Abstract
The first genome-wide association study of dental caries focused on primary teeth in children aged 3 to 12 yr and nominated several novel genes: ACTN2, EDARADD, EPHA7, LPO, MPPED2, MTR, and ZMPSTE24. Here we interrogated 156 single-nucleotide polymorphisms (SNPs) within these candidate genes for evidence of association with dental caries experience in 13 race- and age-stratified samples from 6 independent studies (n = 3600). Analysis was performed separately for each sample, and results were combined across samples via meta-analysis. MPPED2 was significantly associated with caries via meta-analysis across the 5 childhood samples, with 4 SNPs showing significant associations after gene-wise adjustment for multiple comparisons (p < .0026). These results corroborate the previous genome-wide association study, although the functional role of MPPED2 in caries etiology remains unknown. ACTN2 also showed significant association via meta-analysis across childhood samples (p = .0014). Moreover, in adults, genetic association was observed for ACTN2 SNPs in individual samples (p < .0025), but no single SNP was significant via meta-analysis across all 8 adult samples. Given its compelling biological role in organizing ameloblasts during amelogenesis, this study strengthens the hypothesis that ACTN2 influences caries risk. Results for the other candidate genes neither proved nor precluded their associations with dental caries.
Keywords: EDARADD, EPHA7, LPO, MTR, ZMPSTE24, single nucleotide polymorphism
Introduction
Dental caries is one of the most widespread diseases affecting children and adults alike. Among the numerous exogenous and host factors influencing risk of dental caries, genetics plays a substantial role, with heritability estimates of caries experience ranging from 30% to 60% (Shaffer et al., 2012). Furthermore, rapid advances in genotyping capabilities over the past decade have enabled large-scale genetic association studies that seek to catalogue the specific genetic variants contributing to risk of dental caries. The hope for this area of research is that identifying the genetic contributors to dental caries may lead to improvements in prevention, early detection, and treatment.
Several candidate gene studies have investigated the effects of genetic variation in genes chosen a priori based on their known biological functions. For example, enamel genes (i.e., genes having protein products closely involved in processes of amelogenesis) have been studied in several populations (Slayton et al., 2005; Deeley et al., 2008; Patir et al., 2008; Shimizu et al., 2012; Wang et al., 2012b; Gasse et al., 2013; Jeremias et al., 2013). Likewise, genes affecting taste preference (Wendell et al., 2010; Pidamale et al., 2012; Kulkarni et al., 2013), tooth development (Tannure et al., 2012; Wang et al., 2012b), and host defense (Acton et al., 1999; Ozturk et al., 2010; Brancher et al., 2011; Valarini et al., 2012; Briseno-Ruiz et al., 2013) have been studied. Candidate gene studies have had some success in identifying specific variants showing association with dental caries experience. However, a priori candidate genes appear to explain only a fraction of the heritability of dental caries.
The genome-wide association study (GWAS) method is an unbiased and complementary approach to the candidate gene study method, and it has been widely used to identify novel genes for many complex human conditions. To date, a few GWASs have been performed for various dental caries phenotypes, successfully nominating several additional candidate genes for further study (Shaffer et al., 2011; Wang et al., 2012a; Shaffer et al., 2013; Zeng et al., 2013). The first GWAS for dental caries focused on the primary dentition in children aged 3 to 12 yr, and it implicated several novel genes: ACTN2, EDARADD, EPHA7, LPO, MPPED2, MTR, and ZMPSTE24. As a hypothesis-generating method, GWAS results require careful scrutiny and replication in independent samples to distinguish chance results from true associations. This is the aim of the current study. Here, we report results of our follow-up genetic association study seeking to replicate the putative genetic associations identified in the original GWAS of dental caries in white children. Moreover, we test whether the same genes are associated with dental caries in adults and in individuals of different racial backgrounds.
