Abstract
Most species’ sex chromosomes are derived from ancient autosomes and show few signatures of their origins. We studied the sex chromosomes of Drosophila miranda, where a neo-Y chromosome originated only about 1 million years (MY) ago. Whole genome and transcriptome analysis reveals massive degeneration of the neo-Y, that male-beneficial genes on the neo-Y are more likely to undergo accelerated protein-evolution, and that neo-Y genes evolve biased expression towards male-specific tissues, i.e. the shrinking gene content of the neo-Y becomes masculinized. In contrast, while older X chromosomes show a paucity of genes expressed in male tissues, neo-X genes highly expressed in male-specific tissues undergo increased rates of protein evolution if haploid in males. Thus, the response to sex-specific selection can shift at different stages of X differentiation, resulting in masculinization or demasculinization of the X-chromosomal gene content.
X and Y chromosomes follow distinctive evolutionary trajectories after recombination becomes suppressed between ancestral homologous autosomes with a sex-determining function (1). The lack of recombination greatly impairs natural selection on the Y, which loses most of its original genes and often accumulates repetitive DNA (2). However, Y chromosomes are not complete evolutionary dead ends; instead their male-limited transmission favors the gain of male-related genes (masculinization). Low gene density yet enrichment of male-specific genes is shared among many independently evolved ancient Ys (3, 4), but few traces of their evolutionary origins remain, making processes involved in Y degeneration little understood. Conversely, the X still recombines in females, and selection can effectively purge deleterious alleles and incorporate beneficial mutations (2). Unlike autosomes, the X is transmitted more often through females than males, favoring an underrepresentation of male-beneficial genes on the X (demasculinization) (5, 6). Further, almost all X-linked genes are haploid in males (hemizygous) and can fix recessive male-advantageous alleles more easily than autosomes (7), potentially leading to its masculinization. These aspects of X chromosome biology dictate unusual and sometimes opposing patterns of sequence and expression evolution (8), but are difficult to distinguish in ancestral systems.
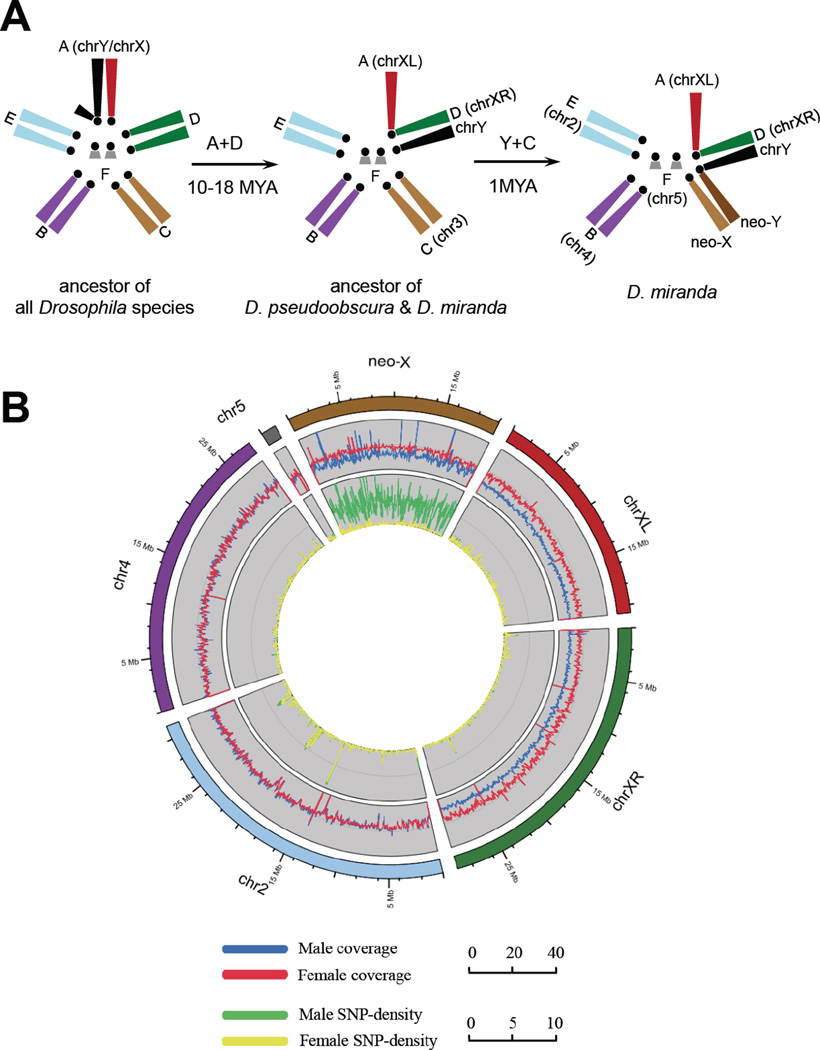
The genome of Drosophila fly species can be divided into a set of homologous chromosomal arms called ‘Muller elements’ (9). Chromosomal fusions between the ancient sex chromosomes (Muller-A element, referred here as ‘chrXL’ and ‘chrY’) with autosomes have repeatedly generated younger secondary sex chromosomes. Male Drosophila lack meiotic recombination (9); thus Y-fused autosomes (neo-Ys) cannot recombine with their homologs (neo-Xs), which sets the stage for sex chromosome differentiation. D. miranda, harbors two such successive fusions that created sex chromosomes of different