Abstract
Advancing pluripotent stem cell technologies for modeling hematopoietic stem cell development and blood therapies requires identifying key regulators of hematopoietic commitment from human pluripotent stem cells (hPSCs). Here, by screening the effect of 27 candidate factors, we reveal two groups of transcriptional regulators capable of inducing distinct hematopoietic programs from hPSCs: panmyeloid (ETV2 and GATA2) and erythro-megakaryocytic (GATA2 and TAL1). In both cases, these transcription factors directly convert hPSCs to endothelium, which subsequently transforms into blood cells with pan-myeloid or erythromegakaryocytic potential. These data demonstrate that two distinct genetic programs regulate the hematopoietic development from hPSCs and that both of these programs specify hPSCs directly to hemogenic endothelial cells. Additionally, this study provides a novel method for the efficient induction of blood and endothelial cells from hPSCs via overexpression of modified mRNA for the selected transcription factors.
Introduction
Human pluripotent stem cells (hPSCs), both embryonic stem cells (hESCs) and induced PSCs (hiPSCs), offer a plentiful source of blood cells for experimentation and therapeutic purposes. Although significant advances have been made in hematopoietic differentiation from hPSCs, a better understanding of the key regulators of hematopoietic commitment is required to achieve the scalability of production of blood cells from hPSCs and to enable de novo generation of hematopoietic stem cells (HSCs).
Transcription factors (TFs) have been recognized as critical regulators of early embryonic development. TFs function as key elements of a gene regulatory network that guide the acquisition of specific properties by particular cell type 1. Several TFs are identified as master regulators of hematopoietic development in the mouse embryo 2-5. Many of them are also involved in the regulation of endothelial development, reflecting a close developmental link between endothelial and hematopoietic cells 6. In fact, recent studies have demonstrated that in the embryo, hematopoietic cells including HSCs arise from endothelial cells with blood-forming potential, hemogenic endothelium 7-9, indicating that blood development proceeds through an endothelial intermediate stage.
To unravel the most essential TFs required for the induction of the blood program from hPSCs, we performed comprehensive gain-of-function screening. Using this approach, we identified two optimal combinations of TFs capable of inducing distinct, robust hematopoietic programs from PSCs: pan-myeloid (ETV2 and GATA2) and erythro-megakaryocytic (GATA2 and TAL1). Interestingly, both TF combinations directly induced hemogenic endothelial cells, which subsequently transformed into blood progenitors with a distinct spectrum of hematopoietic differentiation. These results suggest, firstly the specification to discrete types of hematopoietic progenitors begins at the hemogenic endothelium stage and is regulated by distinct transcriptional programs, and secondly, only a few TFs are sufficient to activate the hematoendothelial program from hPSCs, and trigger in a culture dish the sequence of events observed during blood development in the embryo. Also presented, is a novel approach to induce the efficient production of endothelium and blood from hPSCs using mmRNA.
RESULTS
Selection of candidate genes and screening system design
To induce the hematopoietic program in hPSCs, we first assembled a list of candidate transcriptional regulators involved in mesodermal and angiohematopoietic specification and HSC development through literature review. To prioritize genes for screening, we used molecular profiling data obtained from analysis of the gene expression of hESC-derived mesodermal and vascular progenitors with or without hematopoietic potential we identified in our prior studies 10,11. Based on these data we selected 27 genes (Supplementary Table 1 and Supplementary Fig. 1).
We assumed that the ideal hPSC-based system for a gain-of-function screen for hematopoiesis-inductive factors should meet two major requirements: maintain hPSCs in an undifferentiated state, and support expansion of induced hematopoietic cells. We found that these conditions can be met by maintaining hPSCs as a monolayer on matrigel in a basal growth-factor free mTeSR1 medium supplemented with bFGF and SCF and TPO hematopoietic cytokines. In these conditions, the control hESCs or those transduced with EGFP remained visibly undifferentiated and retained surface markers and gene expression profile characteristic of hPSCs, while hESCs transduced with lineage factors successfully obtained their differentiation phenotypes (Fig. 1a-1e and Supplementary Fig. 2).
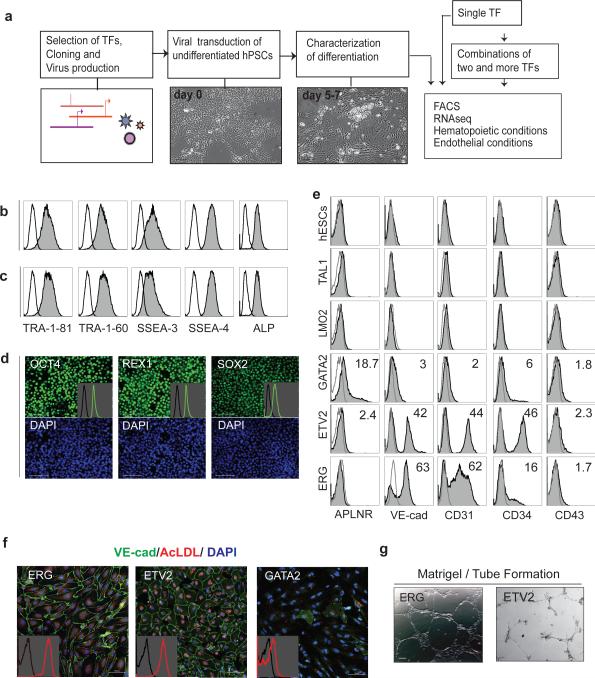
Figure 1. Gain-of-function screening in hPSCs.
(a) Schematic diagram of the screening system; (b-d) Flow cytometric and immunofluorescent analysis of expression of pluripotency markers in H1 hESCs growing on matrigel for 5 days in standard conditions in mTeSR1 medium (b) and basal growth-factor free TeSR1 medium containing 100 ng ml−1 SCF, 50 ng ml−1 TPO, and 20 ng ml−1 bFGF (c and d). Inserts in (d) show analysis of expression of indicated markers by flow cytometry; (e) Flow cytometric analysis of mesodermal, endothelial and hematopoietic markers in control hESCs and hESCs transduced with indicated TFs on day 5 post-transduction; (f,g) ETV2- and ERG-transduced cells acquire endothelial characteristics as shown by positive VE-cadherin immunostaining, AcLDL uptake (f) and formation of endothelial tubes (g). Inserts in (f) shows analysis of AcLDL uptake by flow cytometry. Scale bar, d,f,g, 100 μm.
