Abstract
Hippocampal volume loss has been related to chronic stress as well as genetic factors. Although genetic and environmental variables affecting hippocampal volume have extensively been studied and related to mental illness, limited evidence is available with respect to G × E interactions on hippocampal volume. The present MRI study investigated interaction effects on hippocampal volume between three well-studied functional genetic variants (COMT Val158Met, BDNF Val66Met, 5-HTTLPR) associated with hippocampal volume and a measure of environmental adversity (life events questionnaire) in a large sample of healthy humans (n = 153). All three variants showed significant interactions with environmental adversity with respect to hippocampal volume. Observed effects were additive by nature and driven by both recent as well as early life events. A consecutive analysis of hippocampal subfields revealed a spatially distinct profile for each genetic variant suggesting a specific role of 5-HTTLPR for the subiculum, BDNF Val66Met for CA4/dentate gyrus, and COMT Val158Met for CA2/3 volume changes. The present study underscores the importance of G × E interactions as determinants of hippocampal volume, which is crucial for the neurobiological understanding of stress-related conditions, such as mood disorders or post-traumatic stress disorder (PTSD).
Keywords: COMT, SLC6A4, BDNF, MRI, hippocampus, stress
Introduction
Hippocampal neuroplasticity is critical for cognitive plasticity, novelty learning, and individuality in humans and animals (Garthe et al., 2009; Sahay et al., 2011; Freund et al., 2013; Spalding et al., 2013) and further moderates the adaptation to environmental changes, a process that is intimately linked to hypothalamus-pituitary-adrenal (HPA) axis function (Snyder et al., 2011). Apart from providing negative feedback to the HPA axis, the hippocampus is also a major target of cortisol, which is highlighted by cortisol-mediated hippocampal volume loss after chronic stress exposure (Sapolsky et al., 1990; McEwen, 2001; Brown et al., 2004). The clinical importance of hippocampal volume loss has been shown by numerous studies investigating stress-related and heritable neuropsychiatric disorders, such as major depressive disorder (MDD), post-traumatic stress disorder (PTSD), and schizophrenia (Nelson et al., 1998; MacQueen and Frodl, 2011; Kühn and Gallinat, 2013). In addition to environmental adversity (Gianaros et al., 2007; Dannlowski et al., 2012), genetic variation is known to affect hippocampal volume (MacQueen and Frodl, 2011). Interestingly, several genetic variants known for their direct effects on hippocampal volume have been related to stress susceptibility suggesting the possibility of gene–environment (G × E) interactions at the neural level (Caspi and Moffitt, 2006). This applies specifically to three genetic variants impacting serotonin, dopamine, and neurotrophin signaling. 5-HTTLPR, a functional promoter polymorphism of the serotonin transporter gene (SLC6A4), modulates the relationship between environmental adversity and depression risk (Caspi et al., 2003) resulting in increased cortisol signaling and amygdala response to stressors in S carriers (Hariri et al., 2002; Canli et al., 2006; Miller et al., 2013). Similarly, BDNF Val66Met, a single nucleotide polymorphism (SNP) located in the brain-derived neurotrophic factor gene (BDNF), has been associated with stress susceptibility (Gatt et al., 2009; Alexander et al., 2010). Further, COMT Val158Met, a functional SNP located in the catechol-O-methyltransferase gene (COMT), has been implicated in HPA axis hyper-reactivity (Armbruster et al., 2012), altered μ-opioid neurotransmitter responses to pain stressors (Zubieta et al., 2003), and increased limbic reactivity (Smolka et al., 2005). In line with their effects on the stress system and brain function, these variants have repeatedly been shown to directly affect hippocampal volume (Pezawas et al., 2004; Frodl et al., 2008; Honea et al., 2009), although others failed to show these effects (Dutt et al., 2009; Cole et al., 2011; Molendijk et al., 2012). Such inconsistency is not surprising given the complex interplay between various genes and environmental factors that determine hippocampal volume. The nonconsideration of G × E interactions might be the leading reason for such inconclusive results, especially given that the functional role of the mentioned variants in stress reactivity provides a strong argument for their candidacy in G × E research (Moffitt et al., 2005).
To elucidate potential G × E interactions of these variants on the volume of the hippocampus and its subfields, which considerably vary with respect to their stress sensitivity (McEwen, 2001; Teicher et al., 2012), we conducted an MRI study in a large sample of healthy individuals.
Materials and Methods
Subjects.
Healthy subjects were recruited by online advertisements, announcements on bulletin boards, and word of mouth at two study sites (Division of Biological Psychiatry, Department of Psychiatry and Psychotherapy, Medical University of Vienna, Austria, and Department of Psychology, Dresden University of Technology, Dresden, Germany). All assessments were performed following the same standard procedure, which has been approved by the local ethics committee according to the Declaration of Helsinki (2008): Only right-handed native German speakers of European ancestry aged between 18 and 45 years were invited to participate in this study. Before inclusion, study protocol procedures have been fully explained to study participants before obtaining written informed consent. All participants were further financially compensated for their expenditure of time. At the screening day, subjects underwent a thorough physical examination, including electrocardiography, blood pressure measurement, and routine laboratory testing. Moreover, subjects underwent the Structured Clinical Interview for DSM-IV Axis I disorders (APA, 2000) to ascertain absence of any past or present psychiatric diagnosis except nicotine dependence. The final sample consisted of 153 subjects (21 from the Dresden study site) after exclusion of three subjects due to segmentation failure (female, age 31 years, COMT Val158Met: Met/Met, 5-HTTLPR: SA/LA, BDNF Val66Met: Val/Val; female, age 19 years, COMT Val158Met: Val/Met, 5-HTTLPR: LA/LA,BDNF Val66Met: Val/Met; male, age 27 years, COMT Val158Met: Val/Met, 5-HTTLPR: LA/LA, BDNF Val66Met: Val/Val) following visual quality control of the segmented structural images.
Behavioral measures.
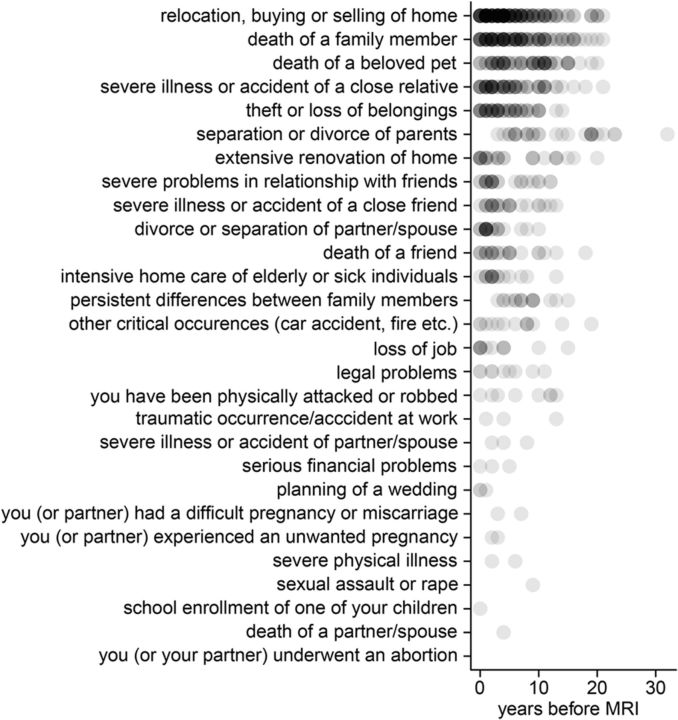
All subjects were asked to complete the life events questionnaire (LEQ), which is a short form of the life history calendar, a data collection method for obtaining reliable retrospective data about life events (LEs) and activities (Caspi et al., 1996; Canli et al., 2006). This self-report questionnaire comprises 28 stressful LEs (Fig. 1). Subjects were asked to indicate whether, when, and how often they had experienced a particular event. The sum of all LEs resulted in the total LEQ score. To test for effects of temperament, the German version of the Temperament and Character Inventory (Version 9) was applied (Cloninger et al., 1993).
Figure 1.
Type and timeline of LEs as reported by the sample (n = 153). LEs are sorted by number of occurrence.
Genotyping.