ages (10) (Fig. 1A). Muller-D became sex-linked before the divergence of D. miranda and D. pseudoobscura roughly 10-18 MY ago, and resembles the ancestral sex chromosomes; the non-recombining Muller-D element is almost completely degenerated and now part of the heterochromatic Y (11), while its recombining counterpart (‘chrXR’) evolved an architecture typical of an X (5, 12). Another fusion unique to D. miranda involves Muller-C element (referred as ‘neo-X’ and ‘neo-Y’) and occurred only about 1 MY ago (13). This very young sex-chromosome system is in the process of evolving from a pair of ordinary autosomes to a pair of heteromorphic sex chromosomes. Cytogenetic studies and investigations of individual genes or genomic regions have revealed that the D. miranda neo-Y is intermediately degenerate (10, 14–16), rendering the neo-X partially hemizygous.
Figure 1.
(A) The reconstructed evolutionary history of D. miranda sex chromosomes. The ancestral sex chromosome chrXL (red) fused to Muller-D element, creating chrXR (green) and the unfused element became part of the heterochromatic ancestral chrY (black). In D. miranda, Muller-C subsequently fused to chrY, creating a neo-Y chromosome (fused Muller-C element, dark brown), and a neo-X (unfused Muller-C element, light brown). (B) Shown are coverage (mapped read counts every 50kb region) and SNP density (sites/kb) derived separately from male and female genomic reads in a 5kb sliding window across the D. miranda genome. The high male SNP density along the neo-X (male: 3.696 vs. female: 0.080 sites/kb) reflects divergence between the neo-X and neo-Y chromosome.
We conducted a whole-genome analysis of the neo-sex chromosomes, integrated with transcriptomes from multiple tissues using next-generation sequencing technology. We sequenced both sexes of an inbred D. miranda strain (MSH22), and assembled scaffolds were anchored onto the D. pseudoobscura genome (17, table S1). We annotated a total of 14819 proteins for the D. miranda genome, using 16133 D. pseudoobscura proteins as queries. We assessed the quality of our assemblies using BAC clone sequences and 454 data (table S3) and validated our chromosomal assignments by comparing male and female mapping coverage along each chromosome (Fig. 1B). Coverage of male and female coincides well along the autosomes, while both chrXL and chrXR show only about half as many reads mapped in males as in females (table S4, Fig. 1B), indicating that their homologous ancient Ys are too degenerate as to show any significant sequence similarity. In contrast, male coverage along the neo-X is about 3/4 that of females, suggesting that parts of the neo-Y are highly diverged from the neo-X. 71.8% of the neo-X sequence can be aligned with the neo-Y, with 1.5% (±0.00093%) nucleotide divergence between aligned regions. Coding regions are under stronger selective constraint and exhibit a higher alignment rate (92.6%) and lower divergence between the neo-X and neo-Y (1.1% ±0.18%).