Materials & Methods
Samples and Phenotypes
Six independent samples were included in this study:
the Center for Oral Health Research in Appalachia (COHRA; n = 1,769), which recruited households from rural Appalachian communities;
the Iowa Head Start (IHS; n = 64) Study, which recruited primarily low-income children through the U.S. Department of Health and Human Services program;
the Iowa Fluoride Study (IFS; n = 136), which recruited children from urban and suburban Iowa;
the Dental Strategies Concentrating on Risk Evaluation (Dental SCORE; n = 502), which recruited adult participants from the Pittsburgh area to study racial and socioeconomic factors leading to disparities in cardiovascular risk;
the Dental Registry and DNA Repository (DRDR; n = 875), which recruited urban adults seeking treatment at the University of Pittsburgh School of Dental Medicine; and
the Center for Education and Drug Abuse Research (CEDAR; n = 241), which included the adolescent offspring of fathers from the Pittsburgh area enrolled in a study of substance use risk factors (Table).
Table.
Characteristics of the Samples
| Sample | n | Female Sex | Age, yr | Caries Prevalencea | dft/DMFTb |
|---|---|---|---|---|---|
| Children | |||||
| COHRA | |||||
| Whites | 608 | 46.7 | 7.3 (3.0-12.0) | 55.4 | 2.3 (0-17) |
| Blacks | 81 | 46.9 | 7.6 (3.2-11.8) | 49.4 | 1.8 (0-8) |
| IHS | |||||
| Whites | 41 | 58.5 | 4.1 (3.2-5.3) | 80.5 | 6.3 (0-20) |
| Blacks | 23 | 52.2 | 4.3 (3.4-5.6) | 82.6 | 5.7 (0-17) |
| IFS whites | 136 | 48.5 | 5.2 (4.4-6.8) | 37.8 | 1.2 (0-16) |
| Adults | |||||
| COHRA | |||||
| Whites | 994 | 62.8 | 34.3 (18.0-75.0) | 96.5 | 10.5 (0-28) |
| Blacks | 86 | 70.9 | 36.2 (18.2-60.8) | 94.2 | 9.3 (9-28) |
| Dental SCORE | |||||
| Whites | 277 | 63.2 | 64.0 (48.0-78.0) | 100.0 | 16.4 (2-28) |
| Blacks | 225 | 72.9 | 61.6 (47.0-79.0) | 100.0 | 14.8 (1-28) |
| DRDR | |||||
| Whites | 702 | 50.0 | 43.0 (18.0-74.8) | 97.9 | 16.6 (0-28) |
| Blacks | 173 | 57.8 | 44.5 (18.0-74.4) | 98.3 | 16.5 (0-28) |
| CEDAR | |||||
| Whites | 173 | 31.2 | 20.4 (15.7-28.6) | 82.1 | 5.4 (0-21) |
| Blacks | 68 | 44.3 | 20.2 (15.6-27.8) | 88.6 | 6.4 (0-16) |
Values expressed as mean (range) or percentage.
COHRA, Center for Oral Health in Appalachia; IHS, Iowa Head Start; IFS, Iowa Fluoride Study; Dental SCORE, Dental Strategies Concentrating on Risk Evaluation; DRDR, Dental Registry and DNA Repository; CEDAR, Center for Education and Drug Abuse Research.
Caries prevalence was defined as dfs ≥ 1 in children or DMFT ≥ 1 in adults.
dft was the measure of caries experience of the primary dentition in children samples; DMFT was the measure of caries experience of the permanent dentition in adult samples.
All study procedures were approved by the institutional review boards of the pertinent universities. Further details of each of the 6 studies are described in the Appendix.
These 6 studies yielded 13 age- and race-stratified samples that were analyzed individually in the current study. Two of these samples (COHRA white children and IFS white children) were included as part of the original GWAS study, whereas 3 additional childhood samples and 8 adult samples were not. Note that a subset of IHS participants were included in the original GWAS; however, the IHS participants included here constitute an independent sample of different individuals. All participants received a full-mouth intraoral examination by a dentist or research dental hygienist to assess dental caries experience (excluding third molars). Dental caries experience in the permanent and primary dentitions was scored via DMFT and dft indices, respectively, which are defined as the number of teeth showing frank decay, missing due to decay, or having fillings/restorations.
