To meet their metabolic needs, starved cells first activate autophagy, but activation in parallel of the general amino acid control pathway increases amino acid uptake, leading to reactivation of mTOR and down-regulation of autophagy.
Abstract
Organisms have evolved elaborate mechanisms to adjust intracellular nutrient levels in response to fluctuating availability of exogenous nutrients. During starvation, cells can enhance amino acid uptake and synthesis through the general amino acid control (GAAC) pathway, whereas nonessential cellular contents are recycled by autophagy. How these two pathways are coordinated in response to starvation is currently unknown. Here we show that the GAAC pathway couples exogenous amino acid availability with autophagy. Starvation caused deactivation of mTOR, which then activated autophagy. In parallel, serum/glutamine starvation activated the GAAC pathway, which up-regulated amino acid transporters, leading to increased amino acid uptake. This elevated the intracellular amino acid level, which in turn reactivated mTOR and suppressed autophagy. Knockdown of activating transcription factor 4, the major transcription factor in the GAAC pathway, or of SLC7A5, a leucine transporter, caused impaired mTOR reactivation and much higher levels of autophagy. Thus, the GAAC pathway modulates autophagy by regulating amino acid uptake and mTOR reactivation during serum/glutamine starvation.
Introduction
Autophagy is a lysosome-based degradation process that helps adjust the cellular response to varying nutrient statuses by degrading and recycling nonessential intracellular contents (Mizushima et al., 1998; Ohsumi, 2001). Amino acids repress autophagy, whereas amino acid starvation stimulates autophagy (Mortimore and Schworer, 1977; Schworer and Mortimore, 1979). Amino acid depletion is linked to activation of autophagy by mTOR, a serine/threonine kinase that integrates signals from various metabolic stimuli (Ma and Blenis, 2009). The intracellular amino acid level is the essential signal for regulating mTOR kinase activity. The kinase activity of mTOR is also controlled by growth factors (Nobukuni et al., 2005; Cohen and Hall, 2009). In many cell types, leucine appears to be the main regulatory amino acid for mTOR (Lynch, 2001). Lowering the leucine concentration abolishes the regulatory effect of amino acids on mTOR. Adding leucine and, to a lesser extent, the other branched-chain amino acids activates mTOR.
During starvation, amino acid levels are maintained by the general amino acid control (GAAC) pathway (Hinnebusch, 2005), which is conserved from yeast to mammals (Sood et al., 2000). In yeast, amino acid starvation elevates translation of the transcription factor GCN4 (Hao et al., 2005), which then causes expression of many genes, including those required for synthesis of all 20 amino acids (Wek et al., 1995). The mammalian counterpart of GCN4 is activating transcription factor 4 (ATF4). ATF4 up-regulates amino acid biosynthesis and controls amino acid transporter genes (Harding et al., 2003; Malmberg and Adams, 2008). Mammalian amino acid homeostasis is more complex because eight essential amino acids cannot be synthesized and must be supplied from outside the cell (Reeds, 2000; Fürst and Stehle, 2004). Leucine is one of these; thus, intracellular leucine homeostasis is likely dependent on exogenous leucine uptake (Nicklin et al., 2009). ATF4 also regulates autophagy by controlling the expression of autophagy genes (Rouschop et al., 2010; Rzymski et al., 2010; B’chir et al., 2013).
Here we report a feedback mechanism that controls the strength of autophagy by regulating amino acid uptake. Glutamine depletion triggers the GAAC pathway, which increases the level of ATF4. The elevated ATF4 level up-regulates amino acid transporters such as SLC7A5, which increase amino acid uptake and elevate intracellular amino acid levels, thus reactivating mTOR and repressing autophagy.
Results and discussion
Amino acid uptake surges during serum/glutamine (S/Gln) starvation
Two starvation protocols are widely used to study autophagy. The harsh protocol uses Dulbecco’s phosphate-buffered saline (DPBS), which lacks serum and all amino acids. The mild protocol is closer to the physiological setting and uses DMEM, which lacks serum and glutamine but contains the other amino acids. DPBS starvation will be referred to herein as serum/amino acid (S/AA) starvation, whereas DMEM starvation will be referred to as S/Gln starvation.
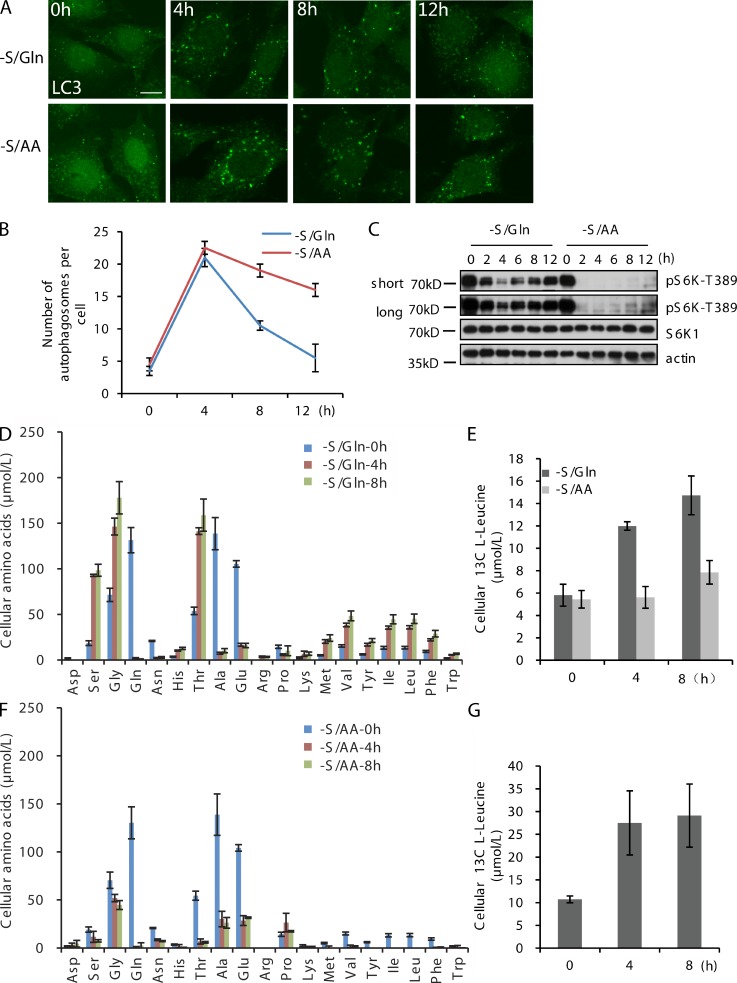
In cells undergoing S/Gln starvation, autophagy is transiently induced. Rapid induction of autophagy by S/Gln starvation is followed by a termination phase, mediated by reactivation of mTOR, in which the number of autophagosomes is rapidly reduced (Yu et al., 2010). We compared the kinetics of autophagy in S/Gln starvation to S/AA starvation in normal rat kidney (NRK) cells and found that S/AA starvation induced persistent autophagy, as indicated by the number of LC3 puncta (Fig. 1, A and B). Accordingly, we observed a much weaker mTOR reactivation in S/AA starvation, as indicated by reduced phosphorylation of the mTOR substrate pS6K1 (Fig. 1 C).