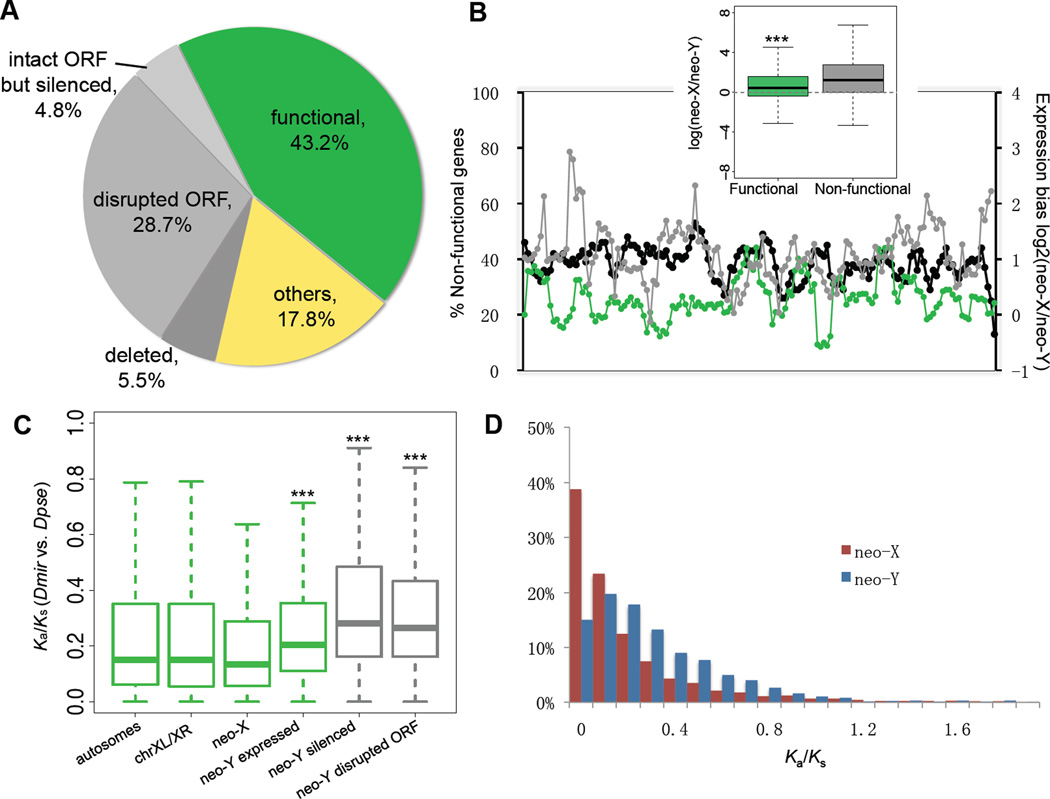
We compared protein-coding regions to gain insights into the process of gene loss of a Y. The neo-X and neo-Y are derived from a gene-rich autosome with initially identical gene sets (10). 2951 genes with intact open reading frames (ORF) could be annotated on the neo-X, while only 1941 intact ORFs were identified on the neo-Y. The remaining 1010 genes that were ancestrally present on the neo-Y (34.2%) are probably non-functional: 848 ORFs are disrupted by premature terminal codons (PTC) and/or frame-shift mutations, and 162 genes are partially or completely deleted from the neo-Y (Fig. 2A, 17). No spatial clustering of non-functional genes was detected on the neo-Y (Fig. 2B).
Figure 2.