ETV2 or ERG alone are sufficient to induce endothelium from hESCs
To test the functional capacity of individual genes, we analyzed their effect on morphology and expression of flowing markers by flow cytometry: APLNR and KDR (mesodermal), VE-cadherin, CD34, CD31 and CD73 (endothelial), CD43 and CD45 (hematopoietic) (Supplementary Table 2). Morphological evaluation of the cultures revealed three major effects of TF overexpression: a change in morphology, thereby indicating differentiation; no change in morphology, thus suggesting that cells remain undifferentiated; and early cell death (Supplementary Table 2). Among the 27 tested factors, only 14 triggered morphologic changes in culture without inducing cell death within first 4-5 days after transduction. Although we could not classify the type of formed cells based on morphology alone in most cases, we noticed that ETV2 or ERG transduced cells acquired the typical morphologic features of endothelial cells (Supplementary Fig. 2). Immunofluorescent and functional analyses revealed that ETV2 and ERG-induced cells expressed VE-cadherin, CD31, and CD34 endothelial markers, uptook AcLDL, and formed vascular tubes in response to VEGF; which is consistent with the endothelial nature of cells (Fig. 1e-1g). Gene expression analysis showed that ETV2 or ERG alone are sufficient to induce the expression of almost the entire set of genes required for angiohematopoietic development, as well as those that are typically expressed in endothelial cells. However, ETV2 and ERG had little effect on the expression of pluripotency genes (Supplementary Fig. 3a,b). None of the selected genes were able to induce formation of round blood cells, though we noted weak expression of CD43 by a very few cells following the transduction of cells with ETV2, GATA1 or GATA2 (Supplementary Table 2). Although FOXF1 and HAND1 TFs are shown to be important for lateral plate/extraembryonic mesoderm development in mouse studies 12,13, we found that they did not upregulate the expression of APLNR or KDR pan-mesodermal markers or the genes known to be expressed in lateral plate mesoderm (Supplementary Fig. 3a). In contrast we noticed that GATA2 transduction triggered the expression of TFPA1, AMOT, APLNR, and some other genes associated with endothelial cells, and repressed ESC-specific genes (Supplementary Fig. 3a,b). Although GATA1 and GATA3 induced expression of some endothelial genes similar to GATA2, they also induced the expression of primitive streak genes, and had a less prominent effect on expression of ESC-specific genes, when compared to GATA2 (Supplementary Fig. 3a). The Principal Component (PC) analysis of global gene expression and global analysis of changes in gene expression profiles using gene expression dynamics investigator (GEDI) revealed that ETV2, ETV6, ERG, EGR1, CEBPA, FOXC2, HHEX, GATA1, GATA2, GATA3, RUNX1B, and RUNX1C caused the most dramatic changes in gene expression, while LMO2 and CBFB TFs, which lack DNA binding activity, and several DNA binding molecules including HES1, LYL1, TAL1, MYB, GFI1 and SPI1 had a minimal effect on gene expression in hESCs (Fig. 2a and Supplementary Fig. 2).
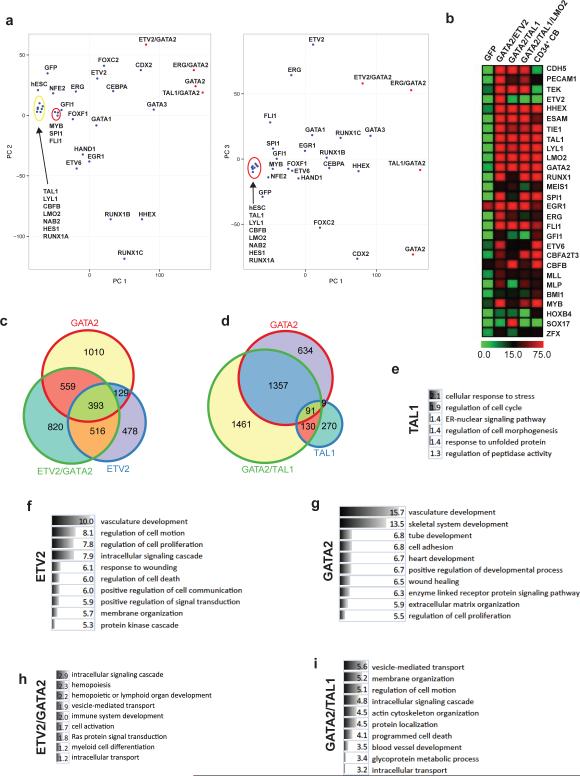
Figure 2. Gene expression profiling of hESCs transduced with transcription factors.
(a) Principal Component (PC) analysis of global gene expression demonstrates the differences in global gene expression in H1 hESCs transduced with indicated genes. The relative distance is collapsed to two PC1/PC2 and PC1/PC3 dimensions. hESC is non-transduced H1 hESCs control. (b) Heat map shows the expression of genes associated with HSC development, expansion and self-renewal in hESCs transduced with blood-inducing combinations and CD34+ cord blood cells. Gene expression is estimated in tpm values. (c-d) The Venn diagrams summarize the number of overlapping and selectively induced genes in hESCs transduced with ETV2, GATA2, TAL1, ETV2/GATA2, and GATA2/TAL1 TFs. Venn diagram shows differentially expressed genes as compared to control hESCs (posterior probability ≥ 0.99 as determined by EBseq analysis). There is a high overlap between ETV2 and GATA2 induced genes. TAL1 alone has little effect on gene expression, however in combination with GATA2, TAL1 causes profound changes in the transcriptome. (e-i) The classification of genes induced by indicated combinations of factors into functional categories defined by Gene Ontology (GO) Term using DAVID program. For GO analysis, each gene set was reduced to include only genes with tpm≥10 in at least 1 sample. ETV2/GATA2 and GATA2/TAL1 show genes uniquely induced by two factors together as compare to single factor-transduced cells. The all (TAL1, ETV2/GATA2) or top ten (ETV2, GATA2, GATA2/TAL1) significantly overrepresented categories with FDR below 0.05 (computed according to Benjamini-Hochberg method) are shown.
ETV2 and GATA2 induce pan-myeloid hematopoiesis from hESCs through the hemogenic endothelium stage
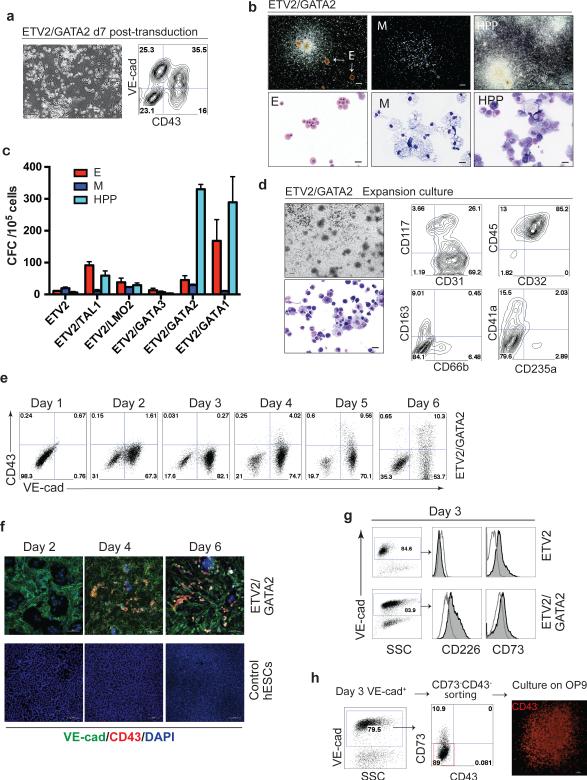
Accumulated knowledge strongly indicates that blood formation in the embryo proceeds through hemogenic endothelial intermediates 7-9. Therefore, we decided to test whether the addition of known hematopoietic factors to ETV2 or ERG endothelium-inductive factors would be sufficient to generate endothelium with hemogenic potential. Given the well-established role of GATA2 and GATA1 factors in hematopoietic development, we selected these TFs as a first choice. In fact, transduction of hESCs with ETV2 and GATA2 led to the formation of round CD43+ blood cells with robust CFC potential, while hESCs transduced with ETV2 alone occasionally produced very few small colonies of macrophages and erythroid cells (Fig. 3a-c). In clonogenic cultures, ETV2/GATA2 transduced cells formed typical erythroid (CFC-E), macrophage (CFC-M), and large (greater than 0. 5mm in diameter) high proliferative potential colonies (CFC-HPP). Most of the HPP colonies were composed of immature granulocytic and monocytic cells (Fig. 3b), but some colonies showed more mature myeloid cells with erythroid and megakaryocytic cells. Cells collected from clonogenic cultures of ETV2/GATA2 transduced hESCs vigorously proliferated in serum-containing medium with hematopoietic cytokines. After 7 days of expansion we were able to obtain 24 million CD43+ cells from one million ETV2/GATA2 transduced undifferentiated H1 hESCs (Supplementary Table 3). Flow cytometric analysis of expansion cultures revealed all types of myeloid cells, including CD31+CD117+ mast cells, CD45+CD32+ myelomonocytic cells, CD163+ macrophages, CD66b+ granulocytes, CD41a+ megakaryocytic, and CD235a+ erythroid cells, thereby indicating that GATA2 and ETV2 induce pan-myeloid hematopoiesis from hESCs (Fig. 3d). GATA1 in combination with ETV2 induced a spectrum of hematopoietic colonies similar to GATA2/ETV2 combination, though we noticed an increase in the number of erythroid colonies with ETV2/GATA1 as compared to ETV2/GATA2 transduced cells (Fig. 3c). Transduction of cells with ETV2 and GATA3, or TAL1 or LMO2 induced far fewer hematopoietic CFCs and CD43+ cells as compared to ETV2 with GATA2, or GATA1 combinations (Fig. 3c and Supplementary Fig. 4a). The addition of other critical hematopoiesis-associated factors, such as TAL1 and LMO2, on top of the ETV2/GATA2 combination, did not substantially change the spectrum of hematopoietic programming (Supplementary Fig. 4b). We also found that hematopoiesis can be initiated by transducing cells with ERG together with GATA2 or TAL1 plus LMO2 (Supplementary Fig. 4b). However, these combinations induced a very few CFCs most of which were erythroid.