Figure 1.
Amino acid uptake surges during S/Gln starvation. (A) CFP-LC3 NRK cells (Yu et al., 2010) were starved with S/Gln (DMEM) or S/AA (DPBS) medium for the indicated times. Cells were stained with antibodies against LC3. Bar, 10 µm. (B) CFP-LC3 NRK cells from A were quantified for LC3 puncta in a blind fashion after starvation (n = 100). (C) CFP-LC3 NRK cells were starved with DMEM or DPBS for the indicated times. Proteins from cell lysates were analyzed by Western blotting for S6K1-T389, S6K1, and β-actin. (D) CFP-LC3 NRK cells were starved with DMEM for the indicated times. The absolute levels of intracellular free amino acids were quantified using LC/MS/MS. (E) CFP-LC3 NRK cells were starved with DMEM and C13-Leucine was added to the cells at the indicated times. After 10 min of incubation, cells were washed, free amino acids were extracted, and the absolute levels of labeled l-Leucine were quantified using LC/MS/MS. (F) CFP-LC3 NRK cells were starved with DPBS for the indicated times. The absolute levels of intracellular free amino acids were quantified using LC/MS/MS. (G) CFP-LC3 NRK cells were treated with DPBS and C13-Leucine was added at the indicated times. After 10 min of incubation, cells were washed, free amino acids were extracted, and the absolute levels of labeled l-Leucine were quantified using LC/MS/MS. Error bars indicate the SD.
Because the difference between DMEM and DPBS is amino acid concentration (DPBS contains no amino acids, whereas DMEM lacks only glutamine), this observation suggested that exogenous amino acids are required for mTOR reactivation.
Using liquid chromatography/tandem mass spectrometry (LC/MS/MS), we found that the concentrations of intracellular free amino acids, including essential amino acids, rapidly increased under S/Gln starvation, but quickly decreased under S/AA starvation (Fig. 1, D and F). This indicated that the increased concentration of intracellular free amino acids under S/Gln starvation depended on the uptake of exogenous amino acids from the medium. Using stable isotope–labeled L-Leu, we observed that S/Gln starvation enhanced the uptake of C13-Leu, whereas S/AA starvation had no obvious effect on C13-Leu uptake (Fig. 1, E and G). These results indicated that S/Gln starvation induced uptake of exogenous amino acids, thus contributing to the increased concentration of intracellular free amino acids.
Glutamine regulates the uptake of exogenous amino acids
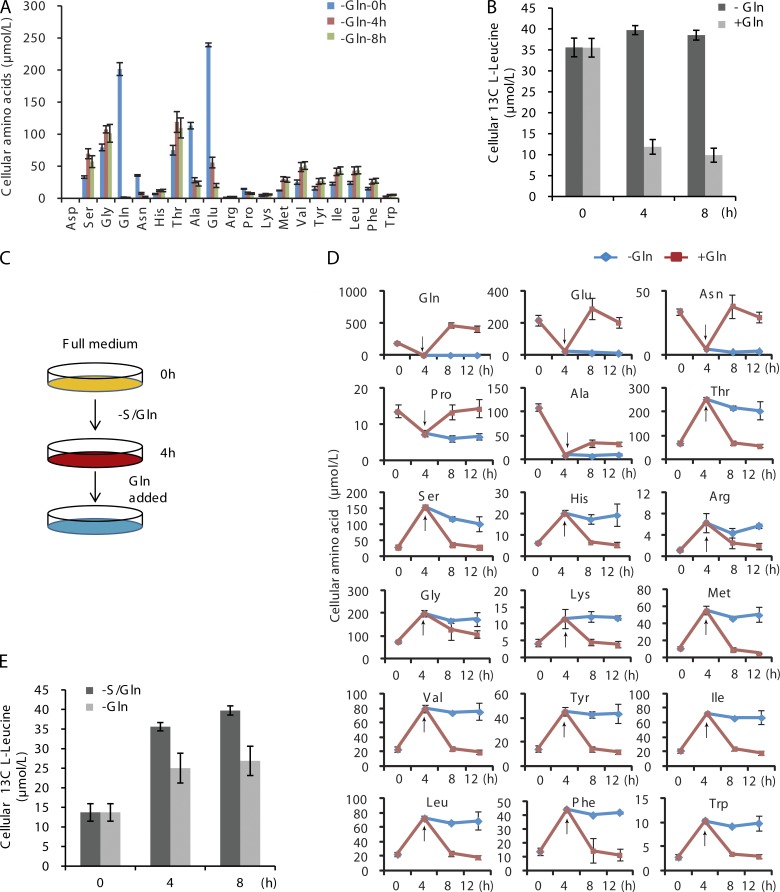
Next, we tested whether serum or glutamine is responsible for triggering amino acid uptake during S/Gln starvation. Glutamine withdrawal, but not serum withdrawal, increases the intracellular free amino acid concentration and the uptake of exogenous amino acids (Fig. 2, A and B). The intracellular levels of Gln, Glu, and Asn are reduced under S/Gln starvation as Glu and Asn are synthesized from Gln. We next starved cells in S/Gln conditions for 4 h, then added glutamine back for an additional 4 h, and finally tested the concentration of intracellular free amino acids and uptake of C13-Leu. Addition of glutamine completely inhibited the increase in intracellular free amino acid levels and the uptake of C13-Leu (Fig. 2, D and E). These results indicated that glutamine deficiency triggers uptake of exogenous amino acids.
