Abstract
Recent studies have identified a growing number of mesoscale protein assemblies in both bacterial and eukaryotic cells. Traditionally, these polymeric assemblies are thought to provide structural support for the cell and thus have been classified as the cytoskeleton. However a new class of macromolecular structure is emerging as an organizer of cellular processes that occur on scales hundreds of times larger than a single protein. We propose two types of self-assembling structures, dynamic globules and crystalline scaffolds, and suggest they provide a means to achieve cell-scale order. We discuss general mechanisms for assembly and regulation. Finally, we discuss assemblies that are found to organize metabolism and what possible mechanisms may serve these metabolic enzyme complexes.
Introduction
A cell’s ability to coordinate the spatial arrangement of its components is crucial for it to achieve functional complexity. Eukaryotes have, partly, achieved this complexity by isolating specific reactions into membrane bound organelles. Similarly, gram-negative prokaryotes use both inner and outer membranes to compartmentalize their cytoplasm and periplasm. However, generating membrane bound compartments de novo is a cumbersome process, and even the most complex cells have a limited number of distinct types of organelles. An alternative strategy is for proteins to organize into distinct assemblies, enabling cells to establish functionally differentiated subcellular regions even in the absence of membrane barriers. While bacteria generally lack membrane-bound organelles, recent evidence suggests that the cytoplasms of both bacterial and eukaryotic cells are highly organized by such protein assemblies [1].
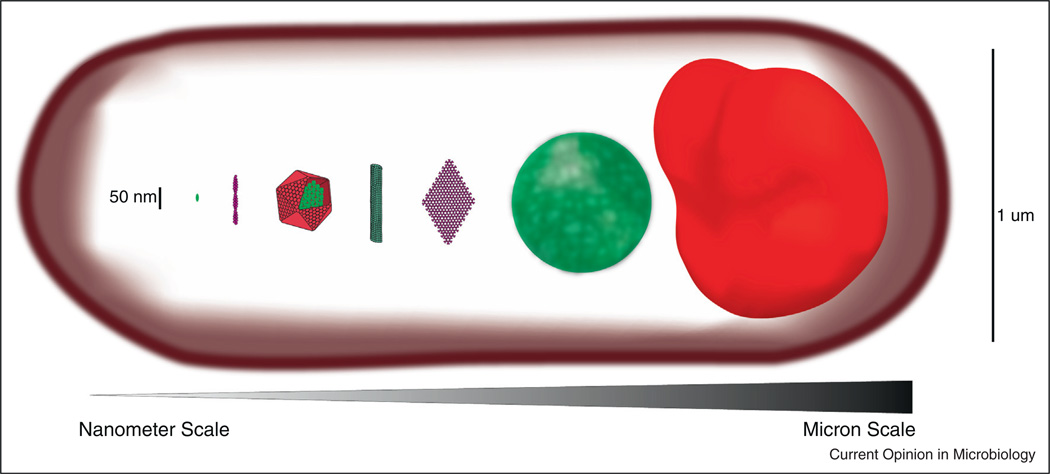
Higher-order protein assemblies exist in a mesoscale between the nanoscale of individual proteins and the microscale of whole cells (Figure 1). To occupy this mesoscale, assemblies are often constructed from one or more types of repeating nanoscale subunit. This feature is advantageous because it enables construction of large structures using only a small number of genes (genetic efficiency) and it is amenable to regulation and quality control by exclusion of malformed subunits [2]. Once assembled, these structures typically extend as polymers that form filaments or sheets, hundreds of times larger than an individual monomer. Multiple polymers can also be densely interconnected (crosslinked) to form dynamic, phase-separated globules or bodies. Here we highlight the recent appreciation that mesoscale protein assemblies are much more common than previously thought and we review some of their emerging forms, how they are regulated, and some of their newly attributed organizing functions. We then focus on how new discoveries about mesoscale assemblies are changing our ability to both understand and manipulate even one of the most well-studied biological networks, metabolism.
Figure 1.
Various mesoscale self-assembling structures drawn to scale in comparison to a single monomer. From left to right, typical scale of a monomeric scaffold like the yeast protein Ste5, an actin filament, a carboxysome shell housing the enzymes of carbon fixation, a microtubule, a chemotaxis array, a low complexity sequence hydrogel, a purinosome. All structures are assembled within a typical E. coli cell (also drawn to scale).
