Abstract
VGX-1027 [(S,R)-3-phenyl-4,5-dihydro-5-isoxasole acetic acid] is a small molecule compound with immunomodulatory properties, which favourably influences the development of immuno-inflammatory and autoimmune diseases in different animal models such as type 1 diabetes mellitus, pleurisy, rheumatoid arthritis and inflammatory bowel disease. However, the precise mechanism of action of VGX-1027 remains to be ascertained. With this aim, we have studied the immunomodulatory effects of VGX-1027 in vitro, using a genome-wide oligonucleotide microarray approach, and in vivo, using the NZB/NZW F1 model of systemic lupus erythematosus. Microarray data revealed that the administration of VGX-1027 profoundly affected the immune response to exogenous antigens, by modulating the expression of genes that are primarily involved in antigen processing and presentation as well as genes that regulate immune activation. When administered in vivo VGX-1027 ameliorated the course of the disease in the NZB/NZW F1 mice, which correlated with higher per cent survival and improved clinical and histopathological signs. The data presented herein support the theory that VGX-1027 modulates immunity, probably by inhibiting inflammatory antigen presentation and so limiting immune cell expansion.
Keywords: autoimmunity, bioinformatics, cell activation
Introduction
VGX-1027 [(S,R)-3-phenyl-4,5-dihydro-5-isoxasole acetic acid] is an isoxazoline compound currently under development for the treatment of human immuno-inflammatory diseases such as type 1 diabetes mellitus and rheumatoid arthritis. VGX-1027 appears to target macrophage function, as it inhibits the lipopolysaccharide (LPS) -induced nuclear factor-κB and p38 mitogen-activated protein kinase signalling pathways.1 Also, VGX-1027 down-regulates tumour necrosis factor-α production from macrophages in response to activators of both Toll-like receptor-4 (e.g. LPS) and Toll-like receptor-2/6 (e.g. zymosan). VGX-1027 is effective in vivo when given both intraperitoneally and orally and it dampens the development of immuno-inflammatory and autoimmune diseases in different animal models such as type 1 diabetes mellitus,2 pleurisy, rheumatoid arthritis1 and inflammatory bowel diseases.3 Acute and sub-acute toxicological studies have showed no toxicity at pharmacological doses in pre-clinical models.3
To further elucidate the mode of action of VGX-1027, which could identify additional therapeutic targets, we have presently studied the immunomodulatory effects of VGX-1027 in vitro, using a genome-wide oligonucleotide microarray screening approach. Pathway enrichment analysis of the microarray data predicted a significant modulation of the systemic lupus erythematosus (SLE) pathway and this prompted us to correlate the in vivo potential of this drug in the NZB/NZW F1 mice model of SLE. We observed a clear amelioration of the disease, which correlated with direct inhibition of cytokine genes as observed in the microarray study.
Materials and methods
Transcriptional profile analysis
Peripheral blood mononuclear cells from three individual healthy donors were obtained from the University of Pennsylvania School of Medicine, Immunology Clinical Core. Cells (5 × 106) were treated with either LPS (5 μg/ml), LPS (5 μg/ml) + VGX-1027 (10 μm) or PBS for 48 hr and, subsequently, RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA) following the manufacturer's instructions.
RNA was hybridized to the Human Gene 1.0 ST array (Affymetrix, Santa Clara, CA). Microarray data were analysed using the web-based utility Babelomics 4.2 and MultiExperiment Viewer.4,5 Data pre-processing and normalization was achieved by performing Robust Multichip Analysis. Principal component analysis (PCA) was conducted on all genes to assign the general variability in the data to a reduced set of variables. Hierarchical clustering was used to determine the relative distance of each sample using Pearson's correlation as similarity comparison. Non-negative Matrix Factorization was used to assign samples to clusters based on their highest metagenes expressions. Gene expression differences were assessed by means of Student's t-test, and genes with P < 0·01 were considered differentially expressed. Functional analysis of microarray data was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool.6
In vivo treatment of lupus-prone NZB/NZW F1 mice and serological, histological and immunological analyses
