The endonuclease AGO3 and mitochondria-associated protein Zucchini together control the dynamic subcellular localization of Armitage between mitochondria and germline granules to regulate secondary piRNA amplification.
Abstract
In Drosophila melanogaster the reciprocal “Ping-Pong” cycle of PIWI-interacting RNA (piRNA)–directed RNA cleavage catalyzed by the endonuclease (or “Slicer”) activities of the PIWI proteins Aubergine (Aub) and Argonaute3 (AGO3) has been proposed to expand the secondary piRNA population. However, the role of AGO3/Aub Slicer activity in piRNA amplification remains to be explored. We show that AGO3 Slicer activity is essential for piRNA amplification and that AGO3 inhibits the homotypic Aub:Aub Ping-Pong process in a Slicer-independent manner. We also find that expression of an AGO3 Slicer mutant causes ectopic accumulation of Armitage, a key component in the primary piRNA pathway, in the Drosophila melanogaster germline granules known as nuage. AGO3 also coexists and interacts with Armitage in the mitochondrial fraction. Furthermore, AGO3 acts in conjunction with the mitochondria-associated protein Zucchini to control the dynamic subcellular localization of Armitage between mitochondria and nuage in a Slicer-dependent fashion. Collectively, our findings uncover a new mechanism that couples mitochondria with nuage to regulate secondary piRNA amplification.
Introduction
Small noncoding RNAs (ncRNAs) play pivotal roles in regulating gene expression via an array of mechanisms (Neilson and Sharp, 2008). PIWI-interacting RNAs (piRNAs), a subset of small ncRNAs (∼24–30 nt in length), associate with PIWI proteins and play conserved roles in silencing the expression of transposable elements to maintain genome integrity and proper germline development (Lin, 2007; Chang et al., 2009; Thomson and Lin, 2009; Juliano et al., 2011).
The Argonaute protein family functions as core components in small RNA–mediated gene regulation pathways (Peters and Meister, 2007; Yin and Lin, 2007; Höck and Meister, 2008; Hutvagner and Simard, 2008). In Drosophila melanogaster there are five well-characterized Argonaute protein family members, which can be grouped into two subfamilies: AGO and PIWI. As members of the AGO subfamily of Argonaute proteins, Argonaute 1 (AGO1) and 2 (AGO2) are known to be involved in miRNA-mediated gene regulation and siRNA-mediated mRNA degradation, respectively (Okamura et al., 2004; Yang et al., 2007). In contrast, Piwi, Aubergine (Aub), and Argonaute 3 (AGO3), members of the PIWI subfamily of proteins, associate with piRNAs, which are derived mainly from transposon-rich clusters, and play an important role in transposon silencing (Saito et al., 2006; Vagin et al., 2006; Brennecke et al., 2007; Gunawardane et al., 2007; Yin and Lin, 2007). Most piRNAs match to the genome in clusters of 20–90 kb in a strand-specific manner, revealing that piRNAs are derived mainly from active transposons and other intergenic regions (Brennecke et al., 2007).
All three PIWI proteins (Piwi, AGO3, and Aub) in Drosophila have endonuclease activity (Gunawardane et al., 2007), and each PIWI protein is associated with a specific set of piRNAs that exhibit certain characteristics, such as piRNA origin and nucleotide/strand bias (Brennecke et al., 2007). Among the three PIWI proteins, Piwi is the only member that is primarily nuclear in both germline and somatic cells in the gonad and it controls piRNA biogenesis in both cell types, whereas AGO3 and Aub are expressed in the cytosolic region in germ cells. Bioinformatic studies have suggested two models for piRNA production, including the primary processing and the feed-forward amplification pathways. Primary piRNAs are produced from piRNA cluster transcripts, which are then processed by multiple factors, including Piwi, Zucchini (Zuc), Armitage (Armi), Shutdown, and Vreteno in the nuclei and cytoplasm (Malone et al., 2009; Olivieri et al., 2010; Saito et al., 2010; Zamparini et al., 2011; Handler et al., 2013). In addition to the primary pathway, the feed-forward amplification model, or “Ping-Pong” model, has been proposed as an important mechanism to amplify piRNA biogenesis in the germline. In this model, Aub and AGO3 associate with distinct sets of piRNAs. piRNA–Aub (or AGO3) protein complexes thus could induce reciprocal Slicer-dependent cleavage of sense and antisense transcripts of transposons, which guide piRNA production (Brennecke et al., 2007; Gunawardane et al., 2007). The Slicer-mediated Ping-Pong model was proposed mainly based on data from deep sequencing and bioinformatic analyses, but whether and how the Slicer activity of AOG3/Aub proteins promotes piRNA biogenesis have not been explored. To address this important question, in this study we generated transgenic flies that express AGO3 Slicer mutant forms in germ cells. We show that the Slicer activity of AGO3 is essential for germline development and piRNA amplification and that AGO3 inhibits the Aub:Aub homotypic Ping-Pong process in a Slicer-independent manner. Importantly, we provide evidence to support that AGO3 acts in concert with the mitochondrion-associated protein Zuc to control dynamic subcellular localization of Armi between mitochondria and nuage. Thus, our findings uncover a mechanism that couples mitochondria and nuage to regulate piRNA amplification.
Results
The Slicer activity of AGO3 is required for transposon silencing
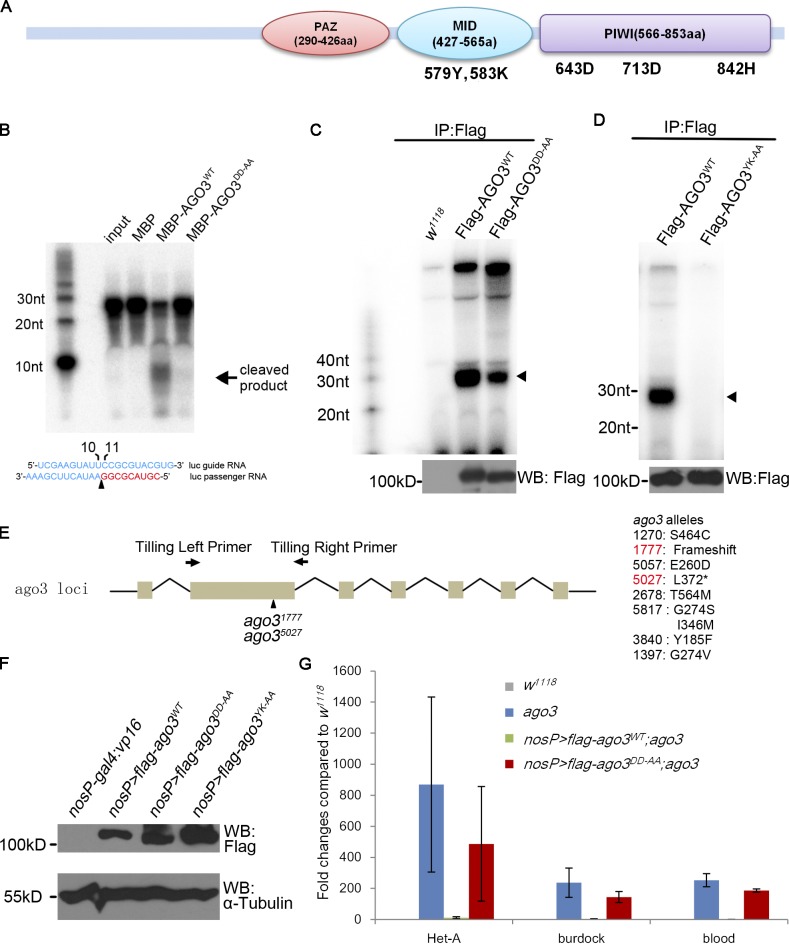
AGO3 contains three typical conserved domains, including the PAZ domain, the MID domain in the middle region, and the PIWI domain in the C terminus (Fig. 1 A). Although the PAZ and MID domains are proposed to bind to the 3′ and 5′ termini of a small RNA, respectively (Jinek and Doudna, 2009; Wang et al., 2009), the PIWI domain possesses the conserved catalytic motif Asp-Asp-His (D-D-H: 643D, 713D, and 842H in AGO3; Fig. 1 A), which exhibits potential Slicer activity (Filipowicz, 2005; Rivas et al., 2005). To verify whether the putative catalytic residues of D-D-H are essential for supporting AGO3 Slicer activity, we performed in vitro Slicer activity assays as described previously (Gunawardane et al., 2007). As shown in Figs. 1 B and S1 A, whereas the wild-type AGO3 protein produced a small 9-nt band, the mutant form of AGO3DD-AA protein, in which both of the aspartic acid (643D and 713D) residues were mutated to alanine in the full-length AGO3, did not, indicating that the 643D and 713D residues are required for the Slicer activity of AGO3 in vitro.
Figure 1.