Genotypes
Participants were genotyped for a custom panel of single-nucleotide polymorphisms (SNPs) using the Illumina Golden Gate platform (San Diego, CA, USA) by the Center for Inherited Disease Research at Johns Hopkins University. This panel was chosen to follow up results from a number of GWAS scans. For this study, we interrogated 156 SNPs across 7 genes (ACTN2, EDARADD, EPHA7, LPO, MPPED2, MTR, and ZMPSTE24) to further investigate associations originally reported by Shaffer et al. (2011). Details regarding the composition of the custom panel, selection of SNPs, and genotype quality assurance are presented in the Appendix. Note that these genes were originally nominated per their proximity to a GWAS hit and plausible biology or experimental evidence relevant to caries etiology or oral health. The originally associated SNP was located within the nominated gene for MPPED2, MTR, and ZMPSTE24 and was physically proximal but not within the other genes (Shaffer et al., 2011). The nominating SNPs in MPPED2 (rs11031093) and MTR (rs11806016) were present in our custom panel.
Statistical Analysis
Dental caries experience in the primary (dft) and permanent (DMFT) dentitions was analyzed separately in participants aged 3 to 12 yr and ≥18 yr, respectively, in all samples except CEDAR, for which “adults” were >15 yr. Analyses were also performed separately in self-reported non-Hispanic whites and blacks (to guard against bias due to population stratification). Linear regression was used to test for genetic association between DMFT/dft, as a quantitative trait, and each SNP under the additive genetic model while modeling the effects of sex and age. Analyses of blacks were also adjusted for the first 4 principal components of ancestry (because individuals of African ancestry potentially exhibit genetic population stratification). As a complementary approach that matches the methods used in the original GWAS, logistic regression was also used to test caries prevalence in children, with each individual coded as affected by dental caries (i.e., dfs ≥ 1) or not (i.e., dfs = 0). Stouffer’s inverse variance weighted method of meta-analysis was used to combine evidence of association across studies. Meta-analysis was performed for whites only, for blacks only, and for all participants. Genetic association and ancestry modeling was performed in PLINK (Purcell et al., 2007). Meta-analysis was performed in METAL (Willer et al., 2010). Discussion of technical issues related to the statistical models employed here is presented in the Appendix.
To assist in the interpretation of our results in light of the issue of multiple comparisons, for each gene we determined the effective number of independent tests—which is less than or equal to the total number of tests due to the linkage disequilibrium (LD; i.e., correlation) among SNPs—using the method by Li and Ji (2005). The threshold for gene-wise significance was set at 0.05, divided by the effective number of independent tests. Note that the gene-wise significance threshold does not control the study-wise error rate.