Female NZB/NZW F1 mice were obtained from Charles River Laboratories (Lecco, Italy) and acclimated for 1 week before the study at the animal house of the Department of Bio-Medical Sciences of the University of Catania (Italy). The mice were maintained under non-specific pathogen-free conditions and studies were performed in accordance with an approved IACUC protocol. Blood samples were obtained for baseline studies, following which the mice were divided into the different experimental groups. The NZB/NZW F1 mice were treated for 20 weeks beginning at 16 weeks of age. VGX-1027 was administered in sterile Na2HPO4 at a dose of 20 mg/kg by daily intraperitoneal injection. Control mice received vehicle alone. Mice were followed for the development of renal disease, as measured by proteinuria, and for survival. Proteinuria was measured using commercially available semi-quantitative strips Albustix (Miles Laboratories, Elkhart, IN), graded as: trace (+/−) = 10 mg/dl; (+) = 30 mg/dl; (++) = 100 mg/dl, (+++) = 300 mg/dl and (++++) = 1000 mg/dl. For statistical analysis the intensity of the colorimetric reaction of each mouse was reported numerically (10 mg/dl = 0·5, 30 mg/dl = 1, 100 mg/dl = 2, 300 mg/dl = 3 and 1000 mg/dl = 4) and the mean value from each experimental group was calculated by dividing the total score by the number of mice in that group.7
After 10 weeks of treatment and at the end of the experimental period, blood was sampled for measurement of autoantibodies. Antibodies to double-stranded DNA (anti-dsDNA antibodies) in the serum were measured by ELISA. At the completion of the study, on week 36, the remaining mice from the different groups were killed by CO2 asphyxiation, blood was sampled by cardiac puncture, and kidney tissues were removed and processed for protein analysis and histology. For histological analyses, the left kidney from each animal was removed and fixed in 10% buffered formalin for subsequent haematoxylin & eosin staining. All histological scoring was performed by an independent medical pathologist. The pathological lesions were graded from 0 to 4 as follows: 0, normal; 1, a small increase of cells in the glomerular mesangium; 2, a larger number of cells in the mesangium; 3, glomerular lobular formation and thickened basement membrane; 4, glomerular crescent formation, sclerosis, tubular atrophy and casts. The score for each animal was calculated by dividing the total score by the number of glomeruli observed.7
Spleens were aseptically isolated and crushed to yield single-cell suspensions. Red blood cells were lysed and lymphomonocytes were used to extract total RNA using Trizol reagent following the manufacturer's instructions (Life Technologies, Monza, Italy). Two micrograms of total RNA was retro-transcribed and cDNA was used for the determination of cytokine by real-time PCR. Primer sequences were: interferon-γ (IFNG) forward: ATGAACGCTACACACTGCATC; IFNG reverse: CCATCCTTTTGCCAGTTCCTC; interleukin-10 (IL10) forward: GCTCTTACTGACTGGCATGAG; IL10 reverse: CGCAGCTCTAGGAGCATGTG; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward: AATGGATTTGGACGCATTGGT GAPDH reverse: TTTGCACTGGTACGTGTTGAT.
Statistical analysis
Data are presented as mean ± SD calculated from the data of at least three independent experiments. Statistical analysis for significant differences was performed according to the Student's t-test for unpaired data or Mann–Whitney U-test. Mantel–Cox log-rank test was used to compare the survival curves of VGX-1027-treated groups with those of vehicle-treated groups. A value of P < 0·05 was considered to be statistically significant.
Results
Microarray analysis
Global transcriptional analysis
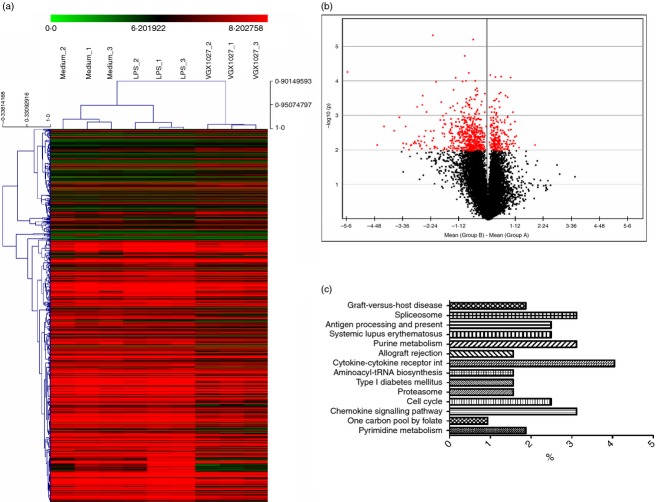
The transcriptional changes associated with VGX-1027 treatment upon LPS stimulation were evaluated using oligonucleotide microarrays. Data indicate that VGX-1027 is able to significantly modify the pro-inflammatory events associated with Toll-like receptor-4 activation. Eigen decomposition yielded nine principal components with the first three of them associated with most of the co-variability among the data (74·9%). Principal components analysis was performed on the entire data set and demonstrates that LPS stimulation of peripheral blood mononuclear cells is associated with a significant modification of the global transcriptome (Fig. 1a). VGX-1027 treatment is also associated with dramatic changes at the transcriptional level, and reveals a complex pattern of gene expression (Fig. 1a). Hierarchical clustering and non-negative matrix factorization confirmed that VGX-1027 is able to modulate gene expression upon LPS stimulation, inducing a distinct phenotype from both medium- and LPS-treated groups (Fig. 1b,c). Treatment with LPS was associated with the modulation of 302 transcripts (Table 1 for the top 50 genes), whereas VGX-1027 induced the modulation of 774 genes relative to LPS (Fig. 2a; Tables 2 and 3 indicate the top 30 down-regulated and up-regulated genes). Most of the genes modulated from VGX-1027 were down-regulated compared with LPS (Fig. 2b for a graphical representation). Functional analysis was performed using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) Database8 on genes with a P < 0·01 and a fold change > 1·5. Functional analysis of the up-regulated genes revealed no enriched pathway whereas the analysis of the down-regulated genes resulted in an over-representation of several pathways (Fig. 2c), with ‘Graft-versus-host disease’, ‘Spliceosome’, ‘Antigen Processing and Presentation’ and ‘Systemic Lupus Erythematosus’ being the most enriched (Fig. 2c).
Figure 1.
