Abstract
MicroRNAs (miRNAs) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic cells to regulate or mostly down-regulate genes by pairing with their complementary base-sequence in related mRNA molecules in the cytoplasm. It has been reported that other than its function in many physiological cell processes, dysregulation of miRNAs plays a role in the development of many diseases. In this short review, the association between miRNAs and some male reproductive disorders is surveyed. Male factor Infertility is a devastating problem from which a notable percentage of couples suffer. However, the molecular mechanism of many infertility disorders has not been clearly elucidated. Since miRNAs have an important role in numerous biological cell processes and cellular dysfunctions, it is of interest to review the related literature on the role of miRNAs in the male reproductive organs. Aberrant expression of specific miRNAs is associated with certain male reproductive dysfunctions. For this reason, assessment of expression of such miRNAs may serve as a suitable molecular biomarker for diagnosis of those male infertility disorders. The presence of a single nucleotide polymorphism (SNP) at the miRNAs’ binding site in its targeted mRNA has been reported to have an association with idiopathic male infertility. Also, a relation with male infertility has been shown with SNP in the genes of the factors necessary for miRNA biogenesis. Therefore, focusing on the role of miRNAs in male reproductive disorders can further elucidate the molecular mechanisms of male infertility and generate the potential for locating efficient biomarkers and therapeutic agents for these disorders.
Keywords: miRNA, Spermatogenesis, Male Infertility, Biomarker, Single Nucleotide Polymorphism
Introduction
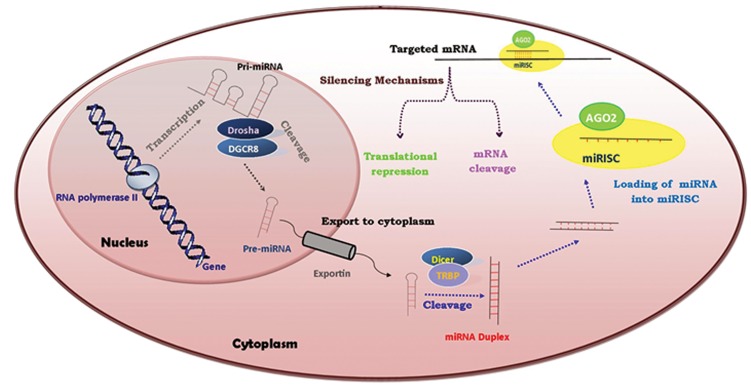
MicroRNAs (miRNAs) are small non-coding single stranded RNA molecules of approximately 22 nucleotides that are physiologically produced in eukaryotic cells (1). After being expressed in the nucleus, miRNA plays a role in regulation of gene expression by pairing with its complementary base-sequence of the targeted mRNA molecule in the cytoplasm. This usually leads to gene silencing through degradation of targeted mRNA or interference with its translation (2, 3). After discovery of miRNAs, as single-stranded non-protein-coding regulatory RNA molecules in C. elegans by Lee et al. (4) in 1993, over 1500 miRNAs have currently been reported to be encoded by the human genome (5-7) which may target around 60% of mammalian genes in various human cell types (8). Figure 1 schematically depicts miRNA biogenesis.
Fig 1.
Biogenesis of miRNA: RNA polymerase II transcribes miRNA gene to produce Pri-miRNA having a stem loop structure. Drosha (a class 2 ribonuclease III) and its RNA-binding partner, DGCR8*, cut the stem loop structure to make pre-miRNA. Exportin 5 transports the pre-miRNA from the nucleus to cytoplasm. In the cytoplasm, Dicer (a RNase III endonuclease) and its RNA-binding partner, TRBP**, cleave pre-miRNA to generate a short double-stranded RNA molecule named miRNA duplex. One strand of this molecule is incorporated into the miRNA-induced silencing complex (miRISC) containig protein Argonaute-2 (AGO2) as a catalytic component to regulate the targeted mRNA. The mechanism of targeted mRNA silencing could be either translational repression or mRNA cleavage.
miRNA; MicroRNA, * DGCR8; DiGeorge syndrome critical region gene and ** TRBP; Transactivating region binding protein.