Results
Characteristics of the samples are shown in the Table; see also the Appendix. Samples represent a range of ages from populations of different risk profiles and hence show considerable variation in caries experience. Two samples included in this follow-up study—COHRA and IFS white children—were part of the original GWAS; that GWAS also included 2 additional samples and a replication sample not included here. Therefore, we have interpreted our results according to 2 benchmarks: whether any samples other than COHRA and IFS white children showed evidence of association and whether the meta-analysis of all samples together showed evidence of association. In addition, we were mindful of whether association was observed in both whites and blacks and in both primary and permanent dentitions.
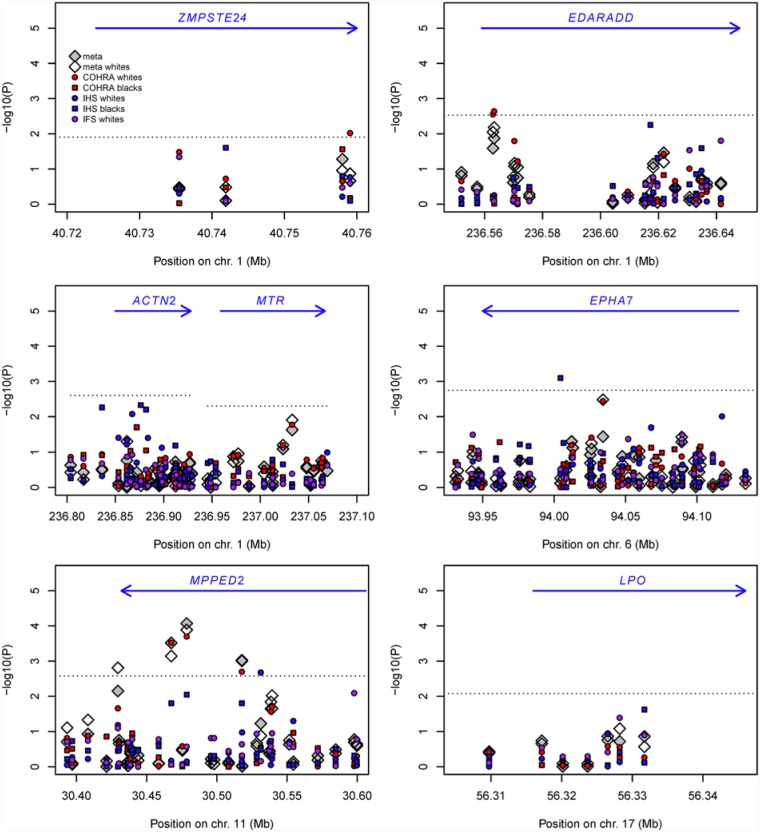
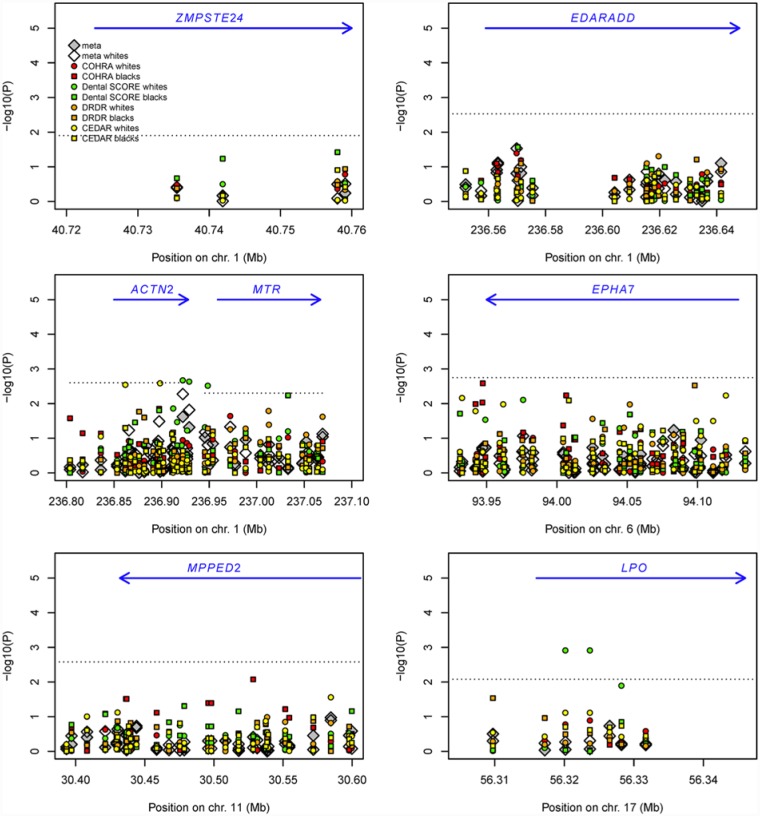
Figures 1 and 2 illustrate evidence of association in children and adults, respectively. Negative log10-transformed p for all SNPs are shown for all samples individually and combined via meta-analysis. Detailed association results for select SNPs from each of the 7 genes are shown in the Appendix Table.
Figure 1.
Genetic association in childhood samples for 7 genes nominated in genome-wide association study of childhood caries. Negative log10-transformed p values are shown for childhood samples: COHRA (red), IHS (blue), and IFS (purple). Circles represent white samples, and squares represent black samples. White diamonds represent meta-analysis across all white childhood samples, and gray diamonds represent meta-analysis across all black and white childhood samples combined. The dotted lines represent the p threshold after adjustment for the number of independent single-nucleotide polymorphisms within a gene. The blue arrows represent the physical location and direction of genes. COHRA, Center for Oral Health in Appalachia; IHS, Iowa Head Start; IFS, Iowa Fluoride Study.
Figure 2.
Genetic association for adult samples for 7 genes nominated in genome-wide association study of childhood caries. Negative log10-transformed p values are shown for childhood samples: COHRA (red), Dental SCORE (green), DRDR (orange), and CEDAR (yellow). Circles represent white samples, and squares represent black samples. White diamonds represent meta-analysis across all white adult samples, and gray diamonds represent meta-analysis across all black and white adult samples combined. The dotted lines represent the p threshold after adjustment for the number of independent single-nucleotide polymorphisms within a gene. The blue arrows represent the physical location and direction of genes. COHRA, Center for Oral Health in Appalachia; Dental SCORE, Dental Strategies Concentrating on Risk Evaluation; DRDR, Dental Registry and DNA Repository; CEDAR, Center for Education and Drug Abuse Research.