AGO3 Slicer activity is essential for transposon repression and piRNA biogenesis in ovaries. (A) Structure diagram of the AGO3 protein showing the essential active sites in the MID and PIWI domains. The PAZ and PIWI domains were analyzed by SMART program (http://smart.embl-heidelberg.de/), and the MID domain was identified between PAZ and PIWI domains. The conserved amino acids were identified via alignment of AGO3 with human AGO2 using CLUSTALW. Although the 579Y and 583K sites in the MID domain are responsible for binding to the 5′ terminus of piRNAs, the 643D, 713D, and 842H sites in the PIWI domain constitute the Slicer motif that is essential for the Slicer activity of AGO3 protein. (B) The DDH motif is essential for AGO3 Slicer activity in vitro. The in vitro assay for Slicer activity was performed according to the method described previously (Gunawardane et al., 2007) with minor modifications (see Materials and methods). Only MBP-AGO3WT, but not MBP itself or MBP-AGO3DD-AA, could cleave target RNA (Luc passenger siRNA) to release 9-nt products. (C and D) Ovaries from uasp-flag-ago3WT; nosP-gal4:vp16 and uasp-flag-ago3DD-AA; nosP-gal4:vp16 (D) or uasp-flag-ago3YK-AA; nosP-gal4:vp16 (E) females were lysed and the supernatants were immunoprecipitated with anti-Flag antibody. RNAs were extracted from the precipitations and then treated with alkaline phosphatase. RNAs were labeled by 32P using T4 polynucleotide kinase for detection of piRNAs (top). A small portion of the precipitations was used to perform Western blot assays with anti-Flag antibody to measure the levels of Flag-tagged AGO3 or its mutants in precipitations (bottom). Arrowhead indicates piRNAs. (E) Generation of ago3 mutants. The ago3 gene region contains 7 exons and the second one is biggest and encodes the major part of the PAZ domain. The tilling primers are in the 5′ terminus of this exon and 5′ terminus of the second intron. All alleles obtained are listed on the right. Two null alleles, ago31777 and ago35027, both result in a stop codon in the second exon. (F) Ovaries from w1118, uasp-flag-ago3WT; nosP-gal4:vp16, uasp-flag-ago3DD-AA; nosP-gal4:vp16, and uasp-flag-ago3YK-AA; nosP-gal4:vp16 females were used to perform Western blot assays to show the abundance of ectopic Flag-AGO3 proteins. α-Tubulin was used as a loading control. (G) Quantitative PCR assays to detect the fold changes in steady-state RNA levels of Het-A, burdock, and blood transposons in ago3 mutant, uasp-flag-ago3WT; nosP-gal4:vp16, ago3, and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 (Normalized to w1118; n = 3; error bars indicate standard errors).
To explore the biological importance of the AGO3 Slicer, we generated a transgenic line, P{uasp-flag-ago3DD-AA}, in which ago3DD-AA was placed under the control of the UASp promoter. Although the AGO3DD-AA protein is unable to cleave the target RNA, it still binds mature piRNAs in ovaries (Fig. 1 C). It has been reported that the first 5′ base of a small RNA stacks with the conserved tyrosine in the MID domain and that the 5′ phosphate also forms a hydrogen bond with the two conserved amino acids, tyrosine and lysine residues in the MID domain (579Y and 583K in AGO3; Fig. 1 A; Song et al., 2004; Schirle and MacRae, 2012). Considering this, we generated another mutant transgene, P{uasp-flag-ago3YK-AA}. In AGO3YK-AA mutant, the conserved tyrosine (579Y) and lysine (583K) residues were both changed to alanine, and this mutant failed to bind piRNAs in ovaries (Fig. 1 D). Unlike the wild-type form of AGO3 (AGO3WT), the germ cell–specific expression of mutant forms of either AGO3DD-AA or AGO3YK-AA driven by the nosP-gal4:vp16 failed to rescue the female sterility caused by the loss of ago3 in ago31777/ago35027 mutant (hereafter referred as ago3 mutant; allelic information shown in Materials and methods and in Fig. 1 E and Fig. S1, B and C), although expression levels of AGO3DD-AA or AGO3YK-AA in germ cells appeared to be comparable with AGO3WT (Fig. 1 F). Additionally, AGO3WT but not AGO3DD-AA or AGO3YK-AA could restore the defects of Oskar or Grk expression in ago3 mutant (Fig. S1, D–F). Collectively, our findings support that the AGO3 Slicer activity is essential for normal germline development.
To obtain direct experimental evidence that supports the role of AGO3 Slicer in the piRNA pathway, we tested whether the AGO3 Slicer influences transposon silencing in ovaries. We performed quantitative RT-PCR assays to measure levels of transposon transcripts in ago3 mutant ovaries expressing AGO3WT and AGO3DD-AA, respectively. In this assay, 15 transposons whose repression is known to be mediated by AGO3 were used. As shown in Fig. 1 G, expression of AGO3WT but not AGO3DD-AA in the ago3 mutant sufficiently suppressed levels of tested transposon transcripts, suggesting that the AGO3 Slicer activity is essential for transposon silencing.
The Slicer activity of AGO3 is required for piRNA amplification
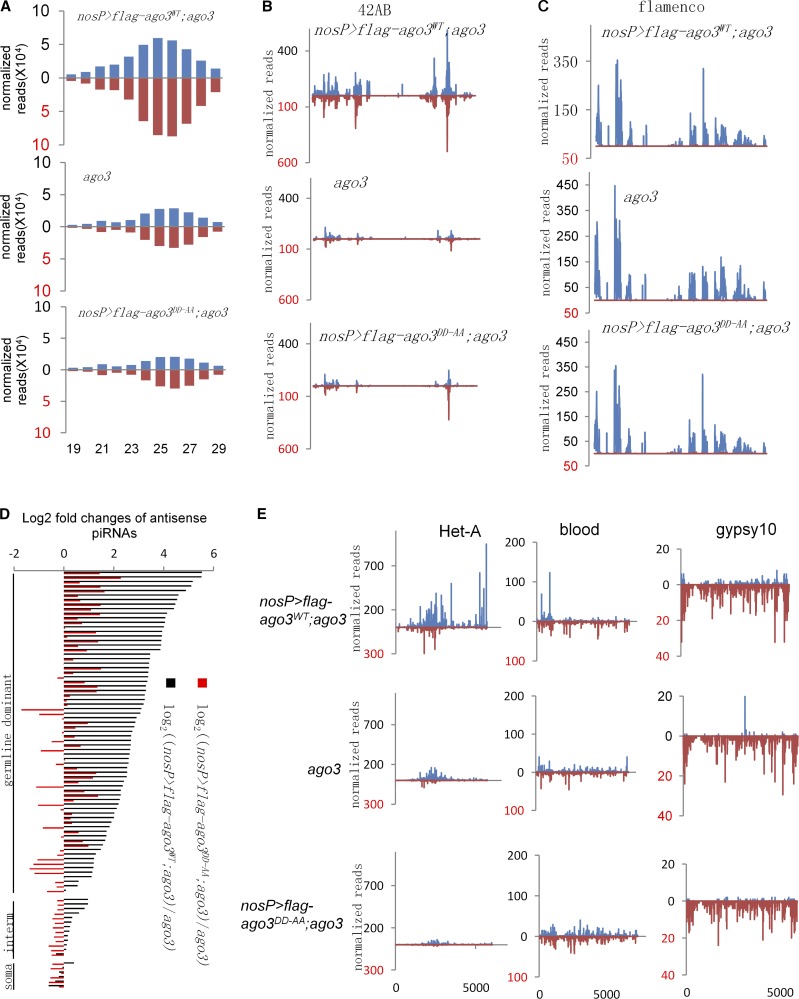
To test the role of AGO3 Slicer activity in regulating piRNA biogenesis, we sequenced piRNAs from ago3 mutant ovaries and ago3 mutant ovaries expressing AGO3WT or AGO3DD-AA. We obtained fully genome-matched non-ncRNA, non-miRNA, 23–29-nt-long small RNAs as piRNAs and normalized the read counts with miRNAs and ncRNAs. We found that expression of AGO3WT in ago3 mutant produced 2.48-fold more 23–29-nt piRNAs, whereas AGO3DD-AA in ago3 only produced 0.83 times as much when compared with control ago3 mutant (Fig. 2 A), indicating that AGO3 Slicer activity is required for piRNA biogenesis in general. We then counted the piRNA reads from two major piRNA clusters, the 42AB site on chromosome 2 and the flamenco site on chromosome X. The 42AB site produced ∼30% of all piRNAs in germline cells, whereas the flamenco transcripts are expressed mainly in soma and produce piRNAs through the primary piRNA pathway (Brennecke et al., 2007). Expression of AGO3WT in ago3 mutant produced much more piRNA from the 42AB site, but not flamenco, than both AGO3DD-AA in ago3 mutant and ago3 mutant alone did (Fig. 2, B and C).
Figure 2.
AGO3 Slicer activity is essential for piRNA biogenesis in ovaries. (A) Length histograms of piRNAs expressed in ago3 mutant, uasp-flag-ago3WT; nosP-gal4:vp16, ago3, and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 ovaries. Blue, sense; red, antisense. (B and C) The normalized abundance of uniquely mapping piRNAs from the 42AB site (B) and flamenco (C) in ago3 mutant, uasp-flag-ago3WT; nosP-gal4:vp16, ago3, and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 ovaries. Blue, sense; red, antisense. (D) The log2 fold changes of antisense piRNA levels mapping to all analyzed transposons in uasp-flag-ago3WT; nosP-gal4:vp16, ago3 (black) and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 (red) compared with ago3 mutant. (E) The normalized abundances of sense and antisense piRNAs mapping to Het-A (left), blood (middle), and gypsy10 (right) were plotted over transposon sequences. Blue, sense; red, antisense.
The majority of piRNAs are derived from transposons and other repeated elements (Brennecke et al., 2007). We then analyzed the piRNA expression from 90 transposon families with at least 500 matching reads in each genotype (Fig. 2 D). Out of 71 germline-dominant families, 68 produced at least two times more antisense piRNAs in uasp-flag-ago3WT, nosP-gal4:vp16, ago3 ovary (1.51–46.4-fold for all 71 families; median fold of 6.71) than in ago3 mutant ovary. Among them, 15 had more than twofold increases in ago3 mutant expressing AGO3DD-AA when compared with ago3 mutant alone. Notably, expression of AGO3DD-AA in ago3 mutant even produced fewer piRNAs than ago3 mutant alone in 22 families (Fig. 2 D).
We next analyzed the specific transposon-derived piRNAs. As shown in Fig. 2 E, typical germline-dominant transposon Het-A has significantly higher levels of both sense and antisense piRNAs in ago3 mutant ovary expressing AGO3WT than in ago3 ovary expressing AGO3DD-AA or in ago3 mutant ovary alone, whereas intermediate, blood, and soma-dominant family gypsy10 had similar levels in those three genotypes. Collectively, our findings suggest that Slicer activity is required for AGO3 to efficiently produce piRNAs in germ cells in general.