Aside from the physiological regulation of gene expression in multiple biological processes such as cell cycle control and differentiation (9), cell growth and apoptosis (10) and embryo development (11), the role of miRNAs in development of many human diseases has been studied and reported. Mutation, dysfunction of biogenesis and dysregulation of miRNAs and their targets may lead to various diseases such as cardiovascular diseases (12), cancers (13), schizophrenia (14), renal function disorders (15), psoriasis (16), muscular disorders (17) and diabetes (18). A comprehensive database of whole miRNA-disease association, miRNA disease spectrum width (DSW), miRNA conservation and miRNA function has been published online at: http://202.38.126.151/hmdd/mirna/md/, entitled "The Human microRNA Disease Database". Among diseases, we have chosen to focus on male infertility with the intent to conduct a literature review of the role of miRNAs in this field.
Infertility is a problem in 10-15% of couples worldwide (19). Approximately, 50% of the infertility cases are attributed to male factors and 60-5% of these cases are idiopathic (20). Of note, the molecular mechanisms of many male infertility disorders are not clear (20). The aim of this review is to pursue the relation between miRNAs and male infertility.
miRNA and spermatogenesis
The presence of miRNA in testis has been verified a few years ago (21). Later, researchers focused on finding the function of miRNAs in germinal tract physiology. Table 1 shows a brief description of the chronological order of discoveries related to miRNAs in testis molecular physiology [for additional information please refer to Papaioannou and Nef (22)].
Table 1.
Discoveries related to microRNAs (miRNAs) in the male reproductive system
| Discoveries | Method of analysis | Year | Reference |
|---|---|---|---|
| Defining expression profiling of miRNA in human testis | miRNA specific oligonucleotide microarrays | 2004 | 21 |
| Report of Mirn122a as a down-regulator of germ cell transition protein 2 (Tnp2) messenger RNA | Real-time, RT-PCR and ribonuclease protection assays | 2005 | 23 |
| Defining of chromatoid bodies as intracellular nerve centers of themiRNA pathway in male germ cells | - | 2006 | 24 |
| In normal spermatogenesis, E2F1 mRNA translation is down regulated by miRNA (from the miR-17-92 cluster) to protect meiotic cells from apoptosis.Expression of miR-17-92 cluster is associated with reduced apoptosis in carcinoma in situ cells of the testis | RT-PCR | 2007 | 25 |
| Determination of miRNA expression profile in mouse testes in sexually immature and mature individuals | Microarray assay andquantitative real-time PCR | 2007 | 26 |
| Describing expression profiling of testis-expressed miRNAs to show some discriminatingly and uniquely expressed miRNA in murine testis | Semi-quantitative RT-PCR analyses | 2007 | 27 |
| Showing the differential pattern of gene expression for the Drosha, Dicer and Argonaute proteins in the germ and somatic cells of mouse testis | Quantitative real-time PCR for mRNA of Drosha, Dicer and Argonaute proteins in the germ and somatic cells of mouse testis | 2008 | 28 |
| Showing the localization of miRNAs (in addition to PIWI associated small RNA) in the nucleolus of Sertoli cells in addition to the chromosome cores, the telomeres and the XY body of spermatocytes | Fluorescence in situ hybridization (FISH), microarray, quantitative real-time PCR | 2008 | 29 |
| Dependency of proliferation of primordial germ cells and spermatogonia to miRNAs | Real-time PCR using a specific knockout mice in which Dicer gene was solely absent in its germ cells | 2008 | 30 |
| Dependency of spermatogenesis to Dicer in testis of mice | Real-time quantitative PCR, microarray | 2009 | 31 |
| Necessity of Drosha (RNase III) for spermatogenesis like Dicer | Generation of postnatal male germ line-specific Drosha or Dicer knock-out mice | 2012 | 32 |
| Identification of seven miRNA molecules discriminately expressed between postnatal gonocytes and spermatogonia to show their possible effect in maintaining germ cell differentiation and or pluripotency in mice | miRNA microarray | 2012 | 33 |
Potential of extracellular miRNAs as biomarkers for infertility
In addition to the presence of miRNA at the cellular level, miRNAs have also been reported to be present in extracellular fluids such as plasma (34), saliva (35, 36), vaginal secretions, menstrual blood and semen (36). Aberrant expression of extracellular miRNA is attributed to different disorders (37). Altered expression of miRNAs, as with altered expression of some testis-specific mRNAs (38-41) has been recently reported in male infertility disorders, hence the possibility of locating potential miRNA biomarkers in male infertility has been proposed.
