764/1798F mutant (GIV-YF), and GIV-FA/YF in which both GEF and tyrosines are mutated. We found that exogenous expression of GIV-WT, but not the mutants in Lx2 cells enhanced phosphorylation of both Akt and SMAD 2/3 proteins (Figure 6A), indicating that both GIV's GEF function and tyrosine phosphorylation are essential. Depletion of endogenous GIV in Lx2 cells suppressed Akt phosphorylation (Figure 6B); an effect reversed by exogenous expression of siRNA resistant GIV-WT, but not GIV-FA (Figure 6B), indicating that GIV's GEF function is required for maximal enhancement of Akt phosphorylation in HSCs. GIV's GEF function was also required for enhancement of Akt phosphorylation in response to a variety of ligands [serum, PDGF-BB, TGF-β1, RANTES, which activates G protein-coupled Chemokine receptors (CCRs), and lipopolysaccharide (LPS) which activates Toll-like receptors, TLR4] that target fibrogenic receptors of diverse classes (Figure 6C, 6D), indicating that GIV enhances the pro-fibrotic PI3K-Akt pathway downstream of multiple receptors via its GEF function. FoxO1, a major downstream target of Akt, whose inactivation and nuclear exclusion triggers HSCs transdifferentiation during liver fibrosis 10, was excluded from the nucleus in ~92% of the cells expressing GIV-WT (Figure 6E-F). By contrast, in cells expressing GIV-FA, FoxO1 was seen in the nucleus in ~50% of the cells, indicating that GIV's GEF function is required for nuclear exclusion and inactivation of FoxO1. Furthermore, compared to cells expressing GIV-WT, those expressing GIV-FA displayed fewer stress fibers (Supplementary Fig. 5), an overall pro-apoptotic phenotype [lower Bcl-2 (Figure 6G) and higher BAX (Figure 6H), higher Caspase 3 activity (Figure 6I) and more abundance of fragmented DNA analyzed by TUNEL (Figure 6J)], lower SMAD 2/3 activation, based on its decreased phosphorylation (Figure 6K), and reduced nuclear localization (Figure 6L-M). Taken together, these results demonstrate that GIV's GEF function is essential for key profibrogenic phenotypes that characterize HSC activation: actin cytoskeletal remodeling, HSCs survival, and enhanced PI3K-Akt-FoxO1 and the TGFβ-SMAD pathways.
Figure 6. GIV's GEF function enhances profibrogenic Akt and SMAD signals in HSCs.
(A) Whole cell lysates of Lx2 cells expressing GIV mutants were analyzed for various signaling pathways by immunoblotting (IB).
(B-D) Lx2 cells treated with control (Scr) or GIV siRNA were transfected with siRNA resistant GIV-WT or the indicated mutants, starved (0% FBS), and then stimulated with serum or various ligands prior to lysis. Equal aliquots of lysates were analyzed for GIV, phospho- and total Akt by immunoblotting (IB).
(E-F) Lx2 cells infected with GIV-WT-HA or GIV-FA-HA adenoviruses were maintained in 0.2% FBS, fixed and stained for HA (GIV, green), FoxO1 (red) and DAPI/DNA (blue) and analyzed by confocal microscopy. Star = nuclear FoxO1. Scale bar = 10 μm. Results display % cells with nuclear FoxO1 (Y axis) (F).
(G, H) Lx2 cells depleted of endogenous GIV were subsequently transfected with various GIV constructs, and RNA extracted of these cells were analyzed for Bcl-2 (G) and BAX (H) mRNA levels by qPCR.
(I, J) Lx2 cells were infected with GIV-WT-HA or GIV-FA-HA adenoviruses, maintained in 0 % FBS, fixed and analyzed for cleaved Caspase 3 (J) by immunofluorescence and for fragmented DNA by TUNEL staining (I). Bar graphs display % cells with cleaved Caspase 3 (J) or fragmented DNA (I; TUNEL) on Y axis.
(K) Lx2 cells depleted of endogenous GIV were subsequently transfected with siRNA-resistant GIV-WT or GIV-FA, stimulated with TGFβ and phospho- (p)SMAD and total (t)SMAD 2/3 were analyzed by immunoblotting (IB). The ratio of pSMAD/tSMAD was ~2.0 fold higher in cells expressing GIV-WT than in those expressing GIV-FA, as determined by band densitometry.
(L, M) Lx2 cells infected with GIV-WT-HA or GIV-FA-HA adenoviruses were maintained in 0.2% FBS, fixed and stained for HA (GIV, green), SMAD2/3 (red) and DAPI/DNA (blue) and analyzed by confocal microscopy. Star = nuclear SMAD2/3. Scale bar = 10 μm. (M) Bar graphs display % cells with nuclear tSMAD2/3 in K. Error bars represent mean ± S.D. n = 3. Statistical significance was assessed with two-tailed Student's t test.
GIV inhibits the anti-fibrotic cAMP signals downstream of multiple receptors
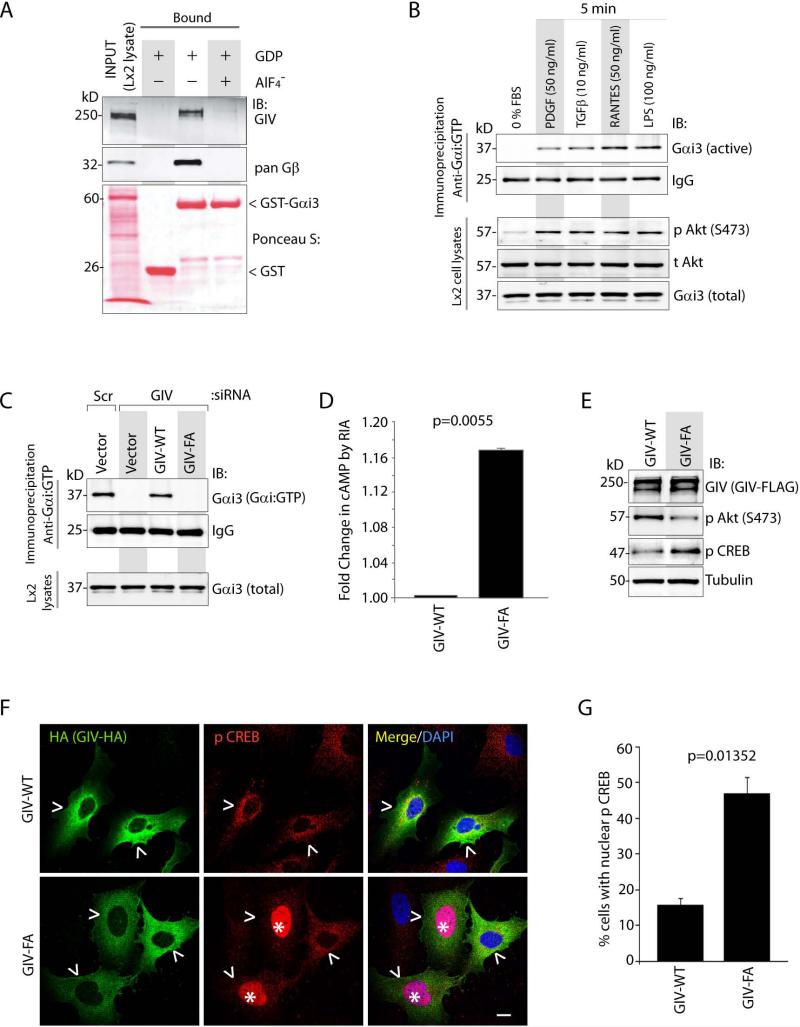
Next we asked if activation of Gαi via GIV's GEF function modulates anti-fibrotic cAMP signals during HSC activation. First, we confirmed that GIV expressed in HSCs bound G proteins as expected for a GEF 16,19, i.e., it preferentially bound inactive Gαi3 (GDP loaded) (Figure 7A), and not to Gαi3 in active conformation (loaded with GDP + AlF −4) in GST pulldown assays. Next we confirmed that various stimuli which require GIV's GEF function to enhance Akt (Figure 6D-E) can also trigger the activation of Gαi3 in Lx2 cells (Figure 7B), as determined by an antibody that specifically recognizes the active GTP-bound conformation of Gαi 37. Depletion of endogenous GIV suppressed Gαi3 activity (Figure 7C); an effect reversed by exogenous expression of siRNA resistant GIV-WT, but not GIV-FA, indicating that GIV's GEF function is required for activation of Gαi3 in HSCs. As expected, loss of Gαi3 activity in HSCs expressing GIV-FA, was accompanied by increased accumulation of cellular cAMP (Figure 7D), and enhanced phosphorylation (Figure 7E) and nuclear translocation (Figure 7F-G) of its downstream target, CREB, indicating that GIV's GEF function is required for inhibition of the cAMP-CREB pathway. These results demonstrate that GIV inhibits the anti-fibrogenic cAMP pathway in HSCs via its GEF function.