In children, the strongest evidence of genetic association was observed for MPPED2. Meta-analyses across child samples for 4 SNPs in this gene yielded significant p values, and multiple samples appeared to drive these signals—notably, COHRA white children and IHS black children. As individual samples, COHRA white children and IHS black children both showed significant associations for 1 or more SNPs in this gene. Other significant associations were observed for COHRA white children in ZMPSTE24 and EDARADD and for IHS black children in EPHA7.
Overall, results for the binary phenotype in children (results not shown) were similar to those for dft. The one exception was ACTN2, for which significant association was observed for the binary phenotype (e.g., rs10925178, p = .0014, in meta-analysis of whites and blacks combined). This signal was driven by COHRA and IFS white children, both of which were included in the initial GWAS but remained significant when other samples were meta-analyzed. SNPs in ACTN2 were also significantly associated with Dental SCORE white adults (p = .002) and showed compelling evidence of association (though not meeting the threshold for gene-wise significance in light of multiple comparisons) in CEDAR whites (p = .003), COHRA blacks (p = .01), Dental SCORE blacks (p = .02), Dental Registry and DNA Repository blacks (p ≤ .03), and CEDAR blacks (p = .05). A particularly interesting finding was that in adults, the effect of SNP rs707204 in ACTN2 differed in direction between whites (p = .03) and blacks (p = .004) and would have been significant overall if the direction of effect was not considered in the meta-analysis.
Other significant associations in adults were observed for Dental SCORE whites in MTR and LPO. However, no SNP yielded significant evidence of association in meta-analysis across all adult samples. Likewise, no SNP was significantly associated with caries in meta-analysis across all adult and children samples.
Discussion
GWAS studies are useful for nominating novel loci for follow-up via hypothesis-driven experiments. In this study, we evaluated nearly 3,600 participants for evidence of genetic association with 7 genes nominated in a previous GWAS of childhood dental caries. Initial nomination of these loci was based loosely on proximity to an associated SNP, as well as corroborating experimental evidence or biologically plausible effects on caries etiology or on the oral environment.
In this follow-up study, we showed that across all samples of children combined, MPPED2 was significantly associated with childhood caries after consideration of the gene-wise issue of multiple comparisons. However, this signal was primarily driven by COHRA white children, who were also included in the original GWAS. Nevertheless, compared with COHRA white children alone, meta-analysis increased evidence of association, indicating that additional samples also contributed to the signal. MPPED2 was not associated with caries in adults, and its possible function in caries etiology is unknown, though one previous report found that MPPED2 was downregulated in oral epithelial cells in response to oral pathogens (Milward et al., 2007).
ACTN2 showed significant evidence of association in meta-analysis across black and white children for the binary caries phenotype but not for dfs index. None of the other genes revealed significant association via meta-analysis. However, individual samples showed evidence of association for some loci. For example, associated SNPs in ACTN2, MTR, and LPO were observed for Dental SCORE white adults. Furthermore, for several genes considered in this study, multiple samples showed some level of evidence but for different specific SNPs (see ACTN2 for CEDAR and Dental SCORE whites and for blacks of all adult samples). In these cases, scientific interpretation is not entirely clear. A liberal definition of replication may allow for different SNPs within a gene to show association among different samples, which could reflect varying patterns of LD across samples. An extreme example of this may be SNP rs707204 in ACTN2, which yielded intriguing evidence of association in both blacks and whites, although in opposite directions. Because of population history and differences in LD patterns between racial groups, opposite alleles at this SNP may occur on the haplotype background (or backgrounds) harboring unobserved caries risk variants.
While SNPs in MPPED2 yielded the smallest p values, ACTN2 is also a very promising gene evaluated in this study. Not only was the statistical evidence encouraging, but the corroborating biology is persuasive: ACTN2 is thought to help regulate and organize ameloblasts during tooth development (Sehic et al., 2010). While this gene was nominated in a GWAS of children, in the present study, stronger evidence was observed for adults. Moreover, whites and blacks both exhibited some level of association. Taken together, these results strengthen the hypothesis that genetic variation in ACTN2 influences dental caries. However, more work is needed to conclusively prove this relationship and to further investigate the issue of heterogeneity across racial groups.