Figure 2.
Glutamine controls amino acid uptake. (A) CFP-LC3 NRK cells were treated with DMEM supplemented with serum (glutamine starvation) for the indicated times. The absolute levels of intracellular free amino acids were quantified. (B) CFP-LC3 NRK cells were treated with DMEM in the absence or presence of serum, and C13-Leucine was added at the indicated times. After 10 min of incubation, cells were extracted and the absolute levels of labeled l-Leucine were quantified. (C) The starvation protocol: cells were grown in complete medium and then pretreated with DMEM (lacking serum and glutamine) for 4 h before addition of glutamine. Amino acid levels were tested at various time points after glutamine was added. (D) CFP-LC3 NRK cells were pretreated with DMEM for 4 h, and then glutamine was added. The absolute levels of intracellular free amino acids were quantified and compared with cells that were starved but not treated with glutamine. Arrows indicate the time when glutamine was added. (E) CFP-LC3 NRK cells were prestarved with DMEM (S/Gln) for 4 h, and then glutamine was added. C13-Leucine was added to the cells at the indicated times. After 10 min of incubation, cells were harvested and the absolute levels of labeled l-Leucine were quantified. Error bars indicate the SD.
SLC7A5 is required for exogenous amino acid uptake
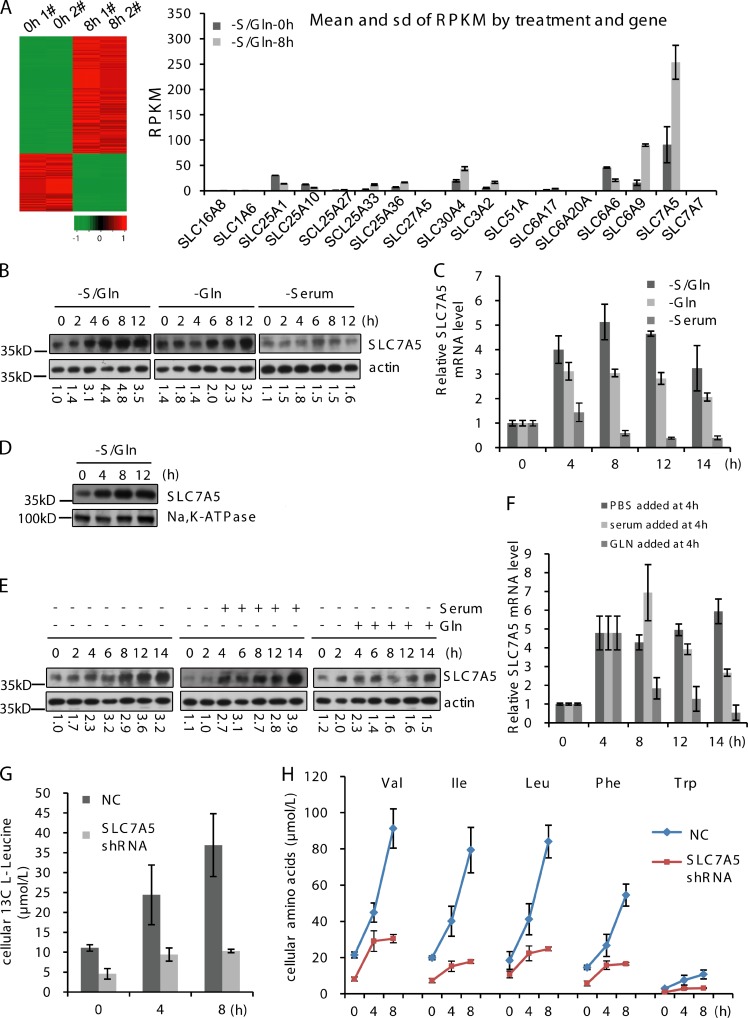
Transcriptome analysis of cells grown in normal or S/Gln starvation conditions revealed that S/Gln starvation dramatically changes the transcription profile (Fig. S1 A), and many amino acid transporter genes are up-regulated. Slc7a5, encoding the Leu transporter (Haase et al., 2007), had the highest induction rate (Fig. 3 A). S/Gln and glutamine starvation, but not serum starvation, induced the up-regulation of SLC7A5 mRNA and protein (Fig. 3, B and C). Moreover, plasma membrane–located SLC7A5 was also elevated under S/Gln starvation (Fig. 3 D). Addition of glutamine, but not serum, blocked the up-regulation of SLC7A5 mRNA and protein induced by S/Gln starvation (Fig. 3, E and F). Thus, transcription of Scl7a5 is induced by glutamine starvation.
Figure 3.
