Centrosome–microtubule interactions during interphase are important for centrosome clustering and cell polarity.
Abstract
Supernumerary centrosomes contribute to spindle defects and aneuploidy at mitosis, but the effects of excess centrosomes during interphase are poorly understood. In this paper, we show that interphase endothelial cells with even one extra centrosome exhibit a cascade of defects, resulting in disrupted cell migration and abnormal blood vessel sprouting. Endothelial cells with supernumerary centrosomes had increased centrosome scattering and reduced microtubule (MT) nucleation capacity that correlated with decreased Golgi integrity and randomized vesicle trafficking, and ablation of excess centrosomes partially rescued these parameters. Mechanistically, tumor endothelial cells with supernumerary centrosomes had less centrosome-localized γ-tubulin, and Plk1 blockade prevented MT growth, whereas overexpression rescued centrosome γ-tubulin levels and centrosome dynamics. These data support a model whereby centrosome–MT interactions during interphase are important for centrosome clustering and cell polarity and further suggest that disruption of interphase cell behavior by supernumerary centrosomes contributes to pathology independent of mitotic effects.
Introduction
The centrosome is the microtubule (MT)-organizing center (MTOC) of the cell, and mutations in centrosome-localized proteins are associated with pathologies such as Huntington disease and lissencephaly (Sathasivam et al., 2001; Tanaka et al., 2004; Badano et al., 2005; Kuijpers and Hoogenraad, 2011). Centrosomes consist of two barrel-shaped centrioles embedded in a protein matrix (pericentriolar material [PCM]; Bettencourt-Dias and Glover, 2007; Bornens, 2012). PCM is organized around the centriole and contains MT nucleation factors, such as γ-tubulin, pericentrin, and NEDD1, and MT nucleation complexes called γ-TuRCs (Kollman et al., 2011; Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). Centrosome MT nucleation capacity increases as cells approach mitosis, and recruitment of MT nucleation proteins is regulated in part by the cell cycle–dependent protein Plk1 (Polo-like kinase 1; Casenghi et al., 2003; Haren et al., 2009; Eot-Houllier et al., 2010). Inhibition, depletion, or mislocalization of Plk1 during mitosis significantly perturbs bipolar spindle formation and leads to mitotic failure, in part through centrosome-mediated defects (Hanisch et al., 2006; Kiyomitsu and Cheeseman, 2012). However, how centrosome-mediated MT nucleation capacity is regulated during interphase is an open question.
A hallmark of tumor cells is the presence of excess (greater than two), or supernumerary, centrosomes (Boveri, 1888, 1901), which disrupt mitotic fidelity and increase aneuploidy (Kwon et al., 2008; Ganem et al., 2009; Silkworth et al., 2009). Endothelial cells of tumor blood vessels also have high frequencies of excess centrosomes (Hida et al., 2004). Tumor endothelial cells (TECs) contribute to vessels that exhibit abnormal morphology and are functionally leaky once they enter a tumor (Carmeliet and Jain, 2011; Aird, 2012). Although cells spend most of their time in interphase, it is not known whether excess centrosomes affect nonmitotic cell processes. Tumor cells with supernumerary centrosomes were overlaid with oocyte extracts containing tubulin monomers; the sections had more MT polymers per cell, but each tumor cell had numerous centrosomes, and neither MT nucleation frequency nor functional observations were reported (Lingle et al., 1998).
Directional cell migration depends on centrosome-derived MTs for Golgi polarization and subsequent vesicle trafficking to the leading edge (Petrie et al., 2009; Kaverina and Straube, 2011; Luxton and Gundersen, 2011). Laser ablation studies reveal a centrosome requirement for initial Golgi organization, but once the MTOC is established, centrosome loss has negligible effects (Miller et al., 2009; Vinogradova et al., 2012). In contrast to centrosome loss, it is unclear whether excess centrosomes impair cell migration.
Here, we show that the presence of even one extra centrosome in endothelial cells leads to a cascade of defects during interphase, resulting in disrupted cell migration and perturbed vessel sprouting. Surprisingly, supernumerary centrosomes had reduced MT nucleations and increased dynamic centrosome movements, leading to Golgi fragmentation and randomized vesicle trafficking. Centrosome ablation to restore normal centrosome numbers partially rescued centrosome dynamics, Golgi morphology, and directional migration. Cells with supernumerary centrosomes had less centrosome-localized γ-tubulin, and Plk1 blockade prevented MT growth, whereas Plk1 overexpression (OE) rescued centrosome dynamics. Thus, centrosome–MT interactions during interphase are important for centrosome clustering, and proper clustering is required for polarized behaviors such as migration. The disruption of interphase cell polarity and migration induced by supernumerary centrosomes may contribute to tissue disorganization and pathology.
Results
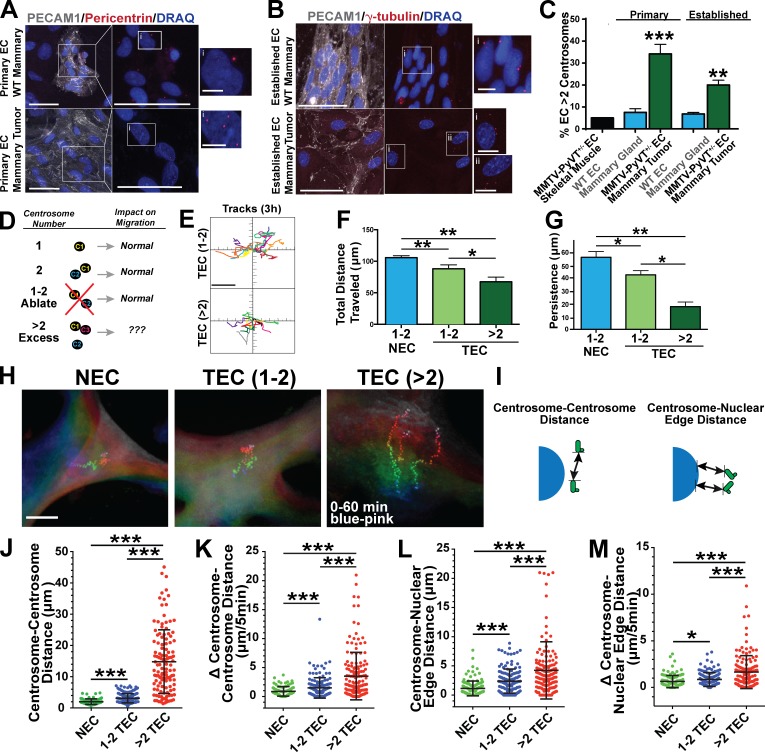
TECs with excess centrosomes have migration defects and centrosome scattering
Endothelial cells derived from tumor blood vessels (TECs) harbor supernumerary centrosomes (greater than two; Hida et al., 2004). To investigate effects of supernumerary centrosomes, we first isolated primary TECs from mammary tumors of PyVT+/− female mice and counted centrosomes. Approximately 34% of TECs from the primary tumors had excess centrosomes, significantly higher than normal endothelial cells (NECs) from mammary tissue of PyVT−/− littermates (Fig. 1, A and C). TECs were established in culture and analyzed for endothelial cell characteristics and general properties (Fig. 1, B and C; and Fig. S1, A–F). Established TECs had ∼20% spontaneous centrosome overamplification, significantly elevated relative to established NECs (Fig. 1, B and C).
Figure 1.
Endothelial cell supernumerary centrosomes disrupt migration and centrosome dynamics. (A) Enriched endothelial cells (EC) from WT mouse mammary tissue and MMTV-PyVT+/− mammary tumors stained for PECAM1 (white, endothelial cells), pericentrin (centrosomes), and DRAQ7 (DNA). (right) Higher magnification of boxed areas on left without PECAM1 channel. Bars: (main images) 50 µm; (insets) 5 µm. (B) Established WT mammary endothelial cell (nontumor [NEC]) and tumor endothelial cell (TEC) stained for PECAM1 (white, endothelial cells), γ-tubulin (centrosomes), and DRAQ7 (DNA). (right) Left images without PECAM1 channel. Bars: (main image) 50 µm; (insets) 10 µm. (C, left bars) Percentage of PECAM1-positive primary endothelial cells with greater than two centrosomes from tissues (MMTV-PyVT+/− skeletal muscle endothelial cells, n = 1 mouse; MMTV-PyVT−/− mammary tissue endothelial cells, n = 10 mice; MMTV-PyVT+/− mammary TECs, n = 15 mice). Statistical comparisons to endothelial cells from control MMTV PyVT−/− mammary tissue. χ2 test; error bars shows means ± SEM. **, P ≤ 0.01; ***, P ≤ 0.001. (right bars) Percentage of established endothelial cells with greater than two centrosomes (WT mammary tissue endothelial cell, four clones and n = 1,082 cells; MMTV-PyVT mammary TEC, seven clones and n = 1,234 cells). χ2 test; error bars show means ± SEM. **, P = 0.013. (D) Diagram of the effect of centrosome number on cell migration. (E) 3-h migration tracks of TECs with one to two centrosomes or greater than two centrosomes; one representative experiment from three repeats. Bar, 40 µm. see also Video 1. (F) Mean total distance traveled of NECs and TECs with either one to two or greater than two centrosomes (NEC, n = 43 cells; TEC 1–2 centrosomes, n = 25 cells; TEC >2 centrosomes, n = 11 cells). Statistics: Student’s t test; error bars show means ± SEM. *, P = 0.02; **, P < 0.01. (G) Mean persistence (linear distance from origin to maximal point of migration) of NECs and TECs with either one to two or greater than two centrosomes (NEC, n = 43 cells; TEC 1–2 centrosomes, n = 25 cells; TEC >2 centrosomes, n = 11 cells). Statistics: Student’s t test; error bars show means ± SEM. *, P = 0.05; **, P < 0.01. (H) Representative endothelial cells with centrosome movements tracked over 1 h. Each track starts with blue colors and ends with pink colors. Note the divergence of tracks over time in TECs with greater than two centrosomes. Bar, 12 µm. see also Video 2. (I) Diagram of the indicated measurements. (J) Centrosome–centrosome distances in indicated groups (NEC, n = 99; TEC 1–2, n = 143; TEC >2, n = 154 centrosome pairs). Statistics: Student’s t test; error bars show means ± SD. ***, P < 0.0001. (K) Change (absolute value) in centrosome–centrosome distance in 5-min time intervals (NEC, n = 99; TEC 1–2, n = 143; TEC >2, n = 154 centrosome pairs). Statistics: Student’s t test; means ± SD. ***, P < 0.0001. (L) Centrosome–nuclear edge distances between the indicated groups (NEC, n = 130 cells; TEC 1–2, n = 142 cells; TEC >2, n = 154 measurements). Statistics: Student’s t test; means ± SD. ***, P < 0.0001. (M) Change (absolute value) in centrosome–nuclear edge distance in 5-min time intervals (NEC, n = 130; TEC 1–2, n = 142; TEC >2, n = 154 measurements). Statistics: Student’s t test; error bars show means ± SD. *, P < 0.05; ***, P < 0.0001.