New organizing functions for macromolecular assemblies
A wealth of macromolecular structures has recently been discovered largely due to new imaging techniques and an increase in our ability to perform high-throughput screens. Genome-wide fluorescent labeling screens in both eukaryotes and prokaryotes have only begun to enumerate the wide assortment of protein assemblies [3,4]. Electron cryotomography (ECT) complemented by genetics has further aided in defining their various shapes. Filaments, rings, sheets, lattices and tubes have all been described with specific functions as diverse as their various forms [5]. However, many of these proteins are characterized as being part of the cytoskeleton with the connotation that they play a structural role in maintaining cell shape in much the same way as our skeletons support our own body shape. On the contrary, we highlight some of the new macromolecular assemblies that serve important organizational roles, beyond simply supporting the shape of the cell.
While classical cytoskeletal structures form linear filament or tubule polymers with rigidly defined repeating structures, newly identified assemblies take on a wider range of shapes and structures including sheets, rings, discs, and gels, which are essential to the organizational functional of these mesoscale assemblies. One newly discovered sheet complex, found in the freshwater bacterium Caulobacter crescentus, is the StpABCD protein complex. The Stp complex localizes to distinct bands that traverse the cross-section of a thin appendage known as a stalk, and ECT imaging has revealed that it forms a sheet that traverses the stalk’s entire diameter [6••]. The Stp complex’s disk-like shape functions to compartmentalize the Caulobacter stalk, thus preventing proteins from diffusing between the stalk and the main cell body. Cells lacking the Stp complex incur the cost of increased protein synthesis to achieve the same effective protein concentrations in the periplasm and cell envelope. Over-expression of StpA, but not StpB, C, or D, leads to a marked increase in the number of complexes per cell, suggesting that the complex assembles through a nucleation-style mechanism [6••]. These findings highlight recurring themes present in many of the emerging macromolecular organizing structures: first, they often form hierarchically through nucleation events, second, they generally accelerate reactions by increasing the effective concentrations of enzymes and their substrates, and third, they assemble into shapes that promote their specific cellular function.
Perturbing classical cytoskeletal proteins often results in gross morphological phenotypes [7,8]. Perturbing macromolecular organizing structures can also result in morphological changes, but often this occurs because they coordinate macroscale processes such as cell division or cell polarity determination, rather than simply providing support for macroscale structures. For example, a macromolecular structure in C. crescentus is formed by the oligomer PopZ, which was discovered based on its polar localization [9] and disruption of division upon overexpression [10]. Purified PopZ forms a lattice-like meshwork of filaments intersecting in 3-way junctions [9]. The PopZ meshwork interacts with a number of other proteins involved in cell cycle progression and chromosome dynamics, thereby serving to organize these processes in relation to the cell pole. A similar screen for cell division defects in the bacterium Streptococcus pneumonia identified SepF (originally named YlmF [11]), which polymerizes into rings that assist FtsZ in constricting the bulky peptidoglycan layer of gram positive bacteria during cell division [12,13]. Such ring structures are important for cell division, as disruption of SepF polymerization stalls septum formation [14]. Despite their different higher-order shapes, both PopZ and SepF serve as organizing scaffolds by binding and localizing multiple proteins and thus coordinating their function both spatially and temporally.
Organizing the transduction of signals is another function well-suited for macromolecular assemblies. Amazingly, cells tend to respond appropriately to a cacophony of signals despite having a comparatively small number of distinct signaling domains [15]. One strategy for coordinating the proteome’s limited number of interaction domains to achieve robust and efficient signaling cascades is to use monomeric scaffolds to organize sets of interacting proteins into signaling complexes [16]. These scaffolds organize small-scale assemblies with low stoichiometries to reduce crosstalk between pathways with structurally similar inputs and facilitate crosstalk when input signals need to be integrated. Larger mesoscale scaffolds can further expand signaling functionality by enabling cells to modulate their signal response dynamics, thereby allowing different cells to use the same components to achieve different signaling outputs in the presence of the same inputs. For example, formation of oligomeric signalisomes in the Toll-like receptor (TLR)-interleukin 1 receptor (IL-1R) superfamilies have been suggested to translate a smoothly – graded input into a digital-like, all or nothing response [17]. Other naturally scaffolded enzymes such as adenylcyclases form micro-domains [18] that are suggested to increase the efficiency of a transduced signal by concentrating intermediate signaling molecules [19•]. Thus, macromolecular assemblies are found to enrich cell-signaling inputs, outputs, and total network integration by organizing the key components into localized centers of orchestrated molecular interaction.