EDARADD and MTR are 2 nearby but less plausible genes originally nominated by the same GWAS hit (i.e., associated locus) as ACTN2. EDARADD is implicated in a single-gene disorder causing dental anomalies and other characteristics. MTR is associated with nonsyndromic cleft lip and palate (Mostowska et al., 2006). Neither of these genes have known roles in caries etiology, per se. Moreover, given their physical proximity and LD patterns, it is difficult to entirely separate the statistical evidence observed for these 2 genes from ACTN2.
LPO, which codes a bactericidal salivary enzyme, was a logical candidate to pursue, but note that it was relatively far (>100 kb) from the original GWAS hit and did not yield evidence of association in this study for any samples except Dental SCORE white adults. Likewise, EPHA7 (involved in murine tooth development; Luukko et al., 2005) and ZMPSTE24 (responsible for mandibuloacral dysplasia, a Mendelian syndrome characterized by craniofacial, dental, skeletal, and epidermal anomalies; Agarwal et al., 2003) showed moderate evidence of association. The latter 2 genes were originally nominated as having gene-by-fluoride interaction effects, although, unfortunately, fluoride data were unavailable for the replication samples reported here. Like previous candidate gene studies, results for these genes did not unequivocally implicate these genes.
In this study, we did not specifically model the effects of exogenous risk factors, such as smoking, oral hygiene, or socioeconomic status, while interrogating the effects of genetic variants, because very few potential risk factors were available across all 6 samples. However, recent studies have suggested that adjustment for covariates of weak effects, which includes most known caries risk factors, may adversely influence genetic association studies (Kuo and Feingold, 2010). Instead, we relied on our large combined sample of nearly 3600 participants to achieve high power to detect SNPs of plausible effect sizes. While incontrovertible evidence of genetic association was not gained for any of these genes and although we cannot determine causality through observation research alone, we feel that this study has strengthened the hypothesis that variation in some of these genes (e.g., ACTN2 and MPPED2) affects risk of dental caries. Understanding the genetic underpinning of disease may influence strategies for prevention, early detection, and treatment.
Supplementary Material
Supplementary Material
Acknowledgments
We would like to express our deep gratitude to the participants and research teams of the 6 parent studies whose contributions made this work possible.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This study was supported by the following National Institutes of Health grants: U01-DE018903, R01-DE014899, R01-DE009551, R01-DE012101, R01-DE018914, P50-DA005605, and R01-DA019157, as well as the National Science Foundation / Department of Defense grant DBI-1263020. The Dental Registry and DNA Repository is supported by the University of Pittsburgh School of Dental Medicine. The Dental SCORE sample is partially supported by the Commonwealth of Pennsylvania Department of Health grant ME-02-384.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Acton RT, Dasanayake AP, Harrison RA, Li Y, Roseman JM, Go RC, et al. (1999). Associations of MHC genes with levels of caries-inducing organisms and caries severity in African-American women. Hum Immunol 60:984-989. [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Fryns JP, Auchus RJ, Garg A. (2003). Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Hum Mol Genet 12:1995-2001. [DOI] [PubMed] [Google Scholar]
- Brancher JA, Pecharki GD, Doetzer AD, Medeiros KG, Cordeiro Junior CA, Sotomaior VS, et al. (2011). Analysis of polymorphisms in the lactotransferrin gene promoter and dental caries. Int J Dent 2011:571726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseno-Ruiz J, Shimizu T, Deeley K, Dizak PM, Ruff TD, Faraco IM, Jr, et al. (2013). Role of TRAV locus in low caries experience. Hum Genet 132:1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley K, Letra A, Rose EK, Brandon CA, Resick JM, Marazita ML, et al. (2008). Possible association of amelogenin to high caries experience in a Guatemalan-Mayan population. Caries Res 42:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse B, Grabar S, Lafont AG, Quinquis L, Opsahl Vital S, Davit-Beal T, et al. (2013). Common SNPs of AmelogeninX (AMELX) and dental caries susceptibility. J Dent Res 92:418-424. [DOI] [PubMed] [Google Scholar]