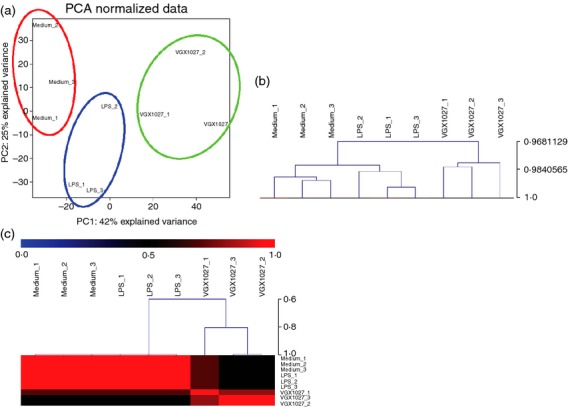
(a) Principal component analysis (PCA) of the transcriptome expressed on either unstimulated peripheral blood mononuclear cells (PBMCs), PBMCs stimulated in vitro with lipopolysaccharide (LPS) or treated with VGX1027 following LPS administration. The PCA was performed on all genes to determine expression trends within the data set. (b) Hierarchical clustering of PBMCs either stimulated with LPS, LPS+VGX-1027 or unstimulated. Pearson's correlation was used as comparison measurement (c) Non-negative matrix factorization of PBMCs either stimulated with LPS, LPS+VGX-1027 or unstimulated.
Table 1.
Top 50 up-regulated genes by lipopolysaccharide
| Gene symbol | P value | Fold change |
|---|---|---|
| CCL7 | 6E-04 | 36·72 |
| IFNG | 5E-03 | 24·42 |
| IL8 | 2E-03 | 14·52 |
| TNFAIP6 | 8E-03 | 13·70 |
| ANKRD1 | 6E-04 | 12·73 |
| MYO1B | 5E-03 | 9·52 |
| CCL3 | 6E-03 | 7·67 |
| IL1B | 3E-03 | 7·35 |
| CCL3L1 | 6E-03 | 7·05 |
| CCL3L1 | 6E-03 | 7·05 |
| CCL3L1 | 6E-03 | 7·05 |
| IL17F | 4E-04 | 6·21 |
| CHRNA6 | 1E-03 | 5·79 |
| GBP6 | 4E-03 | 5·36 |
| IL22 | 2E-03 | 4·50 |
| CCL4 | 7E-03 | 3·80 |
| UBD | 5E-03 | 2·98 |
| IL2RA | 1E-02 | 2·96 |
| UBD | 8E-03 | 2·94 |
| P2RX5 | 1E-02 | 2·58 |
| CD38 | 1E-03 | 2·57 |
| SYTL3 | 1E-02 | 2·40 |
| SLC7A5 | 4E-03 | 2·24 |
| IL2RB | 1E-02 | 2·18 |
| LAMP3 | 2E-03 | 2·16 |
| BATF | 6E-04 | 2·13 |
| FURIN | 4E-03 | 2·03 |
| CTLA4 | 4E-03 | 1·99 |
| MIRN155 | 4E-03 | 1·99 |
| CLU | 5E-03 | 1·93 |
| DUSP4 | 9E-03 | 1·89 |
| KIR2DL3 | 5E-03 | 1·88 |
| HIST2H2AA3 | 7E-03 | 1·88 |
| HIST2H2AA3 | 7E-03 | 1·88 |
| ADAM19 | 5E-03 | 1·88 |
| HIF1A | 5E-03 | 1·86 |
| FEZ1 | 6E-03 | 1·78 |
| IL32 | 3E-03 | 1·78 |
| FAM46C | 1E-02 | 1·73 |
| FBRS | 2E-03 | 1·72 |
| LOC440896 | 4E-03 | 1·71 |
| NFKB2 | 6E-03 | 1·71 |
| JUN | 7E-03 | 1·64 |
| PGLYRP4 | 2E-03 | 1·64 |
| NCDN | 8E-03 | 1·62 |
| KIR2DS2 | 2E-03 | 1·60 |
| GABPB2 | 3E-03 | 1·59 |
| CREM | 5E-03 | 1·58 |
| PTPN1 | 2E-03 | 1·57 |
| PELO | 2E-03 | 1·57 |
Figure 2.
(a) Hierarchical clustering of differentially expressed genes. Each line represents a gene. Gene expression is colour coded from green (lower expression) to red (higher expression). (b) Volcano plot showing in red differentially expressed genes by VGX-1027 compared with lipopolysaccharide (LPS). (c) Enriched pathways by statistically significant down-regulated gene in VGX-1027-treated peripheral blood mononuclear cells compared with LPS-treated cells.
Table 2.