Figure 7. GIV activates Gαi and suppresses the anti-fibrogenic cAMP/PKA/pCREB signals in HSCs.
(A) Lysates of Lx2 cells were used as the source of full-length GIV in pull-down assays with GST or GST-Gαi3 preloaded with GDP alone (inactive) or with AlF −4 (active), and immobilized on glutathione beads. Bound GIV and Gβγ were analyzed by immunoblotting (IB). The GST proteins are shown by Ponceau S stain.
(B) Serum-starved Lx2 cells were stimulated with a variety of ligands prior to lysis. Equal aliquots of lysates were subjected to immunoprecipitation with Gαi:GTP mAb, which recognizes active conformation of the G protein and immunecomplexes were analyzed for Gαi3 by immunoblotting (IB).
(C) Lx2 cells treated with control (Scr) or GIV siRNA were transfected with siRNA resistant GIV-WT or GIV-FA and maintained in 0.2% FBS. Lysates were analyzed for active Gαi3 as in B.
(D) Lx2 cells transfected with GIV-WT or GIV-FA were stimulated with PDGF-BB prior to lysis. Lysates were analyzed for cAMP by RIA and normalized to total protein. Results are displayed as fold change in cAMP (Y axis).
(E) Lx2 cells transfected with GIV-WT or GIV-FA were analyzed for GIV, phospho(p)Akt, phospho(p)CREB and tubulin by immunoblotting (IB). The ratio of pCREB/tubulin was ~2.5 fold higher in cells expressing GIV-FA than in those expressing GIV-WT, as determined by band densitometry.
(F) Lx2 cells infected with GIV-WT-HA or GIV-FA-HA adenoviruses were maintained in 0.2% FBS, fixed and stained for HA (GIV, green), phospho(p)CREB (red) and DAPI/DNA (blue) and analyzed by confocal microscopy. Arrowhead = GIV expressing cell. Star = nuclear p CREB. Scale bar = 10 μm. Results display % cells with nuclear pCREB (Y axis) (G). Error bars represent mean ± S.D. n = 3. Statistical significance was assessed with two-tailed Student's t test.
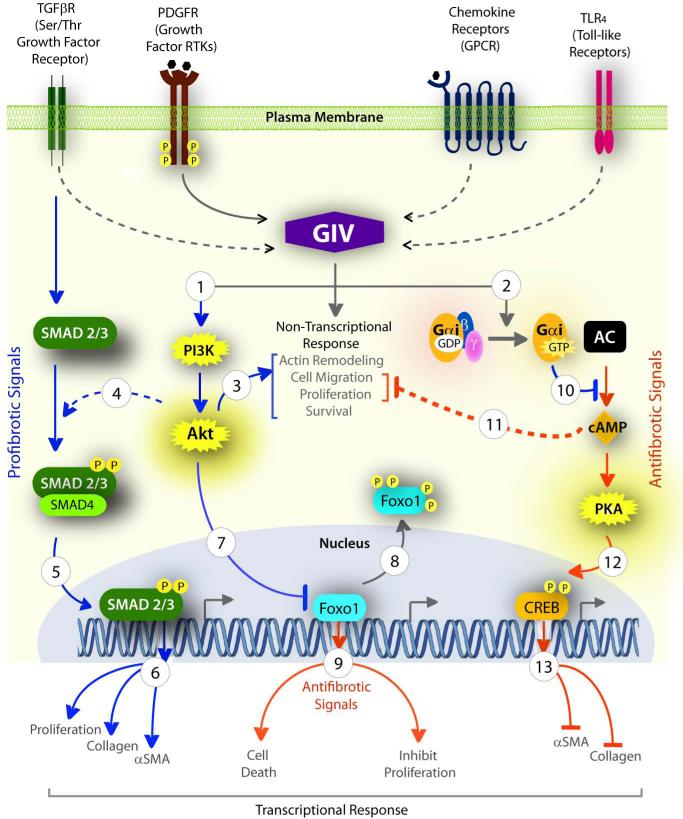
Taken together, we propose (Figure 8) that GIV serves as a central hub which integrates signals downstream of multiple fibrogenic receptors and simultaneously enhances the profibrotic (PI3-KAkt-FoxO1 and TGFβ-SMAD) and inhibits the anti-fibrotic (cAMP-PKA-pCREB) pathways to shift the delicate balance between pro- and anti-fibrogenic signalling to an overall profibrotic signaling program that favors fibrosis. Consequently, phenotypes that are critical for sustained activation of HSCs and collagen synthesis are enhanced through a combination of transcriptional (via FoxO1, SMAD and CREB) and non-transcriptional mechanisms (via PI3K, Akt and cytoskeletal remodeling).
Figure 8. GIV enhances pro-fibrogenic and inhibits anti-fibrogenic signaling pathways in HSCs.
Ligand stimulation of diverse classes of fibrogenic receptors (top) converge upon GIV either via direct interaction (solid arrow) or via unknown mechanism (interrupted arrow) to enhance PI3K-Akt signaling (1) and activate trimeric Gαi (2). GIV-dependent activation of Akt elicits several non-transcriptional responses (3) that lead into activation of HSCs. The GIV-PI3K-Akt pathway also modulates two distinct transcriptional programs-- (i) Potentiation of the profibrotic TGFβ-SMAD pathway by increasing phosphorylation of SMAD2/3 (4) and its translocation into the nucleus (5), and thereby, enhancing proliferation and transcription of collagen and α-SMA (6); (ii) Inhibition of the transcription factor Foxo1 by phosphorylation (7) and by triggering its translocation outside the nucleus (8), thereby antagonizing the anti-fibrotic program (9) that Foxo1 maintains in physiology. Activation of Gi by GIV's GEF function (2) inhibits the production of cAMP (10), a major anti-fibrotic pathway in HSCs by virtue of its ability to inhibit HSC migration and proliferation (11). The cAMP-PKA pathway is also known to inhibit fibrosis at a transcriptional level by phosphorylating CREB (12) which translocates to the nucleus and inhibits transcription of collagen and α-SMA (13). Pro-fibrogenic pathways = blue; Anti-fibrogenic = red.
Expression of GIV in liver biopsies correlates with the degree of fibrosis
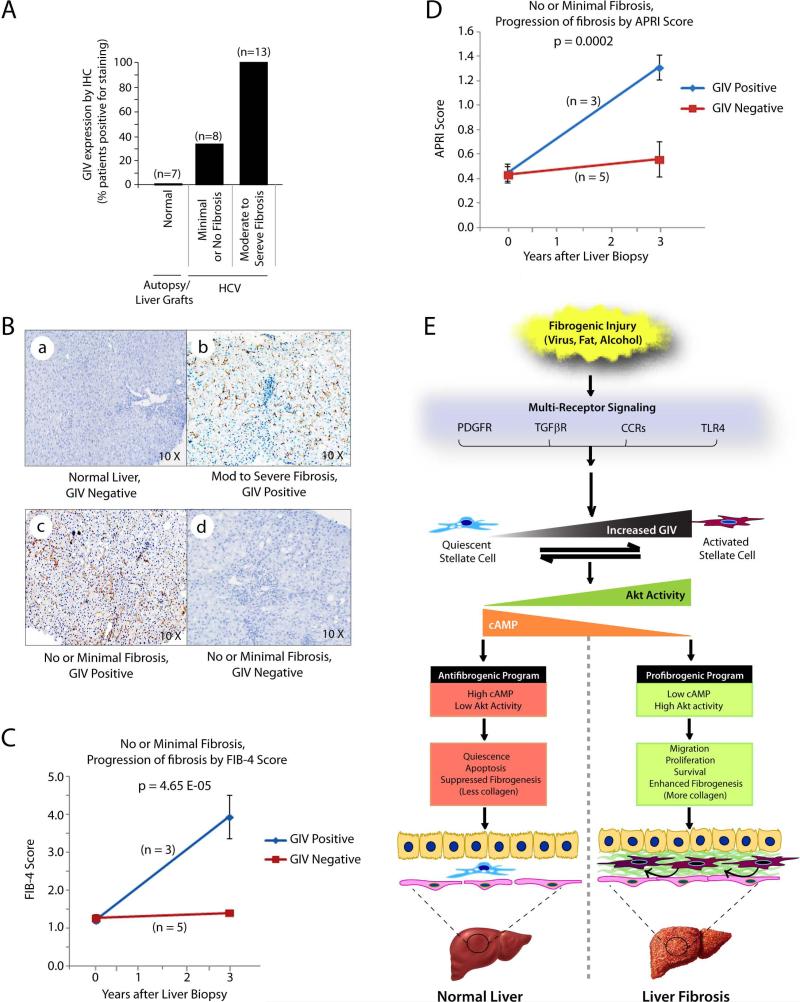
Because GIV is upregulated early during liver fibrosis and its expression is required for HSC activation, we hypothesized that detection of GIV in liver biopsies could indicate sustained activation of HSCs and mark the potential for progression to cirrhosis. We tested this hypothesis in liver biopsies obtained from 21 males with chronic HCV infection because the rate of fibrogenic progression in these patients varies markedly, and the need for a biomarker that differentiates rapid from slow progressors still remains unmet 38. Fibrosis was staged with the Ishak scale (ranging from 0 = no fibrosis to 6 = cirrhosis) 38. As shown previously, GIV was undetectable in normal livers (Figure 9A, 9B-a) and detected in 100% of livers with moderate to severe fibrosis (scores 3-6) (Figure 9A and 9B-b). However, GIV was detected only in ~37% of livers with minimal or no fibrosis (scores = 0-2) (Figure 9A and 9B-c, d). Noteworthy, GIV expression did not show any correlation with age, alcohol intake, degrees of inflammation, or fatty infiltration. These results demonstrate that some, but not all patients express GIV during the early stages of chronic viral hepatitis, even before significant fibrosis has occurred, but with the progression to cirrhosis, all are positive. Of the 8 patients with minimal or no fibrosis, 3 patients whose liver biopsies were positive for GIV progressed to advanced fibrosis, as determined by increasing FIB-4 (Figure 9C) and APRI (Figure 9D) scores 39,40 during a 3 year followup. These results suggest that GIV expression in liver biopsies may predict the progression of fibrosis, and thereby serve as a biomarker for the disease. We propose that detection of GIV in liver biopsies early during HCV infection might distinguish those who progress rapidly to cirrhosis from those who progress slowly or not at all.