Figure 3. Hematopoietic differentiation of hESCs induced by ETV2 and GATA2.
(a) Cell morphology and flow cytometric analysis of ETV2/GATA2-transduced H1 hESCs on day 7 post-transduction. Scale bar, 100 μm. (b) Types of hematopoietic colonies formed by hESCs on day 7 post-transduction with ETV2/GATA2. Erythroid colonies (E); macrophage colonies (M); high proliferative potential myeloid colonies (HPP). Scale bar for CFC-assay, 250 μm; cytospins, 20μm. (c) CFC potential of cells transduced with ETV2 alone and indicated TF combinations. Error bars represent s.e.m. from 3 to 5 independent experiments. (d) Phase-contrast photograph of the culture, Wright-stained cytospin and FACS analysis of ETV2/GATA2-induced hematopoietic cells grown in suspension culture for 14 days in medium supplemented with FBS and 100 ng ml−1 SCF, 10 ng ml−1 IL3, 20 ng ml−1 IL6, 10 ng ml−1 GM-CSF, 20 ng ml−1 G-CSF, and 3 u ml−1 EPO. (e) Kinetic analysis of VE-cadherin and CD43 expression during direct ETV2/GATA2 programing of H1 hESCs by flow cytometry. (f) VE-cadherin and CD43 immunofluorescent staining of untreated control hESCs and hESCs transduced with ETV2/GATA2 at different time points after transduction. Scale bars, 100 μm. (g) Expression of markers of associated with hemogenic and non-hemogenic endothelium by VE-cadherin+ cells emerging on day 3 post-transduction with indicated TFs. Histograms show expression of CD226 and CD73 by VE-cadherin-gated cells. Endothelial cells induced by ETV2 alone have CD226−CD73+ phenotype associated with non-hemogenic endothelial cells, while ETV2/GATA2 induced endothelium have CD226+CD73− phenotype associated with hemogenic endothelium 10. (h) Hematopoietic potential of VE-cadherin+CD43−CD73− endothelial cells isolated from programing cultures. On day 3 after transduction with ETV2/GATA2, VE-cadherin+CD43−CD73− cells were isolated by sorting and cultured on OP9 to assess the hematopoietic potential by counting CD43+ hematopoietic colonies using immunofluorescent staining. Blue and red rectangles show position of gates used for cell sorting. Scale bar, 100 μm.
Based on these observations, we concluded that ETV2 and GATA2 represent the most optimal combination of TFs for the induction of a robust multilineage hematopoiesis from hESCs, and therefore selected this combination for more detailed analysis.
Gene expression profiling revealed that combining ETV2 with GATA2 was sufficient to activate almost the entire spectrum of genes essential for hematopoiesis, including MYB, RUNX1, CBFB, and VAV1 genes critical for definitive hematopoiesis, as well as genes associated with myeloid, lymphoid and erythroid lineages (Fig. 2b and Supplementary Fig. 5a). Compared to ETV2 or GATA2 alone, the ETV2/GATA2 combination uniquely induced the expression of 820 genes enriched in the intracellular signaling cascade, hematopoiesis, immune system development, myeloid differentiation and RAS protein signal transduction functional GO categories (Fig. 2c and 2h), consistent with the induction of functional hematopoietic cells by these factors.
Kinetic analysis of blood formation by ETV2 and GATA2 transduced cells revealed that hematopoietic development from hESCs proceeds through the endothelial stage. As shown in Supplementary Fig. 5b, the ETV2/GATA2 combination quickly induced CDH5 and PECAM1 endothelial genes in hESCs on day 2 post-transduction. The upregulation of CDH5 and PECAM1 was not preceded by substantial and synchronized upregulation of primitive streak and lateral plate mesoderm genes on day 1 post-transduction, thus suggesting that ETV2/GATA2 may directly induce the endothelial program in hESCs bypassing mesodermal stage. The onset of endothelial gene expression was accompanied by induction of the master regulator of definitive hematopoiesis, RUNX1. Microscopic examination and immunofluorescent staining of cultures revealed that 2-3 days after ETV2 and GATA2 transduction, hESCs acquired VE-cadherin expression and typical morphological features of endothelial cells, similar to ETV2 transduced hESCs (Fig. 3e,f). However, in contrast to ETV2 alone, endothelial cells induced after three days of ETV2/GATA2 transduction expressed CD226 and lacked CD73 (Fig. 3g), i.e. displayed phenotypic features that distinguish hemogenic endothelium from nonhemogenic one 10. The following next two days of culture, we observed a transition of endothelial cells into round CD43+ hematopoietic cells, thereby indicating that ETV2 and GATA2 overexpression directly induces formation of endothelial cells with hemogenic properties that subsequently give rise to blood cells (Fig. 3f and Supplementary Movie 1). When VE-cadherin+CD73− cells were collected from ETV2/GATA2 transduced cultures prior to the detection of CD43 expression (day 3) and cultured on OP9, they generated colonies composed of round CD43+ hematopoietic cells (Fig. 4h) at a frequency 501.0±128.4 colonies per 105 plated VE-cadherin+CD43−CD73− cells. In contrast, VE-cadherin+ cells isolated from cultures transduced with ETV2 alone essentially failed to produce CD43+ hematopoietic colonies on OP9 (<2 colonies per 105 plated cells). These findings indicate that VE-cadherin+CD73−CD43− cells induced by ETV2/GATA2 have functional potentials similar to hemogenic endothelium generated from hESC by differentiation on OP9 10
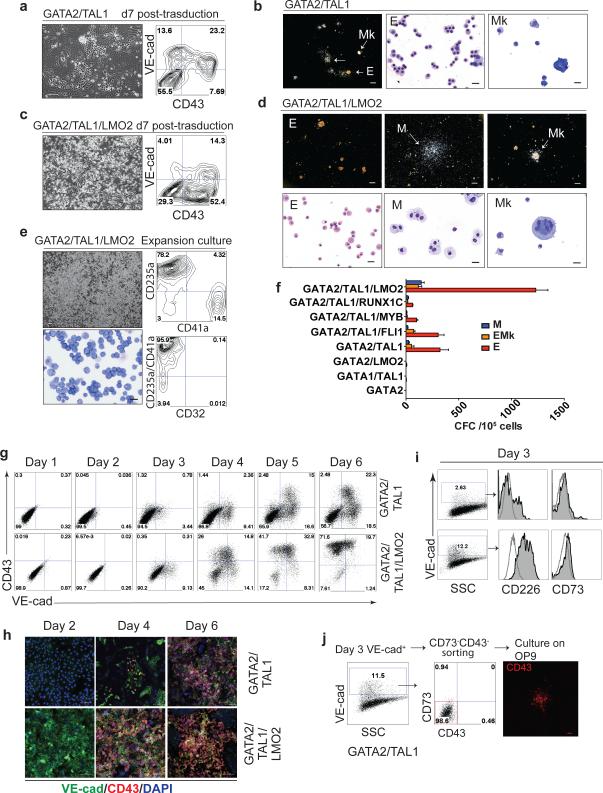
Figure 4. Hematopoietic differentiation of hESCs induced by GATA2 and TAL1.