Although TECs had slightly reduced growth rates compared with NECs (Fig. S1 D), TECs with excess centrosomes underwent mitosis and clustered extra centrosomes at the spindle, as previously described (Kwon et al., 2008), and TECs did not have elevated apoptotic markers absent UV treatment (Fig. S1, E and F). However, the percentage of TECs with excess centrosomes decreased with passage number in culture (Fig. S1 L), although numerous primary isolates had an elevated frequency of centrosome amplification (Fig. 1, A and C). This finding suggests that, in the tumor environment in vivo, TECs with excess centrosomes are either maintained or replenished, while they are selected against in culture.
Because centrosomes form the MTOC that contributes to cell migration, we examined migration in endothelial cell populations with overamplification. TECs had reduced migration in scratch wound and transwell assays compared with NECs (Fig. S1, G and H), and individual cell tracking revealed that TECs had reduced distance traveled and persistence (Fig. S1, I–K). These data indicate that TECs have an elevated frequency of supernumerary centrosomes and migration defects, so we hypothesized that excess centrosomes would disrupt migration (Fig. 1 D). We examined effects of excess centrosomes at the single-cell level via centriole labeling, which allowed us to bin TECs into normal (one to two centrosomes) or excess centrosome (greater than two centrosomes) groups. Individual cell tracking of TECs with excess centrosomes showed significantly reduced migration distance and persistence compared with TECs with one to two centrosomes or NECs (one to two centrosomes; Fig. 1, E–G; and Video 1). Interestingly, TECs with one to two centrosomes had reduced migration compared with NECs, suggesting that centrosome-independent modifications also contribute to TEC migration defects. To test effects of supernumerary centrosomes on migration in primary cells, we induced excess centrosomes in human umbilical vein endothelial cells (HUVECs) using either Cdc14B knockdown (KD) or Plk4 OE (Fig. S1 M). As reported in other cells (Habedanck et al., 2005; Wu et al., 2008), both manipulations promoted significant centrosome overamplification, in the absence of extensive cytokinesis failure (Fig. S1 N). Overall growth was not affected, and centrosome-overamplified HUVECs clustered excess centrosomes at mitosis (Fig. S1 O and not depicted). Cells infected with either construct had reduced migration in scratch wound and transwell assays and increased permeability (Fig. S1, P, Q, and U). Next, we assigned HUVECs into normal (one to two centrosomes) and overamplified (greater than two centrosomes) groups by labeling the centrioles. Similar to TECs, HUVECs containing excess centrosomes had reduced distance traveled and persistence in individual cell-tracking experiments (Fig. S1, R–T). These results indicate that excess centrosomes are associated with perturbed migration in endothelial cells.
After centriole duplication during late G1/S, centrosomes function as a single MTOC by associating in pairs with coordinated movements (Holland et al., 2010; Sluder and Khodjakov, 2010). Interphase centrosomes also connect to the nuclear envelope via attachments between MTs and nuclear membrane proteins (Tapley and Starr, 2013). We noticed that, in TECs with excess centrosomes, centrosomes were more separated from each other and the nucleus than in cells with one to two centrosomes (Fig. 1 H and Video 2). During random cell migration, TECs with supernumerary centrosomes exhibited dynamic centrosome movements and abnormal centrosome positioning, as indicated by significant changes in centrosome–centrosome distance and centrosome–nuclear edge distance over time (Fig. 1, I–M). These results indicate that supernumerary centrosomes are associated with increased dynamic centrosome movements during interphase, and that these movements may prevent normal directional migration.
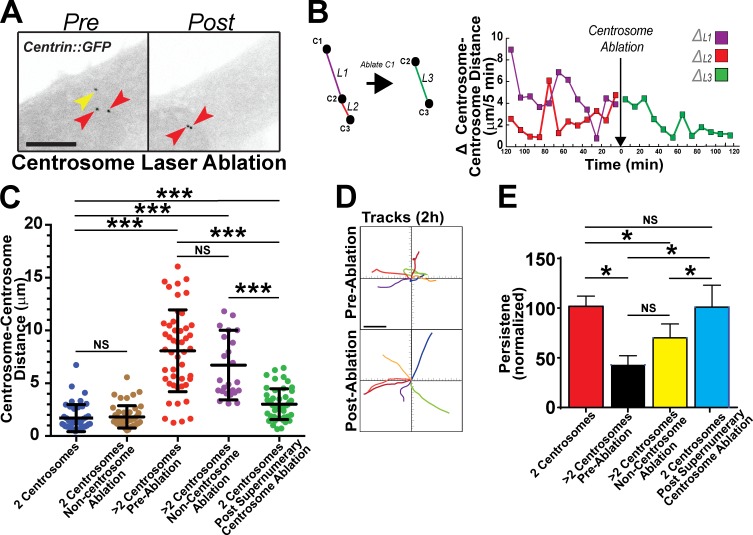
Ablation of excess centrosomes partially rescues centrosome scattering and endothelial cell migration
To determine whether perturbed cell migration and centrosome scattering resulted from supernumerary centrosomes, we removed extra centrosomes in TECs via laser ablation (Magidson et al., 2007). As expected, endothelial cells with excess centrosomes had increased centrosome movements and scattering distances before ablation. It was striking that, after ablation of the excess centrosome, dynamic changes in centrosome separation diminished relatively quickly, and centrosomes came together (Fig. 2 A, Fig. S2 A, and Video 3). We also measured centrosome–centrosome distance, as opposed to changes in distance, over time (Fig. S2 B, diagram) and found a close association between variability in dynamic centrosome movements and centrosome number (Fig. 2, B and C). Cells with supernumerary centrosomes before ablation or after a control, noncentrosomal, ablation had greater variability and larger changes in centrosome–centrosome distance over time. Remarkably, normalization of centrosome number by ablation significantly reduced centrosome–centrosome distance and overall variability, indicating stabilized centrosome associations (Fig. 2 C). Likewise, absolute centrosome–nuclear edge distance was reduced after ablation of excess centrosomes (Fig. S2, C and D). We next analyzed migration tracks of individual cells with excess centrosomes before and after ablation. Although distance traveled was not significantly rescued by ablation, migratory persistence was fully rescued in TECs with excess centrosomes after centrosome ablation (Fig. 2, D and E; Fig. S2 E; and Video 4). Collectively, these results indicate that supernumerary centrosomes promote centrosome scattering and altered directional migration in interphase cells.
Figure 2.
Ablation of supernumerary centrosomes rescues abnormal centrosome dynamics and endothelial cell migration. (A) Representative images of centrin::GFP-expressing TECs before and after ablation of an extra centrosome. Yellow arrow, ablated centrosome; red arrows, remaining centrosomes. Bar, 10 µm. see also Video 3. (B, left) Diagram showing centrosome distances between centrosomes before ablation (purple and red lines) and centrosome distance of the remaining centrosomal pair after ablation (green line). (right) Actual change in centrosome–centrosome distance over time for a single representative TEC before and after centrosome ablation. (C) Individual centrosome–centrosome distances in the indicated groups (n ≥ 6 cells per condition and three independent experiments). Each dot represents individual centrosome–centrosome distances in the indicated cells. Statistics: Student’s t test; means ± SD. ***, P < 0.0001. (D) Migration tracks of TEC before and after centrosome ablation, combined from three experiments. Each color represents a single cell before and after ablation. Bar, 40 µm. see also Video 4. (E) Mean directional persistence of TECs in the indicated groups, labeled as in D (n ≥ 6 cells per condition). Statistics: one-tailed Student’s t test; error bars show means ± SEM; *, P < 0.05.
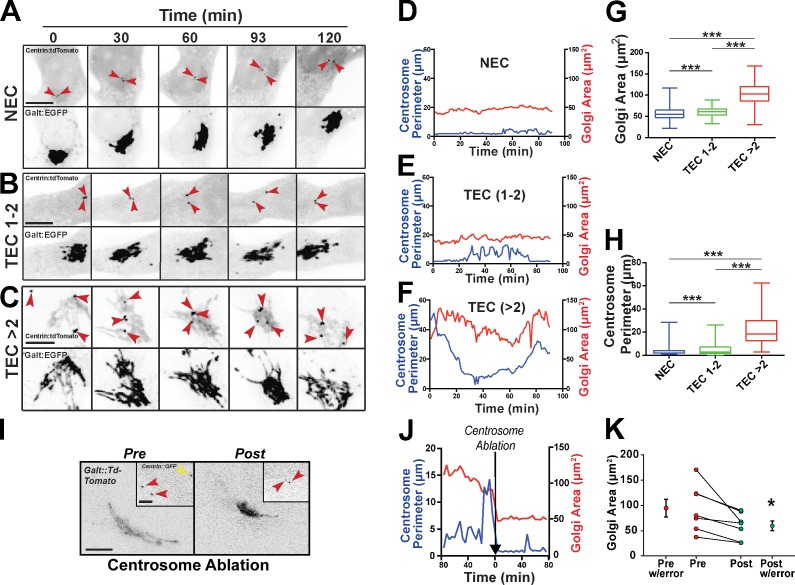
Supernumerary centrosomes disrupt Golgi integrity
We hypothesized that excess centrosomes perturb cell migration by disrupting focal adhesion (FA) dynamics and/or Golgi integrity. TECs with greater than two centrosomes had reduced density of FA (Fig. S3, A and B). However, parameters predicted to be altered upstream of migration defects, such as FA size, length, or orientation, did not differ between TECs with one to two or greater than two centrosomes (Fig. S3 C). Centrosomes also regulate Golgi organization; thus, we next asked whether supernumerary centrosomes affected Golgi morphology. TECs stained for Golgi (GM130, a cis-Golgi protein) and centrosomes (γ-tubulin) had Golgi stacks that were more spread and fragmented in cells with excess centrosomes than cells with one to two centrosomes (Fig. S3, D and E). Because the Golgi uses centrosome-derived MTs to reform after nocodazole treatment and washout, we tested Golgi reformation in this assay and found it significantly impaired in cells with supernumerary centrosomes (Fig. S3, G–I). These data indicate that supernumerary centrosomes perturb interphase Golgi structure and its ability to reform after disruption.
To better define how centrosome movements affect the Golgi, we tracked centrosomes (Fig. 3 F) and Golgi in migrating TECs using time-lapse imaging. Fluctuations in centrosome distance in cells with one to two centrosomes did not significantly affect the Golgi, but TECs with excess centrosomes had elevated centrosome scattering that correlated with increased Golgi area (Fig. 3, A–F). Similar to the fixed image analysis, centrosome perimeter (a measure of centrosome scatter; Fig. S3 J, diagram) and Golgi area were both significantly increased in cells with supernumerary centrosomes (Fig. 3, G and H; and Videos 5 and 6). Additionally, there was moderate correlation between centrosome dynamics and morphological parameters, such as Golgi area (Fig. S3, K and L). Consistent with centrosome scattering causing abnormal Golgi morphology, centrosome ablation significantly rescued Golgi area (Fig. 3, I–K). These results suggest that supernumerary centrosomes cause decreased Golgi compactness and promote fragmentation.