- Jeremias F, Koruyucu M, Kuchler EC, Bayram M, Tuna EB, Deeley K, et al. (2013). Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol 58:1434-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni GV, Chng T, Eny KM, Nielsen D, Wessman C, El-Sohemy A. (2013). Association of GLUT2 and TAS1R2 genotypes with risk for dental caries. Caries Res 47:219-225. [DOI] [PubMed] [Google Scholar]
- Kuo CL, Feingold E. (2010). What’s the best statistic for a simple test of genetic association in a case-control study? Genet Epidemiol 34:246-253. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. (2005). Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95:221-227. [DOI] [PubMed] [Google Scholar]
- Luukko K, Løes S, Kvinnsland IH, Kettunen P. (2005). Expression of ephrin-A ligands and EphA receptors in the developing mouse tooth and its supporting tissues. Cell Tissue Res 319:143-152. [DOI] [PubMed] [Google Scholar]
- Milward MR, Chapple IL, Wright HJ, Millard JL, Matthews JB, Cooper PR. (2007). Differential activation of NF-kappaB and gene expression in oral epithelial cells by periodontal pathogens. Clin Exp Immunol 148:307-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowska A, Hozyasz KK, Jagodzinski PP. (2006). Maternal MTR genotype contributes to the risk of non-syndromic cleft lip and palate in the Polish population. Clin Genet 69:512-517. [DOI] [PubMed] [Google Scholar]
- Ozturk A, Famili P, Vieira AR. (2010). The antimicrobial peptide DEFB1 is associated with caries. J Dent Res 89:631-636. [DOI] [PubMed] [Google Scholar]
- Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, et al. (2008). Enamel formation genes are associated with high caries experience in Turkish children. Caries Res 42:394-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidamale R, Sowmya B, Thomas A, Jose T, Madhusudan KK, Prasad G. (2012). Association between early childhood caries, streptococcus mutans level and genetic sensitivity levels to the bitter taste of, 6-N propylthiouracil among the children below 71 months of age. Dent Res J (Isfahan) 9:730-734. [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehic A, Risnes S, Khan QE, Khuu C, Osmundsen H. (2010). Gene expression and dental enamel structure in developing mouse incisor. Eur J Oral Sci 118:118-130. [DOI] [PubMed] [Google Scholar]
- Shaffer JR, Wang X, Feingold E, Lee M, Begum F, Weeks DE, et al. (2011). Genome-wide association scan for childhood caries implicates novel genes. J Dent Res 90:1457-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, Wang X, Desensi RS, Wendell S, Weyant RJ, Cuenco KT, et al. (2012). Genetic susceptibility to dental caries on pit and fissure and smooth surfaces. Caries Res 46:38-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JR, Feingold E, Wang X, Lee M, Tcuenco K, Weeks DE, et al. (2013). GWAS of dental caries patterns in the permanent dentition. J Dent Res 92:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Ho B, Deeley K, Briseno-Ruiz J, Faraco IM, Jr., Schupack BI, et al. (2012). Enamel formation genes influence enamel microhardness before and after cariogenic challenge. PloS One 7:e45022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayton RL, Cooper ME, Marazita ML. (2005). Tuftelin, mutans streptococci, and dental caries susceptibility. J Dent Res 84:711-714. [DOI] [PubMed] [Google Scholar]
- Tannure PN, Kuchler EC, Lips A, Costa Mde C, Luiz RR, Granjeiro JM, et al. (2012). Genetic variation in MMP20 contributes to higher caries experience. J Dent 40:381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarini N, Maciel SM, Moura SK, Poli-Frederico RC. (2012). Association of dental caries with HLA Class II allele in Brazilian adolescents. Caries Res 46:530-535. [DOI] [PubMed] [Google Scholar]
- Wang X, Shaffer JR, Zeng Z, Begum F, Vieira AR, Noel J, et al. (2012a). Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health 12:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Willing MC, Marazita ML, Wendell S, Warren JJ, Broffitt B, et al. (2012b). Genetic and environmental factors associated with dental caries in children: the Iowa Fluoride Study. Caries Res 46:177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell S, Wang X, Brown M, Cooper ME, DeSensi RS, Weyant RJ, et al. (2010). Taste genes associated with dental caries. J Dent Res 89:1198-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Shaffer JR, Wang X, Feingold E, Weeks DE, Lee M, et al. (2013). Genome-wide association studies of pit-and-fissure- and smooth-surface caries in permanent dentition. J Dent Res 92:432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.