Glutamine starvation up-regulates the expression of amino acid transporters. (A) RNA from cells treated with DMEM was extracted for transcriptome analysis at the indicated times. (B) CFP-LC3 NRK cells were treated with DMEM in the absence or presence of glutamine or serum. Proteins from cell lysates were analyzed by Western blotting for SLC7A5 and β-actin. The values indicate the SLC7A5/actin ratio as the ratio of that in –S/Gln 0-h cells. (C) RNA from CFP-LC3 NRK cells treated with DMEM in the absence or presence of glutamine or serum was extracted and analyzed for endogenous Slc7a5 mRNA expression by quantitative PCR. (D) CFP-LC3 cells were treated with DMEM for the indicated times, and plasma membrane protein was extracted and analyzed by Western blotting for SLC7A5 and Na,K-ATPase. (E) CFP-LC3 NRK cells were pretreated with DMEM for 4 h, and glutamine or serum was added to the cells. Proteins from cell lysates were analyzed by Western blotting for SLC7A5 and β-actin. The values indicate the SLC7A5/actin ratio as the ratio of that in –S/Gln 0-h cells. (F) CFP-LC3 NRK cells were pretreated with DMEM for 4 h, and glutamine or serum was added. RNA was extracted at the indicated times and analyzed for endogenous Slc7a5 mRNA expression by quantitative PCR. (G) CFP-LC3 NRK cells and stable SLC7A5 knockdown cells were treated with DMEM and C13-Leucine was added at the indicated times. After 10 min of incubation, cells were extracted and the absolute levels of labeled l-Leucine were quantified using LC/MS/MS. (H) CFP-LC3 NRK cells and stable SLC7A5 knockdown cells were treated with DMEM for the indicated times. The absolute levels of intracellular free amino acids were quantified using LC/MS/MS. Error bars indicate the SD.
SLC7A5 is a bidirectional amino acid transporter that transports branched side chain amino acids such as l-leucine into cells in exchange for the efflux of intracellular l-glutamine (Yanagida et al., 2001; Meier et al., 2002). Because glutamine starvation up-regulates SLC7A5 expression, we speculated that the up-regulation of SLC7A5 and other amino acid transporters contributed to the increased intracellular concentration of free amino acids under S/Gln starvation. Stable knockdown of SLC7A5 strongly inhibited the increased uptake of C13-Leu, and S/Gln starvation failed to enhance the concentration of a subset of intracellular free amino acids, including Val, Ile, Leu, Phe, and Trp (Fig. 3, G and H). These data indicate that glutamine starvation induces uptake of amino acids through up-regulation of amino acid transporters such as SLC7A5.
Glutamine starvation induces ATF4 up-regulation to control the up-regulation of SLC7A5 and uptake of exogenous amino acids
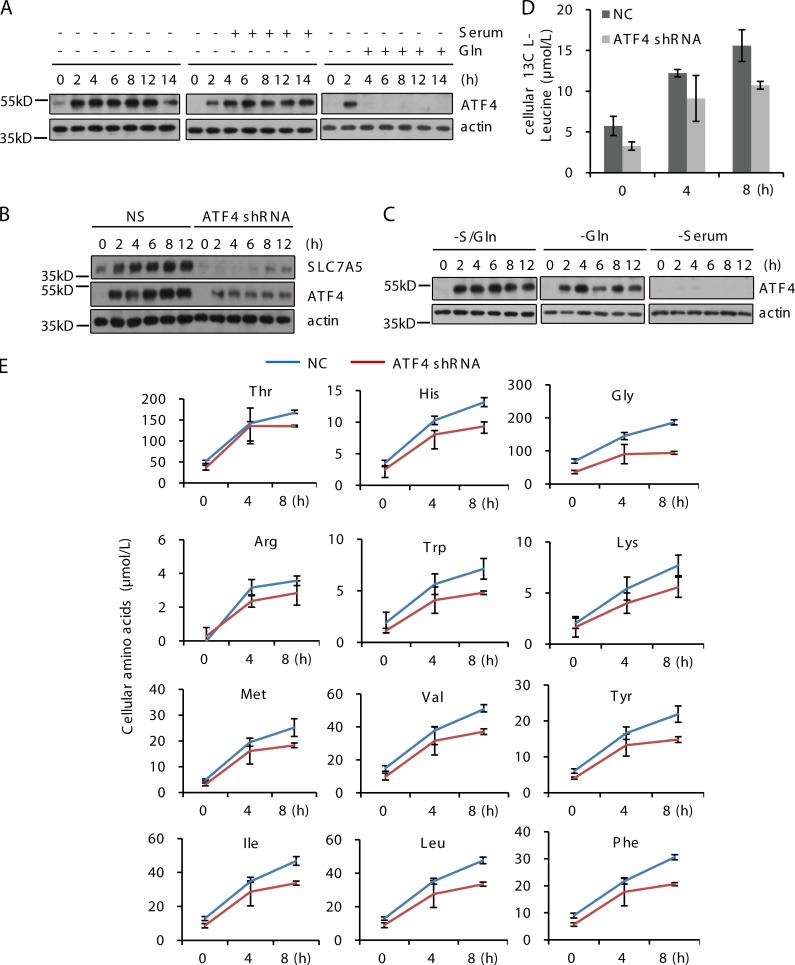
In mammals, the GAAC pathway maintains amino acid homeostasis through the transcription factor ATF4 (Ye et al., 2010). The ATF4 protein level is increased during S/Gln and glutamine starvation, but is unaffected by serum starvation (Fig. 4 A). Adding glutamine back blocked the up-regulation of ATF4 protein after S/Gln starvation (Fig. 4 B). Stable knockdown of ATF4 blocked the induction of SLC7A5 expression and the increased uptake of exogenous amino acids under S/Gln starvation (Fig. 4, C and D). In addition, in the ATF4 stable knockdown cell line, the rise in intracellular free amino acid levels was lower under S/Gln starvation than in control cells (Fig. 4 E). We conclude that glutamine starvation causes activation of the GAAC pathway, which in turn leads to up-regulation of amino acid transporters such as SLC7A5, thereby increasing the uptake of exogenous amino acids and raising the intracellular amino acid levels.
Figure 4.