Phase-separated assemblies as dynamic organizing structures
In addition to the canonical, rigid, self-assembling macromolecular polymers discussed above, an emerging physical class of mesoscale structures form assemblies that undergo a liquid-like phase transition, essentially generating phase-separated microdomains in the absence of membrane barriers. For example, the Caenorhabditis elegans P granule is a structure containing a large number of RNAs and RNA-binding proteins that forms visually distinct globules with physical properties similar to a droplet of oil in water [20]. These phase-separated ‘bodies’ or ‘granules’ are widespread and exist both inside the eukaryotic nucleus such as Cajal Bodies that are thought to organize transcription, and in the cytoplasm such as neuronal granules that are associated with RNA processing (for a longer list see [21]). As with the crystal-line-like structures described above, mesoscale bodies are also thought to accelerate biological processes by concentrating enzymes and substrates into localized reaction volumes [22].
Recent studies suggested that mesoscale bodies can form dynamically through the assembly of physically disordered low complexity (LC) peptide sequences [23•]. LC sequences are common in a wide range of eukaryotic proteins, including many RNA-binding proteins, but their characteristic lack of rigid, well-folded, three-dimensional structures left the function of LC peptides poorly understood. A small molecule, 5-aryl-isoxazole-3-carboxyamide (b-isox), was found to cause multiple LC proteins from a wide variety of eukaryotic organisms and cell types to polymerize into amyloid-like fibers, thereby precipitating the associated protein components of various cellular bodies ranging from P granules to nuclear bodies to neuronal granules [24••]. Further studies with the fused in sarcoma (FUS) LC RNA binding domain demonstrated that they can self-assemble into highly dynamic hydrogels. Under physiological circumstances, LC domain polymer networks may thus form the assembly scaffold for the phase-separated liquid droplets observed in bodies [25].
Another method for forming mesoscale bodies utilizes networks of multivalent binding proteins. For example, Li et al. mixed synthetic proteins composed of repeats of the SRC homology 3 (SH3) domain with synthetic proteins composed of repeats of SH3’s cognate ligand, the proline-rich motif (PRM), and observed the formation of dynamic liquid globules [26•]. Repeats with low valencies (N < 4) did not form globules in concentrations of up to 400 µm (in units of binding domains/volume), while repeats of 4 binding modules or greater assembled into phase separated structures at physiologically relevant concentrations. Thus, changes in monomer valency influence globule formation, which may prove important when considering other synthetic engineering efforts.
In vivo engineering has similarly allowed the understanding of what components or mechanisms are required for granule assembly. Nucleation factors have been shown to be important for the assembly of a nuclear-localized class of cytologically visible puncta known as nuclear bodies. The first type of nuclear body to be created de novo were Cajal Bodies (CB) in HeLa cell lines [27]. Kaiser et al., engineered HeLa cells with both an inducible Escherichia coli LacIGFP-CB component fusion protein and 256 repeats of the cis LacO sequence integrated into its genome. The LacO binding DNA sequences acted as scaffolds that colocalized various candidate CB protein components, thereby locally surpassing the monomer critical concentration necessary for assembly [27]. Later a comprehensive screen of in vivo nuclear body assembly elements revealed that co-localizing sets of functionally relevant RNA molecules was also sufficient for the de novo formation of nuclear bodies [28,29]. Together these findings demonstrate that RNA and DNA can be important nucleation factors with possible structural roles as well. Importantly, they highlight nucleation as an important assembly mechanism.
In vitro experiments have demonstrated the potential for phosphorylation as a widespread mechanism for regulating the assembly of phase-separated structures. In the case of LC protein assembly, phosphorylation of the human FUS protein by recombinant DNA-Protein Kinase prevented the LC-containing peptides from entering into hydrogels [23•]. Phosphorylation can also regulate multivalent interacting protein networks. For example, kidney podocytes form a filtration barrier by assembling a complex of three multivalent signaling proteins: nephrin, NCK, and N-WASP. Nephrin contains three binding sites for a motif on NCK, which in turn contains three binding sites for a motif found six times on N-WASP. As predicted for such multivalent assemblies, nephrin, NCK, and N-WASP form phase transitions. Whereas phosphorylation of the LC-containing FUS protein inhibits assembly, phosphorylation of nephrin increases the proteins’ tendency to phase separate [26]. Phosphorylation may thus provide a general mechanism of regulation by effectively increasing or decreasing monomer valency.