AGO3 inhibits the Aub:Aub Ping-Pong process in a Slicer-independent manner
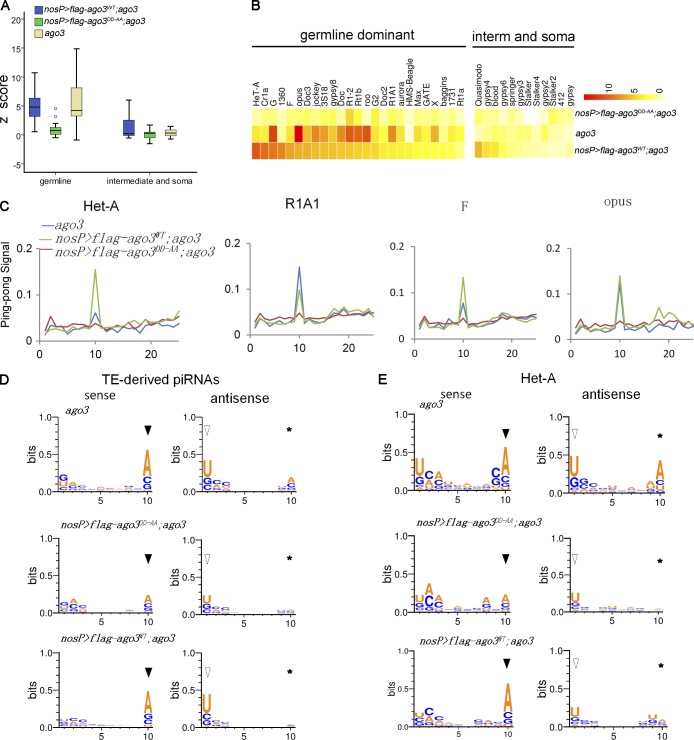
Next, we examined the Ping-Pong features in each genotype to uncover the potential changes in Ping-Pong amplification. We calculated Ping-Pong Z-scores for sense and antisense piRNAs and focused on 37 families (24 germline families and 13 intermediate or somatic families) that had at least 50 piRNAs with 5′ 10-nt complementary Ping-Pong partners (Fig. 3, A–C). In many germline families, AGO3WT expression in ago3 mutant apparently increased the Z-scores compared with ago3 mutant; however, in some families, such as roo, opus, G-element, and Rt1b, ago3 mutant had similar or even higher Z-score values than AGO3WT-expressing ago3 mutant did, indicating that the Ping-Pong cycle might still function in those families in ago3 mutant. It has been reported that, although the loss of ago3 abolishes the AGO3:Aub Ping-Pong cycle, the activity of homotypic Aub:Aub Ping-Pong accordingly increases (Li et al., 2009). Thus, the Ping-Pong–driven piRNAs seen in ago3 mutant are proposed to be the products of the Aub:Aub Ping-Pong cycle. Notably, we found that the Z-scores in ago3 mutant ovary expressing AGO3DD-AA were much lower than in either ago3 mutant ovary expressing AGO3WT or ago3 mutant with respect to all germline families, suggesting that expression of AGO3DD-AA in ago3 mutant significantly suppresses the Aub:Aub Ping-Pong that dominantly exists in ago3 mutant germ cells.
Figure 3.
AGO3 inhibits the Aub:Aub Ping-Pong cycle in a Slicer-independent manner. (A) Box plots illustrating the Ping-Pong Z-score values of piRNAs in each phenotype, with whiskers showing the minimum and maximum numbers in the data group. (B) Heatmap of Z-score values for each transposon family in ago3 mutant, uasp-flag-ago3WT; nosP-gal4:vp16, ago3, and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 ovaries. (C) The Ping-Pong signal showed the probability of 5′ overlap between sense and antisense piRNAs in Het-A, R1A1, F-element, and opus families in ago3 mutant, uasp-flag-ago3WT; nosP-gal4:vp16, ago3, and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 ovaries. (D and E) The nucleotide bias of total TE-derived Ping-Pong piRNAs (D) and Het-A (E) Ping-Pong piRNAs in ago3 mutant, uasp-flag-ago3WT; nosP-gal4:vp16, ago3, and uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 ovaries. Closed arrowhead, open arrowhead, and asterisks indicate sense 10A, antisense 1U, and antisense 10A, respectively.
The primary antisense piRNAs typically begin with U in the 5′ terminus, and the secondary piRNAs produced will bear an A at position 10. Thus, the piRNAs amplified by Ping-Pong in wild-type ovaries usually have nucleotide bias: antisense 5′U and sense 10-nt A. To test whether the Slicer activity of AGO3 affects nucleotide bias, we analyzed the relative nucleotide bias of each position in piRNAs with Ping-Pong partners (Fig. 3, D and E). The Ping-Pong piRNAs in ago3 mutant expressing AGO3WT exhibited strong antisense 1U and sense 10A bias, a feature of Aub:AGO3 Ping-Pong amplification (Li et al., 2009). This feature could still be observed in ago3 mutant ovaries; in addition, ago3 mutant showed a week antisense 10A bias, a feature of Aub:Aub Ping-Pong amplification. Notably, we found that both the antisense 1U/sense 10A and antisense 10A biases were evidently repressed in AGO3DD-AA-expressed ago3 ovaries (Fig. 3, D and E). These findings further argue that Aub:Aub Ping-Pong is repressed in AGO3DD-AA-expressing ago3 ovary.
Nuage localization of AGO3 requires its piRNA binding but not Slicer activity
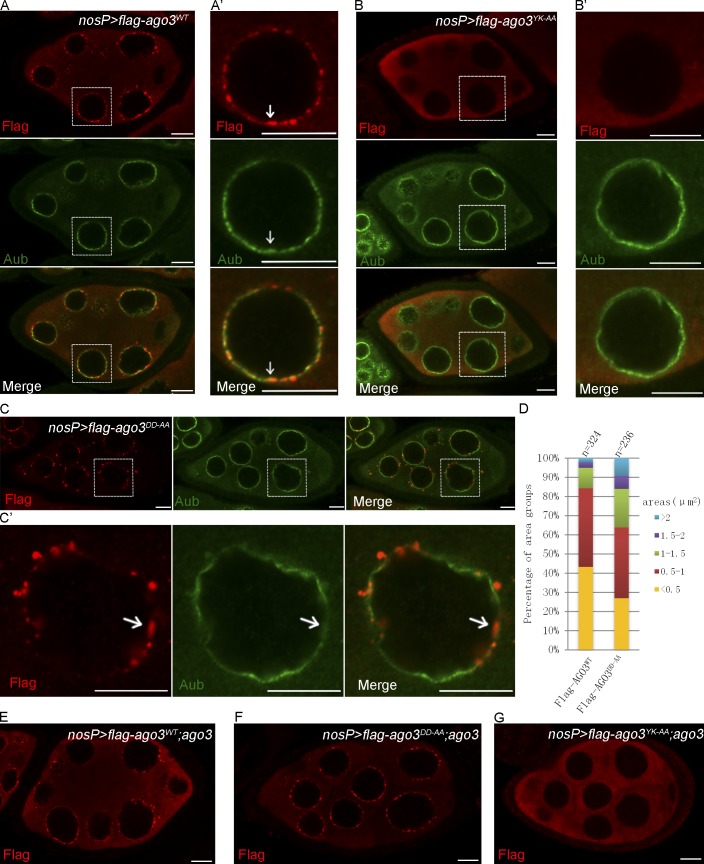
Most piRNA pathway components are localized in the nuage, a heavy electronic density structure around the nurse cell nuclei that includes Vasa, Aub, and AGO3 (Findley et al., 2003; Brennecke et al., 2007; Lim and Kai, 2007; Patil and Kai, 2010). Nuage localization is known to be important for the functions of these components in regulating piRNA biogenesis (Klattenhoff and Theurkauf, 2008; Saxe and Lin, 2011; Siomi et al., 2011). To test the requirements of AGO3 nuage localization, we asked whether the Slicer mutant (AGO3DD-AA) and the piRNA-binding mutant (AGO3YK-AA) affect the subcellular localization of the AGO3 protein. As shown in Fig. 4 (A and A′), in wild-type ovaries, the expression of AGO3WT resulted in the proper localization of AGO3 in the nuage, whereas the AGO3YK-AA almost failed to localize in nuage and evenly distributed in cytoplasm in late-stage nurse cells (Fig. 4, B and B′). Surprisingly, we found that the AGO3DD-AA could be localized in the nuage much like the AGO3WT (Fig. 4, C, C′, and D). To exclude the possibility that endogenous AGO3 might affect the subcellular localization of AGO3 mutants, we investigated the behavior of AGO3 mutant proteins in ago3 mutant ovaries. As shown in Fig. 4 (E and F), AGO3DD-AA displayed a nuage localization pattern in nurse cells, whereas the AGO3YK-AA was occasionally observed in nuage in early nurse cells, but only present in cytoplasm in late-stage nurse cells (Fig. 4 G).
Figure 4.
The Slicer activity of AGO3 is dispensable for its subcellular localization. (A–C′) Ovaries expressing Flag-AGO3WT (A and A′), Flag-AGO3YK-AA (B and B′), and Flag-AGO3DD-AA (C and C′) driven by nosP-gal4:vp16 were stained with anti-Flag (red) and anti-Aub (green) antibodies. A′, B′, and C′ show enlarged images of the boxed portions in A, B, and C, respectively. The arrows indicate the nuage where AGO3 is expressed (A′ and C′) or not (B′). (D) The quantification of the aggregate areas of Flag-AGO3WT and Flag-AGO3DD-AA. The area values were divided into five groups: <0.5, 0.5–1.0, 1.0–1.5, 1.5–2.0, and >2.0 µm2. (E–G) Ovaries from uasp-flag-ago3WT; nosP-gal4:vp16, ago3 (E), uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 (F), and uasp-flag-ago3YK-AA; nosP-gal4:vp16, ago3 (G) were stained with anti-Flag (red) antibody. Bars, 10 µm.