Glutamine starvation induces expression of ATF4, which controls the expression of SLC7A5. (A) CFP-LC3 NRK cells were treated with DMEM in the absence or presence of glutamine or serum. Proteins from cell lysates were analyzed by Western blotting for ATF4 and β-actin. (B) CFP-LC3 NRK cells were pretreated with DMEM for 4 h, and then glutamine or serum was added. Proteins from cell lysates were analyzed for ATF4 and β-actin. (C) CFP-LC3 NRK cells and stable ATF4 knockdown cells were treated with DMEM for the indicated times. Proteins from cell lysates were analyzed for SLC7A5 and β-actin. (D) CFP-LC3 NRK cells and stable ATF4 knockdown cells were treated with DMEM, and C13-Leucine was added to the cells at the indicated times. After 10 min of incubation, cells were extracted and the absolute levels of labeled l-Leucine were quantified using LC/MS/MS. (E) CFP-LC3 NRK cells and stable ATF4 knockdown cells were treated with DMEM for the indicated times. The absolute levels of intracellular free amino acids were quantified using LC/MS/MS. Error bars indicate the SD.
ATF4 and SLC7A5 regulate S/Gln starvation–induced mTOR reactivation and autophagy
Next, we examined the role of the GAAC pathway in mTOR reactivation during S/Gln starvation. We found that mTOR reactivation was tightly controlled by glutamine (Fig. S2, A and B). Serum starvation caused deactivation of mTOR, without noticeable mTOR reactivation, whereas both S/Gln and glutamine starvation caused robust mTOR reactivation. Adding glutamine back blocked S/Gln starvation–induced mTOR reactivation. To examine which exogenous amino acid contributed to mTOR reactivation, we treated cells with different DMEM lacking glutamine and any given single amino acid. We found that exogenous Leu or Arg is required for mTOR reactivation under S/Gln starvation (Fig. S2 C). Thus, uptake of Leu and/or Arg is required for mTOR reactivation, which is consistent with a previous study showing that Leu and Arg are prerequisites for mTOR activity (Lynch, 2001).
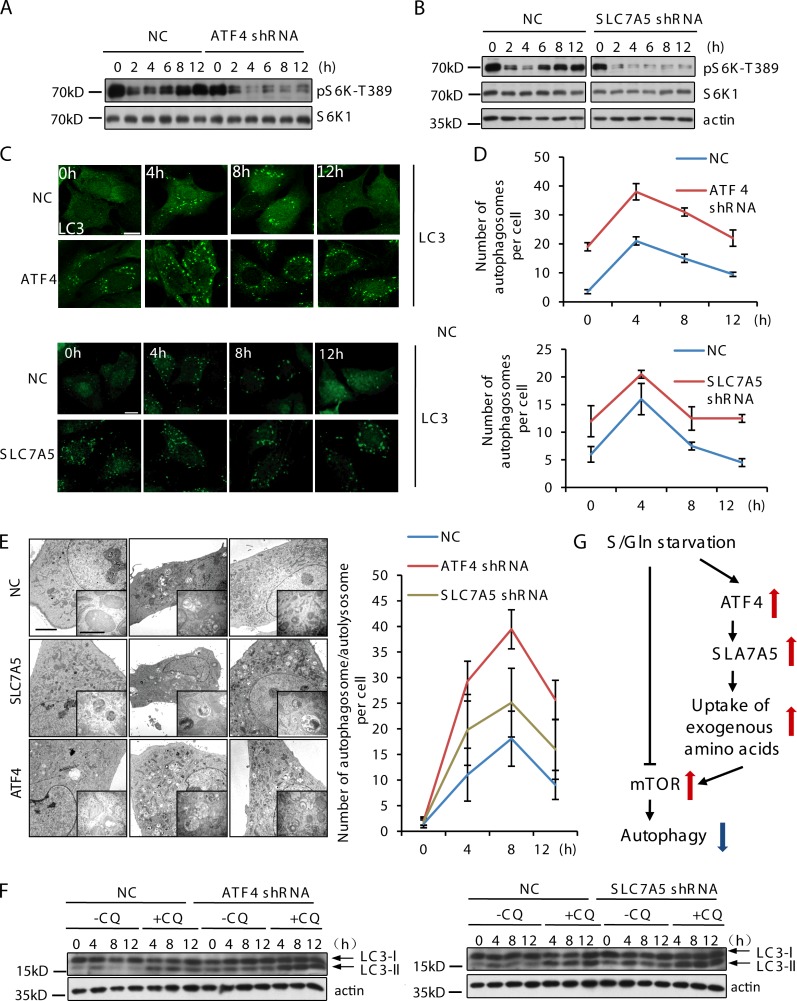
Because S/Gln starvation induces a marked increase in the intracellular Leu level, but only a marginal increase in the Arg level under the same conditions (Fig. S2 D), we speculated that SLC7A5, the major Leu transporter, may regulate mTOR reactivation. Indeed, mTOR reactivation is abolished in SLC7A5 stable knockdown cells and, accordingly, in ATF4 stable knockdown cells (Fig. 5, A and B). Similarly, blocking Leu uptake by treating cells with 2-aminobicyclo-(2, 2, 1)-heptane-2-carboxylic acid, a potent SLC7A5 inhibitor, completely abolished the S/Gln starvation–induced mTOR reactivation (Fig. S2 E). Thus, ATF4-SLC7A5–mediated Leu uptake is responsible for S/Gln starvation–induced mTOR reactivation.
Figure 5.