(a) Cell morphology and flow cytometric analysis of H1 hESCs differentiated by expression of GATA2 and TAL1 on day 7 post-transduction. (b) Types of hematopoietic colonies formed by GATA2 and TAL1 differentiated cells on day 7 post-transduction. Erythroid colonies (E); macrophage colonies (M); megakaryocytic colonies (Mk). Scale bar for CFC-assay, 250μm; for cytospins, 20μm. (c) Cell morphology and flow cytometric analysis of H1 hESCs differentiated by expression of GATA2/TAL1/LMO2 on day 7 post-transduction. (d) Types of hematopoietic colonies formed by GATA2/TAL1/LMO2 differentiated cells on day 7 post-transduction. Scale bar for CFC-assay, 250μm; for cytospins, 20μm. (e) Phenotypic characterization of GATA2/TAL1/LMO2-induced hematopoietic cells grown in SFEM serum-free medium supplemented with 100 ng ml−1 SCF, 50 ng ml−1 TPO, 3 u ml−1 EPO, and 20 ng ml−1 bFGF for 14 days. (f) CFC potential of hESCs transduced with GATA2-based combinations. Error bars represent s.e.m. from 3 experiments. (g) Kinetic analysis of VE-cadherin and CD43 expression by flow cytometry following transduction of H1 hESCs with indicated combinations of TFs. (h) VE-cadherin and CD43 immunofluorescent staining of hESCs transduced with indicated TFs at different time points after transduction. Scale bars, 100 μm. (i) Expression of markers of associated with hemogenic and non-hemogenic endothelium by VE-cadherin+ cells emerging on day 3 post-transduction with GATA2/TAL1 (upper panels) and GATA2/TAL1/LMO2 (lower panels). Histograms show expression of CD226 and CD73 by VE-cadherin-gated cells. Endothelial cells induced by GATA2/TAL1 and GATA2/TAL1/LMO2 shows CD226+CD73− phenotype associated with hemogenic endothelium 10. (j) Hematopoietic potential of VE-cadherin+CD43−CD73− endothelial cells isolated from GATA2/TAL1 programing cultures. On day 3 after transduction with GATA2/TAL1 or ETV2/GATA2, VE-cadherin+CD43−CD73− cells were isolated by sorting and cultured on OP9 to assess the hematopoietic potential by counting CD43+ hematopoietic colonies using immunofluorescent staining. Blue and red rectangles show position of gates used for cell sorting. Scale bar, 100 μm.
GATA2 and TAL1 Induce a Hematopoietic Program Mostly Restricted to Erythromegakaryocytic Cells
Although the basic helix-loop-helix TF TAL1 is a well-known key regulator of hematopoiesis and vasculogenesis 14,15, overexpression of TAL1 alone induced minimal changes in the hESC transcriptome (Fig. 2a,d and Supplementary Fig. 2) and was not able to induce the formation of endothelium or blood cells from hESCs (Fig. 1e). When added to ETV2, TAL1 induced only a few hematopoietic colonies and CD43+ cells (Fig. 3c and Supplementary Fig. 4a), while cotransduction of TAL1 with GATA2 efficiently induced the robust formation of CD43+ hematopoietic progenitors (Fig. 4a,b), similar to the ETV2/GATA2 combination. However, in contrast to ETV2/GATA2 combination, hematopoiesis in GATA2/TAL1 transduced cultures was restricted to erythroid and megakaryocytic cells, with the exception of a few macrophages (Fig. 4b,f). In tandem with TAL1, GATA2 was essential for the induction of hematopoiesis and could not be replaced with GATA1. When hESCs were transduced with the GATA1/ TAL1 combination they slightly upregulated expression of VE-cadherin and CD43 within the first 4-5 days of culture, but failed to produce a significant number of CFCs afterward (Fig. 4f).
Since overexpression of TAL1 and GATA2 induced mostly erythroid and megakaryocytic lineages with very few macrophage CFCs, we tested whether the addition of other factors to these combinations would promote multilineage hematopoiesis. As shown in Fig. 4f, the addition of either MYB or RUNX1C factors that are critical for definitive hematopoiesis to GATA2/TAL1 combination was not able to induce the robust myeloid hematopoiesis as was seen with ETV2/GATA2 transduced cells. Likewise, the addition of another ETS TF FLI1 failed to promote myeloid development, indicating that GATA2 and TAL1 combination “locked” cells to erythroid and megakaryocytic lineages.
The addition of LMO2 to the GATA2/TAL1 combination dramatically increased hematopoiesis, without significant changes in the spectrum of hematopoietic colonies (Fig. 4c,d,f). GATA2/TAL1/LMO2 transduced cells collected from clonogenic cultures robustly expanded in suspension cultures with cytokines. After 7 days of expansion we were able to obtain 33 million CD43+ cells from one million transduced H1 hESCs. Flow cytometric analysis demonstrated that expansion cultures generated almost exclusively CD235a+ erythroid and CD41a+ megakaryocytic cells and no CD32+ myelomonocytic cells (Fig. 4e), thus confirming the restricted differentiation potential of cells generated from hESCs using these TFs.