Top 30 down-regulated genes by VGX1027 relative to LPS
| Gene symbol | P value | Fold change |
|---|---|---|
| CCL8 | 0·000 | −48·54 |
| EPR1 | 0·007 | −21·41 |
| CXCL10 | 0·002 | −17·83 |
| DTL | 0·003 | −13·26 |
| CXCL9 | 0·001 | −11·61 |
| TOP2A | 0·006 | −10·46 |
| KIAA0101 | 0·008 | −10·26 |
| FAM111B | 0·002 | −9·80 |
| HMMR | 0·006 | −9·52 |
| CCNB1 | 0·009 | −8·83 |
| NUF2 | 0·007 | −8·49 |
| TPX2 | 0·007 | −8·28 |
| KIF11 | 0·006 | −7·77 |
| HIST1H2BM | 0·006 | −7·65 |
| MKI67 | 0·005 | −7·55 |
| TYMS | 0·006 | −7·31 |
| BANK1 | 0·001 | −7·09 |
| BUB1B | 0·005 | −6·94 |
| CHEK1 | 0·006 | −6·70 |
| CLC | 0·004 | −6·68 |
| CEP55 | 0·007 | −6·58 |
| RRM2 | 0·007 | −6·52 |
| CDC6 | 0·005 | −6·33 |
| FCGR3A | 0·009 | −6·24 |
| PGDS | 0·000 | −6·18 |
| CPA3 | 0·000 | −6·07 |
| KLRF1 | 0·004 | −5·83 |
| FCGR3A | 0·006 | −5·78 |
| SH2D1B | 0·003 | −5·62 |
| C6orf173 | 0·001 | −5·47 |
Table 3.
Top 30 up-regulated genes by VGX1027 relative to lipopolysaccharide
| Gene symbol | P value | Fold change |
|---|---|---|
| ARMC9 | 0·007 | 3·65 |
| NEFM | 0·008 | 2·34 |
| OR1F2P | 0·004 | 2·17 |
| FBXO2 | 0·006 | 2·12 |
| YPEL2 | 0·001 | 2·09 |
| PI3 | 0·004 | 2·08 |
| KIF5C | 0·007 | 2·03 |
| TUBB4 | 0·005 | 2·03 |
| CACNB3 | 0·006 | 1·99 |
| ETNK2 | 0·001 | 1·98 |
| PTMS | 0·006 | 1·90 |
| BMF | 0·002 | 1·87 |
| LRRC8D | 0·000 | 1·85 |
| FNBP1L | 0·002 | 1·85 |
| CCDC136 | 0·005 | 1·83 |
| STXBP1 | 0·007 | 1·80 |
| DNAJB4 | 0·002 | 1·77 |
| SLC44A1 | 0·008 | 1·75 |
| FLJ40113 | 0·004 | 1·74 |
| RCAN1 | 0·009 | 1·74 |
| TXNRD1 | 0·001 | 1·73 |
| FLJ40125 | 0·008 | 1·72 |
| CDK2AP1 | 0·005 | 1·72 |
| KLF3 | 0·001 | 1·71 |
| SLC38A2 | 0·000 | 1·71 |
| CAMK2N1 | 0·009 | 1·70 |
| SMARCA1 | 0·007 | 1·69 |
| SERPINI1 | 0·004 | 1·67 |
| SLC3A2 | 0·006 | 1·67 |
| GTF2IRD1 | 0·005 | 1·67 |
Analysis of functional categories: ‘Antigen Processing and Presentation’
The genes associated with the ‘Antigen Processing and Presentation’ pathway belonged to the MHC class II molecules HLA-DOB, HLA-DPB1 and HLA-DRA; the Killer Cell Immunoglobulin-like Receptors KIR3DS1, KIR2DS1, KIR2DS2, KIR2DS4 and KIR3DL1, which are involved in the transduction of the activation signals in natural killer cells and T cells;9 and the genes that encode for the three subunits of the 11S regulator of PA28 (PSME2, PSME3), which is the endogenous regulator of the immunoproteasome, involved in the processing of class I MHC peptides10 (Fig. 3a).
Figure 3.
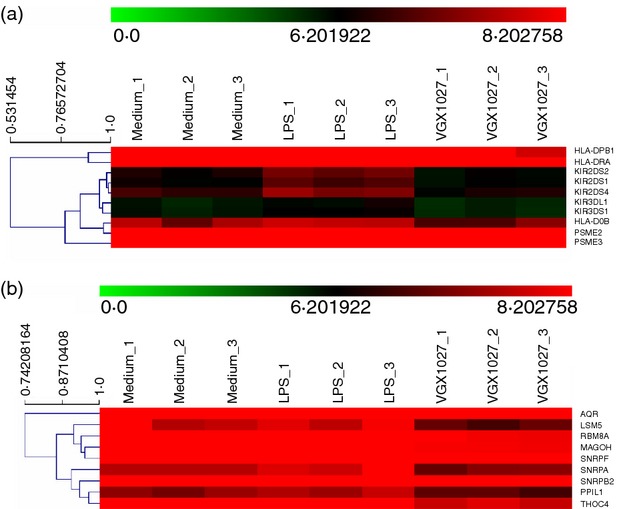
Differential regulation of genes related to the KEGG categories ‘Antigen Processing and Presentation’ (a) or ‘Spliceosome’ (b) is shown as heat map after unsupervised hierarchical clustering. Each line represents a gene. Gene expression is colour coded from green (lower expression) to red (higher expression).