ATF4 and SLC7A5 regulate mTOR reactivation and the strength and duration of autophagy. (A) CFP-LC3 NRK cells and stable ATF4 knockdown cells were treated with DMEM for the indicated times. Proteins from cell lysates were analyzed for S6K1-T389 and S6K1. (B) CFP-LC3 NRK cells and stable SLC7A5 knockdown cells were treated with DMEM for the indicated times. Proteins from cell lysates were analyzed for S6K1-T389, S6K1, and β-actin. RNA was extracted at the indicated times and analyzed for endogenous Slc7a5 mRNA expression by quantitative PCR (right) to determine the knockdown efficiency of SLC7A5 shRNA. (C and D) CFP-LC3 NRK cells and stable ATF4 and SLC7A5 knockdown cells were treated with DMEM for the indicated times, and then stained with antibodies against LC3 to reveal autophagosomes. The mean number of LC3 puncta per cell was assessed in a blind fashion after starvation and quantified. 100 cells from three independent experiments were analyzed. Bar, 10 µm. (E) CFP-LC3 NRK cells and stable ATF4 and SLC7A5 knockdown cells were treated with DMEM for the indicated times, and then fixed for transmission EM. Bars: (main) 2 µm; (inset) 200 nm. (F) CFP-LC3 NRK cells and stable ATF4 and SLC7A5 knockdown cells were treated with DMEM with or without chloroquine (CQ) for the indicated times. Proteins from cell lysates were analyzed by Western blotting for LC3 and β-actin. (G) A model for the role of ATF4 and SLC7A5 in the regulation of mTOR and autophagy under starvation. Error bars indicate the SD.
Next, we investigated the role of ATF4 and SLC7A5 in regulation of autophagy. By examining the formation of LC3 puncta, we found that overexpression of SLC7A5 inhibits the induction of autophagy (Fig. S2, G and H). Conversely, in both ATF4 and SLC7A5 stable knockdown cell lines, numerous LC3 puncta were observed in nutrient-rich conditions, implying that the basal autophagy level was higher in these cells than controls. Moreover, S/Gln starvation further increased the number of LC3 puncta (Fig. 5, C and D). Ulk1 and Atg5 puncta were also more numerous in these cells (Fig. S3, A and B). Transmission EM analysis verified the enhanced formation of autophagosomes/autolysosomes in these cells (Fig. 5 E).
We used an autophagy flux assay to clarify whether ATF4 and SLC7A5 knockdown enhanced autophagy by promoting the induction of autophagy or inhibiting the degradation of autophagosomes. Autophagy flux is indeed much higher in SLC7A5 and ATF4 knockdown cells (Fig. 5 F). Therefore, knockdown of ATF4 and SLC7A5 causes up-regulation of autophagy. We conclude that ATF4-SLC7A5 negatively regulates autophagy by controlling Leu uptake.
In summary, we showed that the GAAC pathway regulates autophagy by controlling the uptake of amino acids. During S/Gln starvation, glutamine deficiency triggered activation of the GAAC pathway, causing up-regulation of ATF4, which then led to up-regulation of amino acid transporters including the Leu transporter SLC7A5. The elevated expression of amino acid transporters caused a surge in the uptake of amino acids, which in turn led to mTOR reactivation and autophagy suppression (Fig. 5 G).
We observed the induction of autophagy and the surge of intracellular amino acid levels after 4 h of S/Gln starvation. At first glance, these two observations appear to be contradictory, as high intracellular amino acid levels will result in the activation of mTOR and suppression of autophagy. It is well established that growth factors that are present in serum regulate mTOR activity through the tuberous sclerosis complex (TSC). Growth factor withdrawal causes activation of the TSC, which in turn inhibits mTOR. In our experimental setting, serum withdrawal appears to play the major role in deactivation of mTOR, as serum starvation can cause sustained deactivation of mTOR in the presence of exogenous amino acids (Fig. S2 A). We speculate that although the intracellular level of amino acids is increased 4 h after S/Gln starvation, the inhibitory effect of TSC is sufficient to cause the deactivation of mTOR. To test this hypothesis, we performed S/Gln starvation in wild-type or TSC−/− mouse embryonic fibroblast (MEF) cells and found that S/Gln starvation causes deactivation of mTOR after 4 h in wild-type MEF cells, but deactivation of mTOR is completely abolished in TSC−/− cells (Fig. S2 F). These data imply that 4 h after S/Gln starvation, the dominant factor that determines mTOR activity is activation of TSC by serum withdrawal, and this can override the effect of increased intracellular amino acid levels. The fact that mTOR reactivation occurs 8 h after starvation indicates that inhibition of mTOR activity by TSC is eventually overcome by increased intracellular amino acids levels.
The possible physiological relevance of this negative feedback mechanism is obvious. As a bulk degradation pathway, autophagy is the last resort to help cells survive during starvation, but self-degradation has a limit past which the cell will die (Schiaffino et al., 2008). Incorporating a negative control mechanism will help cells avoid overactivation of autophagy. The GAAC pathway enhances the cell’s ability to use exogenous amino acids, thus coupling the strength and duration of autophagy induction with the availability of exogenous amino acids. This enables cells to calibrate autophagy to an optimal level, ensuring that they can survive prolonged starvation by using stored material thriftily.
Materials and methods
Reagents and antibodies
Reagents were obtained from the following sources. Monoclonal antibody phospho-T389 S6K1, rabbit antibody S6K, and rabbit antibody ATF4, which could react with antigens from rat, were obtained from Cell Signaling Technology. Rabbit antibody LC3 was obtained from MBL, rabbit antibody β-actin was obtained from Sigma-Aldrich, and rabbit antibody SLC7A5 was obtained from Cosmo Bio Company and could react with antigens from rat. 2-Aminobicyclo-(2, 2, 1)-heptane-2-carboxyl acid was obtained from Sigma-Aldrich. EvaGreen was obtained from Biotium and Trizol from Invitrogen. RNeasy Mini kit was purchased from QIAGEN, RevertAid First Strand cDNA Synthesis kit from Fermentas, BCA Protein Assay kit from Thermo Fisher Scientific, and Fluoromount-G from SouthernBiotech. Western blots were performed as described previously (Yu et al., 2010).
Mammalian cell culture and transfection
CFP-LC3 NRK cells were cultured in DMEM (Life Technologies) supplemented with 10% FBS (5% CO2). Cells were transfected with 200 pmol RNAi or a total of 2 µg DNA by Amaxa nucleofection using solution T and program X-001. Starvation medium was DMEM without the addition of serum and glutamine or DPBS supplemented with 15 mg/liter of phenol red and 4.5 g/liter d-glucose. DMEM deficient in different amino acids was prepared from DPBS by adding nutrients (except for individual amino acids) with the same composition and concentration as found in DMEM. For starvation, the growth medium was removed and cell monolayers were washed twice with starvation medium before adding test medium for the times indicated.