DNA extraction and genotyping were performed at the Department of Laboratory Medicine, Medical University of Vienna, Vienna, Austria. DNA was isolated from EDTA blood samples using the Magna Pure LC DNA Isolation Kit (Roche). A tetra-primer amplification refractory mutation system-PCR (ARMS-PCR) was used for COMT Val158Met (rs4680) genotyping following a previously published protocol (Ruiz-Sanz et al., 2007). All subjects were further genotyped for the SLC6A4 promoter variant (5-HTTLPR), including rs25531 following a previously published protocol (Wendland et al., 2006). Genotyping resulted in 24 SA/SA, 56 SA/LA, 12 SA/LG, 1 SG/LA, 12 LA/LG, and 48 LA/LA carriers. Because the LG allele equals the S allele with regard to 5-HTT expression, the LG allele was grouped together with the S allele (Hu et al., 2006) and is for simplicity referred to as S allele in the course of the manuscript. BDNF Val66Met (rs6265) genotyping was performed using a TaqMan (Applied Biosystems) 5′-nuclease assay. Sequence detection was accomplished in a 384-well format on an ABI 7900HT RT-PCR thermocycler (Applied Biosystems) applying 10 ng genomic DNA in a total volume of 10 μl consisting of 5 μl TaqMan Genotyping Mastermix (Applied Biosystems) and 0.25 μl of a 20× TaqMan Genotyping assay (Assay ID C_11592758_10, Applied Biosystems) containing sequence-specific primers and probes. PCR was performed under the following conditions: initial denaturation step (10 min, 95°C), followed by 40 cycles of DNA denaturation (15 s, 95°C) and oligonucleotide annealing/strand elongation (60 s, 60°C). Evaluation of data was realized using SDS 2.3 sequence detection software (Applied Biosytems). BDNF Val66Met genotyping resulted in 5 Met/Met, 52 Val/Met, and 96 Val/Val carriers. There was no indication for deviation from Hardy-Weinberg equilibrium for COMT Val158Met (p = 0.19), BDNF Val66Met (p = 0.64), 5-HTTLPR (p = 0.50), and rs25531 (p = 0.39).
MRI: acquisition.
3 Tesla (3T) TIM Trio scanners equipped with Siemens 12-channel head coils (Siemens Medical Solutions) were used for structural MRI measurements at both study sites using the same scan protocol and quality control procedures. Head movements were restricted using foam pillows. Structural images were acquired using a 3D magnetization-prepared rapid acquisition with a gradient echo sequence (3D MPRAGE, TR/TE = 2300/4.21 ms, flip angle = 9°, inversion time = 900 ms, and voxel size = 1 × 1 × 1.1 mm). Preprocessing: Anatomical MRI preprocessing was performed on a Linux computer (Red Hat Enterprise Linux 5, x86_64 architecture) using FreeSurfer version 5.1.0 (http://surfer.nmr.mgh.harvard.edu/), a set of automatic tools for morphological operations that require little or no human interventions. Preprocessing included registration to Talairach space, intensity normalization, removal of nonbrain tissue, and segmentation. Hippocampi were delineated by a subcortical segmentation procedure that assigns to each voxel one of 37 anatomical labels based on voxel intensity, intensity of neighboring voxels, and atlas-based prior probabilities (Fischl et al., 2002). Hippocampal subfields were defined by the use of Bayesian inference within a statistical model of image formation around the hippocampus (Van Leemput et al., 2009). Hippocampal volumes as well as subfields were corrected for total intracranial volume using the residual method (Sanfilipo et al., 2004).
Statistics.
To test for interaction effects between genotype and LEs, linear regression models were used with total hippocampal or hippocampal subfield volume as dependent variables and the interactions between COMT Val158Met, BDNF Val66Met, and 5-HTTLPR genotypes and LEs as independent variables. Age, gender, and study site were included as covariates. The following regressors for genotype variables were used: For COMT Val158Met, a linear allele-dose model was assumed by regressing the number of Met alleles (0, 1, 2) given the overwhelming evidence for dosing effects of COMT Val158Met (Smolka et al., 2005; Drabant et al., 2006; Domschke et al., 2012). 5-HTTLPR genotypes were collapsed into risk allele carriers (S allele carriers) and LA homozygotes in analogy to previous imaging genetics studies and coded as 0 (LA homozygotes) and 1 (S carriers) (Canli et al., 2006). Similarly, BDNF Val66Met genotypes were grouped into Met carriers (coded as 0) and Val homozygotes (coded as 1) as in previous studies (Kambeitz et al., 2012). A separate regression model, including the same regressors without the interaction terms, was used to assess potential main effects of genotype and LEs. Where applicable, the resulting p values were corrected for multiple comparisons using the Bonferroni–Holm method (Holm, 1979). Likelihood-ratio tests and Akaike information criterion were used for comparison of nested and non-nested models, respectively. To assess the variance explained by the interaction terms for each hippocampal subfield, we used the lmg metric (R2 partitioned by averaging over orders, provided in R package “relaimpo,” function “calc.relimp”) to break down R2 into shares from the individual regressors in analogy to a previous study (Teicher et al., 2012). All statistical analyses were conducted in R (version 2.15.2; http://cran.r-project.org/).
Results
Demographics
The sample consisted of 153 subjects (81 females, 72 males) with an average age of 23.79 ± 3.03 years (range: 18–43 years; percentiles: 25th: 22 years; 50th: 23 years; 75th: 25 years). Subjects were generally well educated as reflected by the percentage (98%) receiving 12 or more years of schooling. The distributions of 5-HTTLPR, BDNF Val66Met, and COMT Val158Met did not significantly differ between each other (all p > 0.05; Tables 1, 2, and 3). As shown in Tables 1, 2, and 3, genotype groups did not significantly vary with respect to age, gender, smoking status, or study site. Subjects reported on average 4.77 ± 3.14 LEs (range: 0–15 events; percentiles: 25th: 2 events; 50th: 5 events; 75th: 6 events), which occurred on average 5.75 ± 5.07 years before the MRI scan (range: 0–32 years; percentiles: 25th: 2 years; 50th: 4 years; 75th: 9 years). The type and timeline of all LEs in the whole sample are depicted in Figure 1. The number of LEs was significantly correlated with age (ρ = 0.18, p = 0.02). To obtain a measure of stressor intensity and to avoid correlated regressors, we corrected the number of LEs by age, resulting in an average of 0.20 ± 0.13 LEs per year. There was no significant difference with regard to LE between COMT Val158Met, BDNF Val66Met, and 5-HTTLPR groups (all p > 0.05; Tables 1, 2, and 3).
Table 1.
Distributions, sample sizes, means, and SDs according to COMT Val158Met genotypesa
| COMT Val158Met | ValVal | ValMet | MetMet | |
|---|---|---|---|---|
| N = 153 | N = 30 | N = 85 | N = 38 | p |
| Gender (female/male) | 20/10 | 43/42 | 18/20 | 0.23 |
| Age | 23.77 ± 2.37 | 23.84 ± 3.4 | 23.71 ± 2.64 | 0.98 |
| 5-HTTLPR (LALA/S carrier) | 9/21 | 29/56 | 9/29 | 0.50 |
| BDNF Val66Met (Met carrier/ValVal) | 10/20 | 28/57 | 19/19 | 0.17 |
| LE | 0.22 ± 0.14 | 0.18 ± 0.13 | 0.23 ± 0.13 | 0.10 |
| Study site (Vienna/Dresden) | 27/3 | 72/13 | 33/5 | 0.76 |
| Nonsmoker/smoker | 22/8 | 59/25 | 24/14 | 0.63 |
| Novelty seeking | 21.9 ± 5.18 | 21.53 ± 5.13 | 22 ± 5.19 | 0.88 |
| Harm avoidance | 14.23 ± 5.66 | 13.08 ± 5.48 | 12.58 ± 5.68 | 0.46 |
| Hippocampal volume (mm3) | 8113.79 ± 446.72 | 8322.59 ± 771.1 | 8328.82 ± 664.81 | 0.33 |
ap, p value of χ2 test or ANOVA between genotype groups. LE, Life events/year.
Table 2.
Distributions, sample sizes, means, and SDs according to BDNF Val66Met genotypesa
| BDNF Val66Met | ValVal | Met carriers | |
|---|---|---|---|
| N = 153 | N = 96 | N = 57 | p |
| Gender (female/male) | 53/43 | 28/29 | 0.57 |
| Age | 23.7 ± 3.36 | 23.95 ± 2.39 | 0.59 |
| 5-HTTLPR (LALA/S carrier) | 29/67 | 19/38 | 0.82 |
| COMT Val158Met | 19/57/20 | 19/28/10 | 0.17 |
| (MetMet/ValMet/ValVal) | |||
| LE | 0.2 ± 0.13 | 0.2 ± 0.13 | 0.86 |
| Study site (Vienna/Dresden) | 81/15 | 51/6 | 0.52 |
| Nonsmoker/smoker | 62/33 | 43/14 | 0.26 |
| Novelty seeking | 21.68 ± 4.79 | 21.79 ± 5.68 | 0.90 |
| Harm avoidance | 13.23 ± 5.38 | 13.11 ± 5.89 | 0.90 |
| Hippocampal volume (mm3) | 8322.51 ± 735.59 | 8216.98 ± 617.53 | 0.34 |
ap, p value of χ2 test or t test between genotype groups. LE, Life events/year.