Figure 9. GIV expression in liver biopsies may predict fibrogenic progression.
(A) GIV expression was analyzed in liver biopsies from normal subjects and a cohort of HCV-infected patients at various stages of fibrosis by IHC. Staining was scored by two pathologist blinded to the degree of fibrosis using a simple binary scoring system; i.e., positive or negative for GIV. Results are displayed as % patients positive for GIV (Y axis) in each category of fibrosis.
(B) Representative IHC images from A are displayed showing examples of positive and negatively-stained liver samples within different categories of fibrosis. Magnification = 10X.
(C, D) Line plots depict the progression of fibrosis in patients with no or minimal fibrosis, as determined using FIB-4 (C) and APRI (D) scores derived from clinical parameters at the time of liver biopsy and at 3 yr follow-up. p values indicate significant differences in fibrosis scores at 3 yr.
(E) Schematic shows our proposed model for GIV's role in fibrogenic signaling in HSCs during liver fibrosis. Top to Bottom: Fibrogenic injuries of various kinds result in the activation of diverse classes of receptors that trigger upregulation of GIV in HSCs. Increased GIV triggers transformation of quiescent HSCs into activated myofibroblasts by modulating two key signaling pathways: 1) the pro-fibrotic PI3-KAkt pathway and, 2) the anti-fibrotic cAMP-PKA pathway. In quiescent HSCs low levels of GIV expression generate a net antifibrogenic signaling program characterized by low Akt activity and impaired activation of Gαi with resultant high levels of cAMP. In activated HSCs high levels of GIV expression generate a net profibrogenic signaling program characterized by high Akt activity and increased activation of Gαi with resultant low levels of cAMP. Low GIV expression maintains HSCs in quiescent state, triggers apoptosis, and suppresses collagen production in the normal liver, whereas high GIV expression enhances HSC migration, proliferation, and survival, and triggers collagen production, thereby leading to liver fibrosis. n = number of patients. Error bars represent mean ± S.D. Statistical significance was assessed with two-tailed Student's t test.
Discussion
Here we define GIV as a central hub within the fibrogenic signaling network downstream of multiple receptors. By virtue of its ability to activate trimeric G protein Gi, GIV simultaneously modulates proand anti-fibrotic pathways that function antagonistically to balance collagen synthesis and its removal in a delicate equilibrium. Our results provide detailed mechanistic insights into how GIV triggers HSC activation, and supports the model outlined in Figure 9E: In response to a variety of chronic injuries, diverse classes of fibrogenic receptors are activated, which triggers an upregulation of GIV mRNA, translation of GIV protein and its activation in HSCs. Increased expression and activation of GIV's GEF function enhances PI3K-Akt signaling downstream of numerous receptors and blunts cAMP accumulation, both via activation of Gαi. These upstream events trigger subsequent tiers of signals along the pro- and anti-fibrogenic pathways (see Figure 8), skewing the entire network towards an overall profibrotic signaling program that favors fibrogenesis. Consequently, several phenotypes (migration, proliferation, actin remodeling, survival/anti-apoptosis and collagen production) that are critical not only for transdifferentiation of HSCs to an active state but also for a sustained increase in the abundance of activated HSCs in the liver during progressive liver injury, eventually leading to liver fibrosis. Depletion of GIV, or targeted inhibition of its GEF function reverses the profibrogenic program in HSCs, i.e., suppresses Akt activity and increases cellular cAMP, thereby 'resetting' the balance to an anti-fibrogenic program, as exists in normal liver. We conclude that GIV is a key transducer of signals downstream of multiple classes of fibrogenic receptors that trigger liver fibrosis.
Of the many receptors that utilize GIV to transduce profibrotic signals, we found that the fibrogenic RTK PDGFRβ directly binds and phosphorylates GIV. This is in keeping with our previous work on the prototype RTK, EGFR, in which we defined how GIV serves as a signaling platform that links ligand-activated RTKs to G protein signaling. Although the other major pro-fibrotic receptor i.e., TGFβR also requires GIV's GEF function to enhance Akt and SMAD signals and collagen synthesis, surprisingly, we could not identify a direct link (neither binding nor phosphoregulation) between them. Because TGFβ and PDGF pathways crosstalk 41-43, we hypothesize that GIV and its GEF function may enhance Akt signals in response to TGFβ indirectly via activation of PDGFR pathway. Similarly, because the PI3K-Akt pathway can potentiate fibrogenic SMAD signals in cells responding to TGFβ 44, it is possible that GIV and its GEF function enhance SMAD signals via activation of Akt.
We demonstrate that GPCRs, such as CCRs also require GIV's GEF function for enhancement of Akt activity when stimulated with RANTES. We previously demonstrated that GPCRs (e.g., LPAR1 and f-Met-Leu-Pro-R) require GIV and its GEF function for PI3K-Akt signaling and chemotaxis 16,19,45, and that they can trigger tyrosine phosphorylation and activation of GIV via activation of Src-family of non-RTKs 17. We conclude that CCRs modulate pro- and anti-fibrogenic signals via activation of GIV. Finally, we also provide evidence that TLR4, a receptor that is unique to fibrosis in the liver, also requires GIV's GEF function to enhance Akt signals when stimulated with LPS. Our findings are consistent with data showing that TLR4 requires activation of the PI3K-Akt pathway for enhancement of TGFβ-SMAD signaling 46,47, and that cirrhosis-predictive single nucleotide polymorphisms (SNPs) of TLR4 are functionally linked to genes of the PI3K-Akt pathway 48. We conclude that TLR4 activates Akt via GIV, and results shown here provide the first evidence for how GIV may shape the innate immune response by modulating signaling networks initiated by members of the Interleukin-1 (IL-1)/TLR superfamily. Because KCs are the main target of LPS in the liver 49, and these cells also upregulated GIV mRNA after injury, it is possible that the TLR4-GIV-Akt pathway also helps to activate KCs during liver fibrosis.
Besides demonstrating that multiple fibrogenic receptors converge upon GIV, we demonstrate that subsequent activation of Gαi by GIV modulates two functionally antagonistic signaling events (see Figure 8)-- (a) Activation of the profibrotic PI3K-Akt signals that potentiate TGFβ-SMAD signals and inhibit FoxO1, and (b) inhibition of cAMP production, which suppresses the PKA-pCREB pathway; thereby generating an overall profibrotic signaling program that favors fibrosis. Because the PI3K-Akt pathway is strongly implicated in studies of fibrogenic diseases in other organ systems [skin, cardiac, pulmonary (reviewed in 50)], and the cAMP pathway is a well-accepted antifibrotic pathway in various other models of fibrogenesis (cardiac 51, pulmonary 42, kidney 44, and skin 26), and fibrosis in all organs share common receptors/pathways defined here (i.e., TGF-β1 and PDGF 52), we speculate that GIV may also serve as a key mediator in fibrogenic diseases of other organs.