The assembly of mesoscale hydrogels and liquid droplets represents a highly dynamic strategy for spatially localizing cellular processes. These structures share some very general regulatory and assembly characteristics with classical polymers, like phosphorylation-mediated construction and nucleation-dependent assembly. However, phase-separated structures are probably better suited for specific functions that require highly dynamic assembly and disassembly, like the packaging and transport of RNA within a neuron [30]. Granules and other liquid-like assemblies are only beginning to be dissected and could lead to the discovery of exciting new organizing structures which preside over functions that benefit from being localized into distinct subcellular regions.
Coordinating metabolism with mesoscale assemblies
One aspect of cellular physiology for which mesoscale assemblies appear to have particular significance is metabolism. Metabolism is classically thought of as a well-mixed spatially distributed process. However, new studies are identifying large numbers of metabolic enzymes as members of mesoscale assemblies [4,31–33] suggesting that metabolism may benefit from mesoscale spatial organization. The most straightforward function for spatial organization in metabolism is to bring together successive metabolic reactions, thereby channeling the product of one enzyme to the active site of another enzyme. When one considers the extremely low probability of a small molecule diffusing from one active site to a single neighboring active site tens of nanometers away, it becomes clear that simply tethering two sequentially acting enzymes does not result in any changes to pathway flux. However, by co-localizing large numbers of enzymes in a mesoscale assembly, one may be able to dramatically increase flux by increasing the probability that a product will diffuse into one of many possible active sites. Thus, like the macroscale processes of cell division and signalisome-mediated signal transduction, metabolism is a process that lends itself to organization by macromolecular assemblies. We suggest that the physical properties of both highly dynamic globules and more rigid polymers could be useful for coordinating metabolism depending on the specific application.
Increasing pathway flux may be the central function of the recently discovered purinosome, a nutrient-dependent assembly of all six enzymes involved in de novo purine biosynthesis in HeLa cells. Importantly, disrupting purinosome assembly led to a significant decrease in the purine biosynthesis flux [34]. While it is unclear whether the purinosome is a dynamic liquid droplet or a more crystalline-like structure, new findings have hinted at its mechanisms of assembly and regulation. For example, assembly of the entire purinosome complex is coordinated by three core proteins, consistent with a nucleation-like assembly mechanism similar to those controlling Stp complex and Cajal Body formation described above [35]. Furthermore, phosphorylation also appears to be a key regulator of purinosome assembly. Three of the six purine biosynthesis enzymes are phosphorylated by the CK2 kinase [36] and inhibition of the CK2 kinase stimulates purinosome production [37] indicating that phosphorylation represses purinosome assembly.
Allosteric regulation by binding to accessory cofactors has long been known to be important for regulating metabolic enzyme activity, but recent studies suggest that similar mechanisms can also control enzyme assembly. For example, modulating the activity of the CtpS enzyme by adding DON, the glutamine substrate mimic, making active site point mutants, or adding high levels of its pathway end product, CTP, can alter CtpS polymerization in bacteria and yeast [38,39]. Another interesting example comes from morpheeins, which are homoligomeric enzymes that can form a variety of higher order multimers [40]. Interconversion between forms requires that the individual oligomers disassemble and undergo a conformational change, which can be induced by binding a metabolite or other protein. Thus, while protein conformational changes in morpheeins are generally thought to determine enzyme activity [41] they could also determine complex formation by altering binding partners or simply by altering binding site valency.
In the case of enzymes that catalyze easily reversible reactions, enzyme complexes can drive reactions in a favorable direction by locally concentrating substrates and not products. For example, the carboxysome, found in cyanobacteria, is an icosohedral protein cage that locally concentrates carbon dioxide, which in turn increases the forward reaction rate of the kinetically fickle enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) [42]. Similar protein cages serve other functions such as sequestering toxic intermediate metabolites. Salmonella enterica constructs such micro-compartments to catabolize 1,2-propanediol without exposing its DNA to the mutagenic intermediate propionaldehyde [43]. Phase-separated bodies could hypothetically serve a similar function if their local environments enriched or depleted substrates based on their electro-chemical properties. Thus, containment of metabolites by mesoscale protein assemblies enables cells to expand the types of reactions they can effectively perform.