Table 3.
Distributions, sample sizes, means, and SDs according to 5-HTTLPR genotypesa
| 5-HTTLPR | LALA | S carrier | |
|---|---|---|---|
| N = 153 | N = 48 | N = 105 | p |
| Gender (female/male) | 23/25 | 58/47 | 0.50 |
| Age | 23.21 ± 2.76 | 24.06 ± 3.12 | 0.09 |
| COMT Val158Met | 9/29/10 | 29/56/20 | 0.50 |
| (MetMet/ValMet/ValVal) | |||
| BDNF Val66Met (Met carrier/ValVal) | 19/29 | 38/67 | 0.82 |
| LE | 0.21 ± 0.13 | 0.2 ± 0.13 | 0.68 |
| Study site (Vienna/Dresden) | 40/8 | 92/13 | 0.64 |
| Nonsmoker/smoker | 35/13 | 70/34 | 0.61 |
| Novelty seeking | 22.35 ± 5.72 | 21.43 ± 4.83 | 0.33 |
| Harm avoidance | 13.56 ± 5.7 | 13.01 ± 5.51 | 0.58 |
| Hippocampal volume (mm3) | 8310.96 ± 608.05 | 8270.5 ± 732 | 0.72 |
ap, p value of χ2 test or t test between genotype groups. LE, Life events/year.
G × E effects on hippocampal volume
There were no significant main effects of COMT Val158Met, BDNF Val66Met, 5-HTTLPR, or LE on total hippocampal volume (Table 4; Fig. 2). However, each genotype exhibited a significant and independent interaction effect with LE indicating increased negative impact of LE on hippocampal volume in COMT Met homozygotes, BDNF Val homozygotes, or 5-HTTLPR S allele carriers (Table 4; Fig. 2). There was no evidence for statistical epistasis with regard to these interaction effects (LE × Val158Met × 5-HTTLPR: b = −0.13, SE = 0.17, t(140) = −0.76, p = 0.45; LE × Val158Met × Val66Met: b = 0.12, SE = 0.16, t(140) = 0.75, p = 0.45; LE × Val66Met × 5-HTTLPR: b = −0.19, SE = 0.36, t(140) = −0.53, p = 0.60). Given the independence of G × E interaction effects in previous analyses, we performed a post hoc analysis of the combined genetic effect on hippocampal volume. Based on above-mentioned results, we constructed a cumulative risk score (CRS) by summing up genetic risk factors (number of COMT Met alleles, 5-HTTLPR S allele, and BDNF Val/Val genotype) for each individual. CRS was highly predictive for hippocampal volume in interaction with LE (b = −0.41, SE = 0.08, t(146) = −4.99, p < 0.00001; Fig. 3A). Interestingly, this interaction exhibited a gradual effect on hippocampal volume ranging from volume increases to decreases dependent on the load of genetic risk (Fig. 3A). The more parsimonious CRS model explained nearly as much variance as the full model (full model: R2 = 0.19, Table 4; additive model: R2 = 0.18, F(6,146), p < 0.0001) and reduced the Akaike information criterion by 5.96 indicating improved model performance. Moreover, this additive effect was even detectable when analyzing sites separately (Vienna: n = 132, b = −0.42, SE = 0.10, t(126) = −4.10, p < 0.0001; Dresden: n = 21, b = −0.39, SE = 0.15, t(15) = −2.59, p = 0.021). The model fit, however, varied substantially: in the genetically more consistently defined groups, LE explained between 38% and 42% of hippocampal volume variance, notably in opposite directions (CRS = 1: n = 19, b = 0.69, s2 = 42%; CRS = 4: n = 15, b = −0.68, s2 = 38%), whereas only 1% and 5% variance was observed in the intermediate groups. We further performed several post hoc analyses to exclude effects of potential confounding variables on these G × E interaction effects (detailed statistics available upon request). There were no significant main or interaction effects of smoking status (n = 152, all p > 0.6), gender (all p > 0.5), harm avoidance, or novelty seeking (all p > 0.05). Additionally, including all possible covariate × environment and covariate × genotype interactions (similar to model 4 in Keller, 2014) revealed no significant differences to the parsimonious models (full model: F(12,130), p = 0.79, additive model: F(6,140), p = 0.64) and no change in significance or direction of effects. To explore whether these G × E interaction effects were driven by recent or early LEs, we conducted separate analyses restricted to specific developmental periods. Participants reported significantly less LEs during childhood (first 15 years of life) than during the last 5 years before the MRI scan (1.03 ± 1.12 vs 2.52 ± 2.07, t(152) = −8.29, p < 0.001). A restriction to the last 5 years showed a similar, but less significant, pattern compared with the full set of LEs (COMT: b = −0.23, SE = 0.09, t(142) = −2.54, p = 0.01, BDNF: b = −0.39, SE = 0.18, t(142) = −2.11, p = 0.04, SLC6A4: b = −0.32, SE = 0.17, t(142) = −1.89, p = 0.06, additive model: b = −0.33, SE = 0.09, t(142) = −3.76, p < 0.001). Similarly, a restriction to childhood revealed an almost identical, but also less significant, pattern (COMT: b = −0.17, SE = 0.08, t(142) = −2.19, p = 0.03, BDNF: b = −0.26, SE = 0.17, t(142) = −1.58, p = 0.12, SLC6A4: b = −0.28, SE = 0.17, t(142) = −1.63, p = 0.11, additive model: b = −0.25, SE = 0.07, t(142) = −3.34, p = 0.001). Nevertheless, both models explained major parts of variance in total hippocampal volume, but considerably less than the full model (last 5 years: R2 = 0.13, F(10,142) = 2.18, p = 0.023; childhood: R2 = 0.12, F(10,142) = 1.92, p = 0.047; LE: R2 = 0.19, F(10,142) = 3.40, p < 0.001).
Table 4.
Gene × LE interaction effects on hippocampal volumea
| Test for main effects |
Test for interaction effects |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| b | SE | t | p | b | SE | t | p | pcorr | |
| Constant | −0.15 | 0.20 | −0.76 | 0.451 | −0.22 | 0.19 | −1.15 | 0.250 | |
| Age | 0.02 | 0.08 | 0.19 | 0.853 | −0.06 | 0.08 | −0.78 | 0.439 | |
| Gender | 0.26 | 0.17 | 1.57 | 0.118 | 0.37 | 0.16 | 2.37 | 0.019 | |
| Study site | −0.36 | 0.24 | −1.50 | 0.135 | −0.61 | 0.24 | −2.58 | 0.011 | |
| COMT Val158Met | 0.10 | 0.08 | 1.19 | 0.237 | 0.11 | 0.08 | 1.44 | 0.151 | |
| 5-HTTLPR | −0.08 | 0.18 | −0.45 | 0.652 | 0.00 | 0.17 | −0.01 | 0.990 | |
| BDNF Val66Met | 0.21 | 0.17 | 1.27 | 0.207 | 0.22 | 0.16 | 1.42 | 0.158 | |
| LE | 0.03 | 0.08 | 0.37 | 0.709 | 0.70 | 0.19 | 3.78 | <0.001 | |
| LE × COMT Val158Met | −0.27 | 0.08 | −3.51 | <0.001 | 0.002 | ||||
| LE × SLC6A4 5-HTTLPR | −0.43 | 0.17 | −2.56 | 0.012 | 0.012 | ||||
| LE × BDNF Val66Met | −0.55 | 0.17 | −3.21 | 0.002 | 0.003 | ||||
| R2 = 0.05 | F(7,145) = 1.15 | p = 0.335 | R2 = 0.19 | F(10,142) = 3.40 | p < 0.001 | ||||
apcorr, p value corrected for multiple comparisons using the Bonferroni–Holm correction. LE, Life events/year.
Figure 2.