(A) Composition of neo-Y genes with regards to inferred functionality (green: intact ORFs and detectable expression in adult male; grey: disrupted ORF and/or silenced expression; yellow: genes without neo-X expression or without diagnostic SNPs). (B) The chromosomal distribution of non-functional genes across a sliding window size of 20 genes (black line). Average neo-X expression bias within the investigated window was calculated from log ratios of neo-X vs. neo-Y expression for functional (green) and non-functional (grey) genes. Functional neo-Y genes show significantly less neo-X biased expression than non-functional genes (boxplot, P<2.2e-16, Wilcoxon test). (C) Evolutionary rate comparisons (Ka/Ks ratios relative to D. pseudoobscura) among genes on different chromosomes. Wilcoxon tests show significant differences in Ka/Ks ratios between neo-X vs. neo-Y genes, genes with intact neo-Y copies with vs. without expression (P=0.000242) and disrupted vs. intact transcribed neo-Y genes (P=2.971e-12). Different levels of significance are marked as asterisks. (D) The frequency distribution of Ka/Ks ratios of neo-X and neo-Y genes.
Severely disabling mutations have also accumulated in regulatory regions on the neo-Y. We compared allelic expression of 2165 neo-sex linked genes in males that were expressed from the neo-X. 883 genes (40.8%) show similar levels of expression from both chromosomes, while 947 (43.7%) are expressed at a significantly higher level from the neo-X, and 335 genes (15.5%) are neo-Y biased (binomial-test, P<0.05). 220 genes (10.2%) with a transcribed neo-X copy are completely silenced on the neo-Y. A large fraction of neo-Y genes is still transcribed, despite having disrupted ORFs: 83.0% of all neo-Y genes with nonsense mutations are transcribed, yet at a significantly lower level than genes with intact neo-Y ORFs (Fig. 2B, Wilcoxon test, P<2.2e-16). This implies that down-regulated genes either tend to acquire nonsense mutations or that pseudogenes become transcriptionally silenced on the neo-Y, but could also reflect up-regulation of the neo-X copy at non-functional neo-Y genes (dosage compensation) (12). Gene loss is non-random with regard to gene function. Non-functional and down-regulated genes on the neo-Y are significantly enriched for various gene ontology (GO) categories of primary metabolic processes (GO: 0046165, 0016042, GO: 0006094, etc.), while genes involved in regulatory (GO: 0050789, 0048519) or developmental processes (GO: 0032502) tend to maintain non-biased expression and intact ORFs (one-tailed Fisher’s exact test, P<0.01, table S5,6). Thus, while natural selection is impaired on the neo-Y, it tends to maintain haploinsufficient genes (such as regulatory genes), while haplosufficient genes (like metabolic enzymes) are more prone to degeneration (18). Overall, about 40% of the neo-Y genes have lost their functions within 1 MY.
Deleterious mutations with more subtle effects also accumulate on the neo-Y. We calculated pairwise rates of non-synonymous (Ka) and synonymous changes (Ks), and their ratios (ω), using D. pseudoobscura as an outgroup. Genes on the neo-Y evolve significantly faster than their neo-X homologs or genes on other chromosomes at both synonymous and nonsynonymous sites (table S7, Wilcoxon test, P<0.01). Selection to maintain codon usage bias is reduced in D. miranda (19); thus, patterns of synonymous changes (Ks) should largely reflect mutational differences. Although neo-Y genes generally show lower codon bias (table S7), this difference is not statistically significant between the neo-X and neo-Y, or between functional and non-functional neo-Y genes (P>0.05, Wilcoxon test). Thus, the contribution of codon bias selection to elevated Ks patterns appears limited and instead may reflect male-driven evolution (20). Neo-Y genes with disrupted ORFs evolve significantly faster at the protein-level than those with intact ORFs (Fig. 2C, Wilcoxon test, P=2.97e-12) and intact but silenced neo-Y genes evolve significantly faster than those still expressed from the neo-Y (Fig. 2C, Wilcoxon test, P= 0.000242). This suggests that neo-Y genes with disrupted ORFs or silenced expression are subject to little selective constraint, and we classified all these genes as non-functional in subsequent analysis (fig. S4, 17). The distribution of ω at neo-Y genes is shifted towards neutral evolution (ω =1) and the proportion of genes under strong selective constraints (ω <0.1) is greatly reduced compared with neo-X genes (15.02% vs. 38.78%, Fig. 2D). This pattern is consistent with an accumulation of mildly deleterious amino-acid mutations at many neo-Y loci.