Analysis of functional categories: ‘Spliceosome’
The genes associated with the ‘Spliceosome’ pathway include the small nuclear ribonucleoproteins SNRPA, required for splicing; SNRPD1, which belongs to the SNRNP core protein family; SNRPB2, which may play a role in pre-mRNA splicing; and SNRPF, which seems to function in the U7 snRNP complex that is involved in histone 3'-end processing.11 Other genes include RBM8A and MAGOH, which form a heterodimer that remains associated with spliced mRNAs as a tag to indicate where introns had been present;12 AQR, an intron-binding spliceosomal protein required to link pre-mRNA splicing and snoRNP (small nucleolar ribonucleoprotein) biogenesis; PPIL1, member of the cyclophilin family of peptidylprolyl isomerases that are a highly conserved family, involved in protein folding, immunosuppression by cyclosporin A and infection of HIV-1 virions;13 LSM5, important for pre-mRNA splicing;14 and THOC4, a nuclear protein that functions as a molecular chaperone, regulating dimerization, DNA binding, and transcriptional activity of basic region-leucine zipper proteins15 (Fig. 3b).
Analysis of functional categories: ‘Systemic Lupus Erythematosus’
The fourth most enriched pathway modulated by VGX-1027 was ‘Systemic Lupus Erythematosus’. Indeed, VGX-1027 was able to significantly down-regulate the expression of MHC class II molecules, including HLA-DPB1, HLA-DOB, HLA-DRA, important for the presentation of antigens to CD4+ T cells; IFNG, known mediator of cellular immune responses; HIST2H2AB and HIST1H2BM, which have been described as possible antigenic stimulus for SLE autoantibodies;16,17 and FCGR3A, also known as CD16a, which has been found to be expressed by infiltrating monocyte/macrophages in lupus glomeruli18,19 (Fig. 4).
Figure 4.
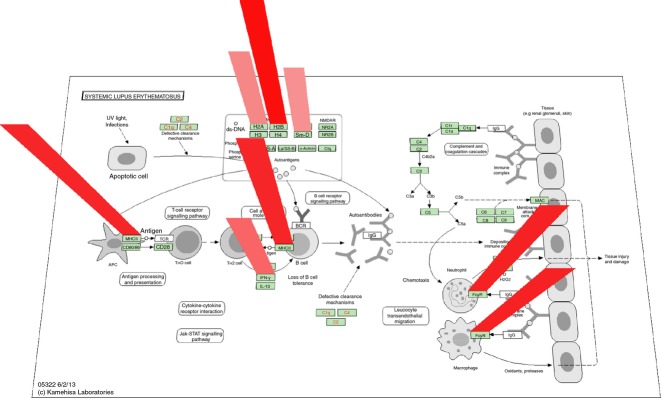
Genes down-regulated by VGX-1027 compared with lipopolysaccharide (LPS) belonging to the ‘Systemic Lupus Erythematosus’ pathway, as revealed by the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool. Gene down-regulation is represented as red columns of progressive height and color intensity.
Modulation of cytokines/chemokines and receptors by VGX-1027
Lipopolysaccharide treatment was associated with a complex pattern of cytokines/cytokine receptors and chemokines/chemokine receptors up-regulation as compared with medium (Table 1). Cytokines up-regulated included IFNG, IL17F, IL1B, IL22, IL32. Chemokines up-regulated included CCL3, CCL4, CCL7, IL8. Among the receptors which resulted upregulated by LPS, we found IL2RA, IL2RB, CX3CR1, CXCR4 and TNFRSF25 (Table 1). VGX-1027 treatment resulted in the significant down-regulation of most of these genes as compared with LPS treatment (Fig. 5 for a graphical representation; Table 2).
Figure 5.
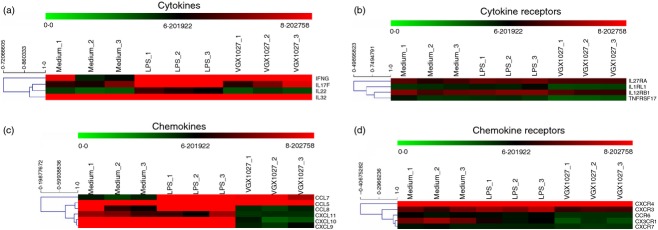
Heatmaps showing the modulation of cytokines (a), cytokines receptors (b), chemokines (c), chemokine receptors (d) by VGX-1027. Gene expression is color coded from green (lower expression) to red (higher expression).
Effects of VGX-1027 in the NZB/NZWF1 mouse model of SLE
VGX-1027 reduces proteinuria and increases survival of NZB/NZW F1 mice
To determine the effects of VGX-1027 on the development and progression of SLE, lupus-prone NZB/NZW F1 were treated daily with 5 mg per mouse VGX-1027 intraperitoneally for 20 weeks, starting at 16 weeks of age. Control mice received an equal volume of vehicle. Control mice started to die at 20 weeks of age. By week 36, 62·5% of them were dead compared with only 33·3% of treated mice (P = 0·0487 by Log-rank test) (Fig. 6a). Improved survival was correlated with inhibition of renal disease. Proteinuria was assessed once a week. The administration of VGX-1027 significantly improved renal disease and the levels of proteinuria in VGX-1027-treated mice remained significantly lower throughout the experimental period (P = 0·0265 by Student's t-test). Eventually, all of the mice developed proteinuria but the delay in disease progression and prolonged survival of VGX-1027-treated animals remained evident (Fig. 6b).
Figure 6.