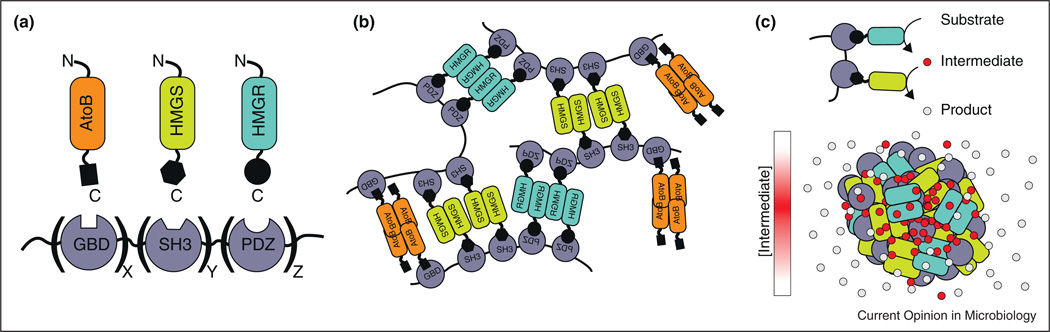
Finally, while we have focused on naturally occurring mesoscale assemblies, recent efforts have begun engineering synthetic assemblies for increased metabolic pathway titers. DNA-based [44] and RNA-based [45•] assemblies have yielded modest increases in the resveratrol, 1,2-propanediol, mevalonate, and H2 pathway titers. A different strategy used artificial scaffolds with multivalent metazoan binding domains to co-recruit the enzymes of the mevalonate biosynthesis pathway, leading to a maximum 77-fold increase in mevalonate titers [46]. One explanation for such a dramatic increase is that the multivalent scaffolds formed mesoscale assemblies similar to the multivalent phase-separated structures discussed above (Figure 2) [19•]. Such networks could possibly form without a scaffold as long as the participating enzymes were oligomeric and bound each other. Furthermore, in addition to controlling pathway titers, such assemblies could be useful for controlling branchpoint fluxes by adjusting enzyme-binding partners in response to particular cellular needs.
Figure 2.
Metabolic enzyme scaffold assembly process. (a) Enzymes involved in mavelonate biosynthesis paired with their cognate metazoan scaffolding domain. (b) Individual metabolic enzymes oligomerize and thus create a network of connected scaffold proteins and metabolic enzymes. (c) Basic two-step metabolic pathway shown scaffolded. When metabolic enzymes oligomerize into assembly the intermediate metabolites are concentrated within the assembly while the product freely diffuses away.
Conclusion
Here we highlight the self-assembling, mesoscale structures as a general mechanism for organizing biological functions. Not only does life utilize multimeric assembly as a rapid and efficient method of bridging the nanoscale to the microscale, but it also enables the coupling of time and space through the kinetics of assembly. Recently discovered examples of polymeric proteins and phase-separated liquid droplets suggest that both eukaryotic and prokaryotic cells abound with these mesoscale assemblies. Such structures serve to organize functions by concentrating components of a process, excluding unnecessary ones, or orienting them in space. Fundamental biological functions such as cell division, cell signaling, and metabolism have all been shown to benefit from this type of organization. Building upon generalizable principles of assembly and regulation such as nucleation and valency, provides a roadmap for probing an assembly’s function.
Just as cells and engineers have learnt to improve cellular function through the construction of mesoscale assemblies, pathogens may exploit such assemblies to rewire cellular pathways for the pathogen’s benefit. Looking forward, we may find that metabolic factories could be used to promote specific metabolic pathways or perhaps drive novel reactions that are needed for the pathogen’s life cycle. In fact, bacterial and viral pathogens have evolved toxic protein scaffolds capable of re-organizing eukaryotic signaling pathways [16,47]. Thus, furthering our understanding of the assembly, regulation, and function of polymeric scaffolds will improve our understanding of basic cell biology, our ability to engineer improved cellular activities, and our capacity to combat cellular disruption by pathogens.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• of outstanding interest
- 1.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. Garland Science; 2008. [Google Scholar]
- 3.Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci U S A. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilhofer M, Jensen GJ. The bacterial cytoskeleton: more than twisted filaments. Curr Opin Cell Biol. 2012 doi: 10.1016/j.ceb.2012.10.019. http://dx.doi.org/10.1016/j.ceb.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–1282. doi: 10.1016/j.cell.2012.10.046. The discovery of the StpABCD complex is shown to compartmentalize the bacterial periplasm and cytoplasm. Competition assays demonstrate the advantage of this subcellular organizing function.