Figure 3.
Excess centrosomes disrupt Golgi integrity. (A–C) Time-lapse fluorescence micrographs of centrosomes (centrin::tdTomato; top) and Golgi (GalT::EGFP; bottom) in freely migrating NECs or TECs with one to two or greater than two centrosomes. Red arrows, individual centrosomes. Bars, 10 µm. (D–F) Plots of centrosome perimeter and Golgi area over time from images in A–C. Each graph is a single experiment from several repeats. (G and H) Box and whisker plot (middle bars, mean; boxes, top and bottom quartiles with error bars [SEMs]) of centrosome perimeter and Golgi area from individual live-cell imaging frames (NEC, n = 1,424; TEC 1–2, n = 775; TEC >2, n = 1,075 frames for both graphs). Statistics: Student’s t test; ***, P < 0.0001. see also Videos 5 and 6. (I) Representative images of both centrosomes (centrin::GFP, insets) and Golgi (GalT::tdTomato) in TECs with greater than two centrosomes before and after centrosome ablation. Red arrows, nonablated centrosomes; yellow arrow, ablated centrosome. Bars: (main images) 10 µm; (insets) 5 µm. (J) Representative plot from a single experiment showing centrosome perimeter and Golgi area over time from images in I, before and after centrosome ablation (black arrow, ablation time point). (K) Golgi area of individual TECs with greater than two centrosomes before and after centrosome ablation (n = 7 cells from two experiments). Statistics: one-way Student’s t test; means ± SD. *, P < 0.05.
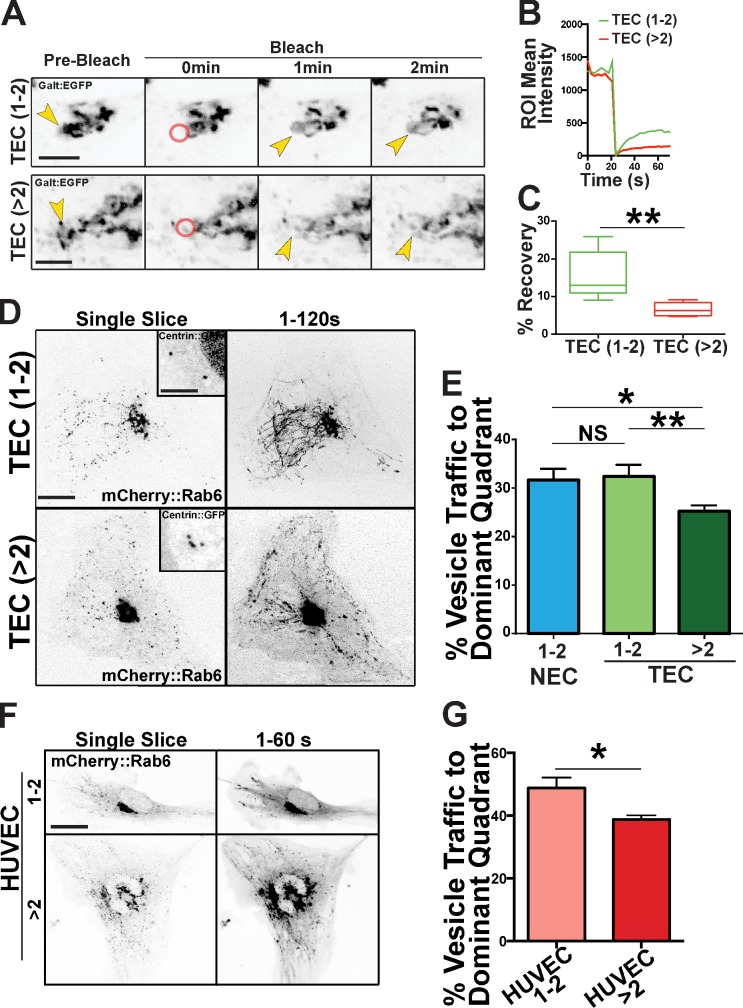
Golgi continuity and vesicle trafficking are altered in cells with excess centrosomes
To determine functional consequences of Golgi changes, we examined Golgi integrity and directed vesicle trafficking. FRAP in the Golgi significantly lagged in TECs with excess centrosomes, indicative of less continuity among the Golgi stacks (Fig. 4, A–C; and Video 7). We next monitored post-Golgi vesicle trafficking by time-lapse imaging of cells labeled with mCherry::Rab6 (endosomes) and centrin::GFP (centrosomes). Among TECs with greater than two centrosomes, there were significantly fewer cells able to localize vesicle trafficking to the cell’s dominant quadrant, the presumed leading edge; in other words, the trajectories of post-Golgi trafficking were disorganized in these cells (Fig. 4, D and E; and Video 8). Similarly, HUVECs with supernumerary centrosomes had more randomized mCherry::Rab6 trafficking compared with controls (Fig. 4, F and G).
Figure 4.
Excess centrosomes disrupt post-Golgi vesicle trafficking. (A) Fluorescence micrographs from time-lapse imaging depicting bleaching of Golgi edge (GalT::GFP) and subsequent recovery (FRAP) between TECs with one to two (top) and greater than two (bottom) centrosomes. Arrows, bleach time point; red circles, bleached areas; ROI, region of interest. Bars, 5 µm. see also Video 7. (B) One representative quantification of FRAP of Golgi marker between TECs with one to two or greater than two centrosomes from two experiments. (C) Box and whisker plots (middle bars, mean; boxes, top and bottom quartiles with error bars [SEMs]) of percentages of fluorescence recovery of Golgi marker 60 s after bleaching of TECs with one to two or greater than two centrosomes (n = 5 cells per group from two experiments). Statistics: Student’s t test; **, P < 0.01. (D) Representative single time point and time-compressed images (1–120 s) of mCherry-Rab6 vesicle trafficking time lapse in TECs with one to two or greater than two centrosomes. Insets are higher magnification of corresponding centrosomes (centrin::GFP). see also Video 8. Bars: (main images) 10 µm; (insets) 10 µm. (E) Mean percentage of vesicle trafficking to the dominant quadrant (see Materials and methods for details; NEC, n = 25; TEC 1–2, n = 17; TEC >2, n = 20 cells; three experiments). Statistics: Student’s t test; *, P < 0.05; **, P < 0.01. (F) Representative images of HUVECs with one to two or greater than two centrosomes expressing Rab6-mCherry. (left) Single slice is one video frame, and 1–60 s are time projections. Bar, 10 µm. (G) Mean percentage of vesicle traffic to the dominant quadrant in HUVECs with one to two or greater than two centrosomes (HUVEC 1–2, n = 10; HUVEC >2, n = 5; two experiments). Statistics: Student’s t test; means ± SEM; *, P < 0.05.
Supernumerary centrosomes promote randomized MT growth and reduced MT nucleations
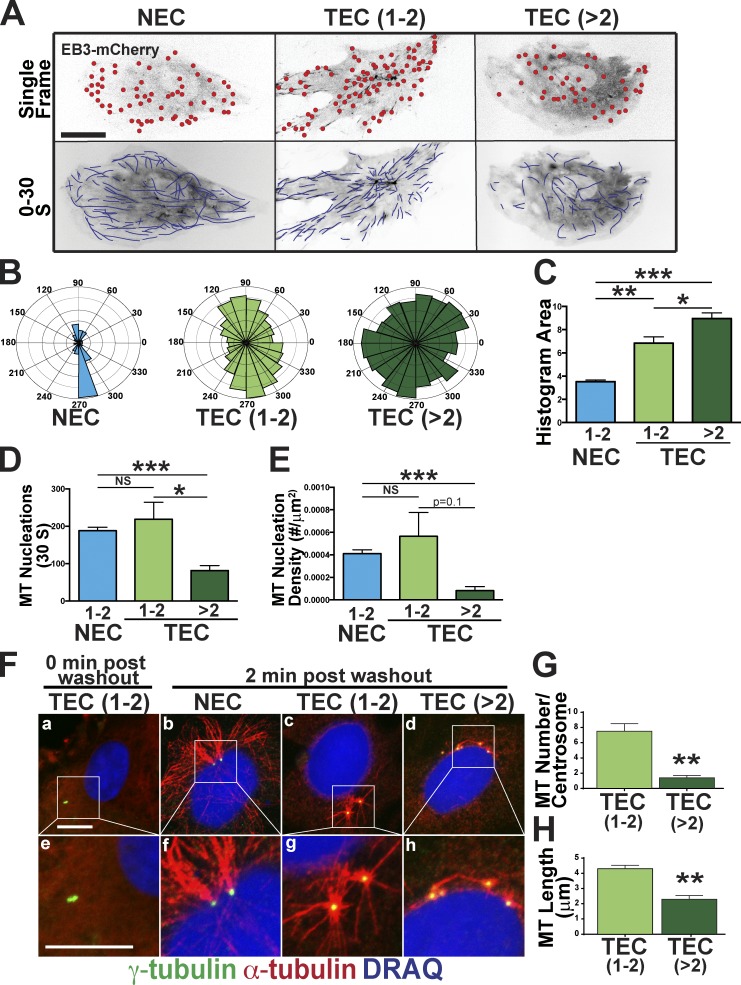
Golgi integrity and vesicle trafficking depend on MTs; thus, we hypothesized that Golgi and trafficking defects were downstream of MT changes. We monitored steady-state MT dynamics using plus-end tip tracking (Applegate et al., 2011). TECs with greater than two centrosomes had more MTs emanating in random directions compared with controls (Fig. 5, A–C; and Video 9). Moreover, in HUVEC with extra centrosomes via Plk4 OE, MT growth polarity was randomized (Fig. S4, A and B). These results suggest that excess centrosomes disrupt post-Golgi vesicle trafficking downstream of randomized MT growth direction.
Figure 5.
Excess centrosomes randomize MT polarity and reduce nucleation capacity. (A) Representative cells expressing centrin::GFP (centrosomes) and EB3::mCherry (growing MT plus ends). (top) Single image, with red circles indicating individual MT tips. (bottom) 30-s stack to show MT paths (blue lines). see also Video 9. (B) Imposition of MT growth directions onto Rose plots to visualize the percentage of growing MTs by angle in the indicated groups. (C) Comparison of MT angle areas from Rose plots in B. (NEC, n = 12 cells; TEC 1–2 centrosomes, n = 11 cells; TEC >2 centrosomes, n = 6 cells). Error bars are SEMs. (D and E) Comparison of number of MT nucleations (D) and nucleation density (E) among groups (NEC, n = 12 cells, TEC 1–2 centrosomes, n = 11 cells; TEC >2 centrosomes, n = 6 cells). (F) MT regrowth after nocodazole washout. Indicated cells stained for MTs (α-tubulin), centrosomes (γ-tubulin), and DNA (DRAQ7). (bottom) Higher magnifications of the insets. (G and H) Mean number of MTs/centrosome (G) and MT length/centrosome (H) in the indicated groups. (TEC 1–2 centrosomes, n = 10 cells; TEC >2 centrosomes, n = 10 cells). Error bars are SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 10 µm.