The other important finding is the invariable upregulation of GIV mRNA and expression of GIV protein after several forms of chronic liver injury, in both humans and mice. Although here we focused on upregulation of GIV in HSCs, it is noteworthy that other cells like ECs and KCs which also contribute to liver fibrosis by promoting sustained activation of HSCs 53 showed significant upregulation of GIV after injury. This was not unexpected because GIV is known to be expressed in vascular endothelial cells15 and we previously showed that GIV is required for macrophage chemotaxis 19. As in the case of HSCs, activation and secretion of cytokines and chemokines by KCs is antagonistically balanced by the PI3K-Akt 7 and the cAMP-pCREB pathway 54. Because GIV modulates those two signaling pathways in HSCs, we suspect that GIV's role in liver fibrosis is not limited to activation of HSCs, but extends also to activation of KCs; all via common signal transduction mechanisms we outline here.
As for mechanism(s) of GIV upregulation, our results indicate that its expression may be regulated at multiple steps. For example, quiescent HSCs have low levels GIV mRNA but no protein is translated, indicating that protein translation is repressed or protein half-life is short in these cells. However, in activated HSCs, high levels of GIV mRNA and full-length protein are seen, indicating that transcriptional induction and/or RNA and/or protein stabilization are responsible for such upregulation. We have recently defined STAT3 as a key transcription factor responsible for upregulation of GIV in response to wounding and during cancer progression 22. Because STAT3 has been widely implicated in perpetuating inflammation/injury response in the liver 55, and liver fibrosis has been likened to would healing 23, it is possible that STAT3 up-regulates GIV after liver injury, thereby linking inflammation to fibrogenesis. Regardless of the mechanism(s) involved, we found that sustained upregulation of GIV is restricted to some forms of injuries, not others: upregulation occurs in all forms of chronic fibrogenic injuries, but not in acute injuries that heal without fibrosis. These results indicate that GIV is not merely a component of an acute phase response in the injured liver, instead, increased expression of GIV during acute injuries may signal a potential for progression to chronicity and fibrosis.
With regard to the clinical relevance of our findings, we provide evidence that GIV could serve both as a therapeutic target against and a biomarker for prognosticating liver fibrosis. Our findings in HSCs expressing the GEF-deficient GIV-FA demonstrate that selective inhibition of the GIV:Gαi interface by a single point mutation is sufficient to alter a multitude of signals and inhibit various phenotypes that are required for sustained activation of HSCs. Based on these we conclude, that the GIV:Gαi interface is an effective and specific target, that is also sensitive to disruption. Furthermore, since accumulation of cellular cAMP can reverse fibrosis 13, and accumulation of cAMP is one of the direct consequences of inhibiting GIV's GEF function, we anticipate that antagonists targeting the GIV:Gαi interface will not only halt, but also cause regression of fibrosis by reinstating anti-fibrotic signaling. Based on our findings that GIV's GEF function is utilized by multiple receptors, inhibiting the GIV:Gαi interface constitutes a novel strategy for externally manipulating multiple fibrogenic receptors via a single target. Because full-length GIV is not expressed in the normal liver, and the majority of cells in the liver (i.e., hepatocytes) do not express GIV even after injury, inhibition of GIV is likely to have fewer off-target effects on major metabolic functions of the liver. We conclude that small molecules against the GIV:Gαi interface have the potential of being both selective and broad-- i.e., selectively inhibit GIV-dependent profibrotic signaling in cells that express full-length GIV, yet broadly downstream of multiple receptors.
With regard to the prognostic potential of GIV, we provide proof-of-concept that detection of GIV in FFPE liver biopsies early during the course of chronic HCV infection (when significant fibrosis is yet to occur) may predict rapid progression to advanced fibrosis. Based on the fact that GIV expression in HSCs precedes collagen synthesis and onset of liver fibrosis, GIV fits the definition of an ideal prognosticator which identifies at-risk patients even at early stages when minimal or no fibrosis is detected by IHC. In addition, our IHC results show that full-length GIV is detected easily and reproducibly with 3 different CT antibodies, and that a simple binary system of scoring (i.e., presence or absence of GIV) is sufficient to distinguish at-risk individuals from others without the need for cumbersome and subjective scoring algorithms. Despite the potential significance of our findings, limitations of this and any future studies on this topic includes the frequent occurrence of sampling error in liver biopsies despite the fact that individual biopsies meet the histologic criteria for adequacy. To circumvent this limitation, here we used two different clinical scoring methods to monitor the progression of fibrosis, FIB-4 and APRI; each score is derived from multiple clinical parameters. We found that both scores concurrently classified the patients into two groups-- non-progressors (fibrosis stages F0 = no fibrosis; F1 = portal fibrosis without bridges; F2 = portal fibrosis with rare bridges) and progressors (fibrosis stages F3 = numerous bridges without cirrhosis; F4 = cirrhosis). Our results indicate that expression of GIV in HSCs by IHC was associated with progression of fibrosis to significant levels within 3 years. We conclude that GIV may serve as a prognostication biomarker for clinical use to identify those patients with chronic liver injury that are at highest risk for progressive fibrosis. Because growing evidence suggests that activated HSCs drive primary liver and biliary cancer progression 56, and because GIV is a bona fide prometastatic protein that imparts aggressiveness to multiple cancers 22,57, it is possible that levels of expression of GIV in HSCs correlates/predicts progression to liver cancer. Further studies on larger cohorts of patients are required to evaluate if GIV expression in liver biopsies independently predicts liver cirrhosis and/or progression to cancer.
In conclusion, here we have defined GIV as a novel signaling hub at the center of the complex multi-receptor signaling system that drives liver fibrosis. Our findings set a new paradigm in which a single multidomain signal transducer centrally integrates and broadly shapes the entire fibrogenic network. As a promising therapeutic target which can serve as a biomarker to predict risk of fibrosis, GIV presents an opportunity for the practice of personalized medicine in patients with fibrogenic diseases of the liver.
Methods
Reagents and antibodies
Unless otherwise indicated, all reagents were of analytical grade and obtained from Sigma-Aldrich. Cell culture media were purchased from Invitrogen. PDGF (Invitrogen), TGFβ1 (Invitrogen), RANTES (R&D Systems), and LPS-E Coli 055:B5 (Sigma) were obtained commercially. BrdU was obtained from Sigma. Recombinant and PDGFRβ was purchased from Cell Signaling. All restriction endonucleases and Escherichia coli strain DH5α were purchased from New England Biolabs. E. coli strain BL21 (DE3), oligofectamine and phalloidin-Texas Red were purchased from Invitrogen. DAPI was purchased from Molecular Probes (Invitrogen). GenejuiceTM transfection reagent was obtained from Novagen (EMD Millipore) and PfuUltra DNA polymerase was purchased from Stratagene. Silencer Negative Control scrambled (Scr) siRNA was purchased from Ambion and and a previously validated 32 GIV siRNA sequence was custom-ordered from Dharmacon. Goat anti-rabbit and goat anti-mouse Alexa Fluor 680 or IRDye 800 F(ab')2 used for Odyssey Infrared Imaging were from Li-Cor Biosciences. Antibodies used in this work include rabbit pAbs against Gαi3 (M-14) and pan-Gβ (Santa Cruz Biotechnology), phospho-Akt Ser 473, phospho-ERK 1/2, phospho-SMAD 2 and phospho-CREB (Cell Signaling) and phospho-Histone H3 (Abcam). Mouse mAbs against hexahistidine (His), FLAG (M2), actin and α-tubulin (Sigma-Aldrich), SMAD 2/3, phospho-PDGFRβ-Tyr716 and GFP (Santa Cruz Biotechnology), BrdU (BD Biosciences), PDGFRβ (Biolegend), α-SMA (DAKO), control IgG (Bio-Rad Laboratories) and anti-active Caspase 3 (Abcam) were obtained commercially. Several anti-GIV antibodies were used in this work. For immunoblotting we used rabbit anti-GIV coiled-coil (GIVcc) (Millipore) and GIV-CT Abs (Santa Cruz Biotechnology). Coiled-coil antibody detects most isoforms of GIV, whereas CT Ab detects exclusively the full length isoform of GIV that contains the domains required for GIV's function as a signal transducer. For immunofluorescence we used rabbit monoclonal anti-GIV-CT (Spring Bioscience) and anti-GIV-CT mAb (Millipore). Three different Anti-GIV-CT Abs were used for IHC detection of GIV (Santa Cruz Biotechnology, IBL America and Spring Bioscience). All these CT antibodies were raised against the last 18 aa of the C-terminus, and were validated in immunoblot assays based on their ability to specifically detect the full length GIV isoform. Anti-Tyr1764-GIV Ab was developed by Spring Bioscience using a proprietary platform for the generation of Rabbit mAb using a peptide encompassing pY1764. It is a diagnostic grade antibody jointly developed by our group, Ventana/Roche and Spring Biosciences and extensively validated in multiple assays prior to use in this project. Anti-Ser1614-GIV was commercially obtained from IBL America, and was previously validated by the Takahashi group32. Anti-Gαi3:GTP (6-F12) was a gift from Dr. Graeme Milligan58.