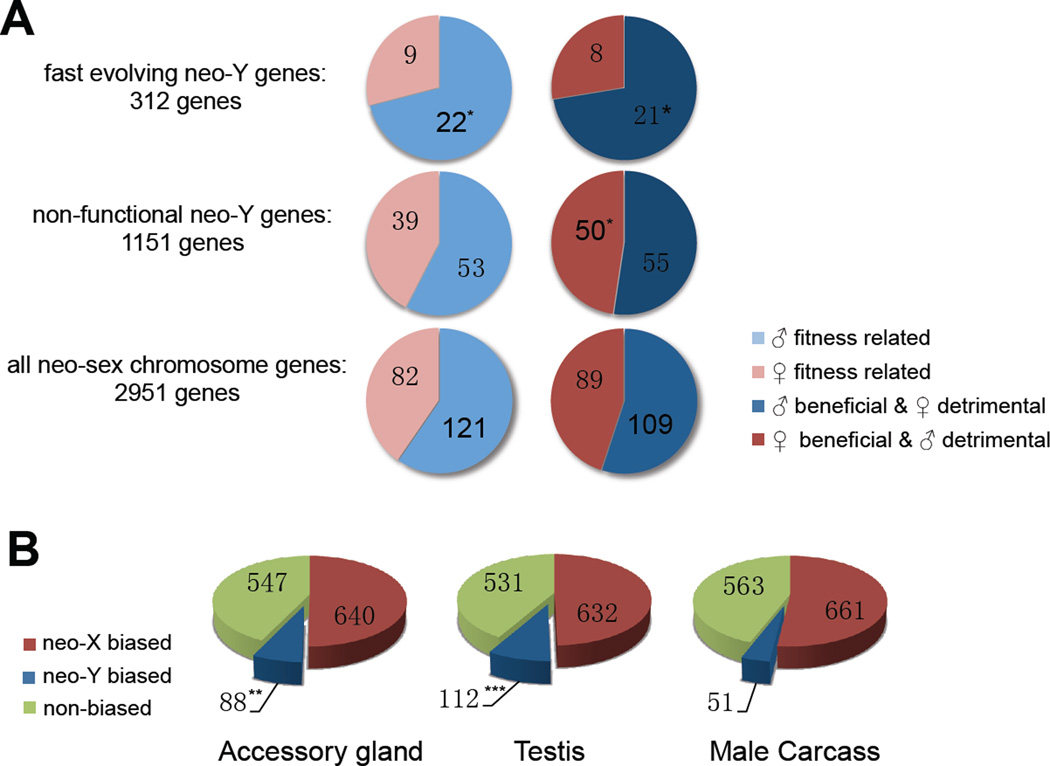
Decay in gene function is the primary but not only force driving early Y evolution. Y-chromosomes are limited to males, and genes found on ancient Ys often have male-specific function (3,4). It is unclear if male-related genes only accumulate on old, gene-poor Ys where adaptive mutations experience little interference from segregating deleterious mutations, or whether masculinization accompanies early stages of Y evolution, and thus contributes to degeneration through hitchhiking effects (fixations of deleterious mutations linked to strongly beneficial alleles) (2). To explore if adaptive evolution for male-function is operating on neo-Y genes, we performed maximum-likelihood analysis of the lineage-specific ω ratio of all functional neo-sex genes (17). We identified 312 transcribed genes evolving significantly faster on the neo-Y lineage, (compared to 66 genes evolving faster on the neo-X lineage, P<0.05, likelihood ratio test), and evaluated if this set of functional neo-Y genes shows characteristics of male-specific selection. Recently acquired male-limited inheritance releases sexually antagonistic male-beneficial/female-detrimental mutations from counter selection in females, and such genes may show increased rates of adaptive evolution on the neo-Y. We classified orthologous D. miranda genes according to a sexual antagonism scheme proposed for D. melanogaster (21, table S8), and found a significant enrichment of male-beneficial/female-detrimental genes or genes correlated with male-specific fitness among the fast-evolving neo-Y genes (Fisher’s exact test, P<0.05, Fig. 3A). These gene categories are not enriched among non-functional genes, indicating that male-beneficial genes are under selective constraint on the neo-Y and that male-specific selection is driving adaptive protein evolution at some neo-Y genes. Genes related to male fitness are more likely to be functional on the neo-Y, while genes classified as female-beneficial/male-detrimental are enriched for genes with disrupted ORFs or silenced expression (Fig. 3A). This suggests that while selection operates to maintain male-beneficial genes, those harming males are actively removed from the neo-Y. Transcriptome analysis of male-specific organs (testis