We noted that, although both AGO3WT and AGO3DD-AA were present in nuage, in contrast to AGO3WT or endogenous AGO3, the AGO3DD-AA protein was often found to form many larger aggregates (Fig. 4 D), which contained both Vasa (Fig. S2, A and A′) and Aub, although the Aub staining was relatively weak (Fig. 4, C and C′). The distinct behavior of AGO3DD-AA in terms of its nuage localization reflects that AGO3 might play roles in regulating the piRNA pathway via Slicer-independent activity.
The Slicer activity of AGO3 disassociates Armi from nuage
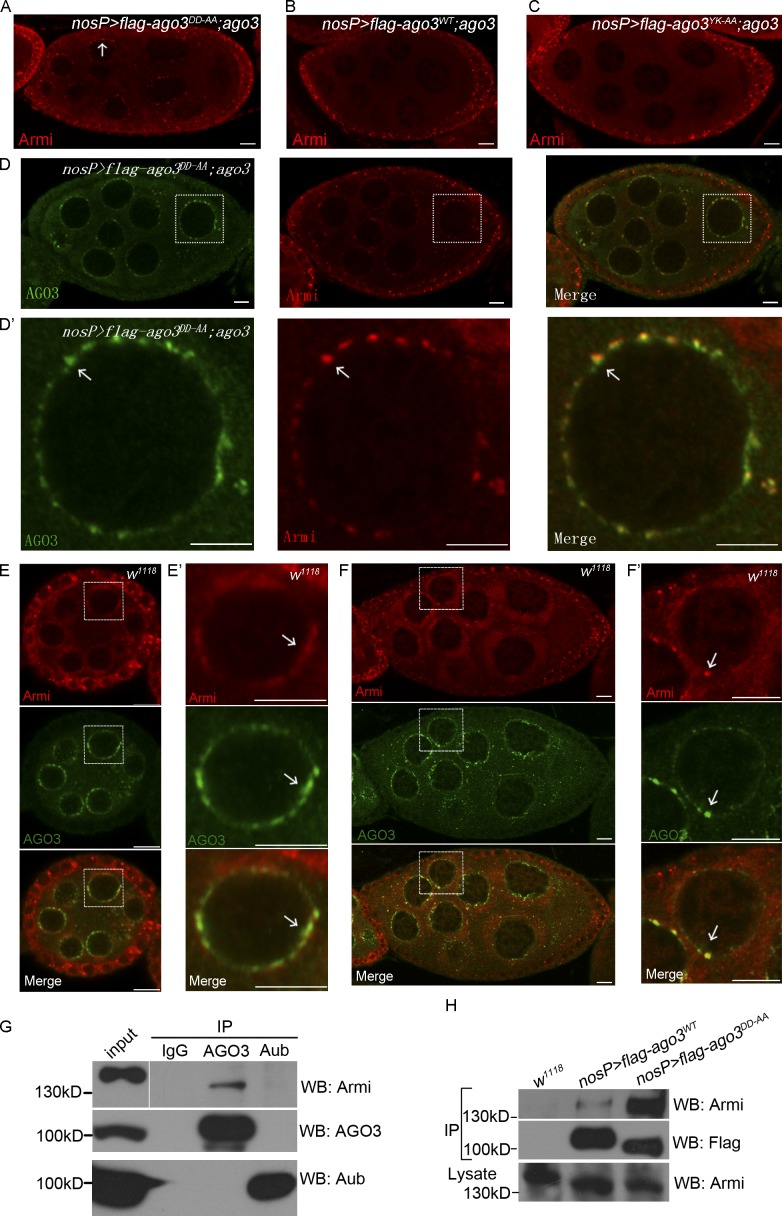
The larger nuage formation induced by ectopic expression of AGO3DD-AA raises a possibility that Slicer activity might contribute AGO3 RISC assembly and nuage formation. To test this, we performed immunostaining assays to determine whether the expression of AGO3DD-AA affects nuage localization of other components in the piRNA pathway in the ago3 mutant. We found that most of the tested nuage components, including Aub, Vasa, Mael, and Tudor, were normally localized in AGO3DD-AA ectopic nuage. Surprisingly, we found that one cytoplasmic protein, Armi, that functions in the primary piRNA processing pathway, was ectopically accumulated in nuage and colocalized with AGO3DD-AA in ago3 mutant ovaries, when compared with ago3 ovaries expressing AGO3WT or AGO3YK-AA (Fig. 5, A–D′). To test the specificity of colocalization of AGO3DD-AA with Armi in nuage, we then examined whether some other nuage components colocalized with AGO3DD-AA/Armi and found that Krimp and Mael were almost fully overlapped with AGO3DD-AA/Armi, and Aub and Tudor were also colocalized, at least in part, with AGO3DD-AA/Armi (Fig. S3, A–E). These results suggest that the AGO3 Slicer mutant promotes nuage accumulation of Armi.
Figure 5.
The Slicer activity of AGO3 disassociates Armi from nuage. (A–C) uasp-flag-ago3WT; nosP-gal4:vp16, ago3 (A), uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 (B), and uasp-flag-ago3YK-AA; nosP-gal4:vp16, ago3 (C) ovaries were stained with anti-Armi (red) antibody. The localization of Armi is indicated by arrows. (D and D′) uasp-flag-ago3DD-AA; nosP-gal4:vp16, ago3 ovary was stained with anti-Armi (red) and anti-AGO3 (green) antibodies. D′ shows the enlarged image of the boxed part in D. The colocalization of AGO3DD-AA–Armi is indicated by arrows. (E–F′) Early (E and E′) and late (F and F′) stage egg chambers in w1118 ovary were stained with anti-Armi (red) and anti-AGO3 (green) antibodies. E′ and F′ show the enlarged image of the boxed part in E and F, respectively. The nuage localization of AGO3–Armi in is indicated by arrows. Bars, 10 µm. (G) Coimmunoprecipitation of Armi with AGO3 in w1118 ovaries. w1118 ovary lysate was immunoprecipitated with IgG, anti-AGO3 or anti-Aub antibodies, and Western blot assays were performed to detect Armi proteins in each immunoprecipitation. (H) Coimmunoprecipitation of Armi with Flag-AGO3WT and Flag-AGO3DD-AA. Ovaries expressing Flag-AGO3WT or Flag-AGO3DD-AA driven by nosP-gal4:vp16 and w1118 ovaries were lysed and immunoprecipitated by anti-Flag antibody. The anti-Armi antibody was used to detect the Armi protein in those immunoprecipitates.
Armi was first identified in Drosophila as a homologue of Arabidopsis thaliana SDE3, an RNA helicase involved in RNAi, which has been shown to be mainly present in both soma and germline (Cook et al., 2004; Tomari et al., 2004). Although it could be detected in nuage with a very small portion, Armi was proposed to be a cytoplasmic regulator in the piRNA pathway and localized to mitochondria (Saito et al., 2010; Handler et al., 2013). Strong colocalization of Armi with the Slicer mutant AGO3DD-AA prompted us to analyze the germ cell expression pattern of Armi in depth. As shown in Fig. 5 (E–F′), Armi was mainly accumulated in Yb bodies in soma, but exhibited stage-specific patterns with relatively low levels in germline. For example, Armi was not only colocalized with AGO3 in nuage but also present in cytosol in the early stage egg chambers, whereas in late egg chambers, Armi proteins were predominantly present in cytosol. To test the behavior of Armi in wild-type ovaries expressing AGO3DD-AA, we used the GFP-tagged AGO3WT or AGO3DD-AA transgenes to detect Armi and AGO3 association and found that Armi was strongly colocalized with expressed GFP-AGO3DD-AA in both nuage and some cytoplasmic foci (Fig. S3, F–G′).
We next explored how Armi was more strongly associated with the AGO3DD-AA than with the wild-type AGO3. Considering that Armi acts in RISC assembly (Tomari et al., 2004), we reasoned that Armi might be a component of AGO3 RISC, and thus has roles in AGO3-mediated piRNA biogenesis. To test this, we performed immunoprecipitation experiments and found that Armi protein could be detected in endogenous AGO3 immunoprecipitates but not in endogenous Aub immunoprecipitates (Fig. 5 G). Interestingly, in our immunoprecipitation assays we found that Armi had a much stronger association with the Slicer mutant AGO3 than with the wild-type AGO3 (Fig. 5 H).
Armi is localized in both mitochondria and nuage
AGO3DD-AA could bind piRNAs, but not cleave its target transcripts, making it likely that the AGO3DD-AA–piRNA–targets complex was blocked at an intermediate state. At this point, Armi could form a more stable complex with AGO3DD-AA in nuage. Thus, our results not only suggest that the Armi nuage localization is tightly controlled by the AGO3 Slicer, but also raises the possibility that Armi might be a shuttling protein between nuage and the cytoplasmic compartment in germ cells. Although Armi is considered a cytoplasmic protein in germ cells, our immunostaining revealed that it was not evenly distributed in cytosol. Previous studies have shown that Armi functions in conjunction with a mitochondrial protein (Zuc) to regulate primary piRNA biogenesis in both germline and soma, because both armi and zuc mutants share a similar phenotype (Haase et al., 2010; Olivieri et al., 2010, 2012; Saito et al., 2010; Handler et al., 2013).