Plasmid Constructs, Mutagenesis and Protein Expression
Cloning of Gαi3 into pGEX-4T-1 (GST-Gαi3), GIV-CT (aa 1,660-1,870) into pET28b (His-GIV CT), and RNA interference-resistant (siRNA rest) GIV was generated by silent mutations32 and cloned into p3XFLAG-CMVTM-14 plasmid (GIV-FLAG) as described previously 16,19,26,45. His-PCT (Podocalyxin Tail) was a gift from Dr. Farquhar MG 59. GIV mutants were generated using QuickChange II (Stratagene) and specific primers (sequence available upon request) following the manufacturer's instructions. All constructs were checked by DNA sequencing.
GST, GST-Gαi3, His-GIV CT and His-PCT fusion constructs were expressed in E. coli strain BL21 (DE3) (Invitrogen) and purified as described previously 16,19,26. Bacterial cultures were induced overnight at 25°C with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Pelleted bacteria from 1 L of culture were re-suspended in 10 ml GST-lysis buffer [25 mM Tris-HCl, pH 7.5, 20 mM NaCl, 1 mM EDTA, 20% (v:v) glycerol, 1% (v:v) Triton X-100, 2X protease inhibitor cocktail (Complete EDTA-free, Roche Diagnostics)] or His-lysis buffer [50 mM NaH2PO4 pH 7.4, 300 mM NaCl, 10 mM imidazole, 1% (v:v) Triton X-100, 2X protease inhibitor cocktail (Complete EDTA-free, Roche Diagnostics)] for GST or His-fused proteins, respectively. After sonication (3 × 30s), lysates were centrifuged at 12,000g at 4°C for 20 min. Solubilized proteins were affinity purified on glutathione-Sepharose 4B beads (GE Healthcare) or HisPur Cobalt Resin (Pierce). Proteins were eluted, dialyzed overnight against PBS and stored at −80 °C.
Mice and fibrosis induction
Expression of collagen type I time was studied using reporter Col-GFP mice (pCol9GFP-HS4,5 transgene) 30. For all other experiments 8- to 12-week-old male C57BL/6 mice (Jackson laboratories) underwent BDL surgery or administration of CCl4 exactly as described previously 7. Sham operation was performed similarly, except that the bile duct was not ligated. Animals were sacrificed 5 or 21 days after BDL. CCl4 (Sigma-Aldrich; diluted 1:3 in corn oil) or vehicle (corn oil) was administered via intraperitoneal route at a dose of 0.5 μl/g body weight, twice per week, for a total of 12 injections. Animals received humane care according to NIH recommendations outlined in the “Guide for the Care and Use of Laboratory Animals.” All animal experiments and cell isolation studies were approved by the UCSD Institutional Animal Care and Use Committees.
Isolation and Culture of Liver Cell Fractions from Mice
Liver cells of 8- to 12-week-old male C57BL/6 mice (Jackson laboratories) were fractionated into hepatocytes, Kupffer cells, endothelial cells and hepatic stellate cells as previously described 60. Perfusion of mouse livers was followed by 3-layer discontinuous density gradient centrifugation with 8.2% and 14.5% Nycodenz (Accurate Chemical and Scientific Corporation). Kupffer cell or endothelial cell fractions were then selected positively by magnetic cell sorting using anti-CD11b antibody or antiliver sinusoidal endothelial cell antibody, respectively (Miltenyi Biotech). The hepatic stellate cell fraction was purified by negative selection for Kupffer cells and endothelial cells by magnetic cell sorting with respective antibodies. These cell fractions were homogenized immediately for RNA extraction. The purity of hepatocytes, HSCs and ECs were examined by phase-contrast microscopy, ultraviolet (UV)–excited autofluorescence (HSC), and uptake of diacetylated low-density lipoprotein (LDL) (EC). The purity of KC was demonstrated by functional analysis by means of phagocytosis of 1- μm latex beads. The purity of each cell type was as follows: hepatocytes, greater than 97%; EC, greater than 98%; KC, greater than 95%; HSC, greater than 96%. When we isolated HSCs for use in various assays throughout the manuscript, purity was confirmed as >98% by immunostaining with anti-desmin antibody. HSCs were cultured on uncoated plastic tissue culture dishes/coverslips in DMEM containing 10% FBS and used as primary cultures only.
RNA isolation and quantitative (qPCR)
Total RNA was isolated using an RNeasy kit (QIAGEN) as per the manufacturer's protocol. First-strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen), followed by ribonuclease H treatment (Invitrogen) prior to performing quantitative real-time PCR. Reactions omitting reverse transcriptase were performed in each experiment as negative controls. Reactions were then run on a real-time PCR system (ABI StepOnePlus; Applied Biosystems). Gene expression was detected with SYBR green (Invitrogen), and relative gene expression was determined by normalizing to GAPDH using the ΔΔT method. Primers used in this work were designed targeting the last two exons of GIV such that they exclusively detect all known full length isoforms of GIV using Invitrogen's Oligo Perfec Designer and evaluated with NetPrimer from Premierbiosoft 26. The sequences of primers used in this work are included in Supplementary Table 1.
Patient samples
Human liver samples used for GIV mRNA analysis were collected at Mount Sinai Hospital. Cirrhosis samples were collected from HCV-infected patients undergoing surgical resection or liver transplantation for hepatocellular carcinoma. Normal liver tissue was collected from patients receiving hepatic resections for non-tumoral diseases, including hepatic adenoma and focal nodula hyperplasia. Liver samples were snap frozen, and RNA was extracted, DNase treated, reverse transcribed, and analyzed by quantitative real-time PCR as described above.
Liver biopsy specimens used in this work for IHC were obtained from 21 patients undergoing percutaneous liver biopsies from year 2009 to 2010 at the VA San Diego Healthcare System. Biopsies were performed using a standard Jamshidi aspiration needle and samples were formalin fixed immediately and embedded in paraffin. Adequacy of the biopsy specimen was established by the presence of at least 6-8 portal triads. In addition, 7 specimens of normal liver were obtained from autopsies performed by the County of San Diego. All tissues used in this work were collected after IRB approval (Project# 04-0056 0001 05 ME X and 101264) and with informed patient consent.
As for limitations of this study, although the biopsies were done according to standard clinical practice and met current criteria for liver biopsy sampling, it is well known that sampling error can occur with liver biopsies despite meeting histologic criteria for individual biopsies. Despite the fact that sampling error is possible this remains the “gold standard” for determination of liver histology.
Immunohistochemistry
GIV IHC assays were developed and performed on a BenchMark XT automated slide stainer (Ventana Medical Systems, Inc.). FFPE samples (4 um section) were deparaffinized, pretreated for antigen retrieval, and followed by inactivation of endogenous peroxidase. Specimens were incubated with anti-GIV rabbit monoclonal (Ventana Medical Systems, Inc.) or polyclonal (IBL, Santa Cruz) antibodies (10 μg/ml) for 16 min at 37 C and the immunolocalized proteins were visualized using OptiView DAB IHC Detection kit. Following detection, all samples were counterstained with Hematoxylin II and Bluing Reagent. There was 100% concordance between all antibodies tested.
Semi-thin Immunofluorescence
Liver specimens were fixed with 4% paraformaldehyde for 4 h at 4°C, cryoprotected and frozen in liquid nitrogen. Semithin sections (0.5 μm) were cut with a Leica Ultracut UCT microtome equipped with a FCS cryoattachment at −100°C. Frozen sections were washed with PBS (3×), washed with 0.15% glycine (2×) and incubated 20 min in blocking buffer (5% normal goat serum, 2% fish skin gelatin and 2 mg/ml of BSA in PBS), 2 h in primary antibodies and 45 min in secondary antibodies. Dilutions of antibodies used were as follows: Mouse anti-GIV-CT (1:30), GFP (1:50); α-SMA (1:200); DAPI (1:1000) and secondary goat anti-mouse (594), goat anti-rabbit (488) and goat anti-rabbit (635) Alexa-conjugated antibodies (1:250). Sections were imaged on a Leica SPE confocal microscope using a 63x oil objective using 488, 561, 633 and 405 laser lines for excitation. The settings were optimized and the final images scanned with line-averaging of 3. All images were processed using Image J software (NIH) and assembled for presentation using Photoshop and Illustrator softwares (Adobe).