In cultures transduced with GATA2 and TAL1, the formation of CD43+ round blood cells was preceded by upregulation of VE-cadherin expression (Fig. 4g,h and Supplementary Movie 2), thus indicating that CD43+ cells generated with these two factors, similar to ETV2 and GATA2 transduced cells, arose from endothelial cells through endothelial-hematopoietic transition. Endothelial cells induced by GATA2 and TAL1 on day three of culture, had both the phenotypic and functional features of hemogenic endothelium, i.e. they expressed CD226, lacked CD73 (Fig. 4i) and were capable of forming of CD43+ blood colonies after culture on OP9 (Fig. 4j) at a frequency of 35.6+ 8.6 colonies per 105 plated VE-cadherin+CD43−CD73− cells. Transduction of hESCs with the GATA2/TAL1/LMO2 combination, accelerated transition of hESCs into round CD43+ hematopoietic cells with early activation of RUNX1 expression (Supplementary Fig. 5b), early appearance of CD43+VE-cadherin-cells and a markedly contracted endothelial stage of development (Fig. 4g,h). Taken together, these observations suggest that GATA2 and TAL1 are critical TFs for specifying hESCs to erythromegakaryocytic progenitors through hemogenic endothelium intermediates.
Induction of hematopoietic program in various hPSCs and by using modified mRNA
To determine whether the identified sets of transcriptional regulators were capable of inducing the hematopoietic program in hPSCs other than H1 hESCs, we overexpressed ETV2/GATA2 or GATA2/TAL1/LMO2 in H9 hESCs and two fibroblast-derived iPSCs. Although yields of blood cells varied between different hPSCs (Supplementary Table 3), the pattern of hematopoietic programming with both combinations of TFs was reliably consistent across all tested cell lines (Fig. 5a-c). ETV2 and GATA2 induced pan-myeloid hematopoiesis, including CD32+ myelomonocytic and CD235a+ and CD41a+ erythroid and megakaryocytic cells, while the TAL1/GATA2/LMO2 combination induced predominantly the erythroid and megakaryocytic cells, but not CD32+ myelomonocytic cells.
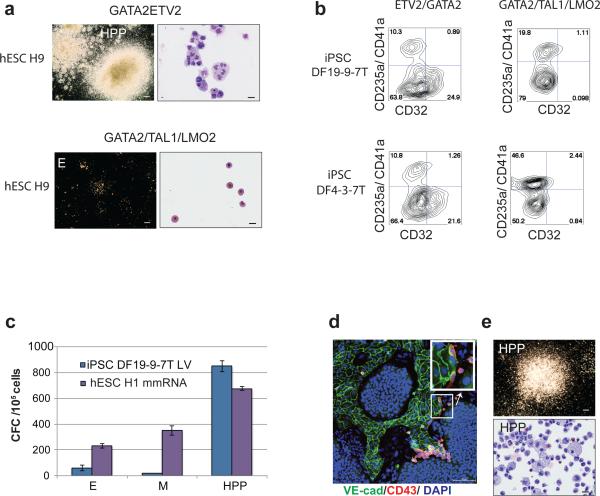
Figure 5. Direct hematopoietic programming of various hPSCs using lentiviral vectors and mmRNA.
(a) CFCs developed from H9 hESCs 7 days after enforced expression of ETV2/GATA2 and GATA2/TAL1/LMO2. (b) FACS analysis of ETV2/GATA2 and GATA2/TAL1/LMO2 transduced iPSC lines DF-19-9-7T and DF-4-3-7T after expansion in suspension cultures in the presence of hematopoietic cytokines (14 day post-transduction). (c) CFC potential of DF19-9-7T hiPSCs transduced with ETV2 and GATA2 lentiviral vectors (LV) and H1 hESCs transfected with ETV2 and GATA2 mmRNA. Error bars represent s.e.m. from 3 experiments. (d) Confocal image of H1 hESC culture five days after transduction with ETV2/GATA2 mmRNA. Insert shows the digital zoom of the area outlined by white rectangle where cells are undergoing endothelial-hematopoietic transition. Scale bar 100 μm. (e) HPP CFCs induced in H1 hESCs by ETV2/GATA2 mmRNA; CFC assay scale bar, 250 μm, cytospins, 20 μm.
To find out whether the induction of blood formation from hPSCs in our system requires continuous or transient expression of transcriptional regulators, we performed direct programming studies using ETV2 and GATA2 modified mRNA (mmRNA). As shown in Figures 5c-e, transfection of H1 hESCs with ETV2 and GATA2 mmRNA induced the hematoendothelial program and robust formation of the same spectrum of CFCs as we observed with lentivirus-transduced cells. Using mmRNA transfection, we were able to generate 11.6 million CD43+ cells from one million H1 hESCs following transfection and 7 day expansion. These studies provide the evidence that brief expression of ETV2 and GATA2 is indeed sufficient to commit the hPSCs to blood fate.
Discussion
Using a gain-of-function genetic screen, we identified ETV2 and GATA2 as the most critical TFs required for induction of the hemogenic endothelium with pan-myeloid potential from hESCs. ETV2 belongs to the ETS family of TFs, which also includes ERG, and plays a critical role in endothelial development16. Gain-of-function experiments in Xenopus and zebrafish embryos have demonstrated that ETV2 and ERG are capable of inducing ectopic endothelial differentiation 17-19. ETV2 and ERG are also essential for establishing definitive hematopoiesis and the proper function of adult HSCs 20-23. We found that ectopic expression of ETV2 and ERG in undifferentiated hESCs upregulates the expression of genes associated with angiohematopoietic development and typical endothelial genes resulting in the formation of endothelial cells. Although overexpression of ETV2 or ERG alone induced expression of endogenous FLI1, GATA2, and TAL1 genes that form the kernel of the gene regulatory network in developing HSCs 24, ETV2 or ERG-induced endothelium was lacking significant blood-forming activity. Overexpressing GATA2 in addition to ETV2 was essential to induce endothelial cells with robust blood forming potential. The ERG-mediated endothelial program was much less susceptible to hemo-inductive effect of GATA2, indicating a unique requirement for synergistic action of ETV2 and GATA2 TFs for optimal activation of pan-myeloid hematopoietic program from hPSCs. Although prior studies in mouse ESCs have shown the synergistic role of Etv2 and Gata2 in hematoendothelial specification from mesoderm 25,26, we have revealed for the first time that these two factors alone are sufficient to directly induce hematoendothelial program from undifferentiated PSCs in humans.
Mouse studies have demonstrated that Tal1 controls the expression of several important hematopoietic regulators, including Runx1, Erg, Gfi1b, and Gata2 among others 27. Tal1 is considered a key component of the regulatory network controlling HSC specification 24. Tal1 also plays an important role in vascular development and is required for embryonic vascular remodeling 15,28 and hemangioblast specification 29,30. Enforced expression of Tal1 in normal zebra fish embryos causes overproduction of endothelial and hematopoietic cells 30. However, enforced TAL1 expression in hESCs induced only minimal changes in the transcriptome and failed to trigger morphologic changes, thereby suggesting that TAL1 alone can't create a TF network to direct the activation of the angiogenic program in hESCs as we observed with ETV2 and ERG. Yet, TAL1 was capable of triggering blood development when coexpressed together with ETV2 or GATA2. ETV2/TAL1 combination induced pan-myeloid hematopoiesis in hESC cultures similar to cultures transduced with ETV2 and GATA2, but with much less efficiency. Enforced expression of TAL1 with GATA2 induced robust hematopoiesis, but it produced mostly erythroid and megakaryocytic cells. Interestingly, hematopoietic development in GATA2/TAL1-transduced cultures proceeded through the hemogenic endothelium stage similar to cultures transduced with ETV2 and GATA2, which generated pan-myeloid CFCs. These observations suggest that two groups transcriptional regulators can trigger activation of TF network leading to formation of hemogenic endothelium with different functional capacities: GATA2/TAL1-induced hemogenic endothelium with restricted erythro-megakaryocytic and macrophage potential, and ETV2/GATA2-induced endothelium with pan-myeloid potential.