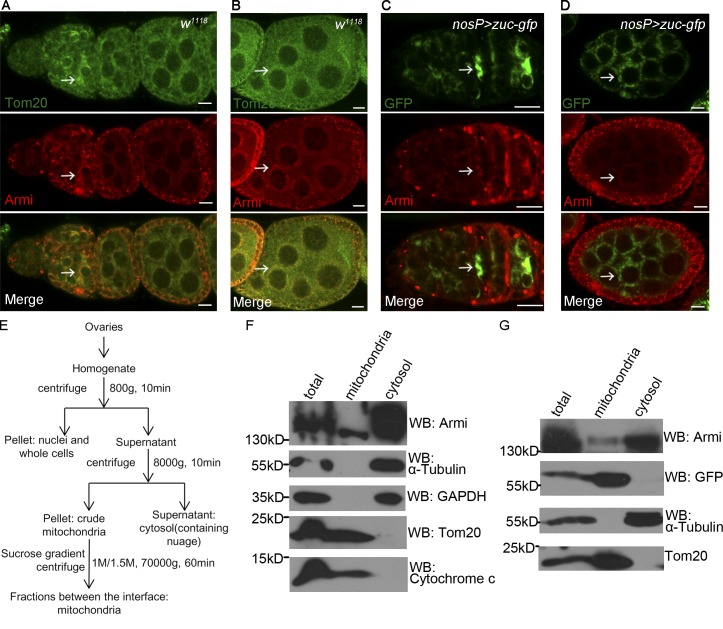
We then sought to confirm whether Armi was localized to mitochondria using multiple methods. We first costained Armi with the mitochondrial outer membrane protein Tom20. In germarium, Armi was highly expressed in region 2 and overlapped with Tom20 (Fig. 6 A). In nurse cells, Armi was also overlapped with Tom20, and both proteins were found localized closely to and around the nuclei (Fig. 6 B). We then tested whether Armi colocalized with Zuc-GFP in ovaries. As shown in Fig. 6 (C and D), Armi was easily detected to be overlapped with Zuc-GFP in both germarium germ cells and egg chamber. Next, we performed biochemical assays by isolation of mitochondria from ovaries and Western blot assays. As shown in Fig. 6 (E and F), the mitochondrion and cytosol fractions could be clearly separated, as indicated by the presence of Tom20 and cytochrome c (Marks for mitochondrion) and α-tubulin and GAPDH (Marks for cytosol), respectively, and a significant portion of Armi was detected in the mitochondrion fraction where it was cofractionated with the mitochondrial protein Zuc (Fig. 6 G). Because the expression of AGO3 Slicer mutant leads to ectopic accumulation of Armi onto nuage and mitochondria are rich close to the perinuclear nuage in ovarian germ cells indicated by the assay of electron microscope (Fig. S4 A), our findings suggest that Armi is a shuttling protein between nuage and mitochondria, and AGO3 Slicer activity might control this shuttling process.
Figure 6.
AGO3 is associated with Armi at mitochondria. (A and B) w1118 ovary was stained with anti-Armi (red) and anti-Tom20 (green). A shows the staining in germarium and early egg chambers. B shows the staining in mid-to-late-stage egg chamber. The mitochondrion localization of Armi with Tom20 is indicated by arrows. (C and D) Ovary expressing Zuc-GFP driven by nosP-gal4:vp16 was stained for Armi (red) and GFP (green). C shows their staining in germarium and early egg chambers. D shows the staining in mid-to-late-stage egg chamber. The mitochondrion localization of Armi with Zuc-GFP is indicated by arrows. Bars, 10 µm. (E) Subcellular fractionation of mitochondria from ovaries. Ovaries were homogenized in a homogenization buffer and then subjected to differential centrifugation and sucrose gradient centrifugation as shown in the diagram. (F) Western blot assays were performed to measure the abundance of Armi, and specific markers in the purified mitochondrion and cytosol fractions of wild-type ovaries. In this assay, the α-tubulin and GAPDH were used as cytosol-specific markers and Tom20 and cytochrome c as mitochondrion-specific markers. (G) Western blot assays were performed to show the abundances of Armi, GFP, Tom20, and α-tubulin in the indicated fractions from ovary expressing Zuc-GFP driven by nosP-gal4:vp16.
AGO3 is associated with Armi in mitochondria
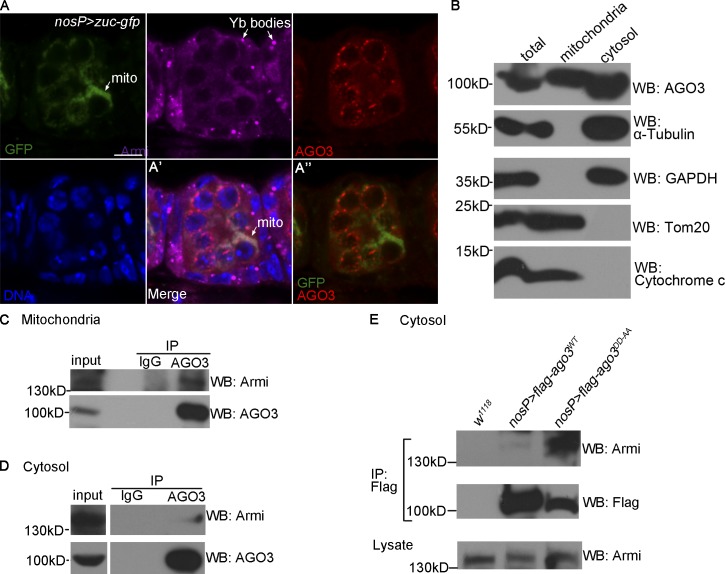
To gain further evidence to support that Armi is a nuage–mitochondria shuttling protein, we sought to test whether AGO3 is present in mitochondria. We performed additional immunostaining assays and found that, although the nuage-localized AGO3 was not overlapped with Tom20 (or Zuc), a faint staining of AGO3 in cytosol seemed to be overlapped with Tom20 (Fig. 7 A and Fig. S4, B and B′). To verify this, we isolated the ovary mitochondrion fraction followed by Western blot assays. As shown in Figs. 7 B and S4 C, we found that a portion of AGO3 could be copurified with Armi and Tom20 in the mitochondrion fraction, suggesting the AGO3 is coexisted with Armi in mitochondrion fraction from ovaries. To rule out the possibility that nuage was cofractionated with mitochondria in our scheme, in addition to Aub, we used Tudor, another typical nuage component, as the marker to perform fractionation followed by Western blot assays. We found that the Tudor protein could never be detected in the mitochondrial fraction (Fig. S4 D). We next asked whether the physical interaction of AGO3–Armi exists in mitochondria, and performed immunoprecipitation using the mitochondrion fraction and the cytosolic fraction without mitochondria, respectively. As shown in Fig. 7 (C and D), the AGO3–Armi interaction was easily detected in mitochondrion fraction, but weakly in the cytosolic fraction. Thus, our biochemistry assays reveal that, in addition to its primary localization at nuage, AGO3 is also localized at mitochondria being associated with Armi. Given that the AGO3 Slicer affects the mitochondrion–nuage localization of Armi, we then performed immunoprecipitation using the ovaries expressing AGO3DD-AA or AGO3WT and found that the AGO3DD-AA–Armi complex was much more abundant than the AGO3WT–Armi complex in the cytosolic fraction (Fig. 7 E). These results indicate that the AGO3 Slicer influences dynamic mitochondria–nuage distribution of AGO3–Armi.
Figure 7.
AGO3 is associated with Armi in mitochondria and nuage. (A) Ovary expressing Zuc-GFP driven by nosP-gal4:vp16 was stained for GFP (green), Armi (pink), AGO3 (red), and Hoechst (blue). A′ shows the merged image of GFP (green), Armi (pink), AGO3 (red), and Hoechst (blue) and A″ shows the merged image of GFP (green) and AGO3 (red). Bar, 10 µm. (B) Western blot assays were performed to measure expression levels of AGO3, α-tubulin, GAPDH, Tom20, and cytochrome c in the indicated fractions of w1118 ovaries. (C and D) Mitochondria (C) and cytosol (D) fractions from w1118 ovaries were used to perform immunoprecipitation with IgG or anti-AGO3 antibody. Armi in those immunoprecipitates was detected by Western blot assays. (E) Cytosol fractions from w1118 ovaries and ovaries expressing Flag-AGO3WT and Flag-AGO3DD-AA driven by nosP-gal4:vp16 were used to perform immunoprecipitation with anti-AGO3 antibody. Western blot assays were performed to detect Armi in those immunoprecipitates.
AGO3 acts in conjunction with Zuc to control the Armi mitochondria–nuage shuttling in a Slicer-dependent manner
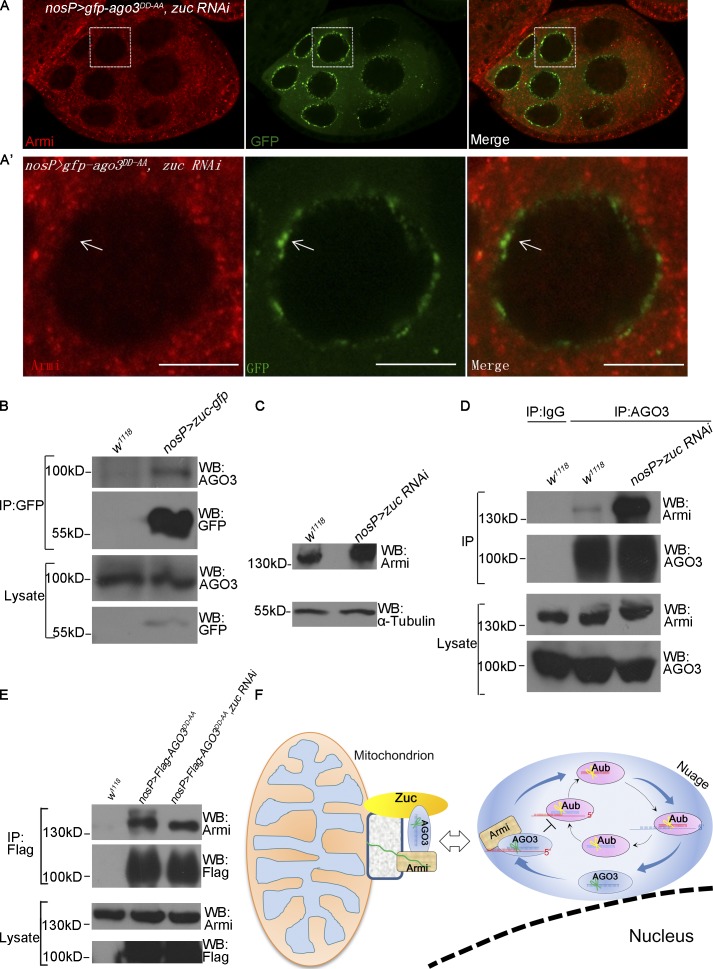
We next attempted to identify the factors affecting nuage localization of Armi besides AGO3. We used the germ cells expressing AGO3DD-AA as a reporter system to perform a candidate knockdown screen. As shown in Fig. S5 (A–E), knockdown of shu, rhi, aub, spnE, and krimp abolished nuage localization of both AGO3DD-AA and Armi, suggesting that these factors indirectly affect Armi–nuage localization by controlling AGO3 expression. Because Zuc is the key mitochondria-associated component in the piRNA pathway, we next determined whether Zuc has a direct role in controlling Armi–nuage localization. As shown in Fig. 6 (C and D), cytoplasmic Armi proteins were evidently colocalized with GFP-tagged Zuc proteins in mitochondria. Next, we tested whether Zuc protein affects the mitochondrion–nuage localization and examined Armi and AGO3DD-AA expression patterns. As shown in Fig. 8 (A and A′), although the nuage localization of AGO3DD-AA exhibited no apparent change in zuc knockdown ovaries, Armi was no longer localized to nuage, suggesting that Zuc supports the nuage localization of Armi and that the ectopic nuage accumulation of Armi by AGO3DD-AA depends on the Zuc activity. In support of this, we found that AGO3 form a complex with Zuc in ovaries (Fig. 8 B).