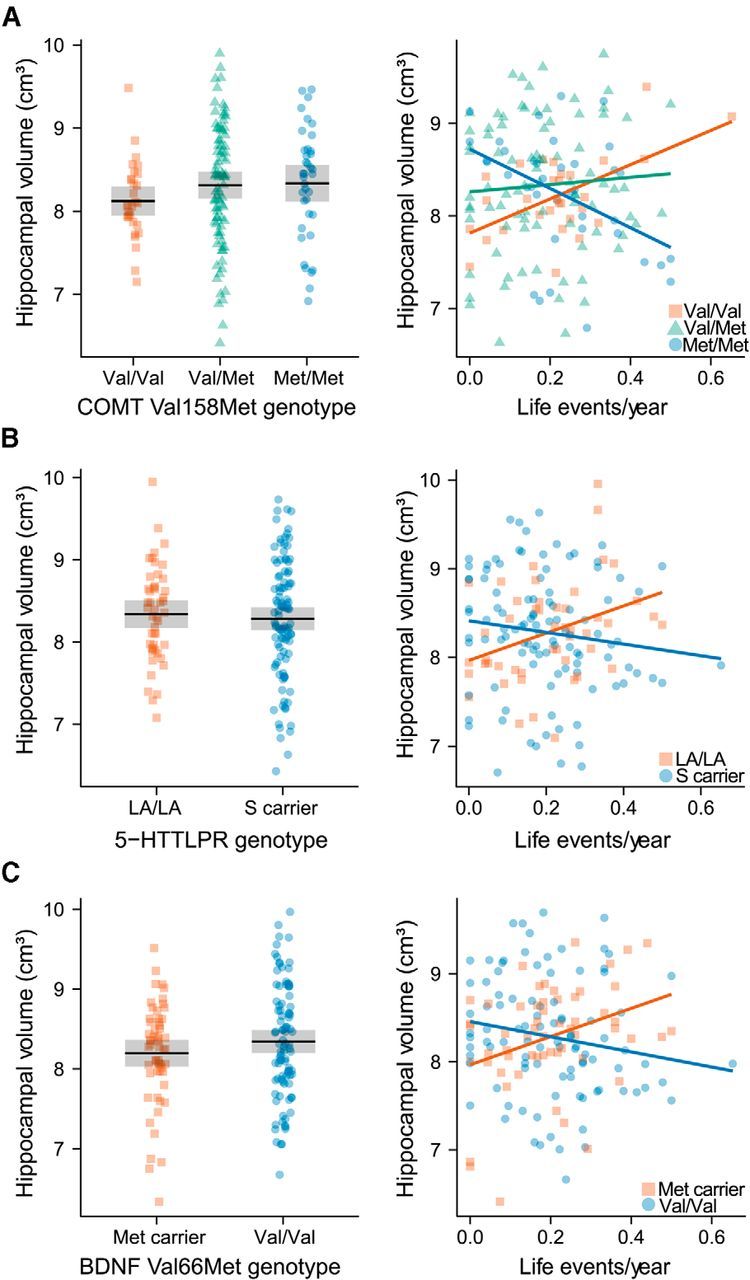
Main effects and gene × LE interaction effects of genetic variation in COMT, SLC6A4, and BDNF on hippocampal volume. Left panel side: Main effects of COMT Val158Met (A), 5-HTTLPR (B), and BDNF Val66Met (C) on hippocampal volume for 153 subjects (n per genotype group is shown in Tables 1, 2, and 3). Hippocampal volumes are corrected for intracranial volume as well as the remaining variables according to the main effects model described in Table 4. The gray box represents the 95% confidence interval. Right panel side: Interaction effects between LEs/year and COMT Val158Met (A), 5-HTTLPR (B), and BDNF Val66Met (C) on hippocampal volume for 153 subjects (n per genotype group is shown in Tables 1, 2, and 3). Hippocampal volumes are corrected for intracranial volume as well as the remaining variables according to the interaction effects model described in Table 4.
Figure 3.
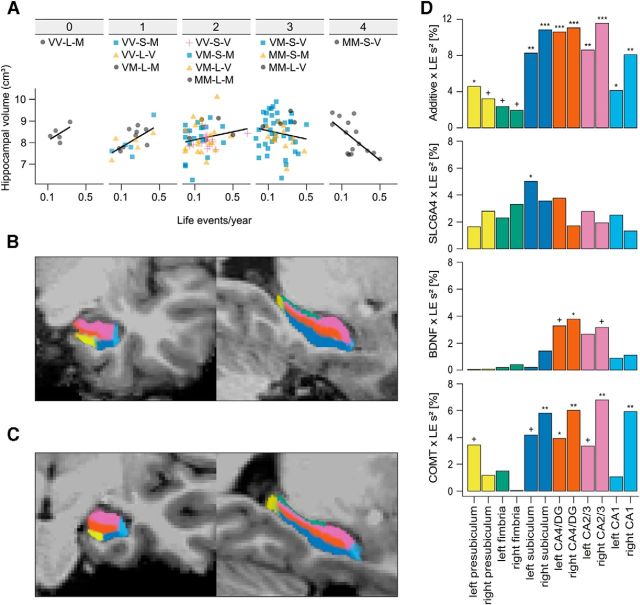
A, Interaction effect between additive genetic risk and LEs/year on hippocampal volume for 153 subjects. Scatter plots between LEs/year and hippocampal volume are shown separately for the number of genetic risk factors (COMT Met alleles, 5-HTTLPR S carrier, BDNF Val/Val carrier). The individual genotype groups are shown above. COMT Val158Met: Val/Val (VV), Val/Met (VM), Met/Met (MM); 5-HTTLPR: LA homozygotes (L), S carrier (S); BDNF Val66Met: Met carrier (M), Val homozygotes (V). B, Hippocampal subfield segmentation of a randomly drawn subject from the Dresden study site. The subfields are color-matched to Figure 3D. C, Hippocampal subfield segmentation of a randomly drawn subject from the Vienna study site. The subfields are color-matched to Figure 3D. D, Percent variance (s2) of hippocampal subfield volumes explained by the interaction effects between LEs/year and COMT Val158Met, BDNF Val66Met, 5-HTTLPR, and the additive risk score for 153 subjects. Bonferroni-Holm-corrected significance of the interaction effect is indicated as follows: +Corrected p < 0.1. *Corrected p < 0.05. **Corrected p < 0.01. ***Corrected p < 0.001. COMT, COMT Val158Met; BDNF, BDNF Val66Met; SLC6A4, 5-HTTLPR; LE, LEs/year.
G × E effects on hippocampal subfield volumes
To study observed G × E interaction effects in specific hippocampal substructures, we applied a novel method for hippocampal subfield segmentation (Fig. 3B,C) allowing for the distinction between presubiculum, subiculum, fimbria, CA1, CA2/3, and CA4/dentate gyrus (Van Leemput et al., 2009). Similar to total hippocampal volume, no main effects of genotype or LE on any of these subfields were present (all p > 0.05). In contrast, G × E interaction analyses revealed spatially distinct effects for single genetic variants that exhibited the same direction as being found for total hippocampal volume (Table 5; Fig. 3D). Interestingly, all genetic variants showed no significant effects at the presubiculum or fimbria, whereas most effects were present in subiculum and Ammon's horn (Table 5; Fig. 3D). In contrast to single G × E effects, the additive model exhibited effects across almost all subfields. Whereas only the right fimbria fell short of significance before multiple comparison correction, nine of the 12 subfields survived Bonferroni-Holm correction, namely, CA1, CA2/3, CA4/dentate gyrus, and subiculum on both hemispheres as well as the left presubiculum. Within these subfields, the interaction effect between LE and CRS explained between 4% and 12% of variance in volume with maximal effects present in right hemispheric regions implicated in neurogenesis and neuroplasticity, such as CA2/3 (12% explained variance) and CA4/dentate gyrus (11% explained variance, Table 5; Fig. 3D).
Table 5.