Although the concept of hemogenic endothelium was developed by observing HSC and blood formation from the aortic endothelium in the AGM region, it has become clear that endothelium in other embryonic and extraembryonic sites possess hemogenic potential. Among these sites are the vitelline and umbilical arteries 31, placenta 32, head vasculature 33, endocardium 34 and nascent yolk sac capillaries 35.
However, not all hemogenic endothelium has the capacity to produce HSCs. In the AGM region, HSCs are restricted to the ventral wall of the dorsal aorta, while hematopoietic progenitors without HSC potential can be found at several other sites within the embryonic aorta 31,36, underscoring that hemogenic potential of the aortic endothelium is variable. Recently, Chen et al. 37 identified at least two distinct types of hemogenic endothelium in the embryo, implying that specification of hematopoietic program may take place at the hemogenic endothelium stage. Further support for this concept came from clonal functional and molecular analyses of hemogenic endothelium undergoing endothelial-hematopoietic transition in mouse embryo which found that the hematopoietic program is already initiated in cells embedded in the endothelial layer. 38 Our present studies have demonstrated that hemogenic endothelium with distinct hematopoietic programs can be induced from hPSCs, and that different groups of TFs are required for the induction of hemogenic endothelium with pan-myeloid or restricted erythromegakaryocytic program. These observations strongly support the concept of heterogeneity of hemogenic endothelium and provide additional evidence that lineage-specific hematopoietic programs from hPSCs can be established at the hemogenic endothelium stage.
Production of hematopoietic cells with long term repopulating potential from hPSCs remains a significant challenge. Although we demonstrated that ETV2/GATA2 combination induces robust formation of multipotential CFCs, we failed to detect meaningful engraftment of these cells in NOD/SCID/IL2Rγ−/− mice, suggesting that TF-induced cells have similar properties to hematopoietic progenitors obtained from hPSCs using conventional differentiation methods. Nevertheless, identification of critical pathways leading to the formation of distinct types of hemogenic endothelium in the current study provided a novel experimental platform to further explore the transcriptional program in hPSCs required for activation of the self-renewal program in hPSC-derived hematopoietic cells. Recently, studies by Pereira et al.39 have shown that the hemogenic program can be induced in mouse fibroblasts via overexpression of Gata2, Gfi1b, cFos, and Etv6, thus indicating the feasibility of generating a hematopoietic hierarchy by bypassing the pluripotency stage. Pursuing both approaches to generating blood cells, PSC-based and direct reprogramming of somatic cells, will be essential to better understand the genetic and epigenetic factors governing hematopoietic fate determination and the self-renewal program, and will contribute substantially to the ultimate goal of developing technologies for in vivo production of HSCs for clinical purposes.
Methods
Cloning of selected genes and virus production
Open Reading Frames (ORFs) of selected genes were amplified from cDNA of H1 hESCs differentiated in coculture with OP9, or from full-length cDNAs clones obtained from Open BioSystems (Pittsburgh, PA) and Gene Copoeia Inc. (Rockville, MD). After sequence verification, ORFs were subcloned into pSIN/EF1α-IRES-Puro lentiviral expression vector. Virus production was carried out by calcium phosphate transfection of Lenti-X 293T cells (Clontech, Mountain View, CA). Packaged lentiviral units were concentrated either on ethylene oxide-sterilized Centricon Plus-70 or Amicon Ultra-15 Centrifugal Filter Units (Millipore, Billerica, MA), or by ultracentrifugation at 33,000rpm for 2.5hr (Beckman Optima XL-A analytical ultracentrifuge, Beckman Coulter, Inc., USA) and re-suspended in 1%BSA in PBS. Amount of Infectious Units (IFU) per mL of viral concentrate was assessed by transduction and puromycin selection of HeLa cells with a working puromycin concentration of 1 μg ml−1. Successful transduction was confirmed by demonstration of virus integration by genomic PCR and transgene mRNA expression by RNAseq analysis.
Cell culture and hPSCs transductions
Irradiated mouse embryonic fibroblasts (MEFs), human embryonic stem cell lines H1 (WA01) and H9 (WA09), fibroblast-derived human induced pluripotent stem cell lines DF-19-9-7T and DF-4-3-7T were obtained from WiCell (Madison, WI). hESCs and hiPSCs were maintained on MEFs in DMEM/F12 basal medium supplemented with 20% of KnockOut Serum Replacement, 0.1 mM non-essential amino acids, 0.1mM β-mercaptoethanol, 1mM L-Glutamine (Gibco, Invitrogen, Carlsbad, CA) and 4 ng ml−1 of bFGF. Prior to lentiviral transduction experiments, pluripotent stem cells were grown on Matrigel (BD Biosciences, Bedford, MA) in feeder-free conditions in chemically defined mTeSR™ 1 medium (StemCell Technologies, Vancouver, Canada) from 2 to 5 passages. hPSCs monolayers at 70% of confluence were washed with PBS and dissociated to a single cell suspension by incubation with StemPro Accutase (Invitrogen, Carlsbad, CA). At all intermediate steps of hPSCs collection, cells were kept on ice in mTeSR™1 medium containing 10 μg ml−1 of Y-27632 ROCK Inhibitor (Stemgent, Cambridge, MA). 0.7-1×106 of dissociated cells were mixed with concentrated viral medium (MOI=1-5) in 1ml of mTeSR™1 containing 10 μg ml−1 Y-27632 ROCK Inhibitor and 6μg ml−1 of polybrene (Sigma). hPSCs suspensions containing virus were plated on six-well matrigel-coated plates, and incubated for 12 hours in standard conditions (37°C, 5% CO2 , >95% humidity). After 12 hours, virus-containing medium was replaced with fresh mTeSR1 medium. On day 1 after transduction, regular mTeSR1 was replaced with basal mTeSR1 medium supplemented with cytokines SCF (100ng ml−1), TPO (50 ng ml−1) and bFGF (20 ng ml−1) (PeproTech, Rocky Hill, NJ). Since the presence of puromycin in differentiation cultures had a little effect on the outcomes of cellular transductions, hPSCs differentiation experiments were performed without antibiotic selection. Cells transduced with a single factor were maintained in indicated conditions from three to seven days, depending on their survival and growth, and collected for analysis (Supplementary Table 2). Cell transduced with different combinations of factors were collected after seven days of culture.