Figure 8.
Zuc is required for Armi shuttling from mitochondria to nuage. (A and A′) uasp-gfp-ago3DD-AA; zuc RNAi/nosP-gal4:vp16 ovary was stained for Armi (red) and GFP (green). A′ shows the enlarged image in the boxed part in A. The arrows indicate nuage localization of GFP-AGO3DD-AA. Bars, 10 µm. (B) Lysates of w1118 ovaries and ovaries expressing Zuc-GFP driven by nosP-gal4:vp16 were immunoprecipitated by anti-GFP antibody. AGO3 levels were detected by Western blot assays. (C) The expression levels of Armi and α-tubulin in w1118 and zucRNAi/nosP-gal4:vp16 ovaries were determined by Western blot assays. (D) w1118 and zuc RNAi/nosP-gal4:vp16 ovaries were lysed and immunoprecipitated with IgG or anti-AGO3 antibody, and Armi was detected by Western blot assays. (E) ovaries from w1118, uasp-flag-ago3DD-AA; nosP-gal4:vp16, and uasp-flag-ago3DD-AA; zuc RNAi/nosP-gal4:vp16 females were lysed and immunoprecipitated with anti-Flag antibodies, and Armi was detected by Western blot. (F) A proposed model describing that AGO3 Slicer functions in the Ping-Pong cycle and acts in concert with Zuc to regulate dynamic subcellular localization of the AGO3–Armi complex between mitochondrion and nuage that contributes to piRNA amplification.
Given that Zuc plays an important role in the primary piRNA pathway, and that assembly of AGO3–Armi–piRNA–targets complex and AGO3 Slicer might affect the behavior of AGO3–Armi, we tested whether knockdown of zuc affects the AGO3–Armi association. We found that Armi protein levels were evidently increased in zuc knockdown ovaries (Fig. 8 C), and a stronger AGO3–Armi association was detected in zuc knockdown ovaries than that in wild-type ovaries (Fig. 8 D). No apparent difference in AGO3DD-AA-Armi association in zuc knockdown and wild-type ovaries was detected (Fig. 8 E). Collectively, our findings suggest that Zuc acts as a key factor to regulate the localization of Armi–AGO3.
Discussion
piRNAs, a subset of small ncRNAs, associate with PIWI proteins and play conserved roles in silencing the expression of transposable elements to maintain genome integrity and proper germline development (Lin, 2007; Yin and Lin, 2007; Chang et al., 2009; Thomson and Lin, 2009; Juliano et al., 2011). Given that all three PIWI proteins in Drosophila have a typical Slicer motif, which is essential for cleaving target RNAs complementary with bound small RNAs in vitro, the Slicer activity in PIWI proteins was proposed to be endonuclease in piRNA biogenesis. However, an elegant study revealed that the Slicer activity of Piwi protein is dispensable for primary piRNA production (Darricarrère et al., 2013). In addition, several studies also reported that the Slicer activities of different members of PIWI family might have distinct functions on piRNA biogenesis or effects on target silencing in other model animals such as worms and mice (De Fazio et al., 2011; Reuter et al., 2011; Bagijn et al., 2012; Di Giacomo et al., 2013). In this study, we explore the mechanism of how piRNAs are amplified in germline and test whether the AGO3 Slicer plays a role in the Ping-Pong model. By generating transgenic flies that express the Slicer mutant form of AGO3, we show that the Slicer activity of AGO3 is essential for the proper germline development and the secondary piRNA amplification, and AGO3 inhibits the Aub:Aub Ping-Pong process in a Slicer-independent manner. Importantly, we provide evidence to support a notion that AGO3 and Zuc coordinately direct dynamic mitochondrion–nuage localization of Armi (Fig. 8 F).
The Ping-Pong selection of piRNAs is independent of AGO3 Slicer activity
Given that both AGO3 and Aub are nuage proteins and that Piwi is also loaded with piRNAs in nuage, the selection of PIWI proteins to anticipate the Ping-Pong cycle is key to piRNA amplifications, but the mechanism is still elusive. It has been shown that the heterotypic Aub:AGO3 Ping-Pong cycle predominates in wild-type germ cells, promoting antisense piRNA production, whereas when levels of AGO3 were decreased (in ago3−/+ ovaries) or completely lost (in ago3−/− ovaries), the homotypic Aub:Aub Ping-Pong increased, indicating that AGO3 proteins in ovaries can inhibit the process of Aub:Aub Ping-Pong (Li et al., 2009; Zhang et al., 2011). Thus, AGO3 protein could play the key role in the Ping-Pong selection of piRNAs. In this study, we show that AGO3 represses Aub:Aub Ping-Pong in a Slicer-independent manner. Compared with wild-type ovaries, piRNA levels in ago3 mutant significantly decreased and Aub:AGO3 Ping-Pong totally collapsed. However, piRNAs from many transposon families still show apparent Ping-Pong signatures, which are believed to be the result of Aub:Aub Ping-Pong. Interestingly, we found that the Aub:Aub Ping-Pong signatures in all families are almost completely lost when Slicer mutant AGO3 was expressed in ago3 mutant germ cells. Collectively, our study provides direct evidence of a role for AGO3 in controlling the piRNA Ping-Pong selection in a Slicer-independent manner (Fig. 8 F).
AGO3 functions in concert with Zuc to regulate the subcellular localization of Armi between mitochondria and nuage
The nuage is a conserved animal germ cell–specific organelle, which is also close to mitochondria indicated by electron micrograph (Lim and Kai, 2007). In Drosophila, several components, such as AGO3, Aub, Vasa, and Krimp, in the piRNA pathway in germ cells localize to nuage, and mutations of these components lead to strong defects in piRNA biogenesis and consequently de-repress the expression of transposons, underscoring that nuage in germ cells functions as a potential processing site of piRNAs, particularly for the Ping-Pong amplification pathway. In addition to these observations, recent studies have also shown that the mitochondrion-associated protein Zuc plays a role in piRNA biogenesis, linking the potential function of mitochondrion-associated proteins to piRNA production (Watanabe et al., 2011; Handler et al., 2013). Earlier studies have proved that Armi acts in the primary piRNA pathway both in the soma and germline. In this study, we show that Armi and AGO3 are mitochondrion-localized proteins as indicated by its biochemical presence in mitochondrion fraction and colocalization with the mitochondrial outer membrane proteins Tom20 and Zuc in germ cells. In addition, we also find that a portion of Armi physically associates and colocalizes with AGO3 in nuage, raising the possibility that Armi might be a shuttling protein between nuage and mitochondria. Interestingly, Armi was preferentially localized to nuage when the Slicer mutant form of AGO3 (AGO3DD-AA) was also present in germ cells, suggesting that the AGO3 Slicer might control the distribution of Armi between mitochondria and nuage. Consistent with this notion, the Slicer mutant form of AGO3 has a stronger physical association with Armi than the wild-type AGO3 does. Thus, in the absence of Slicer activity, AGO3/sense piRNAs likely form a more stable complex with Armi and block the transient disassociation of Armi from nuage to mitochondrion.
Given that both Zuc and Armi are mitochondrion-localized proteins, we also further examined whether Zuc supports the nuage localization of Armi. We found that knockdown of zuc leads to nondetectable localization of Armi in nuage, but does not affects the localization of AGO3DD-AA in nuage, suggesting that the activity of Zuc is essential for the proper subcellular distribution of Armi. Intriguingly, although we could not detect Zuc localization in nuage, we find a physical association of Zuc with AGO3, but not with Armi, in coimmunoprecipitation experiments (Fig. 8 B). Thus, our findings suggest that Zuc might function in concert with AGO3 to control the proper localization of Armi between nuage and mitochondria. Early studies (al-Mukhtar and Webb, 1971; Eddy, 1975) as well as our current work (Fig. S4 A) indicate that mitochondria are enriched in close to the perinuclear nuage in germ cells, emphasizing the functional relationship between mitochondria and nuage in the regulation of piRNA production. Mitochondrion has been recently proposed as a key site for primary piRNA biogenesis. Several studies have identified mitochondrial piRNA factors including Zuc, BmPAPI, GPAT2, and Gasz that were localized in the outer membrane, and, thus, the outer membrane of mitochondria likely provides a scaffold for piRNA precursor process (Pane et al., 2007; Watanabe et al., 2011; Ipsaro et al., 2012; Nishimasu et al., 2012; Handler et al., 2013; Honda et al., 2013; Shiromoto et al., 2013). Identification of the Armi–AGO3 as a shuttling complex between mitochondria and nuage provides novel insights into understanding mechanisms of how primary and secondary piRNAs are coupled in piRNA biogenesis in germ cells.