Whole-cell Immunofluorescence
Lx2 cells were fixed with 3% paraformaldehyde in PBS for 25 min at room temperature, treated with 0.1 M glycine for 10 min, and subsequently blocked/permeabilized with PBS containing 3% BSA and 0.2% Triton X-100 for 30 min at room temperature. Primary and secondary antibodies where incubated for 1 h at room temperature in PBS containing 3% BSA and 0.2% Triton X-100. Cells were imaged on a Leica SPE confocal microscope using a 63× oil objective using 488, 561 and 405 laser lines for excitation. The settings were optimized and the final images scanned with line-averaging of 3. Dilutions of antibodies used were as follows: GIVcc (1:150); GM130 (1:150); phospho-GIV (Tyr1764; 1:500); phospho-PDGFRβ (1:50); PDGFRβ (1:100); mouse anti-GIV-CT (1:100); Phalloidin (1:1000); DAPI (1:2000); secondary goat anti-rabbit (594) and goat anti-mouse (488) Alexa-conjugated antibodies (1:500); anti-phospho-Histone H3 (Ser28) (1:150), anti-BrdU (1:10), SMAD 2/3 (1:100), phospho-CREB (1:250), FoxO1 (1:100) and anti-active Caspase 3 (1:200). All images were processed using Image J software (NIH) and assembled for presentation using Photoshop and Illustrator softwares (Adobe).
For assays investigating nuclear localization of transcription factors, % cells with nuclear localization was quantified by counting any nuclear staining as presence of transcription factor in the nucleus, and used that as a basis for a binary system (i.e., cells with or without nuclear localization of transcription factor). ~200-400 cells per condition were counted from three independent experiments.
Cell Culture, Transfection, Lysis, and Quantitative Immunoblotting
The Lx2 human hepatic stellate cell line was obtained from Scott Friedman (Mount Sinai, NY) and were cultured in DMEM media containing 2% FBS according to previously published guidelines 31. Transfection, lysis and immunoblotting were carried out exactly as described before 19,26,61.
Lx2 cells were transfected using Genejuice (Novagen) for DNA plasmids and Oligofectamine (Invitrogen) for siRNA oligos following the manufacturers’ protocols. For adenovirus infection, cells were incubated with the respective viruses at 50 MOI for 5 h. After, the medium was replaced and cells were maintained for 2-3 days. For assays involving serum starvation, serum concentration was reduced to 0% overnight and for serum reduced treatment 0.2% FBS was used. Whole-cell lysates were prepared after washing cells with cold PBS prior to resuspending and boiling them in sample buffer. Lysates used as a source of proteins in immunoprecipitation or pull-down assays were prepared by resuspending cells in lysis buffer [20 mM HEPES, pH 7.2, 5 mM Mg-acetate, 125 mM K-acetate, 0.4% Triton X-100, 1 mM DTT, supplemented with sodium orthovanadate (500 μM), phosphatase (Sigma) and protease (Roche) inhibitor cocktails], after which they were passed through a 28G needle at 4 °C, and cleared (10,000 × g for 10 min) before use in subsequent experiments.
For immunoblotting, protein samples were separated by SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked with PBS supplemented with 5% nonfat milk (or with 5% BSA when probing for phosphorylated proteins) before incubation with primary antibodies. Infrared imaging with two-color detection and quantification were performed using a Li-Cor Odyssey imaging system. Dilution of primary antibodies were used as follows: Anti-His 1:1000; anti-FLAG 1:500; anti-GIV-CT 1:500; anti-phospho-GIV (Tyr1764) 1:500; anti-phospho-GIV (Ser1416) 1:500; anti-phospho-Akt 1:250; anti-Akt 1:500; anti-phospho-ERK1/2 1:250; anti-Gαi3 1:333; anti-pan-Gβ 1:250; anti-α tubulin 1:2000; anti-actin 1:1000; anti-SMAD2/3 1:250; anti-phospho-PDGFRβ 1:250; anti-PDGFRβ 1:250 and anti-phospho CREB (1:1000). All Odyssey images were processed using Image J software (NIH) and assembled for presentation using Photoshop and Illustrator softwares (Adobe). Uncropped images of immunoblots are shown in Supplementary Fig. 7-9.
Cell Migration and Proliferation Assays
Lx2 cells were transfected with control scrambled (Scr) siRNA or GIV siRNA for ~36 h. Monolayer cultures (100% confluent) were then scratch-wounded using a 20 μl pipette tip, and the cells were subsequently monitored by phase-contrast microscopy over the next 24 h. To quantify cell migration (expressed as percent of wound closure) images were analyzed using Image J software to calculate the difference between the wound area at 0 h and that at 24 h divided by the area at 0 h × 100.
The proliferation rate of Lx2 cells was measured by phospho-Histone H3 (Ser28) (mitotic index) and BrdU incorporation (DNA replication in S phase) as previously described 20. For the mitotic index, cells were grown on coverslips for 24 h, then fixed and analyzed by confocal microscopy. For the BrdU uptake assay, Lx2 cells were grown on coverslips for 24 h and treated with 10 μM BrdU for 30 min. Cells were then fixed with 70% ethanol for 30 min at room temperature, washed with PBS (3×), treated with 2 M HCl for 20 min at room temperature, followed by 0.1 M Na2B407 (pH 8.5) for 2 min at room temperature. Finally, coverslips were washed with PBS (3×) and analyzed by confocal microscopy. For both phospho-Histone and BrdU assays 400-600 cells from 10 randomly chosen fields were counted (DAPI stained) from three independent experiments. Mitotic index was determined by dividing the number of positively stained cells/ the total number of DAPI-stained nuclei × 100. Results are shown as mean ± S.E.M of 10 fields from 3 separate experiments. For the mitotic index in primary murine HSCs, cells were isolated from mouse liver and grown on coverslips for 16 h. Then, mHSCs were infected with GIV-WT or GIV-FA adenoviruses and 24 h later, cells were starved or treated with PDGF-BB (20 ng/ml 16 h) and mitotic index was measured by confocal microscopy.
Assays for measuring cellular apoptosis
Three different approaches were used. Lx2 cells were grown on coverslips and either transfected with control scrambled (Scr) siRNA or GIV siRNA for 48 h or infected with GIV-WT or GIV-FA adenoviruses and 24 h later, cells were starved for another 24 h. To measure Caspase 3 cleavage, cells were fixed and stained with anti-active Caspase 3 Ab (1:200). Measurement of DNA fragmentation by TUNEL assay was performed according to the protocol of In situ Cell Death Detection Kit, TMR red (Roche). 100 cells from randomly chosen fields were counted (DAPI stained) from three independent experiments. The percent of cells with either active Caspase 3 or fragmented DNA was determined by dividing the number of positively stained cells/ the total number of DAPI-stained nuclei × 100. Expression of two bona fide pro- (Bax) and anti- (Bcl-2) apoptotic signaling intermediates was measured by qPCR.
Generation of Adenoviruses Expressing GIV
HA-tagged full length human GIV wild-type (GIV-WT) and GIV-F1685A (GIV-FA) resistant to siRNA against human and murine GIV was cloned into pShuttle-CMV vector 62. The resultant plasmid was linearized with PacI and transformed into electrocompetent AdEasier E.coli (Agilent) using a Gene Pulser Electroporator (Bio-Rad). Correctly recombined clones were chosen based on sequencing and restriction endonuclease analyses, linearized with PacI and transfected into Cre8 cells (a gift from Dr. Ora Weisz, Univ. of Pittsburgh). Adenoviral particles were amplified using HEK-293 cells and purification was done using cesium chloride gradients 62.
In vitro Pulldown Assays and Immunoprecipitation
Purified GST-Gαi3, GST alone, His-GIV CT or His-PCT were immobilized on either glutathione-Sepharose or HisPur Cobalt beads for 90 min at room temperature as described before 17,19,26,63. Lx2 HSCs lysates or phosphorylated PDGFRβ (50 ng) proteins were added to each tube, and binding reactions were carried out for 4 h at 4°C with constant tumbling in binding buffer [50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.4% (v:v) Nonidet P-40, 10 mM MgCl2, 5 mM EDTA, 2 mM DTT, protease inhibitor mixture]. Beads were washed (4×) with 1 mL of wash buffer [4.3 mM Na2HPO4, 1.4 mM KH2PO4 (pH 7.4), 137 mM NaCl, 2.7 mM KCl, 0.1% (v:v) Tween 20, 10 mM MgCl2, 5 mM EDTA, , 2 mM DTT] and boiled in Laemmli's sample buffer. When GST-Gαi3 was used in the assay, both binding buffer and washing buffer were supplemented with 30 μM GDP.