Gene × LE interaction effects on hippocampal subfieldsa
| Subfield | COMT Val158Met |
5-HTTLPR |
BDNF Val66Met |
Additive model |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| s2 | p | pcorr | s2 | p | pcorr | s2 | p | pcorr | s2 | p | pcorr | |
| L presubiculum | 0.03 | 0.020 | 0.099 | 0.02 | 0.094 | 0.285 | 0.00 | 0.696 | 1.000 | 0.05 | 0.007 | 0.036 |
| R presubiculum | 0.01 | 0.163 | 0.484 | 0.03 | 0.036 | 0.251 | 0.00 | 0.610 | 1.000 | 0.03 | 0.023 | 0.069 |
| L fimbria | 0.01 | 0.121 | 0.484 | 0.02 | 0.057 | 0.285 | 0.00 | 0.998 | 1.000 | 0.02 | 0.041 | 0.081 |
| R fimbria | 0.00 | 0.716 | 0.716 | 0.03 | 0.014 | 0.130 | 0.00 | 0.271 | 1.000 | 0.02 | 0.053 | 0.081 |
| L subiculum | 0.04 | 0.007 | 0.051 | 0.05 | 0.004 | 0.048 | 0.00 | 0.421 | 1.000 | 0.08 | <0.001 | 0.002 |
| R subiculum | 0.06 | 0.001 | 0.009 | 0.04 | 0.013 | 0.129 | 0.01 | 0.046 | 0.369 | 0.11 | <0.001 | <0.001 |
| L CA4/DG | 0.04 | 0.005 | 0.041 | 0.04 | 0.012 | 0.129 | 0.03 | 0.007 | 0.078 | 0.11 | <0.001 | <0.001 |
| R CA4/DG | 0.06 | 0.001 | 0.006 | 0.02 | 0.087 | 0.285 | 0.04 | 0.004 | 0.043 | 0.11 | <0.001 | <0.001 |
| L CA2/3 | 0.03 | 0.010 | 0.061 | 0.03 | 0.031 | 0.246 | 0.03 | 0.015 | 0.138 | 0.09 | <0.001 | 0.001 |
| R CA2/3 | 0.07 | <0.001 | 0.004 | 0.02 | 0.068 | 0.285 | 0.03 | 0.007 | 0.078 | 0.12 | <0.001 | <0.001 |
| L CA1 | 0.01 | 0.143 | 0.484 | 0.03 | 0.044 | 0.265 | 0.01 | 0.164 | 0.982 | 0.04 | 0.009 | 0.037 |
| R CA1 | 0.06 | 0.001 | 0.009 | 0.01 | 0.134 | 0.285 | 0.01 | 0.080 | 0.563 | 0.08 | <0.001 | 0.001 |
as2, Variance explained by the interaction effect between LEs and the specified polymorphism; pcorr, p value corrected for multiple comparisons using the Bonferroni–Holm correction; L, left; R, right; CA, cornu ammonis; DG, dentate gyrus.
Discussion
This study provides evidence for the presence of interaction effects between stressful LEs and three functional genetic variants on total hippocampal and hippocampal subfield volumes in healthy individuals in line with previous imaging, animal, and epidemiological evidence (McEwen, 2001; Caspi et al., 2003; Canli et al., 2006; Gianaros et al., 2007). These results highlight the need to model G × E interactions at the intermediate phenotype level to sufficiently map the intimate relationship between environmental and genetic variability (Caspi and Moffitt, 2006).
We observed that the impact of stressful LEs on hippocampal volume is significantly modulated by variation in COMT, BDNF, and SLC6A4 independent of gender, smoking, and temperament. Notably, these effects were driven by both recent and childhood events, suggesting that environmental effects on hippocampal volume occur throughout life and probably last for years (Teicher et al., 2012; Gray et al., 2013).
For COMT Val158Met, we observed a dose-dependent effect of the Met allele resulting in a gradual change from a positive to a negative correlation between LE and hippocampal volume in line with its previously described pleiotropic effects on brain function and behavior (Mier et al., 2010). Previous studies suggested that Met homozygosity strengthens prefrontal cognitive stability (Mier et al., 2010). However, this benefit comes with a trade-off of disadvantageous emotion-related information processing likely because of increased subcortical tonic and increased cortical phasic dopaminergic signaling (Bilder et al., 2004; Mier et al., 2010). Correspondingly, the Met allele has been associated with stress-related phenotypes, such as exaggerated limbic response to unpleasant stimuli (Smolka et al., 2005, 2007), HPA axis hyperreactivity (Armbruster et al., 2012), and increased pain sensitivity (Zubieta et al., 2003). This balance of costs and benefits suggests that each allele can be advantageous depending on the environmental context (“warrior vs worrier” model) in line with our data (Goldman et al., 2005). Interestingly, COMT Val158Met effects were most pronounced in CA1, CA2/3, CA4/dentate gyrus, and subiculum, which is in line with preclinical research suggesting disruptive effects of glucocorticoids and repeated stressors on neurogenesis or neuroplasticity in these hippocampal regions (Sousa et al., 2000; McEwen et al., 2012; MacDougall and Howland, 2013). The specific mechanisms of how dopamine affects hippocampal volume are still obscure but may involve alterations of the dopamine-modulated stress response that translate to changes of HPA axis function (Armbruster et al., 2012; Hernaus et al., 2013). Moreover, given the high hippocampal COMT expression, also direct effects on local dopamine signaling and hippocampal plasticity may be possible (Matsumoto et al., 2003; Lisman et al., 2011; Laatikainen et al., 2012).
G × E interactions on hippocampal volume were also present for BDNF Val66Met in this study. The Val allele, which drove hippocampal volume loss in our data, has shown to promote social defeat stress susceptibility in mouse models because of increased BDNF signaling within the reward circuitry (Krishnan et al., 2007). In humans, Val/Val individuals have been linked to increased neuroticism (Frustaci et al., 2008), diminished antidepressant response (Niitsu et al., 2013), and heightened stress vulnerability (Yu et al., 2012; Jiang et al., 2013). Interestingly, we found maximal effects of BDNF Val66Met in CA4/dentate gyrus, where BDNF loss has been associated with depressive behavior and attenuated antidepressant efficacy (Adachi et al., 2008; Taliaz et al., 2010). However, this region is also highly susceptible to stressor-induced glucocorticoid signaling (Karst and Joels, 2003). It is therefore possible that an exaggerated stress response in Val/Val individuals (because of increased BDNF signaling in the mesolimbic dopamine circuit) diminishes the positive effect of increased BDNF signaling in the hippocampus (Krishnan et al., 2007; Alexander et al., 2010).
Furthermore, hippocampal volume loss was mediated by the S allele of 5-HTTLPR in our analyses in line with previous reports and highlighting the specific role of this allele in stress susceptibility (Hariri et al., 2002; Caspi et al., 2003; Canli et al., 2006; Frodl et al., 2010). 5-HTTLPR has been extensively studied and is known to alter serotonergic neurotransmission as well as brain development (Gaspar et al., 2003; Fisher et al., 2012; Migliarini et al., 2013). Accordingly, the observed effects could be driven by both direct and indirect serotonergic effects, including 5-HTTLPR-mediated alterations of HPA axis activity or direct effects of serotonin on neuroplasticity (Martinowich and Lu, 2008; Klempin et al., 2013; Miller et al., 2013). In this study, 5-HTTLPR exerted maximal effects in the subiculum, which is the principal hippocampal relay for HPA axis control and exhibits neuroplastic changes in response to stress (MacDougall and Howland, 2013). Interestingly, the subiculum is also densely packed with 5-HT1A and 5-HT1B receptors, which are involved in the serotonergic antidepressant response (Boeijinga and Boddeke, 1996; Hall et al., 1997; Cai et al., 2013).
There was no evidence for gene–gene interactions in our data, which may be understandable given that these genes are involved in distinct molecular pathways (Hemani et al., 2014). Instead, most of the individual variance was captured by a simple additive model suggesting that the observed G × E effects on hippocampal volume accumulate similar to other stress-related endophenotypes (Smolka et al., 2007; Stone et al., 2013) or quantitative traits (Hill et al., 2008; Yang et al., 2010). Notably, this additive effect predominantly affected hippocampal subfields that have been reported to be specifically vulnerable to childhood maltreatment, glucocorticoids, or stress in general (Sousa et al., 2000; McEwen et al., 2012; Teicher et al., 2012; MacDougall and Howland, 2013).
Interestingly, subjects at low genetic risk exhibited effects that were diametrically opposite to subjects at high genetic risk in our study. Although mathematically obvious, the biological meaning of this finding is less intuitive. Even so, a significant body of evidence exists that implicates factors promoting well-being in hippocampal growth (Kempermann et al., 1997; Pollak et al., 2008; Erickson et al., 2011; Davidson and McEwen, 2012; Arnone et al., 2013). Moreover, resilience, coping behavior, predictable stress, and low-dose glucocorticoids have been reported to stimulate hippocampal neurogenesis and neuroplasticity (Jeanneteau et al., 2008; Lyons et al., 2010; Schloesser et al., 2010; Delgado y Palacios et al., 2011; Parihar et al., 2011; Chen et al., 2012). It is therefore tempting to speculate that more beneficial stress coping behavior in individuals at lower genetic risk may have led to positive associations in our data. However, it needs to be emphasized that this is highly speculative given the correlative nature of our data and unresolved questions with respect to stress-related hippocampal changes (Czéh and Lucassen, 2007; Petrik et al., 2012). Nonetheless, such diametrically opposite effects may explain the lack of a significant main effect of LEs in several large healthy samples, including this study (Dannlowski et al., 2012; Luby et al., 2012).