The first report of an alteration of miRNA expression in the testis of patients with non-obstructive azoospermia (NOA) has been reported by Lian et al. (42). Of note, that NOA which has a testis-origin, presents a diverse range of defects from hypospermatogenesis and sperm maturation arrest to Sertoli-cell-only-syndrome. Hence, assessment of differential expressions of miRNAs in these infertile individuals is a prerequisite.
Wang et al. (43) examined pooled semen samples obtained from infertile men and compared the results with normal fertile individuals as controls. They found alterations in miRNA profiles by Solexa Sequencing (a sequencing method based on reversible dye-terminators technology and engineered polymerases) in both azoospermia and asthenozoospermia. These authors considered a cut of value of 50-fold higher or lower expression as significant, which was further validated by real time RT-qPCR for 7 miRNAs (miR-34c-5p, miR-122, miR-146b-5p, miR-181a, miR-374b, miR-509-5p, and miR-513a-5p). The level of these 7 miRNAs was significantly lower in azoospermia and higher in asthenozoospermia compared to the control. Another noticeable finding of these authors was the stability of the above miRNAs to different conditions. They attributed this phenomenon to the small size of the miRNAs and their ability to bind with complex organic molecules in cell-free semen samples. Finally, they proposed that these 7 miRNAs might have confirmative molecular diagnostic value for male infertility.
In this regard, Wu et al. (44) evaluated the pattern of expression related to miR-I9b and let-7a in idiopathic infertile individuals with NOA or oligozoospermia by quantitative RT-PCR. They showed that these two miRNAs distinctively expressed at higher levels in infertile cases compared with fertile individuals. Consequently, they concluded that miR-I9b and let-7a were good diagnostic molecular biomarkers for idiopathic infertile cases with NOA or oligozoospermia.
Single nucleotide polymorphism (SNP) and function of miRNA in spermatogenesis
Considering the diverse role of miRNA in spermatogenesis, therefore any polymorphism in related genes might lead to infertility. Zhang et al. (45) were the first researchers who reported an association between miRNA-binding site single nucleotide polymorphism (SNP) and idiopathic male infertility. They systematically surveyed all SNPs in the 3´ UTR of 140 mammal spermatogenesis-related genes and observed that some SNPs within miRNA-binding sites might be related to idiopathic male infertility.
Among 140 surveyed genes, 6 SNPs (two in CYP19, Serpina5 and four in CGA, CPEB1, and CPEB2), involved in down regulation of gene expression in spermatogenesis and meiosis and were predicted to have possibility for altering the binding affinity of miRNA by using bioinformatically specialized algorithms (Pictar, miRanda, Targetscan, and RNAhybrid). The effectiveness of these SNPs in male infertility was analyzed in a case-control study using PCR followed by restriction digestion of the amplified fragments for genotyping. They realized that T substituted for A in rs6631 which lead to diminished binding ability of miR-1302 to its binding site in CGA in vitro, with a subsequent overexpression of CGA. Therefore an association between this special SNP and male infertility was concluded.