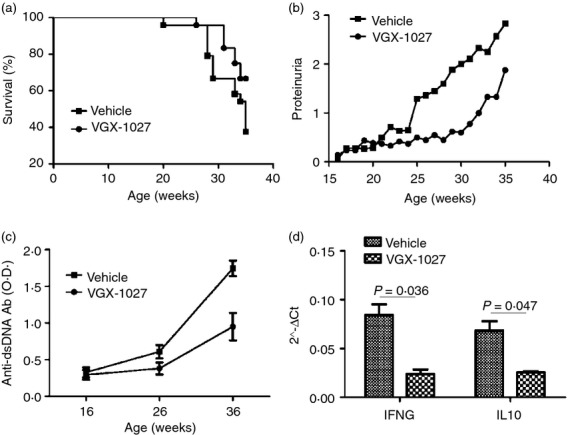
Effects of VGX-1027 administration on survival rate (a), proteinuria (b), serum anti-dsDNA antibodies (c) and cytokine production (d) of NZB/NZW F1 mice (n = 24 per group). Female NZB/NZW F1 mice were treated for 20 weeks beginning at 16 weeks of age with VGX-1027 20 mg/kg by daily intraperitoneal injection. Control mice received vehicle alone. Mice were followed for survival and for the development of renal disease, as measured by proteinuria. Proteinuria was measured using commercially available semi-quantitative strips. After 10 weeks of treatment and at the end of the experimental period, blood was sampled for measurement of autoantibodies using ELISA. Levels of the cytokines interferon-γ and interleukin-10 were determined by real-time PCR from unsorted splenocytes.
VGX-1027 reduced anti-dsDNA autoantibody and cytokine production and nephritis
The presence of anti-dsDNA antibodies is commonly used as a biomarker associated with poor prognosis of SLE and is strongly associated with developing lupus nephritis. The effect of VGX-1027 on anti-dsDNA autoantibody production was evaluated after 10 weeks of treatment and at the end of the experimental period. Only slightly increased levels of anti-dsDNA antibodies were found at 26 weeks of age in both vehicle and VGX-1027-treated animals. In contrast, at the termination of the study, when mice were 36-weeks old, VGX-1027-treated mice showed significantly lower levels of autoantibodies in the serum (P < 0·0001 by analysis of variance) (Fig. 6c).
Real-time PCR analysis of expression for IFNG and IL-10 on spleen cells, revealed a significant down-regulation of IFNG in VGX-1027-treated animals (P = 0·036) and of IL-10 (P = 0·047) (Fig. 6d).
End-stage SLE nephritis was evaluated by histopathology and scored by an independent pathologist for the assessment of total renal damage. Decreases in cellular infiltration/glomerulonephritis were observed in the haematoxylin & eosin-stained renal sections from the VGX-1027-treated group compared with the vehicle treatment group. Mean scores were 1·9 ± 0·8 and 3·1 ± 0·9 for the VGX-1027 and the vehicle control group, respectively (P = 0·01 by Mann–Whitney U-test) (Fig. 7).
Figure 7.
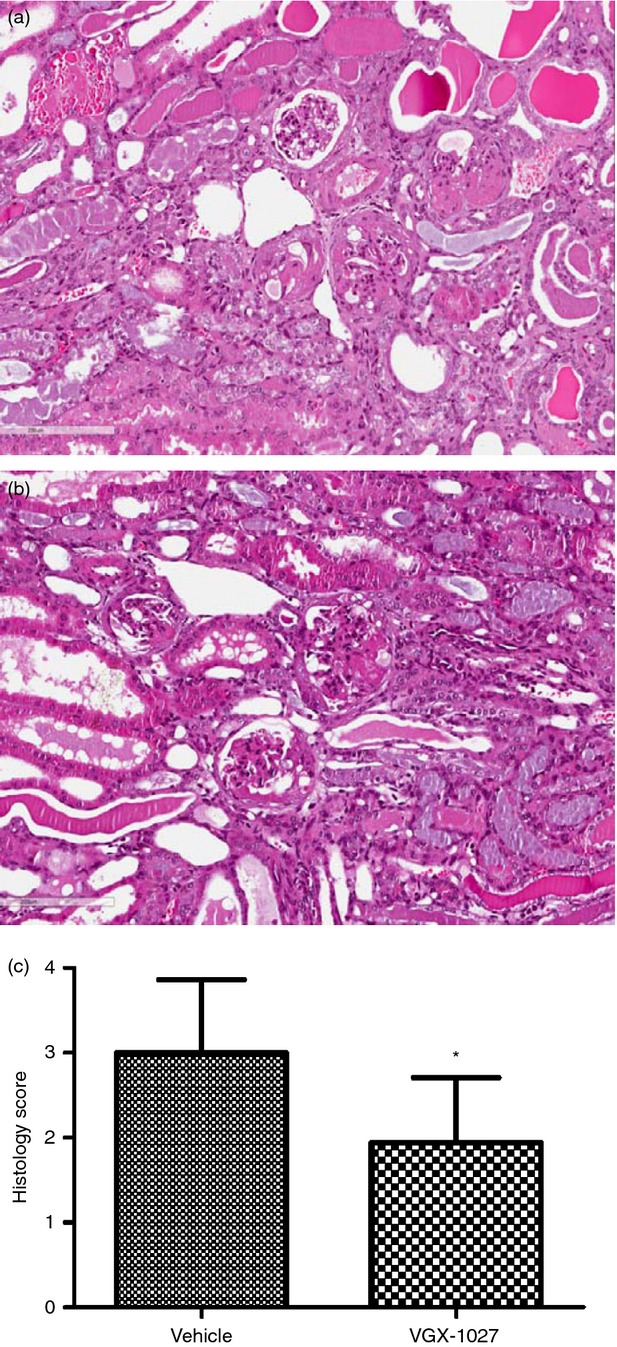
Histopathological analysis of kidney samples from NZB/NZW F1 mice. Female NZB/NZW F1 mice were treated for 20 weeks beginning at 16 weeks of age with VGX-1027 20 mg/kg by daily intraperitoneal injection. Control mice received vehicle alone. At the completion of the study, on week 36, the remaining mice from the different groups were killed and and their kidney tissues were removed and processed for histology. Representative haematoxylin & eosin (H&E) section of a vehicle-treated mouse (a). Representative H&E section of a VGX-1027 treated mouse (b). Histogram showing the H&E score at the completion of the study (c).