The present study is not without limitations. The investigated sample is healthy, highly educated, and fairly homogeneous with regard to age, thereby controlling for major confounders, which might have been advantageous for the detection of subtle effects (Uher and McGuffin, 2008). However, this limits, on the other hand, the generalizability of our results, which especially cannot be extrapolated to patients. Subjects participating in this study reported considerably more recent than early LEs, which may reflect both the difficulty of recalling childhood memories as well as the different landscape of childhood stress (Howe, 2013). The reported diminished significance for early LEs could therefore be attributed to difficulties in memory recall rather than to lower G × E effects on hippocampal volume per se. It would therefore be interesting to further assess these interactions with regard to early life stressors (Teicher et al., 2012). Furthermore, there are other genetic variants in genes, such as FKBP5, CRHR1, NR3C2, or KIBRA, that have been associated with stress vulnerability or hippocampal structure (Mandelli and Serretti, 2013). However, the study of a higher number of variants would unlikely have led to meaningful results given the expected small effects and the requirement for rigorous Type I error control. We therefore focused on three functional variants that have been related to hippocampal structure and, most importantly, stress reactivity by numerous studies providing strong a priori support for the investigated effects (Moffitt et al., 2005). Another limitation is the lack of functional measures of stress responsiveness in our sample, which would have allowed for assessing the potential mediatory effect of HPA axis reactivity. However, the impact of these genotypes on stress reactivity has repeatedly been demonstrated before and was therefore beyond the scope of this study. Finally, pharmacological or behavioral stress tests offer the possibility of standardized stressors that are free of any recall bias but are not suited to study the long-term consequences of real life stressors as intended in our study (Pilger et al., 2014).
In conclusion, this study provides evidence of interaction effects between three well-studied functional genetic variants and environmental adversity on hippocampal anatomy, thereby resembling previous animal, molecular biology, genetic, and imaging work. These results highlight the importance of G × E interactions in imaging studies, which should facilitate a better understanding of the complex interplay between genes and the environment in future research.
Footnotes
This work was supported by the Special Research Project SFB-35 (F3514-B11 and F3506-B11) of the Austrian Science Fund, the Oesterreichische Nationalbank (OeNB 11903), and the Institute for the Study of Affective Neuroscience. U.R. was further supported by an award of the Theodor Körner Fonds. J.-C.D. was supported by the LABEX ANR-11-LABEX-0042 of Université de Lyon, within the program “Investissements d'Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency. We thank Kristin Anacker, Department of Psychology, Dresden University of Technology, Germany, for sharing expertise on the life events questionnaire; Pia Auersperg, Anastasia Gudakovskaja, Viktoria Köller, Elisabeth Kühtreiber, Franziska Mayr, Helge Oswald, Lisa Ott, Sophia Petschnak, Matthias Pilgerstorfer, Sebastian Ribar, and Marie Janina Schwidde for support of study participant recruitment and evaluation; Christian Kasess, Wolfgang Huf, and Roland Boubela for technical advice; and the anonymous reviewers for their suggestion to analyze effects of multiple genetic variants and other thoughtful comments.
The authors declare no competing financial interests.
References
- Adachi M, Barrot M, Autry AE, Theobald D, Monteggia LM. Selective loss of brain-derived neurotrophic factor in the dentate gyrus attenuates antidepressant efficacy. Biol Psychiatry. 2008;63:642–649. doi: 10.1016/j.biopsych.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander N, Osinsky R, Schmitz A, Mueller E, Kuepper Y, Hennig J. The BDNF Val66Met polymorphism affects HPA-axis reactivity to acute stress. Psychoneuroendocrinology. 2010;35:949–953. doi: 10.1016/j.psyneuen.2009.12.008. [DOI] [PubMed] [Google Scholar]
- APA. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- Armbruster D, Mueller A, Strobel A, Lesch KP, Brocke B, Kirschbaum C. Children under stress: COMT genotype and stressful life events predict cortisol increase in an acute social stress paradigm. Int J Neuropsychopharmacol. 2012;15:1229–1239. doi: 10.1017/S1461145711001763. [DOI] [PubMed] [Google Scholar]
- Arnone D, McKie S, Elliott R, Juhasz G, Thomas EJ, Downey D, Williams S, Deakin JF, Anderson IM. State-dependent changes in hippocampal grey matter in depression. Mol Psychiatry. 2012;18:1265–1272. doi: 10.1038/mp.2012.150. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004;29:1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- Boeijinga PH, Boddeke HW. Activation of 5-HT1B receptors suppresses low but not high frequency synaptic transmission in the rat subicular cortex in vitro. Brain Res. 1996;721:59–65. doi: 10.1016/0006-8993(96)00149-7. [DOI] [PubMed] [Google Scholar]
- Brown ES, Woolston DJ, Frol A, Bobadilla L, Khan DA, Hanczyc M, Rush AJ, Fleckenstein J, Babcock E, Cullum CM. Hippocampal volume, spectroscopy, cognition, and mood in patients receiving corticosteroid therapy. Biol Psychiatry. 2004;55:538–545. doi: 10.1016/j.biopsych.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Cai X, Kallarackal AJ, Kvarta MD, Goluskin S, Gaylor K, Bailey AM, Lee HK, Huganir RL, Thompson SM. Local potentiation of excitatory synapses by serotonin and its alteration in rodent models of depression. Nat Neurosci. 2013;16:464–472. doi: 10.1038/nn.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli T, Qiu M, Omura K, Congdon E, Haas BW, Amin Z, Herrmann MJ, Constable RT, Lesch KP. Neural correlates of epigenesis. Proc Natl Acad Sci U S A. 2006;103:16033–16038. doi: 10.1073/pnas.0601674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Moffitt TE. Gene–environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- Caspi A, Moffitt TE, Thornton A, Freedman D, Amell JW, Harrington H, Smeijers J, Silva PA. The life history calendar: a research and clinical assessment method for collecting retrospective event-history data. Int J Method Psych. 1996;6:101–114. doi: 10.1002/(SICI)1234-988X(199607)6:2<101::AID-MPR156>3.3.CO%3B2-E. [DOI] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chen DY, Bambah-Mukku D, Pollonini G, Alberini CM. Glucocorticoid receptors recruit the CaMKIIalpha-BDNF-CREB pathways to mediate memory consolidation. Nat Neurosci. 2012;15:1707–1714. doi: 10.1038/nn.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Svrakic DM, Przybeck TR. A psychobiological model of temperament and character. Arch Gen Psychiatry. 1993;50:975–990. doi: 10.1001/archpsyc.1993.01820240059008. [DOI] [PubMed] [Google Scholar]
- Cole J, Weinberger DR, Mattay VS, Cheng X, Toga AW, Thompson PM, Powell-Smith G, Cohen-Woods S, Simmons A, McGuffin P, Fu CH. No effect of 5HTTLPR or BDNF Val66Met polymorphism on hippocampal morphology in major depression. Genes Brain Behav. 2011;10:756–764. doi: 10.1111/j.1601-183X.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czéh B, Lucassen PJ. What causes the hippocampal volume decrease in depression? Are neurogenesis, glial changes and apoptosis implicated? Eur Arch Psychiatry Clin Neurosci. 2007;257:250–260. doi: 10.1007/s00406-007-0728-0. [DOI] [PubMed] [Google Scholar]
- Dannlowski U, Stuhrmann A, Beutelmann V, Zwanzger P, Lenzen T, Grotegerd D, Domschke K, Hohoff C, Ohrmann P, Bauer J, Lindner C, Postert C, Konrad C, Arolt V, Heindel W, Suslow T, Kugel H. Limbic scars: long-term consequences of childhood maltreatment revealed by functional and structural magnetic resonance imaging. Biol Psychiatry. 2012;71:286–293. doi: 10.1016/j.biopsych.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Davidson RJ, McEwen BS. Social influences on neuroplasticity: stress and interventions to promote well-being. Nat Neurosci. 2012;15:689–695. doi: 10.1038/nn.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado y Palacios R, Campo A, Henningsen K, Verhoye M, Poot D, Dijkstra J, Van Audekerke J, Benveniste H, Sijbers J, Wiborg O, Van der Linden A. Magnetic resonance imaging and spectroscopy reveal differential hippocampal changes in anhedonic and resilient subtypes of the chronic mild stress rat model. Biol Psychiatry. 2011;70:449–457. doi: 10.1016/j.biopsych.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Domschke K, Baune BT, Havlik L, Stuhrmann A, Suslow T, Kugel H, Zwanzger P, Grotegerd D, Sehlmeyer C, Arolt V, Dannlowski U. Catechol-O-methyltransferase gene variation: impact on amygdala response to aversive stimuli. Neuroimage. 2012;60:2222–2229. doi: 10.1016/j.neuroimage.2012.02.039. [DOI] [PubMed] [Google Scholar]