Recently Qin et al. (46) evaluated the relation between seven SNPs in the genes of Drosha (rs10719, rs2291109, rs17409893 and rs642321) and Dicer (rs13078, rs1057035 and rs12323635) and infertility. Performing genotyping by means of real time PCR, these researchers showed that SNPs in rs10719, rs12323635 and rs642321 were related with male infertility in the examined Han Chinese population.
Conclusion
Considering limited number of studies performed in this field, little has been clarified about miRNA-mediated gene regulation in spermatogenesis. In addition, there are few numbers of published studies that focus on the role of miRNAs in male infertility. This review has aimed to elaborate the role of miRNA in the male reproductive system. Any disorder or failure in this system can result in male infertility. Therefore, a dysfunction in miRNA processing as with Dorsha and Dicer can result in azoospermia and infertility. Other anomalies such as dysregulation in expression of certain miRNAs or SNP in the miRNA binding site or SNP in the genes involved in biogenesis of miRNA can be related to infertility. Hence, in this regard, scientists suggest that assessment of miRNA may have future diagnostic value and shed more light on the molecular mechanisms of male infertility. Thus, new paths can be opened for future treatments of male infertility or even in the design of new contraceptive drugs.
Acknowledgments
There is no conflict of interest in this article.
References
- 1.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kusenda B, Mraz M, Mayer J, Pospisilova S. MicroRNA biogenesis, functionality and cancer relevance. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150(2):205–215. doi: 10.5507/bp.2006.029. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38(supple):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 7.Landgraf P, Rusu M, Sewer A, Lovino N, Aravin A, Pfeffer S, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Bot N. miRNAs and cell-cycle control in ESCs. Nat Cell Biol. 2012;14(7):685–685. [Google Scholar]
- 10.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis LM. MicroRNAs in early embryonic development: dissecting the role of miR-290 through miR-295 in the mouse.Presented for the Ph.D. Cambridge: Massachusetts Institute of Technology; 2008. [Google Scholar]
- 12.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6(6):419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Hansen T, Olsen L, Lindow M, Jakobsen KD, Ullum H, Jonsson E, et al. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS One. 2007;2(9):e873–e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JY, Yong TY, Michael MZ, Gleadle JM. Review: The role of microRNAs in kidney disease. Nephrology (Carlton) 2010;15(6):599–608. doi: 10.1111/j.1440-1797.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- 16.Sonkoly E, Wei T, Janson PC, Sääf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2(7):e610–e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104(43):17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes. 2011;60(7):1832–1837. doi: 10.2337/db11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jangravi Z, Alikhani M, Arefnezhad B, Sharifi Tabar M, Taleahmad S, Karamzadeh R, et al. A fresh look at the male-specific region of the human Y chromosome. J Proteome Res. 2013;12(1):6–22. doi: 10.1021/pr300864k. [DOI] [PubMed] [Google Scholar]
- 20.Guzick DS, Overstreet JW, Factor-Litvak P, Brazil CK, Nakajima ST, Coutifaris C, et al. Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med. 2001;345:1388–1393. doi: 10.1056/NEJMoa003005. [DOI] [PubMed] [Google Scholar]
- 21.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaioannou MD, Nef S. microRNAs in the testis: building up male fertility. J Androl. 2010;31(1):26–33. doi: 10.2164/jandrol.109.008128. [DOI] [PubMed] [Google Scholar]
- 23.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73(3):427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 24.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci USA. 2006;103(8):2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novotny GW, Sonne SB, Nielsen JE, Jonstrup SP, Hansen MA, Skakkebaek NE, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007;14(4):879–882. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 26.Yan N, Lu Y, Sun H, Tao D, Zhang S, Liu W, et al. A microarray for microRNA profiling in mouse testis tissues. Reproduction. 2007;134(1):73–79. doi: 10.1530/REP-07-0056. [DOI] [PubMed] [Google Scholar]