Centrosome pairs are clustered by MTs and dynamically repositioned by MT-dependent motor proteins, such as dynein (Robinson et al., 1999; Tanaka et al., 2004). The centrosome scattering and randomized MT directionality led us to hypothesize that supernumerary centrosomes have MT nucleation defects. TECs and HUVECs with supernumerary centrosomes had significantly fewer MT nucleations compared with controls, consistent with lower MT nucleation capacity (Fig. 5, D and E; and Fig. S4, C and D). However, not all MT parameters were altered, as cells with excess centrosomes did not exhibit significant changes in MT polymerization rate or lifetime (Fig. S4, E and F).
To determine whether the whole-cell decrease in MT nucleation was related to defects in centrosome-mediated MT nucleation, we used a regrowth assay in which MTs renucleate after nocodazole-induced MT depolymerization and washout. We observed that, in cells with supernumerary centrosomes, all centrosomes were associated with some α-tubulin, suggesting that they can nucleate MTs. However, TECs with excess centrosomes had significantly decreased MT regrowth potential, producing fewer and shorter MTs per centrosome after washout compared with controls (Fig. 5, F–H). Line scans indicated less α-tubulin around supernumerary centrosomes (Fig. S4, G–I), and HUVECs with excess centrosomes also had decreased MT nucleations (Fig. S4, J–L). Collectively, these results imply that centrosomes in cells with excess centrosomes have reduced MT nucleation capacity.
Supernumerary centrosomes are deficient in recruitment of PCM
Because supernumerary centrosomes have decreased MT nucleations, we thought that stable MTs might be reduced in this scenario. To avoid cell cycle–dependent PCM fluctuations, we arrested TECs in G1/S using a double-thymidine block and then assayed TECs for α-tubulin (all MTs) and acetylated (Ac) tubulin (stable MTs) and compared radial integrated densities around centrosomes. We observed significantly reduced α- and Ac-tubulin associated with supernumerary centrosomes (Fig. S5, A–C), with a positive association between reduced α-tubulin and lower MT acetylation, indicating that the proportion of stable MTs does not change in cells with excess centrosomes (Fig. S5 D).
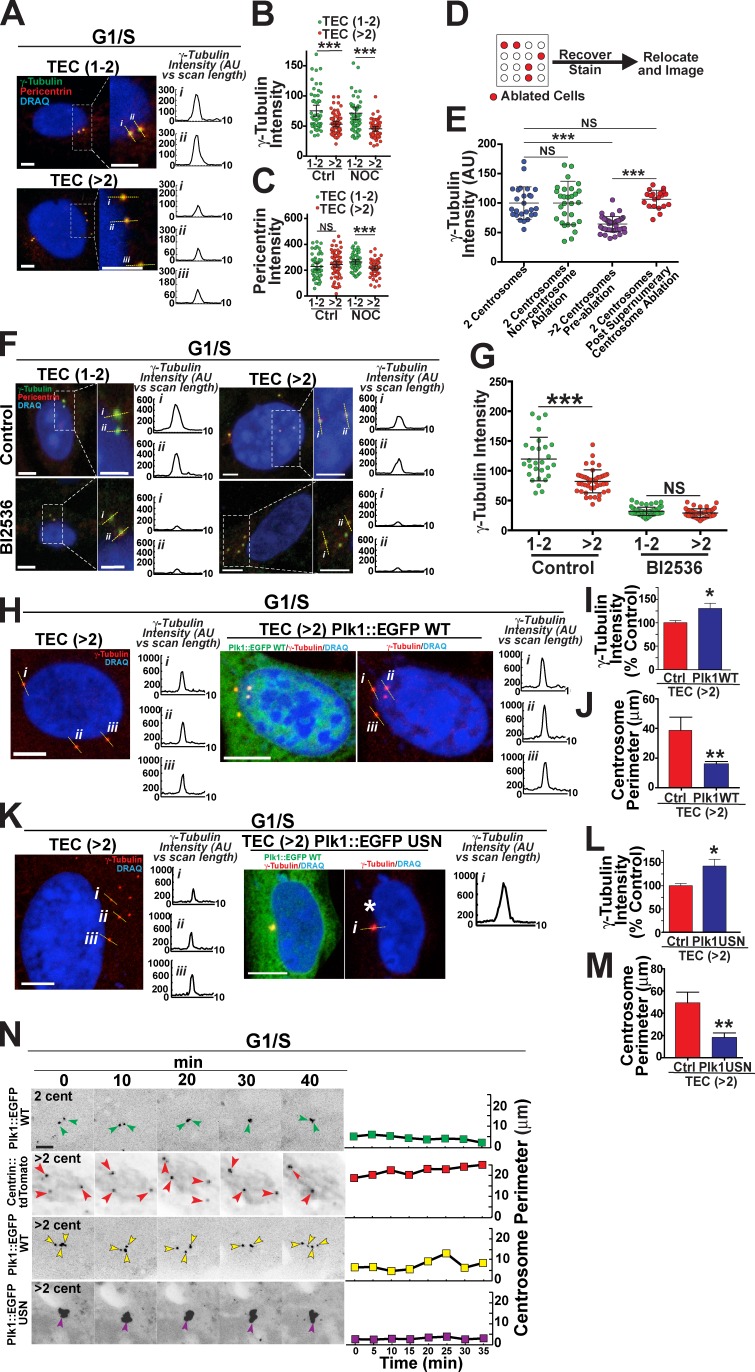
MT nucleation is regulated by recruitment of PCM components; thus, we hypothesized that reduced MT nucleations result from reduced centrosomal recruitment of PCM proteins that promote MT nucleation, such as γ-tubulin and pericentrin. We analyzed radial integrated protein intensities at the centrosomes of untreated TECs that had intact MTs. TECs with supernumerary centrosomes had significantly less γ-tubulin at the centrosome than did cells with one to two centrosomes, whereas centrosome-localized pericentrin levels were not affected (Fig. 6, A–C). To determine the relative amount of γ-tubulin and pericentrin bound to the centrosome in the absence of continual MT-mediated trafficking, we depolymerized MTs via nocodazole and found that levels of centrosomal γ-tubulin and pericentrin were significantly lower in TECs with greater than two centrosomes compared with cells with one to two centrosomes (Fig. 6, B and C). We next performed laser ablation to normalize centrosome numbers in HUVECs with centrosome overamplification. HUVECs expressing Plk4 plated on a grid to identify centrosome-ablated cells after staining showed rescued centrosomal γ-tubulin levels within 90 min after ablation (Fig. 6, D and E). These results show that supernumerary centrosomes have reduced levels of MT-nucleating proteins resulting from their supernumerary status, consistent with their reduced MT nucleation capacity.
Figure 6.
Supernumerary centrosomes have reduced PCM. (A) Representative TECs with the indicated centrosome numbers arrested in G1/S and stained for γ-tubulin, pericentrin, and DNA (DRAQ7). Boxes show areas of greater magnification. Numbered yellow lines, line scans of γ-tubulin intensity shown in graphs to the right. Bars, 5 µm. (B and C) Radial integrated protein intensities of γ-tubulin (B) or pericentrin (C) of TECs of the indicated centrosome numbers in control conditions (left) or after nocodazole (NOC) treatment (right). Scatter plot with mean (middle bars) and 95% confidence intervals are shown for each group (control: TEC 1–2, n = 52 centrosomes; TEC >2, n = 70 centrosomes; nocodazole treatment: TEC 1–2, n = 55 centrosomes; TEC >2, n = 57 centrosomes). Statistics: Student’s t test; ***, P < 0.001. (D) Diagram of setup that allowed centrosome or control-ablated cells to be relocated. (E) Radial integrated protein intensities of γ-tubulin between various groups. Scatter plot with mean (middle bars) and 95% confidence intervals are shown (one to two centrosomes, n = 26 centrosomes; noncentrosome ablation, n = 43 centrosomes; greater than two centrosomes, n = 29 centrosomes; after supernumerary centrosome ablation, n = 20 centrosomes). Statistics: Student’s t test; ***, P < 0.0001. (F) G1/S-arrested TECs with one to two (left) or greater than two (right) centrosomes in control conditions (top) or with BI2536 treatment and stained for γ-tubulin, pericentrin, and DNA (DRAQ7). Boxes show areas of greater magnification. Numbered yellow lines, line scans of γ-tubulin intensity graphed on the right. Bars: (main images) 5 µm; (insets) 2.5 µm. (G) Radial integrated densities of γ-tubulin in TECs of the indicated centrosome numbers in control conditions (left) or after Bl2536 treatment (right). Scatter plot with mean (middle bars) and 95% confidence intervals are shown (control: TEC 1–2, n = 28 centrosomes; TEC >2, n = 47 centrosomes; Bl2536 treatment: TEC 1–2, n = 63 centrosomes; TEC >2, n = 47 centrosomes). Statistics: Student’s t test; ***, P < 0.001. (H) G1/S-arrested TECs containing greater than two centrosomes in control conditions (left) or expressing Plk1::EGFP WT (right) and stained for (γ-tubulin) and DNA (DRAQ7). Numbered yellow lines, line scans of γ-tubulin intensity graphed to the right. Bars, 10 µm. (I) Mean γ-tubulin radial integrated protein density between the indicated groups normalized to controls. (TEC 1–2, n = 27 centrosomes; TEC >2, n = 27 centrosomes). Statistics: Student’s t test; error bars show means ± SEM; *, P < 0.05. (J) Mean centrosome perimeter between indicated groups. (TEC 1–2, n = 12 cells; TEC >2, n = 15 cells). Statistics: Student’s t test; error bars show means ± SEM; **, P < 0.01. (K) G1/S-arrested TECs containing greater than two centrosomes in control conditions (left) or expressing Plk1::EGFP USN (destruction box mutant, right) and stained for (γ-tubulin) and DNA (DRAQ7). Numbered yellow lines, line scans of γ-tubulin intensity graphed to the right. White asterisk denotes cluster of excess centrosomes. Bars, 10 µm. (L) Mean γ-tubulin radial integrated protein density between indicated groups normalized to controls. (TEC 1–2, n = 29 centrosomes; TEC >2, n = 25 centrosomes). Statistics: Student’s t test; error bars show means ± SEM; *, P < 0.05. (M) Mean centrosome perimeter between indicated groups. (TEC 1–2, n = 8 cells; TEC >2, n = 10 cells). Student’s t test; error bars show means ± SEM; **, P < 0.01. (N, left) Time-lapse fluorescence micrographs of G1/S-arrested TECs with one to two or greater than two centrosomes (cent) and expressing Plk1::EGFP WT (first and third rows), centrin::tdTomato (second row), or Plk1::EGFP USN (fourth row). Colored arrows, centrosomes. (right) Plots of centrosome perimeter over time. Bar, 5 µm. AU, arbitrary unit; Ctrl, control.