- Drabant EM, Hariri AR, Meyer-Lindenberg A, Munoz KE, Mattay VS, Kolachana BS, Egan MF, Weinberger DR. Catechol O-methyltransferase val158met genotype and neural mechanisms related to affective arousal and regulation. Arch Gen Psychiatry. 2006;63:1396–1406. doi: 10.1001/archpsyc.63.12.1396. [DOI] [PubMed] [Google Scholar]
- Dutt A, McDonald C, Dempster E, Prata D, Shaikh M, Williams I, Schulze K, Marshall N, Walshe M, Allin M, Collier D, Murray R, Bramon E. The effect of COMT, BDNF, 5-HTT, NRG1 and DTNBP1 genes on hippocampal and lateral ventricular volume in psychosis. Psychol Med. 2009;39:1783–1797. doi: 10.1017/S0033291709990316. [DOI] [PubMed] [Google Scholar]
- Erickson KI, Voss MW, Prakash RS, Basak C, Szabo A, Chaddock L, Kim JS, Heo S, Alves H, White SM, Wojcicki TR, Mailey E, Vieira VJ, Martin SA, Pence BD, Woods JA, McAuley E, Kramer AF. Exercise training increases size of hippocampus and improves memory. Proc Natl Acad Sci U S A. 2011;108:3017–3022. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- Fisher PM, Holst KK, McMahon B, Haahr ME, Madsen K, Gillings N, Baaré WF, Jensen PS, Knudsen GM. 5-HTTLPR status predictive of neocortical 5-HT4 binding assessed with [(11)C]SB207145 PET in humans. Neuroimage. 2012;62:130–136. doi: 10.1016/j.neuroimage.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Freund J, Brandmaier AM, Lewejohann L, Kirste I, Kritzler M, Krüger A, Sachser N, Lindenberger U, Kempermann G. Emergence of individuality in genetically identical mice. Science. 2013;340:756–759. doi: 10.1126/science.1235294. [DOI] [PubMed] [Google Scholar]
- Frodl T, Koutsouleris N, Bottlender R, Born C, Jäger M, Mörgenthaler M, Scheuerecker J, Zill P, Baghai T, Schüle C, Rupprecht R, Bondy B, Reiser M, Möller HJ, Meisenzahl EM. Reduced gray matter brain volumes are associated with variants of the serotonin transporter gene in major depression. Mol Psychiatry. 2008;13:1093–1101. doi: 10.1038/mp.2008.62. [DOI] [PubMed] [Google Scholar]
- Frodl T, Reinhold E, Koutsouleris N, Donohoe G, Bondy B, Reiser M, Möller HJ, Meisenzahl EM. Childhood stress, serotonin transporter gene and brain structures in major depression. Neuropsychopharmacology. 2010;35:1383–1390. doi: 10.1038/npp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frustaci A, Pozzi G, Gianfagna F, Manzoli L, Boccia S. Meta-analysis of the brain-derived neurotrophic factor gene (BDNF) Val66Met polymorphism in anxiety disorders and anxiety-related personality traits. Neuropsychobiology. 2008;58:163–170. doi: 10.1159/000182892. [DOI] [PubMed] [Google Scholar]
- Garthe A, Behr J, Kempermann G. Adult-generated hippocampal neurons allow the flexible use of spatially precise learning strategies. PLoS One. 2009;4:e5464. doi: 10.1371/journal.pone.0005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Cases O, Maroteaux L. The developmental role of serotonin: news from mouse molecular genetics. Nat Rev Neurosci. 2003;4:1002–1012. doi: 10.1038/nrn1256. [DOI] [PubMed] [Google Scholar]
- Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant RA, Schofield PR, Gordon E, Kemp AH, Williams LM. Interactions between BDNF Val66Met polymorphism and early life stress predict brain and arousal pathways to syndromal depression and anxiety. Mol Psychiatry. 2009;14:681–695. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- Gianaros PJ, Jennings JR, Sheu LK, Greer PJ, Kuller LH, Matthews KA. Prospective reports of chronic life stress predict decreased grey matter volume in the hippocampus. Neuroimage. 2007;35:795–803. doi: 10.1016/j.neuroimage.2006.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.175. doi: 10.1038/mp.2013.175. Advance online publication. Retrieved Dec. 17, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall H, Lundkvist C, Halldin C, Farde L, Pike VW, McCarron JA, Fletcher A, Cliffe IA, Barf T, Wikström H, Sedvall G. Autoradiographic localization of 5-HT1A receptors in the post-mortem human brain using [3H]WAY-100635 and [11C]way-100635. Brain Res. 1997;745:96–108. doi: 10.1016/S0006-8993(96)01131-6. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Hemani G, Shakhbazov K, Westra HJ, Esko T, Henders AK, McRae AF, Yang J, Gibson G, Martin NG, Metspalu A, Franke L, Montgomery GW, Visscher PM, Powell JE. Detection and replication of epistasis influencing transcription in humans. Nature. 2014;508:249–253. doi: 10.1038/nature13005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hernaus D, Collip D, Lataster J, Ceccarini J, Kenis G, Booij L, Pruessner J, Van Laere K, van Winkel R, van Os J, Myin-Germeys I. COMT Val158Met genotype selectively alters prefrontal [18F]fallypride displacement and subjective feelings of stress in response to a psychosocial stress challenge. PLoS One. 2013;8:e65662. doi: 10.1371/journal.pone.0065662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Honea R, Verchinski BA, Pezawas L, Kolachana BS, Callicott JH, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Impact of interacting functional variants in COMT on regional gray matter volume in human brain. Neuroimage. 2009;45:44–51. doi: 10.1016/j.neuroimage.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe ML. Memory development: implications for adults recalling childhood experiences in the courtroom. Nat Rev Neurosci. 2013;14:869–876. doi: 10.1038/nrn3627. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, Goldman D. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneteau F, Garabedian MJ, Chao MV. Activation of Trk neurotrophin receptors by glucocorticoids provides a neuroprotective effect. Proc Natl Acad Sci U S A. 2008;105:4862–4867. doi: 10.1073/pnas.0709102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Brummett BH, Babyak MA, Siegler IC, Williams RB. Brain-derived neurotrophic factor (BDNF) Val66Met and adulthood chronic stress interact to affect depressive symptoms. J Psychiatr Res. 2013;47:233–239. doi: 10.1016/j.jpsychires.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambeitz JP, Bhattacharyya S, Kambeitz-Ilankovic LM, Valli I, Collier DA, McGuire P. Effect of BDNF val(66)met polymorphism on declarative memory and its neural substrate: a meta-analysis. Neurosci Biobehav Rev. 2012;36:2165–2177. doi: 10.1016/j.neubiorev.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Karst H, Joëls M. Effect of chronic stress on synaptic currents in rat hippocampal dentate gyrus neurons. J Neurophysiol. 2003;89:625–633. doi: 10.1152/jn.00691.2002. [DOI] [PubMed] [Google Scholar]
- Keller MC. Gene × environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Kühn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- Klempin F, Beis D, Mosienko V, Kempermann G, Bader M, Alenina N. Serotonin is required for exercise-induced adult hippocampal neurogenesis. J Neurosci. 2013;33:8270–8275. doi: 10.1523/JNEUROSCI.5855-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, Laplant Q, Graham A, Lutter M, Lagace DC, Ghose S, Reister R, Tannous P, Green TA, Neve RL, Chakravarty S, Kumar A, Eisch AJ, Self DW, Lee FS, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Kühn S, Gallinat J. Gray matter correlates of posttraumatic stress disorder: a quantitative meta-analysis. Biol Psychiatry. 2013;73:70–74. doi: 10.1016/j.biopsych.2012.06.029. [DOI] [PubMed] [Google Scholar]
- Laatikainen LM, Sharp T, Bannerman DM, Harrison PJ, Tunbridge EM. Modulation of hippocampal dopamine metabolism and hippocampal-dependent cognitive function by catechol-O-methyltransferase inhibition. J Psychopharmacol. 2012;26:1561–1568. doi: 10.1177/0269881112454228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Grace AA, Duzel E. A neoHebbian framework for episodic memory: role of dopamine-dependent late LTP. Trends Neurosci. 2011;34:536–547. doi: 10.1016/j.tins.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby JL, Barch DM, Belden A, Gaffrey MS, Tillman R, Babb C, Nishino T, Suzuki H, Botteron KN. Maternal support in early childhood predicts larger hippocampal volumes at school age. Proc Natl Acad Sci U S A. 2012;109:2854–2859. doi: 10.1073/pnas.1118003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DM, Buckmaster PS, Lee AG, Wu C, Mitra R, Duffey LM, Buckmaster CL, Her S, Patel PD, Schatzberg AF. Stress coping stimulates hippocampal neurogenesis in adult monkeys. Proc Natl Acad Sci U S A. 2010;107:14823–14827. doi: 10.1073/pnas.0914568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall MJ, Howland JG. Acute stress, but not corticosterone, disrupts short- and long-term synaptic plasticity in rat dorsal subiculum via glucocorticoid receptor activation. Cereb Cortex. 2013;23:2611–2619. doi: 10.1093/cercor/bhs247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen G, Frodl T. The hippocampus in major depression: evidence for the convergence of the bench and bedside in psychiatric research? Mol Psychiatry. 2011;16:252–264. doi: 10.1038/mp.2010.80. [DOI] [PubMed] [Google Scholar]