and accessory gland) vs. male somatic carcass tissues (removing these organs) provides additional evidence for masculinization. In all tissues, the majority of genes show neo-X biased expression. However, testis and accessory glands harbor about twice as many genes with neo-Y biased expression compared to male somatic carcass (Fig. 3B, P<0.05, Fisher’s exact test), caused by up-regulation of the neo-Y alleles (fig. S5, 17). Also, functional neo-Y genes in D. miranda have evolved significantly increased expression specificity in accessory glands (relative to male somatic carcass tissues) compared to their orthologous D. pseudoobscura genes (fig. S6,7, 17). Further, genes with significant neo-Y biased expression are enriched for male reproductive GO terms including ‘insemination’, ‘copulation’ and ‘reproductive process’ (GO: 0007320, 0007620, 0022414) (table S11). Thus, although most neo-Y genes undergo degeneration, a subset acquires or improves male-related functions. The absence of sexual conflicts on a male-limited chromosome enables male-specific adaptation, and may significantly contribute to Y degeneration through the hitchhiking effect (22).
Figure 3.
(A) Sex-specific fitness effects and sexual antagonism of neo-sex genes (light-blue: male-fitness related; light-red: female-fitness related; dark-blue: male-beneficial/female-detrimental; dark-red: female-beneficial/male-detrimental). Significance is evaluated by comparing all neo-sex genes to either fast evolving or non-functional neo-Y genes (‘*’ P-value <0.05, ‘**’ P-value <0.01). (B) The number of neo-X biased (red), neo-Y biased (blue) and non-biased (green) genes in different tissues of male D. miranda.
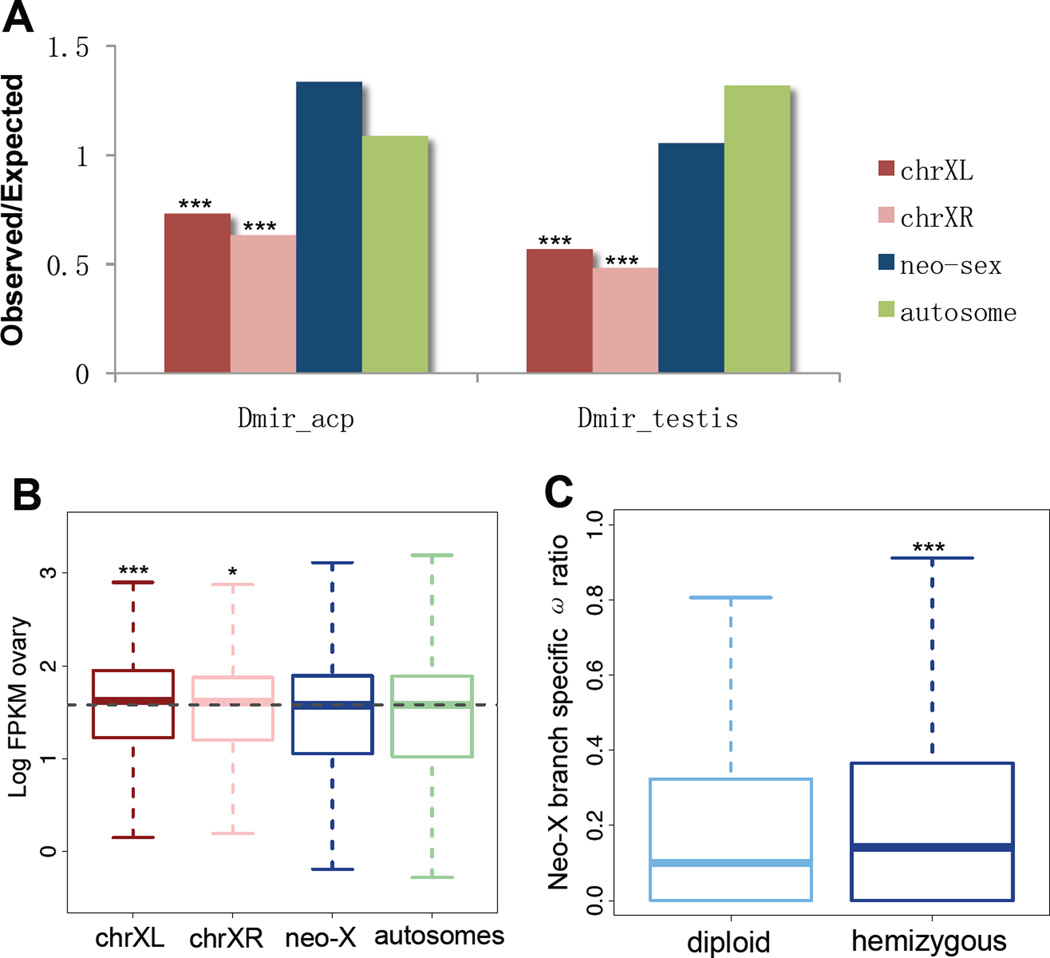
Evolutionary forces on an evolving X chromosome can operate in opposite directions. Female-biased transmission will favor female-specific genes and disfavor male-beneficial genes. Old X chromosomes in Drosophila contain a deficiency of genes expressed in male-specific tissue (5, 6). In D. miranda, both chrXL and chrXR show a clear under-representation of testis and accessory gland genes (Fig. 4A, Fisher’s exact test, P<0.05), and ovary expression is higher for genes located on chrXL and chrXR (Fig. 4B, P<0.05, 17). Thus, an X chromosome becomes fully