For immunoprecipitation, cell lysates (~2 mg of protein) were incubated for 4 h at 4°C with 2 μg anti-PDGFR mAb or pre-immune control mouse IgG. Protein G Sepharose beads (GE Healthcare) were added and incubated at 4°C for an additional 60 min. Beads were washed and bound immune complexes were eluted by boiling in Laemmli's sample buffer.
For immunoprecipitation of active Gαi3, cell lysates were incubated for 1 h at 4°C with the conformational Gαi3: GTP mouse antibody (1 μg) 58 or pre-immune control mouse IgG. Protein G Sepharose beads (GE Healthcare) were added and incubated at 4°C for additional 45 min. Beads were washed and immune complexes were eluted by boiling in Laemmli's sample buffer as previously described 59,61,62.
In vitro kinase assay
In vitro kinase assays were perfomed using purified His-GIV-CT or His-PCT proteins (~5 μg) and commercially obtained PDGFRβ kinase (Cell Signaling). Reactions were started by addition of 1000 μM ATP and carried out at 25°C for 60 min in 30 μl kinase buffer [60 mM Hepes (pH 7.5), 5 mM MgCl2, 5 mM MnCl2, 3 μM Na3OV4]. Reactions were stopped by addition of Laemmli sample buffer and boiling.
Measurement of cAMP
Lx2 cells were transfected with GIV-WT or GIV-FA, serum starved (0% FBS, 16 h) and incubated with isobutylmethylxanthine (IBMX, 200 μM, 20 min) followed by PDGF (20 ng/ml, 10 min) and Forskolin (10 μM, 10 min). Reactions were terminated by aspiration of media and addition of 150 μl of ice-cold TCA 7.5% (w/v). cAMP content in TCA extracts was determined by radioimmunoassay (RIA) and normalized to protein [(determined using a dyebinding protein assay (Bio-Rad)] 64. Data is expressed as fold change over Forskolin stimulation.
Data Analysis and Statistics
All experiments were repeated at least three times, and results were presented either as one representative experiment or as average ± S.D. Statistical significance was assessed with two-tailed Student's t test.
Supplementary Material
Acknowledgements
We thank Marilyn Farquhar (UCSD) and Mikel Garcia-Marcos (Boston U) for helpful comments and critiques during the preparation of this manuscript, Katsumi Miyai (Pathology, UCSD) for generously giving us access to liver autopsy samples used in this work, Timo Meerloo for help with the preparation of semi-thin cryosections used to perform the immunofluorescence analysis, and Robert Esteban (Ventana Inc.) for optimization of IHC staining of liver sections. This work was supported by NIH grants CA160911 and DK099226, CAMS (Burroughs Wellcome Fund), and CSDA (Doris Duke Charitable Foundation) Awards to P.G. I.L-S was supported by American Heart Association (AHA #14POST20050025); Y.M. was supported by the Sarah Rogers Fellowship (UCSD). D.A.B was supported by P42ES010337, S.B.H. by Veterans Affairs Research Service, E.S by R01AA020172, R01DK085252 and P42ES010337, and F.M. by NIH HL091061.
Footnotes
Author Contributions:
I.L-S. and P.G. designed, performed and analyzed most of the experiments in this work and also wrote the paper. Y.D and Y.M. performed experiments and analyzed data. Liver fractionation and chronic injury of mice were carried out by Y-S.R and S.D.M and supervised by E.S. and D.A.B.. A.M, S.S and K.S performed IHC at Ventana. N.A. and F.M. carried out RIA assay for cAMP. S.B.H. contributed human liver samples used in this study. P.G. funded the project.
Competing financial interest
The authors declare no conflict of interest.
References
- 1.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinzani M, Gesualdo L, Sabbah GM, Abboud HE. Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver fat-storing cells. J Clin Invest. 1989;84:1786–1793. doi: 10.1172/JCI114363. doi:10.1172/JCI114363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Frontiers in bioscience : a journal and virtual library. 2002;7:d793–807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 4.Svegliati-Baroni G, et al. Bile acids induce hepatic stellate cell proliferation via activation of the epidermal growth factor receptor. Gastroenterology. 2005;128:1042–1055. doi: 10.1053/j.gastro.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Yoshiji H, et al. Vascular endothelial growth factor and receptor interaction is a prerequisite for murine hepatic fibrogenesis. Gut. 2003;52:1347–1354. doi: 10.1136/gut.52.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svegliati-Baroni G, et al. Insulin and insulin-like growth factor-1 stimulate proliferation and type I collagen accumulation by human hepatic stellate cells: differential effects on signal transduction pathways. Hepatology (Baltimore, Md. 1999;29:1743–1751. doi: 10.1002/hep.510290632. [DOI] [PubMed] [Google Scholar]
- 7.Seki E, et al. CCR1 and CCR5 promote hepatic fibrosis in mice. J Clin Invest. 2009;119:1858–1870. doi: 10.1172/JCI37444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow EK, et al. TLR agonists regulate PDGF-B production and cell proliferation through TGF-beta/type I IFN crosstalk. The EMBO journal. 2005;24:4071–4081. doi: 10.1038/sj.emboj.7600867. doi:10.1038/sj.emboj.7600867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons CJ, Takashima M, Rippe RA. Molecular mechanisms of hepatic fibrogenesis. Journal of gastroenterology and hepatology. 2007;22(Suppl 1):S79–84. doi: 10.1111/j.1440-1746.2006.04659.x. [DOI] [PubMed] [Google Scholar]
- 10.Adachi M, et al. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology. 2007;132:1434–1446. doi: 10.1053/j.gastro.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Son G, Hines IN, Lindquist J, Schrum LW, Rippe RA. Inhibition of phosphatidylinositol 3-kinase signaling in hepatic stellate cells blocks the progression of hepatic fibrosis. Hepatology (Baltimore, Md. 2009;50:1512–1523. doi: 10.1002/hep.23186. doi:10.1002/hep.23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solis-Herruzo JA, et al. G proteins are involved in the suppression of collagen alpha 1 (I) gene expression in cultured rat hepatic stellate cells. Cellular signalling. 1998;10:173–183. doi: 10.1016/s0898-6568(97)00036-3. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Sun SQ, Hassid A, Ostrom RS. cAMP inhibits transforming growth factor-beta-stimulated collagen synthesis via inhibition of extracellular signal-regulated kinase 1/2 and Smad signaling in cardiac fibroblasts. Molecular pharmacology. 2006;70:1992–2003. doi: 10.1124/mol.106.028951. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh AK, Yuan W, Mori Y, Varga J. Smad-dependent stimulation of type I collagen gene expression in human skin fibroblasts by TGF-beta involves functional cooperation with p300/CBP transcriptional coactivators. Oncogene. 2000;19:3546–3555. doi: 10.1038/sj.onc.1203693. doi:10.1038/sj.onc.1203693. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura T, et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nature cell biology. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. doi:10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, et al. Tyrosine phosphorylation of the Galpha-interacting protein GIV promotes activation of phosphoinositide 3-kinase during cell migration. Sci Signal. 2011;4:ra64. doi: 10.1126/scisignal.2002049. doi:10.1126/scisignal.2002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang P, et al. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer research. 2008;68:1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. The Journal of cell biology. 2008;182:381–393. doi: 10.1083/jcb.200712066. doi:10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Sanchez I, et al. Protein kinase C-theta (PKCtheta) phosphorylates and inhibits the guanine exchange factor, GIV/Girdin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5510–5515. doi: 10.1073/pnas.1303392110. doi:10.1073/pnas.1303392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh P, Garcia-Marcos M, Farquhar MG. GIV/Girdin is a rheostat that fine-tunes growth factor signals during tumor progression. Cell adhesion & migration. 2011;5:237–248. doi: 10.4161/cam.5.3.15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunkel Y, et al. STAT3 protein up-regulates Galpha-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. The Journal of biological chemistry. 2012;287:41667–41683. doi: 10.1074/jbc.M112.390781. doi:10.1074/jbc.M112.390781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albanis E, Friedman SL. Hepatic fibrosis. Pathogenesis and principles of therapy. Clinics in liver disease. 2001;5:315–334. v–vi. doi: 10.1016/s1089-3261(05)70168-9. [DOI] [PubMed] [Google Scholar]
- 24.Schuppan D, Krebs A, Bauer M, Hahn EG. Hepatitis C and liver fibrosis. Cell death and differentiation. 2003;10(Suppl 1):S59–67. doi: 10.1038/sj.cdd.4401163. doi:10.1038/sj.cdd.4401163. [DOI] [PubMed] [Google Scholar]