- Mandelli L, Serretti A. Gene environment interaction studies in depression and suicidal behavior: an update. Neurosci Biobehav Rev. 2013;37:2375–2397. doi: 10.1016/j.neubiorev.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Lu B. Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology. 2008;33:73–83. doi: 10.1038/sj.npp.1301571. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Weickert CS, Akil M, Lipska BK, Hyde TM, Herman MM, Kleinman JE, Weinberger DR. Catechol O-methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience. 2003;116:127–137. doi: 10.1016/S0306-4522(02)00556-0. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Plasticity of the hippocampus: adaptation to chronic stress and allostatic load. Ann N Y Acad Sci. 2001;933:265–277. doi: 10.1111/j.1749-6632.2001.tb05830.x. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62:3–12. doi: 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mier D, Kirsch P, Meyer-Lindenberg A. Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Mol Psychiatry. 2010;15:918–927. doi: 10.1038/mp.2009.36. [DOI] [PubMed] [Google Scholar]
- Migliarini S, Pacini G, Pelosi B, Lunardi G, Pasqualetti M. Lack of brain serotonin affects postnatal development and serotonergic neuronal circuitry formation. Mol Psychiatry. 2013;18:1106–1118. doi: 10.1038/mp.2012.128. [DOI] [PubMed] [Google Scholar]
- Miller R, Wankerl M, Stalder T, Kirschbaum C, Alexander N. The serotonin transporter gene-linked polymorphic region (5-HTTLPR) and cortisol stress reactivity: a meta-analysis. Mol Psychiatry. 2013;18:1018–1024. doi: 10.1038/mp.2012.124. [DOI] [PubMed] [Google Scholar]
- Moffitt TE, Caspi A, Rutter M. Strategy for investigating interactions between measured genes and measured environments. Arch Gen Psychiatry. 2005;62:473–481. doi: 10.1001/archpsyc.62.5.473. [DOI] [PubMed] [Google Scholar]
- Molendijk ML, Bus BA, Spinhoven P, Kaimatzoglou A, Oude Voshaar RC, Penninx BW, van IJzendoorn MH, Elzinga BM. A systematic review and meta-analysis on the association between BDNF val(66)met and hippocampal volume: a genuine effect or a winners curse? Am J Med Genetics. 2012;159B:731–740. doi: 10.1002/ajmg.b.32078. [DOI] [PubMed] [Google Scholar]
- Nelson MD, Saykin AJ, Flashman LA, Riordan HJ. Hippocampal volume reduction in schizophrenia as assessed by magnetic resonance imaging: a meta-analytic study. Arch Gen Psychiatry. 1998;55:433–440. doi: 10.1001/archpsyc.55.5.433. [DOI] [PubMed] [Google Scholar]
- Niitsu T, Fabbri C, Bentini F, Serretti A. Pharmacogenetics in major depression: a comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45:183–194. doi: 10.1016/j.pnpbp.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Parihar VK, Hattiangady B, Kuruba R, Shuai B, Shetty AK. Predictable chronic mild stress improves mood, hippocampal neurogenesis and memory. Mol Psychiatry. 2011;16:171–183. doi: 10.1038/mp.2009.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik D, Lagace DC, Eisch AJ. The neurogenesis hypothesis of affective and anxiety disorders: are we mistaking the scaffolding for the building? Neuropharmacology. 2012;62:21–34. doi: 10.1016/j.neuropharm.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilger A, Haslacher H, Ponocny-Seliger E, Perkmann T, Böhm K, Budinsky A, Girard A, Klien K, Jordakieva G, Pezawas L, Wagner O, Godnic-Cvar J, Winker R. Affective and inflammatory responses among orchestra musicians in performance situation. Brain Behav Immun. 2014;37:23–29. doi: 10.1016/j.bbi.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Pollak DD, Monje FJ, Zuckerman L, Denny CA, Drew MR, Kandel ER. An animal model of a behavioral intervention for depression. Neuron. 2008;60:149–161. doi: 10.1016/j.neuron.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sanz JI, Aurrekoetxea I, Ruiz del Agua A, Ruiz-Larrea MB. Detection of catechol-O-methyltransferase Val158Met polymorphism by a simple one-step tetra-primer amplification refractory mutation system-PCR. Mol Cell Probes. 2007;21:202–207. doi: 10.1016/j.mcp.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O'Carroll CM, Kheirbek MA, Burghardt NS, Fenton AA, Dranovsky A, Hen R. Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature. 2011;472:466–470. doi: 10.1038/nature09817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilipo MP, Benedict RH, Zivadinov R, Bakshi R. Correction for intracranial volume in analysis of whole brain atrophy in multiple sclerosis: the proportion vs residual method. Neuroimage. 2004;22:1732–1743. doi: 10.1016/j.neuroimage.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Uno H, Rebert CS, Finch CE. Hippocampal damage associated with prolonged glucocorticoid exposure in primates. J Neurosci. 1990;10:2897–2902. doi: 10.1523/JNEUROSCI.10-09-02897.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloesser RJ, Lehmann M, Martinowich K, Manji HK, Herkenham M. Environmental enrichment requires adult neurogenesis to facilitate the recovery from psychosocial stress. Mol Psychiatry. 2010;15:1152–1163. doi: 10.1038/mp.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MN, Schumann G, Wrase J, Grüsser SM, Flor H, Mann K, Braus DF, Goldman D, Büchel C, Heinz A. Catechol-O-methyltransferase val158met genotype affects processing of emotional stimuli in the amygdala and prefrontal cortex. J Neurosci. 2005;25:836–842. doi: 10.1523/JNEUROSCI.1792-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MN, Bühler M, Schumann G, Klein S, Hu XZ, Moayer M, Zimmer A, Wrase J, Flor H, Mann K, Braus DF, Goldman D, Heinz A. Gene-gene effects on central processing of aversive stimuli. Mol Psychiatry. 2007;12:307–317. doi: 10.1038/sj.mp.4001946. [DOI] [PubMed] [Google Scholar]
- Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476:458–461. doi: 10.1038/nature10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa N, Lukoyanov NV, Madeira MD, Almeida OF, Paula-Barbosa MM. Reorganization of the morphology of hippocampal neurites and synapses after stress-induced damage correlates with behavioral improvement. Neuroscience. 2000;97:253–266. doi: 10.1016/S0306-4522(00)00050-6. [DOI] [PubMed] [Google Scholar]
- Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Boström E, Westerlund I, Vial C, Buchholz BA, Possnert G, Mash DC, Druid H, Frisén J. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone LB, McGeary JE, Palmer RH, Gibb BE. Identifying genetic predictors of depression risk: 5-HTTLPR and BDNF Val66Met polymorphisms are associated with rumination and co-rumination in adolescents. Front Genet. 2013;4:246. doi: 10.3389/fgene.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaz D, Stall N, Dar DE, Zangen A. Knockdown of brain-derived neurotrophic factor in specific brain sites precipitates behaviors associated with depression and reduces neurogenesis. Mol Psychiatry. 2010;15:80–92. doi: 10.1038/mp.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher MH, Anderson CM, Polcari A. Childhood maltreatment is associated with reduced volume in the hippocampal subfields CA3, dentate gyrus, and subiculum. Proc Natl Acad Sci U S A. 2012;109:E563–E572. doi: 10.1073/pnas.1115396109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the aetiology of mental illness: review and methodological analysis. Mol Psychiatry. 2008;13:131–146. doi: 10.1038/sj.mp.4002067. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19:549–557. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Simultaneous genotyping of four functional loci of human SLC6A4, with a reappraisal of 5-HTTLPR and rs25531. Mol Psychiatry. 2006;11:224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wang DD, Wang Y, Liu T, Lee FS, Chen ZY. Variant brain-derived neurotrophic factor Val66Met polymorphism alters vulnerability to stress and response to antidepressants. J Neurosci. 2012;32:4092–4101. doi: 10.1523/JNEUROSCI.5048-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta JK, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, Koeppe RA, Stohler CS, Goldman D. COMT val158met genotype affects μ-opioid neurotransmitter responses to a pain stressor. Science. 2003;299:1240–1243. doi: 10.1126/science.1078546. [DOI] [PubMed] [Google Scholar]