Constructs
SLC7A5 and ATF4 shRNA were synthesized by GenePharma. The target of SLC7A5 shRNA was 5′-GCAATATCACACTGATCAA-3′; the target of ATF4 shRNA was 5′-GGGCTGAAGAAAGCCTAGGTCTCTT-3′.
Real-time quantitative PCR
CFP-LC3 NRK cells were treated as described in Mammalian cell culture and transfection, and then the cells were lysed and RNA was extracted using TRIzol (Invitrogen). cDNA was generated by reverse transcription using a RevertAid First Strand cDNA Synthesis kit. Real-time quantitative PCR was performed using cDNA with EvaGreen on a real-time PCR system (Mx3000PTM; Agilent Technologies). The reaction was performed in a volume of 50 µl in triplicate, according to the manufacturer’s instructions. The relative abundance of Slc7a5 RNA was calculated using the ΔΔCt method. Data were normalized to β-actin cDNA. Primers were designed using Primer blast on the National Center for Biotechnology Information website. All primers were determined to be 95–100% efficient and all exhibited only one dissociation peak. Sequences are as follows: SLC7A5 sense, 5′-GCACTGTGCTACGCGGAGCT-3′; SLC7A5 antisense, 5′-AGCCGTGAGTAGTAGCACGCAG-3′; β-actin sense, 5′-CACCCGCGAGTACAACCTTC-3′; β-actin antisense, 5′-CCCATACCCACCATCACACC -3′.
Immunofluorescence
Cells were plated on glass coverslips in 6-well tissue culture plates. After treatment as described in Mammalian cell culture and transfection, the cells were washed with PBS, fixed in 4% paraformaldehyde for 10 min, and permeabilized in 0.1% saponin for 20 min. Fixed cells were blocked with 0.1% saponin and 10% FBS in PBS for 30 min, stained with 10 µg/ml of antibody in blocking buffer for 1 h, and washed with PBS three times every 5 min. Cells were then stained with secondary antibody (Alexa Fluor 488 Dye; Life Technologies) in PBS for 1 h and washed with PBS three times. Slides were mounted on glass coverslips using Fluoromount-G and imaged. Images were acquired using a confocal microscope (FV-1000; Olympus) that was equipped with Uplansapo 60×/1.35 oil and WHN 20×/22. Imaging medium was IMMERSIN OIL TYPE-E (Olympus). Image acquisition and processing were performed using FV10-ASW 3.1 software (Applied Precision). The mean environmental temperature during experimental acquisitions was 22°C. Images were copied to Illustrator (Adobe) and cut for the final figures.
LC/MS/MS measurement of intracellular free amino acids
CFP-LC3 NRK cells were cultured in a 15-cm dish with 20 ml of media for each sample. After treatment as described in Mammalian cell culture and transfection, the growth medium was discarded and cells were rapidly washed two times with 10 ml of precooled PBS. 5 ml MeOH prechilled to the appropriate temperature (−40°C or on dry ice) was added to the culture dish to disrupt the cells. Dishes were then lightly scraped with a rubber-tipped cell scraper and the cell suspensions were transferred to 15-ml conical tubes. The dishes were washed two times with 4 ml MeOH and the sample was transferred to liquid nitrogen for 10 min, thawed in an ice bath for 10 min, and briefly vortexed. This freeze–thaw cycle was repeated three times for complete cell disruption. The sample was centrifuged at 4°C and 3,000 g for 30 min and the supernatant was transferred to a new tube. The pellet was lysed in 1 ml of lysis buffer and the protein concentration was quantitated using a BCA Protein Assay kit according to the manufacturer’s instructions. After sample normalization based on total protein levels, absolute levels of amino acids in the supernatants were quantified by amine reactive isotope-coded tags (iTRAQ Reagents; Applied Biosystems) in combination with LC/MS/MS (Beijing Amino Acid Medical Research Co.).
Measurement of leucine uptake
Leucine uptake was determined as previously described (Nicklin et al., 2009). In brief, C13-leucine was added to the cells. After 10 min of incubation, cells were washed twice with cold PBS and 5 ml MeOH prechilled to the appropriate temperature (−40°C or on dry ice) was added to the culture dish to disrupt the cells. Dishes were then lightly scraped with a rubber-tipped cell scraper and the cell suspensions were transferred to 15-ml conical tubes. The dishes were washed two times with 4 ml MeOH and the sample was transferred to liquid nitrogen for 10 min, thawed in an ice bath for 10 min, and briefly vortexed. This freeze–thaw cycle was repeated three times for complete cell disruption. The sample was centrifuged at 4°C and 3,000 g for 30 min and the supernatant was transferred to a new tube. The pellet was lysed in 1 ml of lysis buffer and the protein concentration was quantitated using a BCA Protein Assay kit according to the manufacturer’s instructions. After sample normalization based on total protein levels, absolute levels of amino acids in the supernatants were quantified by amine reactive isotope-coded tags in combination with LC/MS/MS.
Transcriptome analysis
Total RNA was extracted using an RNeasy Mini kit prepared for reverse transcription according to the manufacturer’s protocols. The target cDNA was sheared into 80–130-bp fragments and a cDNA library was prepared as described previously (Tang et al., 2010). RNA sequencing was performed by the Biodynamics Optical Imaging Center at Peking University. Transcriptome analyses of the data were performed by the Biodynamics Optical Imaging Center as described previously (Tang et al., 2010). The quality of the RNA sequence data were analyzed. These analyses showed that our RNA sequence data are highly reproducible, reliable, and accurate.
Plasma membrane protein extraction
CFP-LC3 NRK cells were treated as described in Mammalian cell culture and transfection and the plasma membrane protein extraction was performed according to the instruction of a Pierce Cell Surface Protein Isolation kit (Thermo Fisher Scientific).
