Abstract
Pseudopilins form the central pseudopilus of the sophisticated bacterial type 2 secretion systems. The crystallization of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus was greatly accelerated by the use of nanobodies, which are the smallest antigen-binding fragments derived from heavy-chain only camelid antibodies. Seven anti-EpsI:EpsJ nanobodies were generated and co-crystallization of EpsI:EpsJ nano-body complexes yielded several crystal forms very rapidly. In the structure solved, the nanobodies are arranged in planes throughout the crystal lattice, linking layers of EpsI:EpsJ heterodimers. The EpsI:EpsJ dimer observed confirms a right-handed architecture of the pseudopilus, but, compared to a previous structure of the EpsI:EpsJ heterodimer, EpsI differs 6° in orientation with respect to EpsJ; one loop of EpsJ is shifted by ~5 Å due to interactions with the nanobody; and a second loop of EpsJ underwent a major change of 17 Å without contacts with the nanobody. Clearly, nanobodies accelerate dramatically the crystallization of recalcitrant protein complexes and can reveal conformational flexibility not observed before.
Keywords: General secretory pathway, Pseudopilins, Single-chain antibody, Crystallization chaperones
1. Introduction
Of all the bottlenecks of structure determinations through X-ray crystallography, arguably, the most critical is the actual production of crystals. Thus far, numerous techniques have been developed to circumvent this major obstacle. Natural partner proteins can greatly improve the probability of obtaining crystals by stabilizing the protein of interest, and by creating additional crystal contact surfaces. However, not all proteins have natural partners with whom they interact strongly, or these partners are not yet known, therefore alternative binders has been explored, including “Designed Ankyrin Repeat Proteins” (DARPins) (Huber et al., 2007; Stumpp and Amstutz, 2007), and a diversity of antibody domains, in particular Fab's (Kovari et al., 1995) and single-chain Fv's (Essen et al., 2003; Hunte and Michel, 2002).
The occurrence of bona fide antibodies devoid of light chains in camelidae (Hamers-Casterman et al., 1993) is at the origin of major new developments in antibody technology (Muyldermans et al., 2001). These so-called heavy-chain antibodies bind antigens solely with one single variable domain, referred to as VHH or nanobody (Nb). The single-domain antigen-binding fragments are smaller (~12–15 kDa) and have several advantages compared to their larger antibody counterparts in terms of stability (Perez et al., 2001; van der Linden et al., 1999), expression yield, protease resistance, solubility (Whitlow et al., 1993) and cost (Wolfson, 2006).
The nanobodies in the crystal structures available so far exhibit the classical immunoglobulin fold, with a scaffold of nine anti-parallel β-strands forming two sandwiching β-sheets. At the time of this study, there are structures reported of 22 protein camelid nano-body complexes (De Genst et al., 2004, 2005, 2006; Decanniere et al., 1999, 2001; Desmyter et al., 2001, 2002, 1996; Dolk et al., 2005; Dumoulin et al., 2003; Koide et al., 2007; Loris et al., 2003; Spinelli et al., 2006; Tegoni et al., 1999; Tereshko et al., 2008; Transue et al., 1998). Of all the protein-nanobody complexes, only two proteins had no previous available structure prior to solving the complex with the nanobody: MazE and phage p2 RBP (Loris et al., 2003; Spinelli et al., 2006). While the purpose of the VHH of the VHH:phage p2 RBP structure was to identify the receptor-binding site, the VHH:MazE structure, in which only 44 of the 98 amino acids of MazE were ordered, is the only case reported in which the nanobody was used for stabilization and crystallization of a novel protein.
The nanobody antigen-binding loops have a more diverse repertoire than the canonical antigen-binding loops seen in traditional human and mouse antibodies (Decanniere et al., 2000). Each nanobody has three hypervariable loops, called complementarity determining regions (CDRs), which are apposed to each other and often interact with the antigen. For nanobodies, the CDR3 commonly makes the most contacts with the antigen which is likely due to its exceptional length (16–18 amino acids versus typically 9 amino acids in mouse and 12 amino acids in human antibodies) and sequence variability (Muyldermans et al., 2001; Revets et al., 2005). Interestingly, not all three CDRs need to interact with the antigen for binding to occur. The current study focuses on the complex of a nanobody with a heterodimer from a protein secretion system. Many pathogenic bacteria secrete a diversity of proteins, including bacterial toxins, from the periplasm into the extracellular milieu via an intricate, two-membrane spanning, multi-protein machinery called the “Type 2 Secretion System” (T2SS) or the “General Secretory Pathway” (Cianciotto, 2005; Filloux, 2004; Overbye et al., 1993; Sandkvist et al., 1997; Tauschek et al., 2002). The T2SS is also referred to as the “Extracellular Protein Secretion” (Eps) system in Vibrio species (Sandkvist et al., 1997). In Vibrio cholerae, the causative agent of cholera, the T2SS protein machinery translocates the ~85 kDa cholera toxin and several unrelated proteins in their folded state into the lumen of the gut (Bortoli-German et al., 1994; Cianciotto, 2005; Connell et al., 1995; Hardie et al., 1995; Hirst and Holmgren, 1987; Pugsley, 1992; Sandkvist, 2001b).