Nucleofection of human pluripotent stem cells with modified mRNAs
ETV2 and GATA2 modified messenger RNAs (mmRNAs) were kindly provided by NHLBI Progenitor Biology Consortium RNA Core (http://www.progenitorcells.org/content/requesting-rnacore-service). Nucleofection of H1 hESCs with modified messenger RNAs (mmRNAs) was performed using Amaxa Human Stem Cell Nucleofector® Kit 2 (Lonza). Prior to nucleofection, cells were washed with PBS and dissociated to a single cell suspension using StemPro Accutase (Invitrogen, Carlsbad, CA) as described above. For one nucleofection reaction 2×106 cells were resuspended in 100 μl of nucleofection reagent containing mmRNA (1.75 μg of both GATA2 and ETV2; 3.5 μg in total), transferred immediately to nucleofection cuvette and transfected using B-016 program on the Amaxa Nucleofector II (Lonza). After the procedure, cells were resuspended in 500 μl of mTeSR1 medium with Y-27632 ROCK inhibitor and transfered to matrigel coated six-well plates containing 2ml of mTeSR1 medium. After a 24 hours incubation, cells were collected and transfected a second time as described above. 24 hours after the second transfection, medium was change to growth factor-free basal TeSR1 medium containing SCF (100 ng ml−1), TPO (50 ng ml−1) and bFGF (20 ng ml−1).
Immunostaining procedures
Expression of cell-surface proteins was assessed by routine flow cytometry protocol (FACSCalibur, BD Biosciences). For intracellular staining by FACS, cells were fixed for 10 minutes at 37°C in BD Cytofix™ buffer (BD Biosciences), followed by permeabilzation for 30 minutes on ice in Perm Buffer III (BD Biosciences). After washing, cells were stained at 4°C for 2 hours with fluorescence-conjugated antibodies. For in situ staining, cell cultures were fixed with 4% paraformaldehyde, permeabilized with 0.05% of Triton X-100, and incubated overnight at 4°C with primary antibodies, followed by staining with the secondary fluorochrome-labeled antibodies. Intranuclear staining of pluripotency markers was performed by permeabilization with ice-cold 0.2% Triton X-100 in PBS. All antibodies used in this study are listed in Supplementary Table 4. Images acquired with an inverted Olympus IX71 microscope or Nikon Eclipse Ti-E configured with an A1R confocal system.
Hematopoietic Colony-forming Assay
Hematopoietic clonogenic assays were performed using cells collected at day 7 after lentiviral transduction and serum-containing methylcellulose media (MethoCult) supplemented with SCF, G-CSF, GM-CSF, IL3, IL6, and EPO (StemCell Technologies) according to the manufacture's protocol. Wright staining (Sigma-Aldrich) was used to evaluate the morphology of cells within colonies on cytospins.
Endothelial Assays
To assess endothelial potential, cells were collected on day 7 post-transduction and placed on fibronectin-coated six-well plates (hFibronectin, BD) supplemented with complete Endothelial Cell Medium ECM (ScienCell, Carlsbad, CA). Once cells achieve monolayer, they were evaluated for VE-cadherin expression by immunofluorescence, Ac-LDL uptake, and tube formation. For AcLDL uptake assay, cells growing in a monolayer were incubated with 10μm/ml of Alexa-594- or Alexa-488-conjugated AcLDL (Invitrogen) for 4 hours at 37°C followed by fluorescent microscopy or flow cytometric analysis. For vascular tube formation, 2×104 cells were resuspended in ECM medium (ScienCell) supplemented with 40 ng ml−1 VEGF
(PeproTech, Rocky Hill, NJ) and placed on matrigel matrix. Cells were incubated at 37°C, 5% CO2 for 18-24 hours when tube formation was observed.
Evaluation of hemogenic potential of induced endothelium
Cells transduced with TFs were collected at day 3 post-transduction and labeled with VE-cadherin, CD43, and CD73 antibodies. VE-cadherin+CD43−CD73− cells were sorted using FACSAria™ cell sorter and cultured in a 6 well plate (3×104 cell/per well) on OP9 monolayer in αMEM basal medium supplemented with 10% FBS (HyClone), 100mM monothioglycerol (MTG) and 50 μg ml−1 of ascorbic acid. After 2 weeks of culture, the formation of blood colonies in cultures was assessed using CD43 immunoflurorescent staining.
RNA-seq and Bioinformatics Analysis
Cells transduced with single TF were collected on day 3-7 post-transduction as indicated in Supplementary Table S2. Cells transduced with different TF combinations were collected on day 7 after transduction. Cord blood CD34+ cells were obtained from Cincinnati Children's Hospital Medical Center (CCHMC). Total RNA was isolated using RNeasy Micro Kit (Qiagen). Treatment with DNaseI was performed on the column according to the manufacture's protocol. Purity and integrity of RNA was estimated by the capillary electrophoresis on the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Samples were then prepared for sequencing using the Illumina TruSeq RNA Sample Preparation Kit v2 (RS-122-2001), according to the manufacturer's protocol. Final sample libraries were quantitated with the Life Technologies Qubit fluorometer and sequenced on the Illumina HiSeq 2500 (SY-401-1003-PRE). Base-calling and demultiplexing were done with the Illumina Genome Analyzer Casava Software, version 1.8.2. After quality assessment and filtering for adapter molecules and other sequencing artifacts, the remaining sequencing reads were aligned to 19084 RefSeq genes extracted from the Illumina iGenomes annotation, selecting only “NM_” designated genes. Bowtie v 0.12.9 was used, allowing two mismatches in a 28 bp seed, and excluding reads with more than 200 alignments.40 RSEM v 1.2.3 was used to estimate isoform or gene relative expression levels in units of “transcripts per million” (tpm), as well as “expected counts” (the non-normalized absolute number of reads assigned by RSEM to each isoform/gene).41,42 To determine differentially expressed genes between selected treated samples and H1 hESC controls, the expected counts data were analyzed using EBSeq. Genes with a posterior probability ≥ 0.99 were considered differentially expressed. Venn diagrams were constructed showing the overlap of gene sets identified in each of the treatment samples as “Up-regulated” with respect to the H1 controls. For GO analysis, each gene set was further reduced to include only genes with tpm≥10 in at least 1 sample. Enrichment of functional categories in the list of responsive genes was computed using the DAVID webservice (http://david.abcc.ncifcrf.gov/) 43. To visualize the significance of the overrepresentation, negative log10 of FDRs were plotted as bars, where FDR is computed by DAVID according to the Benjamini-Hotchberg method 44. To visualize the gene-expression levels, a heat-map was composed using MultiExperiment Viewer v4.2 (http://www.tm4.org). The visualization of global transcriptional signature of cells after transduction with single factor was performed using the Gene Expression Dynamics Inspector (GEDI) plots 45.
Time-lapse microscopy
To capture the endothelial-hematopoietic transition, the time-lapse movies were recorded using Nikon Eclipse Ti-E configured with an A1R confocal system and motorized stage (Nikon Instruments Inc. Melville, NY). Cell cultures were washed thoroughly to remove debris, and VE-cadherin-FITC and CD43-PE antibodies were added to a final concentration of 100 ng ml−1. Presented movies were made on day 3.5 post-transduction for GATA2/TAL1-induced cells and on day 4 for GATA2/ETV2-induced cells. Images were acquired using Nikon Elements (NIS-element C) imaging software for every 5 minutes with CFI Plan Fluor DLL 20x NA 05 WD 2.1 MM objective (Nikon Instruments Inc. Melville, NY). To convert time-lapse serial images to movies, the Quick-time movies and ImageJ (NIMH, Bethesda, MD) software were applied.
Supplementary Material
Acknowledgments
We thank Mitchell Probasco for cell sorting and Matt Raymond for editorial assistance. This work was supported by funds from the National Institute of Health (U01HL099773, U01HL100407, U01HL099997 and P51 RR000167) and The Charlotte Geyer Foundation.