demasculinized and feminized within 10-18 MY in Drosophila (5). No chromosome-wide changes in overall expression patterns in sex-tissues are observed on the neo-X (Fig. 4A,B), and its origin may be too recent for a large turnover of gene content to have taken place. Young X-linked genes in Drosophila tend to be male-biased (23), possibly due to the fixation of recessive, male-beneficial mutations, and demasculinization appears to happen over longer evolutionary time periods. In D. miranda, about half of the neo-X genes have no functional neo-Y homologs (they are hemizygous) and might show different evolutionary dynamics than those with functional neo-Y copies (i.e. diploid neo-X genes). We find that hemizygous genes evolve significantly faster at their neo-X branch than diploid ones (median ω ratios: 0.1416 vs. 0.0997, Wilcoxon test, P= 0.0005187; Fig. 4C). If male-beneficial adaptation drives elevated protein evolution at hemizygous neo-X genes, this pattern should be more pronounced for genes expressed highly in a male-specific tissue. Indeed, hemizygous (but not diploid) neo-X genes show a significant positive correlation between their absolute expression levels in accessory gland and ω ratios (F-statistic comparing diploid vs. hemizygous neo-X genes, P<0.05; 17) and a similar trend is observed for expression levels in testis but not somatic genes (fig. S1, 1, 17). Thus, masculinization occurs on hemizygous neo-X loci, while demasculinization and feminization dominate on chrXL and chrXR. Gene loss, gain and movement appear as the dominant mechanisms for depleting male genes on the X (5), and differences in dominance and rates could contribute to the observed temporal dynamics of masculinization and feminization/demasculinization. Recessive male-beneficial amino acid substitutions might accumulate relatively quickly in hemizgyous neo-X genes, while gene content turnover removing male genes might proceed slowly over longer time periods. Also, demasculinization might result from cellular processes that only operate on older X chromosomes, such as dosage compensation or silencing of the X during spermatogenesis (24, 25). Overall, a variety, and sometimes opposing, evolutionary forces operate on evolving sex chromosomes due to common sexual conflicts.