Reduced centrosomal PCM localization in cells with supernumerary centrosomes may result from defects in recruitment via MT motor proteins. Dynein stabilizes interphase MTs by shuttling MT nucleation proteins, and dynein alterations perturb MT architecture (Koonce et al., 1999). We hypothesized that stabilization of existing MTs around supernumerary centrosomes would provide time for dynein to deposit PCM in spite of the lower MT abundance. To test this, we stabilized MTs with taxol, which fully rescued γ-tubulin and pericentrin levels of supernumerary centrosomes (Fig. S5, E and F). We next asked whether dynein dysfunction caused reduced protein levels on supernumerary centrosomes, and we predicted that dynein inhibition would reduce control centrosome-localized γ-tubulin and/or pericentrin levels to levels seen in cells with excess centrosomes. However, centrosomal γ-tubulin and pericentrin in control cells exposed to the dynein inhibitor ciliobrevin D remained significantly elevated compared with cells with excess centrosomes, suggesting that reduced centrosomal γ-tubulin levels do not directly result from dynein-mediated defects (Fig. S5, E and F; compare Fig. 6 C and Fig. S5 F). However, consistent with a previous study (Levy and Holzbaur, 2008), dynein inhibition caused centrosomes to drift apart, supporting the hypothesis that dynein is responsible for centrosome clustering (Fig. S5 G).
We hypothesized that one or more centrosome-localized regulatory factors were reduced around supernumerary centrosomes. Plk1 is a centrosome-associated kinase that regulates recruitment of γ-tubulin (Eot-Houllier et al., 2010). As predicted, antibody staining for Plk1 in TECs showed colocalization with γ-tubulin (Fig. S5 H). In interphase-arrested TECs, pharmacological inhibition of Plk1 profoundly reduced MT growth after nocodazole washout (Fig. S5 I), and Plk1 inhibition also caused a significant reduction in centrosome-localized γ-tubulin (Fig. 6, F and G). These results suggest that Plk1 is upstream of γ-tubulin recruitment to the centrosome in interphase cells, and it may be a limiting factor for PCM recruitment.
We predicted that if Plk1 regulates interphase γ-tubulin recruitment, elevated levels of Plk1 would alter centrosome dynamics. We transiently expressed wild-type (WT) Plk1 (Plk1::EGFP WT) or Plk1 with mutations that inactivate the ubiquitination site and thus stabilized the protein (Plk1::EGFP ubiquitin site null [USN]; Golsteyn et al., 1994; Lindon and Pines, 2004; Elowe et al., 2007; Neef et al., 2007) in G1/S-arrested TECs. Both conditions increased α-tubulin around centrosomes after nocodazole washout, suggesting elevated centrosomal MT nucleation capacity (Fig. S5 J). G1/S-arrested TECs with excess centrosomes and overexpressing WT Plk1 had more centrosome-localized γ-tubulin and significantly reduced centrosome scattering compared with cells with excess centrosomes and no Plk1 manipulation (Fig. 6, H–J). Similarly, G1/S-arrested TECs expressing Plk1 USN had higher centrosome-localized γ-tubulin and reduced scattering of supernumerary centrosomes (Fig. 6, K–M). Live imaging of TEC with elevated Plk1 levels also showed enhanced centrosome clustering compared with nontransfected controls (Fig. 6 N). Together, these data show that increasing interphase Plk1 levels in cells with supernumerary centrosomes rescues centrosomal γ-tubulin levels and reduces centrosome scattering, suggesting that the increased centrosome scattering associated with supernumerary centrosomes results from decreased recruitment of γ-tubulin downstream of perturbed Plk1.
3D migration and blood vessel sprouting is compromised in endothelial cells harboring supernumerary centrosomes
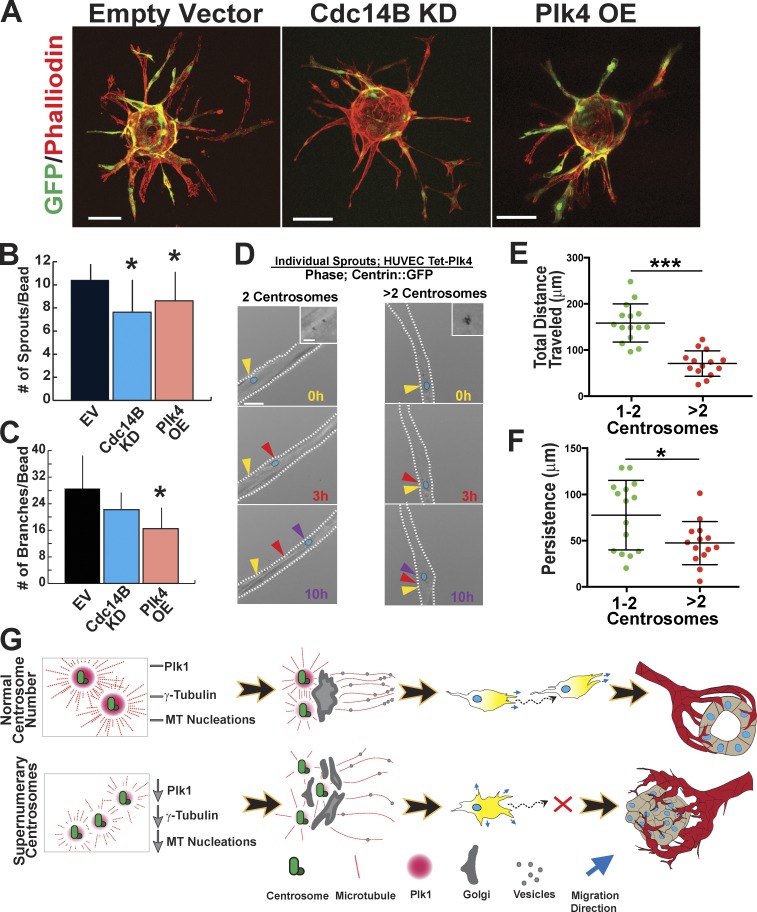
Sprouting angiogenesis is essentially collective cell migration to form a blood vessel network. Thus, we hypothesized that the centrosome scattering and loss of migratory polarity produced by supernumerary centrosomes would perturb vascular sprouting. Using a 3D vessel sprouting assay, we analyzed sprouting parameters of HUVECs with an elevated frequency of supernumerary centrosomes, via either Cdc14B KD or Plk4 OE. Both manipulations produced fewer primary sprouts compared with controls, and Plk4 OE also reduced branching (Fig. 7, A–C). To investigate the contribution of individual cell movements to vessel morphology, we tracked nuclei of normal and centrosome-overamplified cells. Similar to 2D migration results, endothelial cells with excess centrosomes showed significantly reduced total distance traveled and persistence within growing sprouts (Fig. 7, D–F; and Video 10). Thus, primary endothelial cells with excess centrosomes have perturbed migration as blood vessels sprout, suggesting that interphase effects of supernumerary centrosomes extend from basic cellular processes to tissue organization.
Figure 7.
Centrosome overamplification affects 3D migration and blood vessel sprouting. (A) HUVECs on microcarrier beads in sprouting angiogenesis assay infected with the indicated viral constructs (green) and stained with phalloidin to delineate sprout structures. Bars, 100 µm. (B and C) Mean of sprouts/bead (B) and branches/bead (C) in the indicated groups in a sprouting angiogenesis assay (empty vector [EV], n = 20 beads; Cdc14B KD, n = 20 beads; Plk4 OE, n = 8 beads). Error bars show SDs. Statistical comparisons versus empty vector. *, P < 0.05. (D) Live-imaging micrographs with fluorescence (centrin::GFP) overlaid on phase (see also Video 10). Arrowheads, nucleus position of a given cell at the indicated time points; blue ovals, nucleus. Bar, 20 µm. (insets) Fluorescence of centrosomes at higher magnification. Bar, 2 µm. White dotted lines, sprout borders. (E and F) Scatter plot with mean (middle bars) and 95% confidence intervals of total distance traveled (E) and persistence (F) of individual nuclei tracked within sprouts (1–2 centrosomes, n = 15 cells; >2 centrosomes, n = 14 cells for both graphs). *, P < 0.05; ***, P < 0.0001. (G) Model depicting mechanism of supernumerary centrosome effects on Golgi organization, migratory polarity, and blood vessel formation. Endothelial cells with supernumerary centrosomes may have reduced Plk1/centrosome, leading to reduced levels of γ-tubulin and fewer MT nucleations/centrosome. Reduced MT nucleations compromise centrosome clustering at the MTOC, which leads to disorganized Golgi that randomizes vesicle trafficking and directional migration. In blood vessels, this abnormal migration perturbs vessel morphogenesis and integrity.
Discussion
Excess centrosomes promote chromosome missegregation at mitosis, but whether excess centrosomes affect cell behaviors before mitosis has not been explored. Our data show that even one extra centrosome induces centrosome scattering and disrupts cell migration during interphase and reveal a novel requirement for precise centrosome numbers (one to two) before mitosis. A model consistent with our data (Fig. 7 G) suggests that supernumerary centrosomes promote imbalances in centrosome-localized proteins that regulate MT nucleations, leading to centrosome scattering and abnormal directional migration. Elevated growth factor signaling promotes endothelial cell centrosome overamplification (Taylor et al., 2010), suggesting that centrosome overamplification may be more prevalent than previously appreciated in pathologies with aberrant growth factor signaling (Chen et al., 2013). Elucidation of how excess centrosomes affect nonmitotic cell behaviors also sheds light on mechanisms that regulate normal interphase centrosome functions, and the importance of centrosome clustering during interphase.
Excess centrosomes and cell migration
Centrosomes are MTOCs, so how do supernumerary centrosomes affect MT-dependent interphase cell behaviors? Our work shows that endothelial cells with excess centrosomes have reduced directional migration and fewer MT nucleations per centrosome, indicating that even one excess centrosome significantly lowers the MT nucleation capacity of all centrosomes. Consistent with our results, reduced migration was correlated with drug-induced loss or overstabilization of MTs in HUVECs (Myers et al., 2011). The dramatic rescue of directed cell migration upon ablation of excess centrosomes in our work strongly suggests that MT defects are downstream of supernumerary centrosomes.