Footnotes
Competing financial interests
J.A.T. owns stock in, serves on the Board of Directors of, and serves as Chief Scientific Officer of Cellular Dynamics International. I.S. owns stock in Cellular Dynamics International and Cynata. MV owns stock in Cynata. The remaining authors declare no competing financial interests.
Authors Contribution
IE conducted and analyzed experiments, interpreted experimental data, made figures and contributed to paper writing. VB conducted and analyzed experiments, performed GO analysis. AK performed time-lapse studies and analyzed data. PL, JL, and LT assisted with cloning and establishing library of TFs. MV analyzed molecular profiles of hematoendothelial subsets to prioritize genes for screening and contributed to initial design of gain-of-function screening system. RS and SW performed bioinformatics analysis of RNAseq data. MK contributed to design of GATA2/TAL1/LMO2 studies. EY and JC provided modified mRNA and assisted in developing optimal protocol for hESC transduction with mmRNA. JT contributed to concept development and directed molecular profiling studies. IS developed concept, led and supervised all aspects of the studies, analyzed and interpreted data, and wrote the paper.
Accession codes
The RNAseq data has been deposited in Gene Expression Omnibus under accession number GSE57395.
References
- 1.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. doi:10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godin I, Cumano A. The hare and the tortoise: an embryonic haematopoietic race. Nat Rev Immunol. 2002;2:593–604. doi: 10.1038/nri857. [DOI] [PubMed] [Google Scholar]
- 3.Lessard J, Faubert A, Sauvageau G. Genetic programs regulating HSC specification, maintenance and expansion. Oncogene. 2004;23:7199–7209. doi: 10.1038/sj.onc.1207940. [DOI] [PubMed] [Google Scholar]
- 4.Teitell MA, Mikkola HK. Transcriptional activators, repressors, and epigenetic modifiers controlling hematopoietic stem cell development. Pediatr Res. 2006;59:33R–39R. doi: 10.1203/01.pdr.0000205155.26315.c7. doi:10.1203/01.pdr.0000205155.26315.c7. [DOI] [PubMed] [Google Scholar]
- 5.Wilson NK, Calero-Nieto FJ, Ferreira R, Gottgens B. Transcriptional regulation of haematopoietic transcription factors. Stem Cell Res Ther. 2011;2:6. doi: 10.1186/scrt47. doi:10.1186/scrt47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson IJ, et al. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum Mol Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. Epub 2005 Jan 2013. [DOI] [PubMed] [Google Scholar]
- 7.Boisset JC, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. doi:nature08764 [pii] 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 8.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffredo T, Gautier R, Brajeul V, Dieterlen-Lievre F. Tracing the progeny of the aortic hemangioblast in the avian embryo. Dev Biol. 2000;224:204–214. doi: 10.1006/dbio.2000.9799. doi:10.1006/dbio.2000.9799 S0012-1606(00)99799-9 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Choi KD, et al. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:553–567. doi: 10.1016/j.celrep.2012.08.002. doi:10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vodyanik MA, et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 2010;7:718–729. doi: 10.1016/j.stem.2010.11.011. doi:S1934-5909(10)00633-8 [pii] 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 13.Barnes RM, Firulli BA, Conway SJ, Vincentz JW, Firulli AB. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev Dyn. 2010;239:3086–3097. doi: 10.1002/dvdy.22428. doi:10.1002/dvdy.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robb L, et al. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. Embo J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 15.Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meadows SM, Myers CT, Krieg PA. Regulation of endothelial cell development by ETS transcription factors. Semin Cell Dev Biol. 2011;22:976–984. doi: 10.1016/j.semcdb.2011.09.009. doi:10.1016/j.semcdb.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–433. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. doi:10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 18.Wong KS, Proulx K, Rost MS, Sumanas S. Identification of vasculature-specific genes by microarray analysis of Etsrp/Etv2 overexpressing zebrafish embryos. Dev Dyn. 2009;238:1836–1850. doi: 10.1002/dvdy.21990. doi:10.1002/dvdy.21990. [DOI] [PubMed] [Google Scholar]
- 19.Neuhaus H, Muller F, Hollemann T. Xenopus er71 is involved in vascular development. Dev Dyn. 2010;239:3436–3445. doi: 10.1002/dvdy.22487. doi:10.1002/dvdy.22487. [DOI] [PubMed] [Google Scholar]
- 20.Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115:5338–5346. doi: 10.1182/blood-2009-09-244640. doi:10.1182/blood-2009-09-244640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D, Kim T, Lim DS. The Er71 is an important regulator of hematopoietic stem cells in adult mice. Stem Cells. 2011;29:539–548. doi: 10.1002/stem.597. doi:10.1002/stem.597. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka H, et al. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. doi:10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- 23.Loughran SJ, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nature immunology. 2008;9:810–819. doi: 10.1038/ni.1617. doi:10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 24.Pimanda JE, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. Epub 12007 Oct 17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, et al. Enhanced hemangioblast generation and improved vascular repair and regeneration from embryonic stem cells by defined transcription factors. Stem cell reports. 2013;1:166–182. doi: 10.1016/j.stemcr.2013.06.005. doi:10.1016/j.stemcr.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi X, et al. Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev Biol. 2014;389:208–218. doi: 10.1016/j.ydbio.2014.02.018. doi:10.1016/j.ydbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson NK, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. doi:blood-2009-01-200048 [pii] 10.1182/blood-2009-01-200048 [doi] [DOI] [PubMed] [Google Scholar]
- 28.Elefanty AG, Begley CG, Hartley L, Papaevangeliou B, Robb L. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood. 1999;94:3754–3763. [PubMed] [Google Scholar]
- 29.D'Souza SL, Elefanty AG, Keller G. SCL/Tal-1 is essential for hematopoietic commitment of the hemangioblast but not for its development. Blood. 2005;105:3862–3870. doi: 10.1182/blood-2004-09-3611. Epub 2005 Jan 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gering M, Yamada Y, Rabbitts TH, Patient RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. doi: 10.1242/dev.00875. [DOI] [PubMed] [Google Scholar]
- 31.Yokomizo T, Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–3661. doi: 10.1242/dev.051094. doi:dev.051094 [pii] 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, et al. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell. 2012;11:663–675. doi: 10.1016/j.stem.2012.07.004. doi:10.1016/j.stem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Nakano H, et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat Commun. 2013;4:1564. doi: 10.1038/ncomms2569. doi:10.1038/ncomms2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Ferkowicz MJ, Johnson SA, Shelley WC, Yoder MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14:44–54. doi: 10.1089/scd.2005.14.44. [DOI] [PubMed] [Google Scholar]
- 36.Taoudi S, Medvinsky A. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc Natl Acad Sci U S A. 2007;104:9399–9403. doi: 10.1073/pnas.0700984104. doi:0700984104 [pii] 10.1073/pnas.0700984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen MJ, et al. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. doi:10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swiers G, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat Commun. 2013;4:2924. doi: 10.1038/ncomms3924. doi:10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira CF, et al. Induction of a Hemogenic Program in Mouse Fibroblasts. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.05.024. doi:10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:gb-2009-10-3-r25 [pii] 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. doi:10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. doi:btp692 [pii] 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang da W, et al. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 45.Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.