- 25.Weiler-Normann C, Herkel J, Lohse AW. Mouse models of liver fibrosis. Zeitschrift fur Gastroenterologie. 2007;45:43–50. doi: 10.1055/s-2006-927387. doi:10.1055/s-2006-927387. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh P, et al. A G{alpha}i-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Molecular biology of the cell. 2010;21:2338–2354. doi: 10.1091/mbc.E10-01-0028. doi:10.1091/mbc.E10-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Kuncio GS, Zern MA. In: Liver Growth and Repair. Strain Alastair, Mae Diehl Anne., editors. Springer Netherlands; 1998. pp. 558–576. [Google Scholar]
- 28.Moreira RK. Hepatic stellate cells and liver fibrosis. Archives of pathology & laboratory medicine. 2007;131:1728–1734. doi: 10.5858/2007-131-1728-HSCALF. doi:10.1043/1543-2165(2007)131[1728:HSCALF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 29.Mederacke I, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nature communications. 2013;4:2823. doi: 10.1038/ncomms3823. doi:10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yata Y, et al. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology (Baltimore, Md. 2003;37:267–276. doi: 10.1053/jhep.2003.50067. doi:10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 31.Xu L, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. doi:10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enomoto A, et al. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Developmental cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Molecular human reproduction. 1998;4:1099–1109. doi: 10.1093/molehr/4.12.1099. [DOI] [PubMed] [Google Scholar]
- 34.Wright MC, et al. Gliotoxin stimulates the apoptosis of human and rat hepatic stellate cells and enhances the resolution of liver fibrosis in rats. Gastroenterology. 2001;121:685–698. doi: 10.1053/gast.2001.27188. [DOI] [PubMed] [Google Scholar]
- 35.Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. doi:10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong M, Iwaisako K, Jiang C, Kisseleva T. Cell signals influencing hepatic fibrosis. International journal of hepatology. 2012;2012:158547. doi: 10.1155/2012/158547. doi:10.1155/2012/158547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane JR, Powney B, Wise A, Rees S, Milligan G. G protein coupling and ligand selectivity of the D2L and D3 dopamine receptors. The Journal of pharmacology and experimental therapeutics. 2008;325:319–330. doi: 10.1124/jpet.107.134296. doi:10.1124/jpet.107.134296. [DOI] [PubMed] [Google Scholar]
- 38.Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. International journal of medical sciences. 2006;3:47–52. doi: 10.7150/ijms.3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallet-Pichard A, et al. FIB-4: an inexpensive and accurate marker of fibrosis in HCV infection. comparison with liver biopsy and fibrotest. Hepatology (Baltimore, Md. 2007;46:32–36. doi: 10.1002/hep.21669. doi:10.1002/hep.21669. [DOI] [PubMed] [Google Scholar]
- 40.Chou R, Wasson N. Blood tests to diagnose fibrosis or cirrhosis in patients with chronic hepatitis C virus infection. Annals of internal medicine. 2013;159:372. doi: 10.7326/0003-4819-159-5-201309030-00021. doi:10.7326/0003-4819-159-5-201309030-00021. [DOI] [PubMed] [Google Scholar]
- 41.Shah R, et al. TGF-beta1 Up-Regulates the Expression of PDGF-beta Receptor mRNA and Induces a Delayed PI3K-, AKT-, and p70 -Dependent Proliferative Response in Activated Hepatic Stellate Cells. Alcoholism, clinical and experimental research. 2013 doi: 10.1111/acer.12167. doi:10.1111/acer.12167. [DOI] [PubMed] [Google Scholar]
- 42.Messadi DV, et al. Effect of TGF-beta 1 on PDGF receptors expression in human scar fibroblasts. Frontiers in bioscience : a journal and virtual library. 1998;3:a16–22. doi: 10.2741/A246. [DOI] [PubMed] [Google Scholar]
- 43.Ikuno Y, Kazlauskas A. TGFbeta1-dependent contraction of fibroblasts is mediated by the PDGFalpha receptor. Investigative ophthalmology & visual science. 2002;43:41–46. [PubMed] [Google Scholar]
- 44.Das F, et al. Akt kinase targets association of CBP with SMAD 3 to regulate TGFbeta-induced expression of plasminogen activator inhibitor-1. Journal of cellular physiology. 2008;214:513–527. doi: 10.1002/jcp.21236. doi:10.1002/jcp.21236. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Marcos M, Ghosh P, Ear J, Farquhar MG. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. The Journal of biological chemistry. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. doi:10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia Y, Turek JJ. Altered NF-kappaB gene expression and collagen formation induced by polyunsaturated fatty acids. The Journal of nutritional biochemistry. 2005;16:500–506. doi: 10.1016/j.jnutbio.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 47.He Z, Zhu Y, Jiang H. Toll-like receptor 4 mediates lipopolysaccharide-induced collagen secretion by phosphoinositide3-kinase-Akt pathway in fibroblasts during acute lung injury. Journal of receptor and signal transduction research. 2009;29:119–125. doi: 10.1080/10799890902845690. [DOI] [PubMed] [Google Scholar]
- 48.Guo J, et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology (Baltimore, Md. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwabe RF, Seki E, Brenner DA. Toll-like receptor signaling in the liver. Gastroenterology. 2006;130:1886–1900. doi: 10.1053/j.gastro.2006.01.038. doi:10.1053/j.gastro.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 50.Kisseleva T, Brenner DA. Mechanisms of fibrogenesis. Exp Biol Med (Maywood) 2008;233:109–122. doi: 10.3181/0707-MR-190. doi:10.3181/0707-MR-190. [DOI] [PubMed] [Google Scholar]
- 51.Swaney JS, et al. Inhibition of cardiac myofibroblast formation and collagen synthesis by activation and overexpression of adenylyl cyclase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:437–442. doi: 10.1073/pnas.0408704102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kisseleva T, Brenner DA. Fibrogenesis of parenchymal organs. Proceedings of the American Thoracic Society. 2008;5:338–342. doi: 10.1513/pats.200711-168DR. doi:10.1513/pats.200711-168DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolios G, Valatas V, Kouroumalis E. Role of Kupffer cells in the pathogenesis of liver disease. World journal of gastroenterology : WJG. 2006;12:7413–7420. doi: 10.3748/wjg.v12.i46.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chao W, Liu HL, Zhou WG, Hanahan DJ, Olson MS. Regulation of platelet-activating factor receptor and platelet-activating factor receptor-mediated biological responses by cAMP in rat Kupffer cells. The Journal of biological chemistry. 1990;265:17576–17583. [PubMed] [Google Scholar]
- 55.Wang H, Lafdil F, Kong X, Gao B. Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. International journal of biological sciences. 2011;7:536–550. doi: 10.7150/ijbs.7.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coulouarn C, et al. Hepatocyte-stellate cell cross-talk in the liver engenders a permissive inflammatory microenvironment that drives progression in hepatocellular carcinoma. Cancer research. 2012;72:2533–2542. doi: 10.1158/0008-5472.CAN-11-3317. doi:10.1158/0008-5472.CAN-11-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Marcos M, et al. Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. Faseb J. 2011;25:590–599. doi: 10.1096/fj.10-167304. doi:10.1096/fj.10-167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lane JR, et al. Antibodies that identify only the active conformation of G(i) family G protein alpha subunits. Faseb J. 2008;22:1924–1932. doi: 10.1096/fj.07-100388. doi:10.1096/fj.07-100388. [DOI] [PubMed] [Google Scholar]
- 59.Fukasawa H, et al. Phosphorylation of podocalyxin (Ser415) Prevents RhoA and ezrin activation and disrupts its interaction with the actin cytoskeleton. The American journal of pathology. 2011;179:2254–2265. doi: 10.1016/j.ajpath.2011.07.046. doi:10.1016/j.ajpath.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taura K, et al. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology. 2008;135:1729–1738. doi: 10.1053/j.gastro.2008.07.065. doi:10.1053/j.gastro.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Molecular biology of the cell. 2011;22:673–686. doi: 10.1091/mbc.E10-08-0738. doi:10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nature protocols. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. doi:10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Marcos M, et al. Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1961–1966. doi: 10.1073/pnas.1120538109. doi:10.1073/pnas.1120538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ostrom RS, et al. Receptor number and caveolar co-localization determine receptor coupling efficiency to adenylyl cyclase. The Journal of biological chemistry. 2001;276:42063–42069. doi: 10.1074/jbc.M105348200. doi:10.1074/jbc.M105348200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.