Figure 4.
(A) The observed/expected ratio of genes highly expressed (top 500; for different cutoffs see fig. S7) in testis or accessory glands. (B) Log-based absolute expression levels (FPKM) from ovary for each chromosome (see also fig. S7). (C) The ω ratio on the neo-X branch at hemizygous and diploid neo-X genes.
Supplementary Material
Acknowledgments
We thank R. Nielsen, M. Eisen, X. Xun and L. Tian for help and discussion. Funded by NIH grants (R01GM076007 and R01GM093182) and a Packard Fellowship to D.B. All DNA/RNA-seq reads are deposited at http://www.ncbi.nlm.nih.gov/sra under the accession number SRA048115. The genome assembly and annotation is available on NCBI under BioProject ID PRJNA77213
Footnotes
Supporting Online Material
Materials and Methods
figs. S1 to S11
table S1 to S12
References (26-42)
References and Notes
- 1.Bull JJ. Evolution of sex determining mechanisms. Benjamin Cummings; Menlo Park, CA: 1983. [Google Scholar]
- 2.Charlesworth B, Charlesworth D. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1563. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koerich LB, Wang X, Clark AG, Carvalho AB. Nature. 2008 Dec 18;456:949. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skaletsky H, et al. Nature. 2003 Jun 19;423:825. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 5.Sturgill D, Zhang Y, Parisi M, Oliver B. Nature. 2007 Nov 8;450:238. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betran E, Thornton K, Long M. Genome Res. 2002 Dec;12:1854. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth B, Coyne J, Barton N. Am. Nat. 1987;130:113. [Google Scholar]
- 8.Vicoso B, Charlesworth B. Nat. Rev. Genet. 2006;7:645. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 9.Muller HJ. In: The New Systematics. Huxley J, editor. Oxford: Clarendon Press; 1940. pp. 185–268. [Google Scholar]
- 10.Steinemann M, Steinemann S S. Genetica. 1998;102-103:409. [PubMed] [Google Scholar]
- 11.Carvalho AB, Clark AG. Science. 2005 Jan 7;307:108. doi: 10.1126/science.1101675. 2005. [DOI] [PubMed] [Google Scholar]
- 12.Marín I, Franke A, Bashaw GJ, Baker BS. Nature. 1996 Sep;383:160. doi: 10.1038/383160a0. [DOI] [PubMed] [Google Scholar]
- 13.Bachtrog D, Charlesworth B. Nature. 2002;416:323. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- 14.Steinemann M, Steinemann S, Lottspeich F. Proc Natl Acad Sci U S A. 1993 Jun 15;60:5737. doi: 10.1073/pnas.90.12.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachtrog D. Nat. Genet. 2003;34:215. doi: 10.1038/ng1164. [DOI] [PubMed] [Google Scholar]
- 16.Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genome Biol. 2008 Feb 12;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Supporting Online Material. [Google Scholar]
- 18.Kondrashov FA, Koonin EV. Trends Genet. 2004 Jul;20:287. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Bachtrog D. Genetics. 2003 Nov;165:1221. doi: 10.1093/genetics/165.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachtrog D. Mol. Biol. Evol. 2008 Apr;25:617. doi: 10.1093/molbev/msn020. [DOI] [PubMed] [Google Scholar]
- 21.Innocenti P, Morrow EH. PLoS Biol. 2010;8:e1000335. doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachtrog D. Nat. Genet. 2004;36:518. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- 23.Zhang YE, Vibranovski MD, Krinsky BH, Long M. Genome Res. 2010 Nov;20:1526. doi: 10.1101/gr.107334.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachtrog D, Toda N, Lockton S. Curr Biology. 2010 Aug 24;20:1476. doi: 10.1016/j.cub.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lifschytz E, Lindsley D. Proc Natl Acad Sci U S A . 1972;69:182. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.