Discussion
VGX-1027 is a small molecule compound with already known immunomodulatory properties. This compound is able to reduce the secretion of both the pro-inflammatory cytokines IL-1β and tumour necrosis factor-α, and the anti-inflammatory cytokine IL-10 from murine macrophages stimulated in vitro with LPS, and it has also been shown to down-regulate the activation of the nuclear factor-κB and p38 mitogen-activated protein kinase pathways along with an up-regulation of the extracellular signal-regulated kinase pathway. However, the precise mechanism of action of VGX-1027 remains to be ascertained, given the complexity of the cross-talk between the innate and adaptive immune cells. This prompted us to exploit the genome-wide microarray approach to investigate the overall effect of VGX-1027 on a mixed population of immune cells upon exposure to the Toll-like receptor-4 ligand, LPS. Microarray data revealed that the administration of VGX-1027 profoundly affected the immune response to exogenous antigens, by strongly modulating the expression of genes that are primarily involved in antigen processing and presentation and immune activation. The data obtained are consistent with previous findings, which revealed that VGX-1027 primarily acts at the level of antigen-presenting cells such as macrophages and dendritic cells,1,3 activation of nuclear factor-κB and the release of cytokines, alarming the organism and coordinating appropriate defence mechanisms. The results from the functional analysis of modulated genes prompted us to evaluate the in vivo role of VGX-1027 in the NZB/NZW F1 model of SLE.
In agreement with the in vitro observations and functional analysis of significantly down-regulated genes, the NZB/NZW F1 mouse model benefited from VGX-1027 treatment as its administration prolonged treatment with VGX-1027 significantly increased survival and reduced kidney pathology, as shown by the proteinuria levels and histopathology.
VGX-1027 also inhibited other pathways involved in the pathogenesis of murine SLE including the production of IFNG and IL-10. This is in accordance with the essential role played by endogenous IL-10 in the NZB/NZW F1 model20 as well as with the capacity of VGX-1027 to down-regulate the secretion of this cytokine from murine macrophages in vitro.1
From the therapeutic point of view, SLE is a complex autoimmune disease with heterogeneous clinical manifestations and a course that has hindered the successful development of new drugs for the last 30 years.21 The standard of care for SLE patients with severe steroid-refractory inflammatory disease is the administration of alkylating agents.22 Hence, in spite of recent approval of the anti-BLYSS monoclonal antibody (Belimumab) for a fraction of patients,23 SLE remains an unmet medical need that urges for new therapeutic options. Additional and more innovative approaches are warranted in patients whose disease remains active despite available treatments. The use of drugs acting specifically on a critical step in the autoimmune process is another alternative. This may include agents inhibiting interactions between B and T lymphocytes, specific monoclonal antibodies against B cells, or antagonists of cytokines essential for SLE pathogenesis.22 The emerging evidence of a pathogenetic role for IL-10 in SLE24–26 supported by the beneficial response of SLE patients to specific inhibitors of IL-10 along with the ability of VGX-1027 to inhibit the function and the production of these cytokines and to markedly ameliorate SLE-like syndrome in NZB/NZWF1 clearly qualifies VGX-1027 as a drug candidate with relevant immunopharmacological profile that strongly warrants being tested in Phase II studies in SLE patients.
Disclosures
FN is cofounder and shareholder of Onconox that has out-licensed VGX1027 to Inovio Pharmaceuticals; KM is shareholder of Inovio Pharmaceuticals; JJK is CEO and shareholder of Inovio Pharmaceuticals; NYS is vice president and shareholder of Inovio Pharmaceuticals.