Statistical tests
Statistical analysis was performed on data from three independent experiments. The error bars in the figures represent SD; the n value is specified in the legends.
Online supplemental material
Fig. S1 shows the change in gene expression in cells undergoing S/Gln starvation. Fig. S2 shows that SLC7A5 regulates mTOR reactivation and autophagy by controlling the uptake of exogenous amino acids. Fig. S3 shows that knockdown of SLC7A5 and ATF4 affects the strength and duration of autophagy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201403009/DC1.
Supplementary Material
Acknowledgments
We are grateful to Nikon instruments (Shanghai), Tokai Hit Co., Ltd., and the Tsinghua Cell Biology Core Facility for technical support.
This research was supported by 973 Program (2010CB833704 and 2011CB910100) and National Science Foundation (grants 31125018, 31030043, and 31321003 to L. Yu).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ATF4
- activating transcription factor 4
- DPBS
- Dulbecco’s phosphate-buffered saline
- GAAC
- general amino acid control
- LC/MS/MS
- liquid chromatography/tandem mass spectrometry
- MEF
- mouse embryonic fibroblast
- NRK
- normal rat kidney
- S/AA
- serum/amino acid
- S/Gln
- serum/glutamine
- TSC
- tuberous sclerosis complex
References
- B’chir, W., Maurin A.C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., and Bruhat A.. 2013. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41:7683–7699 10.1093/nar/gkt563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A., and Hall M.N.. 2009. An amino acid shuffle activates mTORC1. Cell. 136:399–400 10.1016/j.cell.2009.01.021 [DOI] [PubMed] [Google Scholar]
- Fürst, P., and Stehle P.. 2004. What are the essential elements needed for the determination of amino acid requirements in humans? J. Nutr. 134:1558S–1565S [DOI] [PubMed] [Google Scholar]
- Haase, C., Bergmann R., Fuechtner F., Hoepping A., and Pietzsch J.. 2007. L-type amino acid transporters LAT1 and LAT4 in cancer: uptake of 3-O-methyl-6-18F-fluoro-L-dopa in human adenocarcinoma and squamous cell carcinoma in vitro and in vivo. J. Nucl. Med. 48:2063–2071 10.2967/jnumed.107.043620 [DOI] [PubMed] [Google Scholar]
- Hao, S., Sharp J.W., Ross-Inta C.M., McDaniel B.J., Anthony T.G., Wek R.C., Cavener D.R., McGrath B.C., Rudell J.B., Koehnle T.J., and Gietzen D.W.. 2005. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 307:1776–1778 10.1126/science.1104882 [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G.2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407–450 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- Lynch, C.J.2001. Role of leucine in the regulation of mTOR by amino acids: revelations from structure-activity studies. J. Nutr. 131:861S–865S [DOI] [PubMed] [Google Scholar]
- Ma, X.M., and Blenis J.. 2009. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10:307–318 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- Malmberg, S.E., and Adams C.M.. 2008. Insulin signaling and the general amino acid control response. Two distinct pathways to amino acid synthesis and uptake. J. Biol. Chem. 283:19229–19234 10.1074/jbc.M801331200 [DOI] [PubMed] [Google Scholar]
- Meier, C., Ristic Z., Klauser S., and Verrey F.. 2002. Activation of system L heterodimeric amino acid exchangers by intracellular substrates. EMBO J. 21:580–589 10.1093/emboj/21.4.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., and Ohsumi Y.. 1998. A protein conjugation system essential for autophagy. Nature. 395:395–398 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- Mortimore, G.E., and Schworer C.M.. 1977. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 270:174–176 10.1038/270174a0 [DOI] [PubMed] [Google Scholar]
- Nicklin, P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 136:521–534 10.1016/j.cell.2008.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni, T., Joaquin M., Roccio M., Dann S.G., Kim S.Y., Gulati P., Byfield M.P., Backer J.M., Natt F., Bos J.L., et al. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 102:14238–14243 10.1073/pnas.0506925102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi, Y.2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2:211–216 10.1038/35056522 [DOI] [PubMed] [Google Scholar]
- Reeds, P.J.2000. Dispensable and indispensable amino acids for humans. J. Nutr. 130:1835S–1840S [DOI] [PubMed] [Google Scholar]
- Rouschop, K.M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J.W., et al. 2010. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120:127–141 10.1172/JCI40027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzymski, T., Milani M., Pike L., Buffa F., Mellor H.R., Winchester L., Pires I., Hammond E., Ragoussis I., and Harris A.L.. 2010. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 29:4424–4435 10.1038/onc.2010.191 [DOI] [PubMed] [Google Scholar]
- Schiaffino, S., Mammucari C., and Sandri M.. 2008. The role of autophagy in neonatal tissues: just a response to amino acid starvation? Autophagy. 4:727–730 [DOI] [PubMed] [Google Scholar]
- Schworer, C.M., and Mortimore G.E.. 1979. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc. Natl. Acad. Sci. USA. 76:3169–3173 10.1073/pnas.76.7.3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood, R., Porter A.C., Olsen D.A., Cavener D.R., and Wek R.C.. 2000. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics. 154:787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., and Surani M.A.. 2010. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 6:468–478 10.1016/j.stem.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek, S.A., Zhu S., and Wek R.C.. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15:4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida, O., Kanai Y., Chairoungdua A., Kim D.K., Segawa H., Nii T., Cha S.H., Matsuo H., Fukushima J., Fukasawa Y., et al. 2001. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta. 1514:291–302 10.1016/S0005-2736(01)00384-4 [DOI] [PubMed] [Google Scholar]
- Ye, J., Kumanova M., Hart L.S., Sloane K., Zhang H., De Panis D.N., Bobrovnikova-Marjon E., Diehl J.A., Ron D., and Koumenis C.. 2010. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29:2082–2096 10.1038/emboj.2010.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465:942–946 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.