Although reduced MT nucleation capacity could directly affect directional migration, we also found defects in MT-dependent Golgi organization and vesicle trafficking, indicating disrupted polarization. Centrosome-derived MTs are largely responsible for Golgi coalescence, whereas Golgi-derived MTs primarily provide tracks for vesicle transport (Vinogradova et al., 2012). In our work, Golgi integrity was disrupted, and polarized vesicle trafficking was randomized by supernumerary centrosomes, suggesting that residual MT nucleation capacity of excess centrosomes exerts directional force on the Golgi. Increased centrosome scattering correlated with fragmented Golgi, and both parameters were rescued by centrosome ablation. In line with our results, decreased Golgi integrity disturbs polarized vesicle delivery to the leading front (Yadav et al., 2009; Hurtado et al., 2011). Moreover, polarized transport of regulators of small GTPases that affect migration, such as Rac1, is MT dependent (Mikhailov and Gundersen, 1998; Waterman-Storer et al., 1999). Consistent with this idea, MT plus-end trajectories were polarized in endothelial cells with one to two centrosomes but randomized in cells with supernumerary centrosomes.
Dynamic scattering of excess centrosomes
Our work points to insufficient centrosome clustering in endothelial cells with excess centrosomes as a defect that promotes abnormal cell behaviors. Centrosomes cluster and connect to the nucleus after duplication during S phase (Holland et al., 2010; Sluder and Khodjakov, 2010; Tapley and Starr, 2013). Centrosome clustering in mitotic cells with supernumerary centrosomes occurs, allowing for bipolar spindle formation and mitotic progression (Kwon et al., 2008; Ganem et al., 2009). Although it was possible that excess centrosomes also clustered during interphase, we found that excess interphase centrosomes exhibit dynamic scattering that is rescued by laser ablation that also rescues Golgi and migration defects, suggesting that centrosome scattering causes interphase defects in endothelial cells with excess centrosomes. Interphase clustering is primarily achieved through intracentrosomal MTs and dynein pulling forces (Koonce et al., 1999). We show that centrosome overamplification results in lax centrosome clustering linked to reduced MT nucleation capacity, although dynein blockade did not change the relative levels of γ-tubulin on centrosomes. Interestingly, during mitosis, excess centrosomes clustered, allowing for bipolar spindle pole formation and progression through the cell cycle. However, endothelial cells with extra centrosomes were eventually lost upon passage, suggesting some loss of fidelity.
As cells approach mitosis, centrosomes recruit factors that enhance MT nucleation capacity and set up the spindle (Casenghi et al., 2003; Haren et al., 2009; Eot-Houllier et al., 2010). Specific changes include recruitment and activation of Plk1, which promotes recruitment of γ-tubulin and pericentrin and increased centrosome MT nucleation capacity (Elowe et al., 2007). We hypothesized that parallel mechanisms regulate interphase centrosome MT nucleations and are disrupted in cells with supernumerary centrosomes. Consistent with this hypothesis, cells with supernumerary centrosomes had reduced centrosome-localized γ-tubulin and pericentrin, and γ-tubulin levels were rescued by ablation of excess centrosomes. Moreover, our data indicate that Plk1 regulates centrosome function during interphase because Plk1 blockade reduced MT growth and centrosome-localized γ-tubulin levels, mimicking the effects of supernumerary centrosomes, whereas overexpression restored γ-tubulin levels and reduced centrosome scattering. It may be that a limited pool of Plk1 is active during interphase, and this pool does not change with centrosome number. In this scenario, supernumerary centrosomes would effectively dilute the active Plk1 available at each centrosome and promote reduced MT nucleations. This model is supported by a report that supernumerary centrosomes nucleated supernumerary cilia, but each cilium had reduced signaling capacity (Mahjoub and Stearns, 2012). Moreover, other factors likely also affect centrosome clustering, because we observed differences in MT-nucleating capacity between normal and TECs independent of centrosome number.
Conclusions
The centrosome-clustering defects we observed indicate that interphase endothelial cell centrosomes are less buffered to perturbations than mitotic centrosomes, and that they require precise levels of PCM components to function properly, such that relatively subtle changes have significant effects on centrosome clustering and polarized cell behaviors. The effects of supernumerary centrosomes on centrosome dynamics, MT regulation, and directional migration have implications for tissue organization. Populations of endothelial cells harboring excess centrosomes do not sprout efficiently, and endothelial cells with excess centrosomes have reduced migration within sprouts. Given that ∼30% of TECs have supernumerary centrosomes and do not express tumor markers, they probably originate from normal vessels. The tumor likely provides an environment conducive to centrosome overamplification, perhaps via elevated growth factor signaling (Taylor et al., 2010). Thus, abnormal directional migration downstream of centrosome overamplification is likely to be a novel mechanism whereby TECs sustain migration defects that contribute to the documented abnormalities of tumor vessels. Indeed, it is tempting to speculate that bone fide tumor cells, which exhibit even more profound centrosome overamplification, also alter their responses to environmental cues downstream of supernumerary centrosomes.
Materials and methods
Cell isolation, culture, and viral transduction
HUVECs and human lung fibroblasts were grown under standard conditions. Lentivirus expressing a GFP reporter (GiPZ; GE Healthcare), shRNA against Cdc14B (GiPZ), or cytomegalovirus (CMV)-driven full-length human Plk4 (pcDNA3.1 3×myc-A; gift from E. Nigg, University of Basel, Basel, Switzerland) were first cloned into pCR8/GW/TOPO (Invitrogen) and then inserted into several lentiviral backbones using a conventional Gateway LR reaction (Life Technologies): pLenti CMV GFP DEST (736; 19732; Addgene), pLenti CMV neomycin (Neo) DEST (705-1; 17392; Addgene), and pLIX 402 (41394; Addgene). Human full-length Plk1 was cloned from a pEGFP-C1 construct (37406; Addgene) and inserted into a pEGFP-N1 backbone (Takara Bio Inc.). USN Plk1 was generated by introducing the mutations R337A and L340A using fusion PCR. Centrioles were visualized with a centrin::EGFP (human centrin-2)-expressing lentivirus (pLentiLox 7.0 CMV), a centrin::dsRed lentivirus (pLentiLox 7.0 CMV), or a centrin::tdTomato lentivirus (pLentiLox CMV). For MT tracking, lentivirus (pLenti CMV Neo DEST (705-1)) expressing full-length EB3::mCherry (gift from V. Small, Institute of Molecular Biotechnology, Vienna, Austria) or full-length EB1::EGFP was generated by subcloning into pCR8/GW/TOPO and then cloning into lentiviral destination vector (pLenti CMV Neo DEST (705-1)). For Golgi visualization, a CMV-driven galactosyltransferase (GalT)::EGFP was used (gift from J. Lippincott-Schwartz, National Institutes of Health, Bethesda, MD). Vesicles were marked with full-length Rab6a::EGFP (pEGFP-N1 CMV; gift from J. Lippincott-Schwartz). HUVECs were incubated in media with virus and 1 µg/ml polybrene (EMD Millipore) for 24 h and then in media for 24 h.
Isolation of TECs
Mouse cells were isolated via magnetic bead-assisted cell sorting (MACS; Miltenyi Biotec) as previously described (Dudley et al., 2008), with modifications. Female MMTV-PyVT (mouse mammary tumor virus–polyoma middle T antigen) transgenic mice and WT littermates were sacrificed when tumors were 1–1.5 cm2. Tissue was minced and incubated in 2 µg/ml collagenase (Worthington Biochemical Corporation) for 75 min and then passed through a 70-µm strainer. Cells were washed in PBS, resuspended in autobuffer (autoMACS), and incubated with rat anti–mouse polyclonal platelet endothelial cell adhesion molecule (PECAM) conjugate to phycoerythrin (PE) antibody (1:1,000; BD) and then with anti-PE microbeads (Miltenyi Biotec). Labeled cells were passed through an LS column (Miltenyi Biotec) on a magnetized stand and washed 3× with autobuffer (autoMACS) before elution into MACS buffer. Cells were pelleted, plated onto gelatin-coated Petri dishes in DMEM media containing 20% FBS and a cell growth kit (EGM-2 SingleQuot; Lonza), and maintained at 10% CO2. Some MACS-isolated cells were incubated with media containing polyomavirus middle T–expressing virus for 3 d and then selected for resistance to neomycin (Balconi et al., 2000). Endothelial cell colonies were identified by DiI-acLDL (1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate) uptake (L-23380; Invitrogen) and picked for expansion. For FACS analysis, cells were detached with Accutase, washed, and resuspended in PBS/0.1% BSA containing rat anti–mouse polyclonal PECAM-PE–conjugated antibody (1:1,000; BD). After three washes, cells were analyzed using a flow cytometer (FACSCalibur; BD).
Centrosome counts
HUVECs were stained with rabbit polyclonal anti-pericentrin (1:1,000; Abcam) or mouse monoclonal anti–γ-tubulin (GTU88 clone; 1:1,000; Sigma-Aldrich) to visualize centrosomes, mouse monoclonal anti–α-tubulin Alexa Fluor 555 conjugate (1:1,000; EMD Millipore) to visualize MTs, and DRAQ7 or DAPI to visualize nuclei. Individual centrosomes were scored as either γ-tubulin– or pericentrin-positive dots adjacent to the nucleus with distinct separation from neighboring centrosomes. PECAM-enriched mouse cell isolates were incubated for a maximum of 72 h, and these cells or established mouse endothelial cells were fixed and stained for γ-tubulin, rat monoclonal anti-CD31 (1:1,000; Invitrogen), and DNA (DRAQ7; BioStatus), and centrosome counts were performed. Images were acquired using a confocal microscope (LSM 5 Pascal; Carl Zeiss) and a 63×, 1.4 NA oil objective at RT.
Scratch wound, transwell, and random cell migration assays
Scratch wound assays were performed by plating 104 cells/well and growing to confluence. A linear scratch was made down the middle of the well, and media were exchanged. Cells bordering the wounded area were imaged at 20× magnification at t = 0 and t = 5 h. The relative closure for each condition was calculated using the difference in distances between the opposing sides of the wound before and after incubation, with 10 measurements/area.
Transwell assays were performed using a Transwell kit (12-well at 8 µm; Corning) with the membrane seeded with 1 × 104 cells. After 24 h, cells were serum starved with Opti-MEM media (Gibco) overnight, and then, the upper chamber (containing cells) was transferred to a well containing DMEM with 20% FBS and incubated for 5 h. Cells were fixed and stained using the stain set (Protocol Hema 3; Thermo Fisher Scientific), and the number of cells on the bottom of the transwell was counted.
Random cell migration was performed using cells that were sparsely plated on 35-mm glass-bottom dishes (MatTek Corporation) coated with 2 µg/ml fibronectin. Cells were imaged at 5-min intervals for 3 h using an imaging system (BioStation IM; Nikon), with a 20×, 0.45 NA phase objective. Tracks were analyzed with mTrackJ software (Meijering Laboratory).
Growth curves and mitotic cell assessment
Cells were plated at a density of 103 cells per well in triplicate and then trypsinized and counted every 2 d. To identify mitotic cells, fixed cultures were incubated with rabbit anti–phosphohistone-3 Alexa Fluor 555 conjugate (1:1,000; Cell Signaling Technology) and DRAQ7 (1:1,000; Abcam; 20 min at RT) was used to visualize nuclei.