Materials and methods
Fly stocks and transgenes
All fly stocks were maintained in standard conditions. Knockdown lines are from Bloomington or Tsinghua Stock Center. Transgenes were constructed by standard methods. The transgenic DNA constructs uasp-flag-ago3WT, uasp-flag-ago3DD-AA, uasp-flag-ago3YK-AA, uasp-gfp-ago3WT, and uasp-gfp-ago3DD-AA were generated by inserting wild-type or mutant ago3 coding sequences tagged with 3× flag or GFP at its N terminus into UASp vectors.
To generate ago3 mutant, we used tilling methods (http://tilling.fhcrc.org) and a pair of primers (5′-ACGACGGATGAATCCAAGGGAGTTTTT-3′ and 5′-TGAAATACCATTGGTTTGCCGAATTTGA-3′) to amplify a fragment 1,532 bp in length that almost covers the second exon of the ago3 gene. We screened 6,000 lines carrying the third chromosome of ethyl methanesulfonate mutations, and each mutant was analyzed by sequencing of genomic PCR amplicons. We isolated eight ago3 alleles (Fig. 1 E), and, among them, two alleles, ago31777 and ago35027, are similar to a previous study (Li et al., 2009). Both ago31777 and ago35027 alleles carry either a point mutation or small deletion that results in premature stop codons. Western blot and immunostaining assays revealed that both alleles are null for the ago3 gene because no AGO3 protein was detected in ago31777/ago35027mutant ovaries (Fig. S1, B and C).
Antibody production
Anti-AGO3, -Aub, -Armi, -Mael, -Tudor, -Krimp, and -Osk antibodies were generated by immunizing rabbits or mice with the recombinant protein His6-AGO3 (1–289 aa), His6-Aub (1–273 aa), His-Armi (1–148 aa), His-Mael (1–315 aa), His-Tudor (701–983 aa), His-Krimp (1–318 aa), and His-Osk produced in Escherichia coli, respectively.
Immunohistochemistry and microscopy
Ovaries were prepared for reaction with antibodies as described previously (Chen et al., 2009). In brief, 3–5-d-old ovaries were dissected in PBS and fixed in fixation buffer (4% formaldehyde, 0.3% Tween 20, and 1×PBS) for 20 min. After permeabilization with 1% Triton X-100/1×PBS for 30 min, the ovaries were blocked with blocking buffer (1.5% BSA, 0.3% Tween 20, and 1×PBS) for 30 min. Then the ovaries were incubated with primary antibodies at appropriate concentrations in blocking buffer at 4°C overnight. After washing three times, the ovaries were incubated with secondary antibodies in blocking buffer at room temperature for 3 h. The ovaries then were washed three times and mounted with Vectashield (Vector Laboratories). Rabbit polyclonal anti-Vasa (Santa Cruz Biotechnology, Inc.) and anti-Tom20 (Santa Cruz Biotechnology, Inc.) antibodies were used at 1:1,000 and 1:300 dilution, respectively. Rabbit and mouse polyclonal anti-AGO3 and mouse anti-Aub, mouse anti-Armi, mouse anti-Vasa, mouse anti-Krimp, mouse anti-Tudor, and mouse anti-Mael antibodies were all used at 1:500 dilutions. Mouse polyclonal anti-Osk was used at 1:1,000, and mouse monoclonal anti-Grk (Developmental Studies Hybridoma Bank) was used at 1:1,000 dilution. Rabbit anti–γ-H2Av (EMD Millipore) was used at 1:1,000. Secondary antibodies used were goat anti–mouse Alexa 555, goat anti–rabbit Alexa 488, and goat anti–rabbit Alexa 647 (Molecular Probes), all at 1:1,000 dilution. All samples were examined by microscope (Axio Observer; Carl Zeiss) using a Fluar 40×/1.30 oil objective, and all images were captured by Zen2008 software using the LSM710 system (Carl Zeiss) at room temperature and processed using Photoshop (Adobe) and ImageJ (National Institutes of Health).
For statistical assay of AGO3 aggregates in Fig. 4 D, images of good quality were processed using the Analyze particles function of ImageJ to calculate the foci areas, and for accurate statistics, only perinuclear foci with areas >0.3 µm2 were included.
Small RNA cloning and bioinformatics analysis
Total RNA was extracted from ovaries using TriZol (Invitrogen) according to the manufacturer’s instructions. After 2S rRNA depletion, small RNAs ranging from 15 to 35 nt in length were purified from 15% 7M-urea PAGE. The small libraries were constructed using Ion Total RNA-Seq kit (Ion Torrent) according to the manufacturer’s instructions for small RNA library. The libraries were sequenced by Personal Genome Machine (Ion Torrent) using an Ion PGM 200 Sequencing kit (Ion Torrent) following the manufacturer’s instructions.
The sequencing reads were mapped to the fly genome (release 5.0), and perfect matches were used for the following analysis. After filtering miRNAs and ncRNAs, small RNAs were mapped to piRNA clusters (Brennecke et al., 2007) and transposon element canonical sequences (http://www.fruitfly.org/p_disrupt/TE.html). For piRNA cluster mapping, we considered genome-unique mappers without mismatch, but for TE mappings, we considered all mappers with up to three mismatches. For plotting mapping piRNAs over clusters, we used a window size of 250 nt and a step size of 25 nt, and for plotting over TE, we used a window and step size of 1 nt. The Z-score values were calculated by the probability of a 10-nt overlap minus background probability (1–9, 11–25-nt overlap) divided by its standard deviation. The calculation and plot of nucleotide bias were performed using weblogo3.0. Figs. 2 and 3 represent the data of one array of high-throughput sequencing.
Mitochondria isolation
Ovaries were dissected and collected in cold PBS and then homogenized with homogenization buffer (5 mM Hepes-K, pH 7.4, 0.3 M mannitol, 5 mM KCl, and 2 mM EDTA) on ice with a 1-ml glass homogenizer (Dounce; loose for 12 times and then tight for 12 times). The homogenate was centrifuge at 800 g at 4°C for 10 min and the supernatant was collected and then centrifuged at 8,000 g at 4°C for 10 min. The supernatant was collected as a cytosol fraction and the pellet was resuspended with homogenization buffer and collected as crude mitochondria. For further purification, the crude pellet was suspended in 0.8-M sucrose solution (10 mM Hepes-K, pH 7.4, 0.8 M sucrose, and 1 mM EDTA) and loaded onto the top of the 1.0-M/1.5-M noncontinuous sucrose gradient. After centrifugation at 75,000 g at 4°C for 60 min, the fraction in the interface of the gradient was collected and then diluted to 0.3 M sucrose, followed by centrifugation to get the pellets as purified mitochondria.
When performing immunoprecipitation with the cytosol fraction and resuspended mitochondria, Triton X-100 was added to 1% and NaCl to 150 mM and then lysed on ice. After centrifugation, the supernatants were incubated with specific antibodies at 4°C overnight and then with protein A/G beads for 2 h. The beads were washed extensively with lysis buffer three times. The immunoprecipitates were eluted with SDS sample buffer and analyzed by Western blot.
In vitro AGO3 Slicer activity assay
The in vitro assay for AGO3 Slicer activity was performed according to the method described previously (Gunawardane et al., 2007) with minor modification. The MBP, MBP-AGO3WT, and MBP-AGO3DD-AA proteins were produced in E. coli and incubated with single-strand Luc guide siRNA for 90 min at 29°C, and then a 32P-labeled single-strand Luc passenger siRNA was added and incubated for another 90 min. The products were analyzed in urea-containing PAGE and X-film was developed.
Immunoprecipitation and Western blot
Ovaries were dissected in cold PBS and then homogenized with lysis buffer (30 mM Hepes, pH 7.4, 150 mM sodium chloride, 2 mM magnesium acetate, 2 mM DTT, and 0.1% NP-40) with protease inhibitors. The supernatant was incubated with specific antibodies overnight at 4°C, and then protein A/G beads were added and incubated for another 2 h at 4°C. The beads were washed extensively with lysis buffer three times. The immunoprecipitates were eluted with SDS sample buffer and analyzed by Western blot. Rabbit anti-AGO3, anti-GFP (Invitrogen), and Flag beads (M2; Sigma-Aldrich) were used for immunoprecipitation. Mouse anti-AGO3 (1:1,000), -Aub (1:1,000), -Armi (1:1,000), -Flag (1:3,000; M2; Sigma-Aldrich), and -GFP (1:3,000; Abmart) and rabbit anti–α-tubulin (1:1,000; Santa Cruz Biotechnology, Inc.) and anti-Tom20 (1:1,000; Santa Cruz Biotechnology, Inc.) were used for Western blot.
Real-time PCR analysis
Total RNA was extracted from ovaries using TriZol (Invitrogen) according to the manufacturer’s instructions, and first-strand cDNA synthesis was performed with an iScript cDNA Synthesis kit (Bio-Rad Laboratories). Real-time PCR was performed via standard methods using SYBR Premix Ex Taq II (Takara Bio Inc.), and each sample was repeated in triplicate. The relative enrichment was calculated by normalizing the quantity of rp49. Primers used were as follows: rp49-s, 5′-ATGACCATCCGCCCAGCATAC-3′; rp49-as, 5′-CTGCATGAGCAGGACCTCCAG-3′; Het-A-s, 5′-ATCCTTCACCGTCATCACCTTCCT-3′; Het-A-as, 5′-GGTGCGTTTAGGTGAGTGTGTGTT-3′; blood-s, 5′-TGCCACAGTACCTGATTTCG-3′; blood-as, 5′-GATTCGCCTTTTACGTTTGC-3′; burdock-s, 5′-GTAACAAGACGAGACAGAACC-3′; burdock-as, 5′-CTTTATGAACGACCCTCCCT-3′.
Accession numbers
Sequence data has been deposited to Gene Expression Omnibus (accession no. GSE49436).