References
- 1.Stojanovic I, Cuzzocrea S, Mangano K, et al. In vitro ex vivo and in vivo immunopharmacological activities of the isoxazoline compound VGX-1027: modulation of cytokine synthesis and prevention of both organ-specific and systemic autoimmune diseases in murine models. Clin Immunol. 2007;123:311–23. doi: 10.1016/j.clim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Stosic-Grujicic S, Cvetkovic I, Mangano K, et al. A potent immunomodulatory compound, (S,R)-3-Phenyl-4,5-dihydro-5-isoxazole acetic acid, prevents spontaneous and accelerated forms of autoimmune diabetes in NOD mice and inhibits the immunoinflammatory diabetes induced by multiple low doses of streptozotocin in CBA/H mice. J Pharmacol Exp Ther. 2007;320:1038–49. doi: 10.1124/jpet.106.109272. [DOI] [PubMed] [Google Scholar]
- 3.Mangano K, Sardesai N, D'Alcamo M, et al. In vitro inhibition of enterobacteria-reactive CD4+ CD25– T cells and suppression of immunoinflammatory colitis in mice by the novel immunomodulatory agent VGX-1027. Eur J Pharmacol. 2008;586:313–21. doi: 10.1016/j.ejphar.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Medina I, Carbonell J, Pulido L, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38:W210–3. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 6.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 7.Nicoletti F, Di Marco R, Zaccone P, et al. Dichotomic effects of IFN-gamma on the development of systemic lupus erythematosus-like syndrome in MRL-lpr/lpr mice. Eur J Immunol. 2000;30:438–47. doi: 10.1002/1521-4141(200002)30:2<438::AID-IMMU438>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malmberg KJ, Michaëlsson J, Parham P, et al. Killer cell immunoglobulin-like receptor workshop: insights into evolution, genetics, function, and translation. Immunity. 2011;35:653–7. doi: 10.1016/j.immuni.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Whitby FG, Masters EI, Kramer L, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–20. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 11.Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–59. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 12.Zhao XF, Nowak NJ, Shows TB, et al. MAGOH interacts with a novel RNA-binding protein. Genomics. 2000;63:145–8. doi: 10.1006/geno.1999.6064. [DOI] [PubMed] [Google Scholar]
- 13.Mann SS, Pettenati MJ, von Kap-herr C, et al. Reassignment of peptidyl prolyl isomerase-like 1 gene (PPIL1) to human chromosome region 6p21.1 by radiation hybrid mapping and fluorescence in situ hybridization. Cytogenet Cell Genet. 1999;83:228–9. doi: 10.1159/000015186. [DOI] [PubMed] [Google Scholar]
- 14.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, et al. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2003;8:1489–501. [PMC free article] [PubMed] [Google Scholar]
- 15.Virbasius CM, Wagner S, Green MR. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol Cell. 1999;4:219–28. doi: 10.1016/s1097-2765(00)80369-x. [DOI] [PubMed] [Google Scholar]
- 16.Sui M, Lin Q, Xu Z, et al. Simultaneous positivity for anti-DNA, anti-nucleosome and anti-histone antibodies is a marker for more severe lupus nephritis. J Clin Immunol. 2013;33:378–87. doi: 10.1007/s10875-012-9825-6. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Bertucci AM, Ramsey-Goldman R, et al. Major pathogenic steps in human lupus can be effectively suppressed by nucleosomal histone peptide epitope-induced regulatory immunity. Clin Immunol. 2013;149:365–78. doi: 10.1016/j.clim.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriques A, Inês L, Carvalheiro T, et al. Functional characterization of peripheral blood dendritic cells and monocytes in systemic lupus erythematosus. Rheumatol Int. 2012;32:863–9. doi: 10.1007/s00296-010-1709-6. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimoto S, Nakatani K, Iwano M, et al. Elevated levels of fractalkine expression and accumulation of CD16+ monocytes in glomeruli of active lupus nephritis. Am J Kidney Dis. 2007;50:47–58. doi: 10.1053/j.ajkd.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Ishida H, Muchamuel S, Sakaguchi S, et al. Continuous administration of anti-interleukin 10 antibodies delays onset of autoimmunity in NZB/W F1 mice. J Exp Med. 1994;179:305–10. doi: 10.1084/jem.179.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenberg R. Why can't we find a new treatment for SLE? J Autoimmun. 2009;32:223–30. doi: 10.1016/j.jaut.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zandman-Goddard G, Shoenfeld Y. Novel approaches to therapy for SLE. Clin Rev Allergy Immunol. 2003;25:105–12. doi: 10.1385/CRIAI:25:1:105. [DOI] [PubMed] [Google Scholar]
- 23.Lutalo PM, D'Cruz DP. Belimumab for the management of systemic lupus erythematosus. Expert Opin Biol Ther. 2012;12:957–63. doi: 10.1517/14712598.2012.682980. [DOI] [PubMed] [Google Scholar]
- 24.Llorente L, Zou W, Levy Y, et al. Role of interleukin 10 in the B lymphocyte hyperactivity and autoantibody production of human systemic lupus erythematosus. J Exp Med. 1995;181:839–44. doi: 10.1084/jem.181.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross JT, Benton HP. The roles of interleukin-6 and interleukin-10 in B cell hyperactivity in systemic lupus erythematosus. Inflamm Res. 1999;48:255–61. doi: 10.1007/s000110050456. [DOI] [PubMed] [Google Scholar]
- 26.González-Amaro R, Portales-Pérez D, Baranda L, et al. Role of IL-10 in the abnormalities of early cell activation events of lymphocytes from patients with systemic lupus erythematosus. J Autoimmun. 1998;11:395–402. doi: 10.1006/jaut.1998.0228. [DOI] [PubMed] [Google Scholar]