- 7.Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2010;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB Complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massidda O. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol. 2003;185:6209–6214. doi: 10.1128/JB.185.20.6209-6214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamoen LW, Meile J-C, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol. 2006;59:989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 13.Monahan LG, Robinson A, Harry EJ. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol Microbiol. 2009;74:1004–1017. doi: 10.1111/j.1365-2958.2009.06914.x. [DOI] [PubMed] [Google Scholar]
- 14.Gundogdu ME, Kawai Y, Pavlendova N, Ogasawara N, Errington J, Scheffers D-J, Hamoen LW. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 2011;30:617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawson T. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 16.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the toll-like receptor-interleukin-1 receptor superfamily. Science Signaling. 2012;5 doi: 10.1126/scisignal.2003124. re3-re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baillie GS. Compartmentalized signalling: spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 2009;276:1790–1799. doi: 10.1111/j.1742-4658.2009.06926.x. [DOI] [PubMed] [Google Scholar]
- 19. Lee H, DeLoache WC, Dueber JE. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14:242–251. doi: 10.1016/j.ymben.2011.09.003. The assembly of metabolic enzyme complexes is discussed as a way of creating subcellular microdomains with relatively high concentrations of intermediates similar to the microdomains observed in Ca+ and CAMP signaling.
- 20.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 21.Spector DL. SnapShot: cellular bodies. Cell. 2006;127:1071.e1–1071.e2. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. A companion study to [24] here they demonstrate the possibility of phosphorylation as a means of dynamically regulating the components within a hydrogel.
- 24. Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. This study used a small molecule to precipitate the components of RNA granules and determined their shared structural component to be the Low Complexity peptide sequence, which they then show to be sufficient for the formation of dynamic hydrogels.
- 25.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 26. Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2013;483:336–340. doi: 10.1038/nature10879. The authors synthetically engineer various multivalent assemblies, demonstrate that they resemble phase-separated gels, and show how phosphorylation could be used to regulate their assembly.
- 27.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 28.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nature. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 29.Rajendra TK, Praveen K, Matera AG. Genetic analysis of nuclear bodies: from nondeterministic chaos to deterministic order. Cold Spring Harb Symp Quant Biol. 2011;75:365–374. doi: 10.1101/sqb.2010.75.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 31.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 32.Chen AH, Silver PA. Designing biological compartmentalization. Trends Cell Biol. 2012;22:662–670. doi: 10.1016/j.tcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Agapakis CM, Boyle PM, Silver PA. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat Chem Biol. 2012;8:527–535. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- 34.An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci U S A. 2010;107:12872–12876. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng Y, Gam J, French JB, Zhao H, An S, Benkovic SJ. Mapping protein–protein proximity in the purinosome. J Biol Chem. 2012;287:36201–36207. doi: 10.1074/jbc.M112.407056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen J. PhD thesis. University of San Francisco; 2008. Development and Application of Technologies to Study Individual Kinase Substrate Relationships. [Google Scholar]
- 37.An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J Biol Chem. 2010;285:11093–11099. doi: 10.1074/jbc.M110.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nature. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selwood T, Jaffe EK. Dynamic dissociating homo-oligomers and the control of protein function. Arch Biochem Biophys. 2012;519:131–143. doi: 10.1016/j.abb.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mouilleron S, Badet-Denisot MA, Pecqueur L, Madiona K, Assrir N, Badet B, Golinelli-Pimpaneau B. Structural basis for morpheein-type allosteric regulation of Escherichia coli Glucosamine-6-phosphate synthase: equilibrium between inactive hexamer and active dimer. J Biol Chem. 2012;287:34533–34546. doi: 10.1074/jbc.M112.380378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Micro. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 43.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnsek J, Tomsic N, Avbelj M, Gaber R, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr888. http://dx.doi.org/10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. Synthetic RNA scaffolds are designed and used to colocalize enzymes involved in H2 biosynthesis. Large increases in pathway titers are observed.
- 46.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 47.Lesser CF, Leong JM. Bacterial scaffolds assemble novel higher-order complexes to reengineer eukaryotic cell processes. Sci Signal. 2011;4:e32. doi: 10.1126/scisignal.2002252. [DOI] [PMC free article] [PubMed] [Google Scholar]