Online supplemental material
Fig. S1 shows the expression levels of recombinant proteins used in Fig. 1 B, the AGO3 expressions in ago3 mutant, and the Grk and Osk expression and localization in the ovaries of wild type, ago3 mutant, and ago3 mutant flies expressing different forms of AGO3 proteins. Fig. S2 shows the colocalization of AGO3DD-AA with Vasa. Fig. S3 shows the localizations of Armi, AGO3, Mael, Krimp, Tudor, and Aub in nosP>flag-ago3DD-AA; ago3 ovaries, as well as the localization of GFP-AGO3WT or GFP-AGO3DD-AA with Armi. Fig. S4 shows the localization of AGO3 protein in mitochondria. Fig. S5 shows the localization of Armi and GFP-AGO3DD-AA in shu, rhi, aub, spnE, and krimp knockdown ovaries. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201401002/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201401002.dv.
Supplementary Material
Acknowledgments
The authors would like to thank the core facilities of the State Key Laboratory of Reproductive Biology in the Institute of Zoology, Chinese Academy of Sciences. The authors would also like to thank H. Siomi and C. Strauss for critical reading of the manuscript.
D. Chen is supported by the National Basic Research Program of China (2013CB945002), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010306), and Natural Science Foundation of China (91019022, 31130036, and 31329004). P. Jin is supported by National Institutes of Health grants R01 NS051630 and R21 NS067461.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ncRNA
- noncoding RNA
- piRNA
- PIWI-interacting RNA
References
- al-Mukhtar, K.A., and Webb A.C.. 1971. An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. J. Embryol. Exp. Morphol. 26:195–217 [PubMed] [Google Scholar]
- Bagijn, M.P., Goldstein L.D., Sapetschnig A., Weick E.M., Bouasker S., Lehrbach N.J., Simard M.J., and Miska E.A.. 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 337:574–578 10.1126/science.1220952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., and Hannon G.J.. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 128:1089–1103 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Chang, S., Wen S., Chen D., and Jin P.. 2009. Small regulatory RNAs in neurodevelopmental disorders. Hum. Mol. Genet. 18(R1):R18–R26 10.1093/hmg/ddp072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Wang Q., Huang H., Xia L., Jiang X., Kan L., Sun Q., and Chen D.. 2009. Effete-mediated degradation of Cyclin A is essential for the maintenance of germline stem cells in Drosophila. Development. 136:4133–4142 10.1242/dev.039032 [DOI] [PubMed] [Google Scholar]
- Cook, H.A., Koppetsch B.S., Wu J., and Theurkauf W.E.. 2004. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 116:817–829 10.1016/S0092-8674(04)00250-8 [DOI] [PubMed] [Google Scholar]
- Darricarrère, N., Liu N., Watanabe T., and Lin H.. 2013. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc. Natl. Acad. Sci. USA. 110:1297–1302 10.1073/pnas.1213283110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio, S., Bartonicek N., Di Giacomo M., Abreu-Goodger C., Sankar A., Funaya C., Antony C., Moreira P.N., Enright A.J., and O’Carroll D.. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 480:259–263 10.1038/nature10547 [DOI] [PubMed] [Google Scholar]
- Di Giacomo, M., Comazzetto S., Saini H., De Fazio S., Carrieri C., Morgan M., Vasiliauskaite L., Benes V., Enright A.J., and O’Carroll D.. 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol. Cell. 50:601–608 10.1016/j.molcel.2013.04.026 [DOI] [PubMed] [Google Scholar]
- Eddy, E.M.1975. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 43:229–280 10.1016/S0074-7696(08)60070-4 [DOI] [PubMed] [Google Scholar]
- Filipowicz, W.2005. RNAi: the nuts and bolts of the RISC machine. Cell. 122:17–20 10.1016/j.cell.2005.06.023 [DOI] [PubMed] [Google Scholar]
- Findley, S.D., Tamanaha M., Clegg N.J., and Ruohola-Baker H.. 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 130:859–871 10.1242/dev.00310 [DOI] [PubMed] [Google Scholar]
- Gunawardane, L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., and Siomi M.C.. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 315:1587–1590 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- Haase, A.D., Fenoglio S., Muerdter F., Guzzardo P.M., Czech B., Pappin D.J., Chen C., Gordon A., and Hannon G.J.. 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 24:2499–2504 10.1101/gad.1968110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler, D., Meixner K., Pizka M., Lauss K., Schmied C., Gruber F.S., and Brennecke J.. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell. 50:762–777 10.1016/j.molcel.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck, J., and Meister G.. 2008. The Argonaute protein family. Genome Biol. 9:210 10.1186/gb-2008-9-2-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, S., Kirino Y., Maragkakis M., Alexiou P., Ohtaki A., Murali R., Mourelatos Z., and Kirino Y.. 2013. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA. 19:1405–1418 10.1261/rna.040428.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., and Simard M.J.. 2008. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9:22–32 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- Ipsaro, J.J., Haase A.D., Knott S.R., Joshua-Tor L., and Hannon G.J.. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 491:279–283 10.1038/nature11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M., and Doudna J.A.. 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature. 457:405–412 10.1038/nature07755 [DOI] [PubMed] [Google Scholar]
- Juliano, C., Wang J., and Lin H.. 2011. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 45:447–469 10.1146/annurev-genet-110410-132541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff, C., and Theurkauf W.. 2008. Biogenesis and germline functions of piRNAs. Development. 135:3–9 10.1242/dev.006486 [DOI] [PubMed] [Google Scholar]
- Li, C., Vagin V.V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M.D., Syrzycka M., Honda B.M., et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 137:509–521 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, A.K., and Kai T.. 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 104:6714–6719 10.1073/pnas.0701920104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H.2007. piRNAs in the germ line. Science. 316:397 10.1126/science.1137543 [DOI] [PubMed] [Google Scholar]
- Malone, C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., and Hannon G.J.. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 137:522–535 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson, J.R., and Sharp P.A.. 2008. Small RNA regulators of gene expression. Cell. 134:899–902 10.1016/j.cell.2008.09.006 [DOI] [PubMed] [Google Scholar]
- Nishimasu, H., Ishizu H., Saito K., Fukuhara S., Kamatani M.K., Bonnefond L., Matsumoto N., Nishizawa T., Nakanaga K., Aoki J., et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 491:284–287 10.1038/nature11509 [DOI] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka A., Siomi H., and Siomi M.C.. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18:1655–1666 10.1101/gad.1210204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri, D., Sykora M.M., Sachidanandam R., Mechtler K., and Brennecke J.. 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 29:3301–3317 10.1038/emboj.2010.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri, D., Senti K.A., Subramanian S., Sachidanandam R., and Brennecke J.. 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell. 47:954–969 10.1016/j.molcel.2012.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane, A., Wehr K., and Schüpbach T.. 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell. 12:851–862 10.1016/j.devcel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, V.S., and Kai T.. 2010. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 20:724–730 10.1016/j.cub.2010.02.046 [DOI] [PubMed] [Google Scholar]
- Peters, L., and Meister G.. 2007. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 26:611–623 10.1016/j.molcel.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Reuter, M., Berninger P., Chuma S., Shah H., Hosokawa M., Funaya C., Antony C., Sachidanandam R., and Pillai R.S.. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 480:264–267 10.1038/nature10672 [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., and Joshua-Tor L.. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12:340–349 10.1038/nsmb918 [DOI] [PubMed] [Google Scholar]
- Saito, K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., and Siomi M.C.. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20:2214–2222 10.1101/gad.1454806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Ishizu H., Komai M., Kotani H., Kawamura Y., Nishida K.M., Siomi H., and Siomi M.C.. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24:2493–2498 10.1101/gad.1989510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe, J.P., and Lin H.. 2011. Small noncoding RNAs in the germline. Cold Spring Harb. Perspect. Biol. 3:a002717 10.1101/cshperspect.a002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle, N.T., and MacRae I.J.. 2012. The crystal structure of human Argonaute2. Science. 336:1037–1040 10.1126/science.1221551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiromoto, Y., Kuramochi-Miyagawa S., Daiba A., Chuma S., Katanaya A., Katsumata A., Nishimura K., Ohtaka M., Nakanishi M., Nakamura T., et al. 2013. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA. 19:803–810 10.1261/rna.038521.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, M.C., Sato K., Pezic D., and Aravin A.A.. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12:246–258 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith S.K., Hannon G.J., and Joshua-Tor L.. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 305:1434–1437 10.1126/science.1102514 [DOI] [PubMed] [Google Scholar]
- Thomson, T., and Lin H.. 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25:355–376 10.1146/annurev.cellbio.24.110707.175327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari, Y., Du T., Haley B., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., and Zamore P.D.. 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 116:831–841 10.1016/S0092-8674(04)00218-1 [DOI] [PubMed] [Google Scholar]
- Vagin, V.V., Sigova A., Li C., Seitz H., Gvozdev V., and Zamore P.D.. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 313:320–324 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., and Patel D.J.. 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 461:754–761 10.1038/nature08434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Chuma S., Yamamoto Y., Kuramochi-Miyagawa S., Totoki Y., Toyoda A., Hoki Y., Fujiyama A., Shibata T., Sado T., et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell. 20:364–375 10.1016/j.devcel.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Chen D., Duan R., Xia L., Wang J., Qurashi A., Jin P., and Chen D.. 2007. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 134:4265–4272 10.1242/dev.009159 [DOI] [PubMed] [Google Scholar]
- Yin, H., and Lin H.. 2007. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 450:304–308 10.1038/nature06263 [DOI] [PubMed] [Google Scholar]
- Zamparini, A.L., Davis M.Y., Malone C.D., Vieira E., Zavadil J., Sachidanandam R., Hannon G.J., and Lehmann R.. 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 138:4039–4050 10.1242/dev.069187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Xu J., Koppetsch B.S., Wang J., Tipping C., Ma S., Weng Z., Theurkauf W.E., and Zamore P.D.. 2011. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell. 44:572–584 10.1016/j.molcel.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.