RT-PCR
Total RNA was isolated with TRIZOL reagent (Invitrogen), and 1 µg RNA was used to synthesize cDNA using a cDNA synthesis kit (iScript; Bio-Rad Laboratories, Inc.). 2 µl of the threefold diluted cDNA was used as a template for PCR using Taq PCR Core kit (QIAGEN). The sample was denatured at 94°C for 3 min and then subjected to 26 cycles of denaturation at 94°C for 30 s, annealing at 52–62°C for 30 s, and extension at 72°C for 30 s, with the final extension at 72°C for 10 min. Products were resolved on 1.5% agarose gels.
Western blot analysis
Western blot analysis was performed as previously described (Kappas et al., 2008). In brief, cells were lysed in radioimmunoprecipitation assay buffer containing protease inhibitor cocktail (Sigma-Aldrich). 50 µg protein was separated on a 10% SDS-polyacrylamide gel and transferred to a polyvinylidene fluoride membrane (GE Healthcare). Primary antibodies used were rabbit anti-Cdc14B (1:200; Invitrogen), rabbit anti-Plk4 (1:250; Sigma-Aldrich), rabbit anti–cleaved caspase (1:1,000; Abcam), goat anti-actin (1:5,000; Santa Cruz Biotechnology, Inc.), and goat anti-GAPDH (1:5,000; Abcam). The HRP-tagged secondary antibodies included anti–rabbit IgG (1:1,000; GE Healthcare) and anti–goat IgG (1:1,000; Santa Cruz Biotechnology, Inc.), which were visualized using enhanced chemiluminescence (GE Healthcare).
Sprouting angiogenesis assay
The sprouting assay was performed as previously described (Nakatsu et al., 2007). In brief, HUVECs were mixed with Cytodex beads, incubated with agitation at 20-min intervals for 4 h, and then plated overnight. HUVEC-coated beads were washed in PBS and resuspended in PBS with 2 mg/ml fibrinogen, thrombin was added to form fibrin, and human lung fibroblasts were seeded on top of the fibrin plug. Media were changed every 2 d, and cultures were fixed at day 7 with 4% PFA and stained with phalloidin (Invitrogen) to visualize F-actin or live imaged on days 5–6. Images were acquired using a confocal microscope (LSM 5 Pascal) and 10× air objective at RT. Sprout and branch analyses were performed using the BoneJ plugin for ImageJ software (National Institutes of Health). For live imaging, HUVECs were co-infected with tetracycline-inducible Plk4 and centrin::GFP lentivirus and then incubated with media containing 500 ng/ml doxycycline from day 0 to 2 of the sprouting assay. Live imaging was performed on days 5–6 using a live-imaging system (FV1200; Olympus) at 37°C with 5% CO2 and glass-bottom 35-mm dishes. Confocal stacks were acquired every 20 min with a U Plan S Apochromat 40×, 1.25 NA silicon objective (Olympus).
Centrosome ablation
TECs expressing centrin::GFP were plated on glass-bottom dishes (MatTek Corporation) coated with 2 µg/ml fibronectin or HUVEC on a gridded micropattern. Cells were live imaged in an environmental chamber on an FV1000 at 10-min intervals for 2–3 h before centrosome ablation. Laser ablation was performed as previously described (Magidson et al., 2007). In brief, ablations were performed using a Ti:sapphire laser (Olympus) at 12% power at 900 nm for 10 s and a 60×, 1.42 NA Plan Apochromat objective. In some cells, a third centrosome was ablated, and controls consisted of a similar ablation distant from the centrosomes. For each cell, a comprehensive z stack was acquired before and immediately after the laser pulse to confirm centrosome ablation. After ablation, cells were monitored for 2–3 h. HUVECs on micropattern grids were fixed and stained for γ-tubulin (mouse monoclonal anti–γ-tubulin; GTU88 clone; 1:1,000) 90 min after ablation, and coordinates were used to identify cells that had experienced centrosome ablation.
MT regrowth and MT dynamics
Cells plated on glass-bottom fibronectin-coated plates were incubated in 5 µg/ml nocodazole (in DMSO; Sigma-Aldrich) for 2 h at 37°C. After 5× washes in cold PBS, cells were warmed to 37°C and fixed at various intervals with 100% MeOH. MTs and centrosomes were visualized by immunofluorescence using mouse anti–α-tubulin Alexa Fluor 555 conjugate (1:250) and mouse anti–γ-tubulin (1:5,000; Sigma-Aldrich). Images were acquired using a confocal microscope (LSM 5 Pascal) and 63×, 1.4 NA oil objective at RT. Line scans were generated using a 10-µm line drawn through the centroid of the MT-nucleating center and the centroid of the nucleus.
For MT tip tracking, cells were imaged on an inverted spinning-disk confocal microscope (TE2000E; Nikon) using a 60×, 1.2 NA oil immersion objective (Carl Zeiss). Images were captured using a camera (OrcaER; Hamamatsu Photonics) and SimplePCI software (Hamamatsu Photonics). Excitation was provided by a ∼100-mW Krypton/Argon laser and scan head (Yokogawa Electric Corporation). Acquisitions of EB1/3 videos used a cooled charge-couple device operated by a 16-bit mode at 1–2-s intervals for 1 min. MT dynamics were analyzed using the plusTipTracker software program (Jaqaman et al., 2008) and processed as previously described (Myers et al., 2011). In brief, plusTipTracker is a MATLAB (MathWorks)-based software that detects EB1/3 fluorescently tagged proteins and monitors temporal and spatial dynamics. The first 30 s of each video were analyzed to reduce effects of photobleaching. Tracking parameters were as follows: maximum gap length of 12 frames; minimum track length of three frames; search radium range of 5–10 pixels; maximum forward angle of 25°; maximum backward angle of 8°; maximum shrinkage factor of 1.0; and fluctuation radius of 2 pixels. For all groups, the entire cell was analyzed.
Centrosome and Golgi dynamics
TECs and HUVECs were infected with GalT::EGFP- and centrin::tdTomato-expressing lentivirus overnight. Cells were plated on poly-l-lysine to reduce migration out of frame. HUVECs were also infected with Plk4 to induce centrosome overamplification. Time-lapse videos of Golgi and centrosome dynamics were acquired on a microscope (CellVoyager 1000; Yokogawa Electric Corporation) at 37°C and 5% CO2 with a 60×, 1.4 NA oil objective and electron-multiplying charge-coupled device camera (Rolera; QImaging). Z stacks were acquired over 4–6 h. Centrosome spread is the perimeter of a polygon containing all the centrosomes. Centrosome–centrosome distance was determined by measuring the intracentrosome length between adjacent centrioles. Golgi spread, or area, was calculated using the Analyze Particles function of ImageJ. FRAP analysis was performed by photobleaching a portion of the Golgi with a 488-nm laser and then monitoring fluorescence recovery over 120 s, using a live-imaging system (FV1200) with a 60×, 1.35 NA oil objective and SIM module at 37°C and 5% CO2 (Vinogradova et al., 2012).
Vesicle trafficking
TECs were transfected with Rab6a::GFP and centrin::tdTomato. TEC transfections were performed using HUVEC OLD Nucleofector kit obtained from Amaxa. HUVECs were transfected with program A-034 (Amaxa), and TECs were transfected with M-003 (Amaxa). Cells were plated on dishes (MatTek Corporation) coated with 0.1% gelatin. Images were taken every 2 s for 2 min in a single plane. The images were then time projected and divided into eight radial quadrants with the origin at the Golgi. These quadrants, excluding the Golgi area and extending to the cell border, were outlined using the spline tool on ImageJ, and the fluorescence intensity was measured and background corrected. For comparisons, the intensity percentage in each quadrant was determined. Live imaging was at 37°C and 5% CO2 on an imaging system (FV1200) with a 60×, 1.4 NA oil objective in culture media.
FAs
Measurement of FA dynamics was performed as previously described (Berginski et al., 2011). In brief, TECs were held in nocodazole for 90 min. After washout, cells were fixed at 10-min intervals and stained with rabbit polyclonal anti-vinculin (1:1,000; Sigma-Aldrich) and mouse monoclonal anti–γ-tubulin (1:5,000; Sigma-Aldrich) antibodies. A single plane that most clearly showed FAs was selected and analyzed using the Focal Adhesion Analyzer Server for area, orientation, and length. Images were acquired using a confocal microscope (LSM 5 Pascal) and 63×, 1.4 NA oil objective at RT.
PCM protein quantification
Nonsaturated 1,024 × 1,024–pixel images of cells were acquired on a confocal microscope (LSM 5 Pascal) with a 63×, 1.4 NA objective. After summed z projection of 0.5-µm stacks and background subtraction, the radial integrated density of individual centrosomes was measured by placing a circle (0.8-µm diameter) around centrosome centroids and measuring the pixel intensity. The summed intensity projections of the images were used to quantify the radial integrated fluorescence intensity using ImageJ. Centrosomes that significantly overlapped were excluded from the analysis. Line scans through centrosomes were generated for visual comparison and were not used for quantitative assessments.
Microscope image acquisition and processing
Unless otherwise indicated, fixed samples were mounted using Vectashield and imaged with a confocal microscope (LSM 5 Pascal) with a 63×, 1.4 NA or a 40×, NA 1.3 Plan Neofluar oil objective at RT. Unless otherwise indicated, all fluorescent secondary antibodies were Alexa Fluor (Invitrogen) diluted at 1:250. Image processing and analysis were performed using ImageJ. Illustrator (CS6; Adobe) was used to adjust input levels of the entire image, so the main range of signals spanned the entire output grayscale, and to adjust brightness and contrast.
Statistics
Error bars represent SEMs, except where noted. χ2 analysis was used for centrosome count comparisons, one-tailed Student’s t test was used for pre- versus postablation comparisons, and two-tailed Student’s t test was used for the remainder of the statistical analyses.
Online supplemental material
Fig. S1 characterizes TECs and HUVECs with supernumerary centrosomes. Fig. S2 shows the effects of centrosome ablation on centrosome dynamics and migration. Fig. S3 shows how excess centrosomes affect FAs and Golgi. Fig. S4 shows excess centrosome-mediated changes in MT dynamics in TECs and HUVECs. Fig. S5 shows effects of excess centrosomes on PCM. Video 1 shows TECs with excess centrosomes and blunted migration. Video 2 shows HUVECs with excess centrosomes and blunted migration. Video 3 shows that ablation of excess centrosome restores centrosome clustering. Video 4 shows that ablation of excess centrosome restores directional migration. Videos 5 and 6 show Golgi integrity reduced in cells with excess (two [Video 5] or greater than two [Video 6]) centrosomes. Video 7 shows extended FRAP in Golgi of cells with excess centrosomes. Video 8 shows randomized vesicle trafficking in cells with excess centrosomes. Video 9 shows some changed MT tip-tracking outputs in cells with excess centrosomes. Video 10 shows normal migration of HUVEC with one to two centrosomes and reduced migration of HUVECs with greater than two centrosomes within blood vessel sprouts. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201311013/DC1.