- 27.Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311(2):592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Gonzalez E, Lopez-Casas PP, del Mazo J. The expression patterns of genes involved in the RNAi pathways are tissuedependent and differ in the germ and somatic cells of mouse testis. Biochim Biophys Acta. 2008;1779(5):306–311. doi: 10.1016/j.bbagrm.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Marcon E, Babak T, Chua G, Hughes T, Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16(2):243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3(3):e1738–e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papaioannou MD, Pitetti JL, Ro S, Park C, Aubry F, Schaad O, et al. Sertoli cell Dicer is essential for spermatogenesis in mice. Dev Biol. 2009;326(1):250–259. doi: 10.1016/j.ydbio.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, et al. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem. 2012;287(30):25173–25190. doi: 10.1074/jbc.M112.362053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McIver SC, Stanger SJ, Santarelli DM, Roman SD, Nixon B, McLaughlin EA. A unique combination of male germ cell miRNAs coordinates gonocyte differentiation. PLoS One. 2012;7(4):e35553–e35553. doi: 10.1371/journal.pone.0035553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5(8):1961–1965. [PubMed] [Google Scholar]
- 35.Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004;83(3):199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- 36.Hanson EK, Lubenow H, Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem. 2009;387(2):303–314. doi: 10.1016/j.ab.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 37.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5(10):e13735–e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aghajanpour S, Ghaedi K, Salamian A, Deemeh MR, Tavalaee M, Moshtaghian J, et al. Quantitative expression of phospholipase C zeta, as an index to assess fertilization potential of a semen sample. Hum Reprod. 2011;26(11):2950–2956. doi: 10.1093/humrep/der285. [DOI] [PubMed] [Google Scholar]
- 39.Tseden K, Topaloglu O, Meinhardt A, Dev A, Adham I, Müller C, et al. Premature translation of transition protein 2 mRNA causes sperm abnormalities and male infertility. Mol Reprod Dev. 2007;74(3):273–279. doi: 10.1002/mrd.20570. [DOI] [PubMed] [Google Scholar]
- 40.Motiei M, Tavalaee M, Rabiei F, Hajihosseini R, Nasr-Esfahani MH. Evaluation of HSPA2 in fertile and infertile individuals. Andrologia. 2013;45(1):66–72. doi: 10.1111/j.1439-0272.2012.01315.x. [DOI] [PubMed] [Google Scholar]
- 41.Yatsenko AN, Georgiadis AP, Murthy LJ, Lamb DJ, Matzuk MM. UBE2B mRNA alterations are associated with severe oligozoospermia in infertile men. Mol Hum Reprod. 2013;19(6):388–394. doi: 10.1093/molehr/gat008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang Cet al. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7:13–13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Yang C, Chen X, Yao B, Yang C, Zhu C, et al. Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem. 2011;57(12):1722–1731. doi: 10.1373/clinchem.2011.169714. [DOI] [PubMed] [Google Scholar]
- 44.Wu W, Hu Z, Qin Y, Dong J, Dai J, Lu C, et al. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol Hum Reprod. 2012;18(10):489–497. doi: 10.1093/molehr/gas022. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H, Liu Y, Su D, Yang Y, Bai G, Tao D, et al. A single nucleotide polymorphism in a miR-1302 binding site in CGA increases the risk of idiopathic male infertility. Fertil Steril. 2011;96(1):34–39. doi: 10.1016/j.fertnstert.2011.04.053. [DOI] [PubMed] [Google Scholar]
- 46.Qin Y, Xia Y, Wu W, Han X, Lu C, Ji G, et al. Genetic variants in microRNA biogenesis pathway genes are associated with semen quality in a Han-Chinese population. Reprod Biomed Online. 2012;24(4):454–461. doi: 10.1016/j.rbmo.2012.01.006. [DOI] [PubMed] [Google Scholar]