Supplementary Material
Acknowledgments
We thank members of the Bautch laboratory for discussion and Jim Bear, Bob Goldstein, and Mark Peifer for critical reading and comments on the manuscript. We thank the University of North Carolina (UNC) Hooker Microscopy Facility, Rob Peterson and the UNC Olympus Center, and Tal Kafri and Ping Zhang in the UNC Lenti-shRNA Core Facility.
This work was supported by grants to V.L. Bautch (National Institutes of Health R01 HL116719 and HL43174), an Innovation Award from the Lineberger Comprehensive Cancer Center, and fellowship support for E.J. Kushner (5-T32-CA09156-36 and American Heart Association 11POST7220000).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Ac
- acetylated
- CMV
- cytomegalovirus
- FA
- focal adhesion
- GalT
- galactosyltransferase
- HUVEC
- human umbilical vein endothelial cell
- KD
- knockdown
- MACS
- magnetic bead-assisted cell sorting
- MT
- microtubule
- MTOC
- MT-organizing center
- NEC
- normal endothelial cell
- OE
- overexpression
- PCM
- pericentriolar material
- PE
- phycoerythrin
- PECAM
- platelet endothelial cell adhesion molecule
- TEC
- tumor endothelial cell
- USN
- ubiquitin site null
- WT
- wild type
References
- Aird, W.C.2012. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2:a006429 10.1101/cshperspect.a006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate, K.T., Besson S., Matov A., Bagonis M.H., Jaqaman K., and Danuser G.. 2011. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176:168–184 10.1016/j.jsb.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano, J.L., Teslovich T.M., and Katsanis N.. 2005. The centrosome in human genetic disease. Nat. Rev. Genet. 6:194–205 10.1038/nrg1557 [DOI] [PubMed] [Google Scholar]
- Balconi, G., Spagnuolo R., and Dejana E.. 2000. Development of endothelial cell lines from embryonic stem cells: A tool for studying genetically manipulated endothelial cells in vitro. Arterioscler. Thromb. Vasc. Biol. 20:1443–1451 10.1161/01.ATV.20.6.1443 [DOI] [PubMed] [Google Scholar]
- Berginski, M.E., Vitriol E.A., Hahn K.M., and Gomez S.M.. 2011. High-resolution quantification of focal adhesion spatiotemporal dynamics in living cells. PLoS ONE. 6:e22025 10.1371/journal.pone.0022025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., and Glover D.M.. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Bornens, M.2012. The centrosome in cells and organisms. Science. 335:422–426 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Boveri, T.1888. Zellen-Studien II: Die Befruchtung und Teilung des Eies von Ascaris megalocephala. Gustav Fischer Verlag, Jena, Germany: 198 pp [Google Scholar]
- Boveri, T.1901. Uber die natur der centrosomen. Jena Z Med. Naturw. 28:1–220 [Google Scholar]
- Carmeliet, P., and Jain R.K.. 2011. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat. Rev. Drug Discov. 10:417–427 10.1038/nrd3455 [DOI] [PubMed] [Google Scholar]
- Casenghi, M., Meraldi P., Weinhart U., Duncan P.I., Körner R., and Nigg E.A.. 2003. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell. 5:113–125 10.1016/S1534-5807(03)00193-X [DOI] [PubMed] [Google Scholar]
- Chen, H., Huang Z., Dutta K., Blais S., Neubert T.A., Li X., Cowburn D., Traaseth N.J., and Mohammadi M.. 2013. Cracking the molecular origin of intrinsic tyrosine kinase activity through analysis of pathogenic gain-of-function mutations. Cell Reports. 4:376–384 10.1016/j.celrep.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, A.C., Khan Z.A., Shih S.-C., Kang S.-Y., Zwaans B.M.M., Bischoff J., and Klagsbrun M.. 2008. Calcification of multipotent prostate tumor endothelium. Cancer Cell. 14:201–211 10.1016/j.ccr.2008.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe, S., Hümmer S., Uldschmid A., Li X., and Nigg E.A.. 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21:2205–2219 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eot-Houllier, G., Venoux M., Vidal-Eychenié S., Hoang M.T., Giorgi D., and Rouquier S.. 2010. Plk1 regulates both ASAP localization and its role in spindle pole integrity. J. Biol. Chem. 285:29556–29568 10.1074/jbc.M110.144220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn, R.M., Schultz S.J., Bartek J., Ziemiecki A., Ried T., and Nigg E.A.. 1994. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J. Cell Sci. 107:1509–1517 [DOI] [PubMed] [Google Scholar]
- Habedanck, R., Stierhof Y.D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hanisch, A., Wehner A., Nigg E.A., and Silljé H.H.W.. 2006. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol. Biol. Cell. 17:448–459 10.1091/mbc.E05-08-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Stearns T., and Lüders J.. 2009. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE. 4:e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida, K., Hida Y., Amin D.N., Flint A.F., Panigrahy D., Morton C.C., and Klagsbrun M.. 2004. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 64:8249–8255 10.1158/0008-5472.CAN-04-1567 [DOI] [PubMed] [Google Scholar]
- Holland, A.J., Lan W., and Cleveland D.W.. 2010. Centriole duplication: A lesson in self-control. Cell Cycle. 9:2731–2736 10.4161/cc.9.14.12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado, L., Caballero C., Gavilan M.P., Cardenas J., Bornens M., and Rios R.M.. 2011. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 193:917–933 10.1083/jcb.201011014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman, K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappas, N.C., Zeng G., Chappell J.C., Kearney J.B., Hazarika S., Kallianos K.G., Patterson C., Annex B.H., and Bautch V.L.. 2008. The VEGF receptor Flt-1 spatially modulates Flk-1 signaling and blood vessel branching. J. Cell Biol. 181:847–858 10.1083/jcb.200709114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina, I., and Straube A.. 2011. Regulation of cell migration by dynamic microtubules. Semin. Cell Dev. Biol. 22:968–974 10.1016/j.semcdb.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu, T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, J.M., Merdes A., Mourey L., and Agard D.A.. 2011. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12:709–721 10.1038/nrm3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce, M.P., Köhler J., Neujahr R., Schwartz J.-M., Tikhonenko I., and Gerisch G.. 1999. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 18:6786–6792 10.1093/emboj/18.23.6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers, M., and Hoogenraad C.C.. 2011. Centrosomes, microtubules and neuronal development. Mol. Cell. Neurosci. 48:349–358 10.1016/j.mcn.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Kwon, M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., and Pellman D.. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22:2189–2203 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo, S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Levy, J.R., and Holzbaur E.L.F.. 2008. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121:3187–3195 10.1242/jcs.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon, C., and Pines J.. 2004. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol. 164:233–241 10.1083/jcb.200309035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle, W.L., Lutz W.H., Ingle J.N., Maihle N.J., and Salisbury J.L.. 1998. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA. 95:2950–2955 10.1073/pnas.95.6.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton, G.W.G., and Gundersen G.G.. 2011. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23:579–588 10.1016/j.ceb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson, V., Lončarek J., Hergert P., Rieder C.L., and Khodjakov A.. 2007. Laser microsurgery in the GFP era: a cell biologist’s perspective. Methods Cell Biol. 82:239–266 10.1016/S0091-679X(06)82007-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub, M.R., and Stearns T.. 2012. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr. Biol. 22:1628–1634 10.1016/j.cub.2012.06.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella, V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov, A., and Gundersen G.G.. 1998. Relationship between microtubule dynamics and lamellipodium formation revealed by direct imaging of microtubules in cells treated with nocodazole or taxol. Cell Motil. Cytoskeleton. 41:325–340 [DOI] [PubMed] [Google Scholar]
- Miller, P.M., Folkmann A.W., Maia A.R., Efimova N., Efimov A., and Kaverina I.. 2009. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 11:1069–1080 10.1038/ncb1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, K.A., Applegate K.T., Danuser G., Fischer R.S., and Waterman C.M.. 2011. Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J. Cell Biol. 192:321–334 10.1083/jcb.201006009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu, M.N., Davis J., and Hughes C.C.. 2007. Optimized fibrin gel bead assay for the study of angiogenesis. J. Vis. Exp. 3:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef, R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., and Barr F.A.. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Petrie, R.J., Doyle A.D., and Yamada K.M.. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10:538–549 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Wojcik E.J., Sanders M.A., McGrail M., and Hays T.S.. 1999. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146:597–608 10.1083/jcb.146.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam, K., Woodman B., Mahal A., Bertaux F., Wanker E.E., Shima D.T., and Bates G.P.. 2001. Centrosome disorganization in fibroblast cultures derived from R6/2 Huntington’s disease (HD) transgenic mice and HD patients. Hum. Mol. Genet. 10:2425–2435 10.1093/hmg/10.21.2425 [DOI] [PubMed] [Google Scholar]
- Silkworth, W.T., Nardi I.K., Scholl L.M., and Cimini D.. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder, G., and Khodjakov A.. 2010. Centriole duplication: analogue control in a digital age. Cell Biol. Int. 34:1239–1245 10.1042/CBI20100612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen, K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Serneo F.F., Higgins C., Gambello M.J., Wynshaw-Boris A., and Gleeson J.G.. 2004. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165:709–721 10.1083/jcb.200309025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley, E.C., and Starr D.A.. 2013. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr. Opin. Cell Biol. 25:57–62 10.1016/j.ceb.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.M., Nevis K.R., Park H.L., Rogers G.C., Rogers S.L., Cook J.G., and Bautch V.L.. 2010. Angiogenic factor signaling regulates centrosome duplication in endothelial cells of developing blood vessels. Blood. 116:3108–3117 10.1182/blood-2010-01-266197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova, T., Paul R., Grimaldi A.D., Loncarek J., Miller P.M., Yampolsky D., Magidson V., Khodjakov A., Mogilner A., and Kaverina I.. 2012. Concerted effort of centrosomal and Golgi-derived microtubules is required for proper Golgi complex assembly but not for maintenance. Mol. Biol. Cell. 23:820–833 10.1091/mbc.E11-06-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., Worthylake R.A., Liu B.P., Burridge K., and Salmon E.D.. 1999. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat. Cell Biol. 1:45–50 10.1038/9018 [DOI] [PubMed] [Google Scholar]
- Wu, J., Cho H.P., Rhee D.B., Johnson D.K., Dunlap J., Liu Y., and Wang Y.. 2008. Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. J. Cell Biol. 181:475–483 10.1083/jcb.200710127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., Puri S., and Linstedt A.D.. 2009. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol. Biol. Cell. 20:1728–1736 10.1091/mbc.E08-10-1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.