In Vibrio species the T2SS is assembled from 11 different proteins, many of these being present in multiple copies (Filloux, 2004; Sandkvist, 2001a; Sandkvist et al., 2000). The T2SS can be thought of as consisting of three major components: (i) the “secretin” EpsD, which forms a protein-conducting pore in the outer membrane; (ii) the “pseudopilins” in the periplasm (EpsG, EpsH, EpsI, EpsJ, and EpsK) assembled into a filamentous “pseudopilus”; and (iii) the “inner membrane platform”. In our attempts to enhance the understanding of the architecture and functioning of the T2SS machinery we have solved previously crystal structures of several T2SS proteins (Johnson et al., 2006; Korotkov and Hol, 2008; Korotkov et al., 2006; Yanez et al., 2008a, 2008b).
Importantly, unraveling three-dimensional structures of the T2SS secretion machinery also assists in understanding aspects of the Type 4 Pilus Biogenesis (T4PB) system (Filloux, 2004; Hobbs and Mattick, 1993; Peabody et al., 2003). The pilins and pseudopilins share a highly variable Type 4a Pilin fold which consists of a long N-terminal α-helix, a variable region, and a conserved C-terminal β-sheet (Craig and Li, 2008). In the T2SS, EpsG is called the “major” pseudopilin because it is the predominant protein component of the pseudopilus. EpsH, EpsI, EpsJ and EpsK are present in lesser amounts and are therefore known as the “minor” pseudopilins. These proteins are thought to form the piston-like pseudopilus that pushes the secreted proteins out of the bacteria and/or may function as a plug for the outer membrane pore (Filloux et al., 1998; Mattick and Alm, 1995; Sandkvist, 2001a; Sauvonnet et al., 2000).
The EpsI:EpsJ heterodimer had previously proven to be very difficult to crystallize, requiring a combination of species variation, surface entropy reduction mutagenesis, and N- and C-terminal truncations (Yanez et al., 2008b). Here we report how nanobodies assisted in obtaining crystals almost immediately of a variant of the heterodimer which had never given a crystal hit before. It appears that the nanobody promoted crystal formation by forming layers of nanobodies interspersed by layers of EpsI:EpsJ heterodimers.
2. Materials and methods
2.1. Cloning, expression, and purification of EpsI and EpsJ
Soluble domains of Vibrio vulnificus EpsI and EpsJ, with the first 24 residues of both chains truncated, were obtained as previously described in Yanez et al. (2008b). Briefly, EpsI:EpsJ, with the EpsI containing an N-terminal TEV-cleavable hexahistidine (His6) tag, were cloned into pCDF-NT, a modified pCDFDuet-1 vector (Novagen). The proteins were co-expressed in Escherichia coli BL21(DE3) (Novagen) cells that were grown at 30 °C until A600 = 0.6, induced with 0.5 mM IPTG. After 4 h of expression at 25 °C, the cells were harvested, lysed using French press, and centrifuged at 20,000g for 40 min to clarify the lysate. The EpsI:EpsJ protein complex was purified with IMAC using Ni-NTA resin (Qiagen), TEV His6 cleavage, followed by an additional pass through the Ni-NTA resin. Finally a Superdex 75 HR10/30 size-exclusion column (GE Health-care), equilibrated in 20 mM Tris-HCl (pH 7.8), 250 mM NaCl, 1 mM EDTA was utilized as a final polishing step.
2.2. Nanobody generation
A llama (Lama glama) was injected several times with in total 2 mg of purified EpsI:EpsJ complex. The immunization, library construction and selection have been performed following standard procedures (Conrath et al., 2001), with minor modifications: total RNA was extracted from the peripheral blood lymphocytes (Chom czynski and Sacchi, 1987), 50 lg of total RNA was used to prepare cDNA using SuperScript II (Invitrogen) and a dN6 primer according to the manufactures instruction. Finally, all selected nanobody genes were cloned in a pHEN6 vector for expression with a Histag in E. coli (Conrath et al., 2001).
2.3. Solid-phase ELISA
Maxisorb 96-well plates (Nunc) were coated with EpsI:EpsJ complex preparations overnight at 4 °C at 1 μg/ml in sodium bicarbonate buffer pH 8.2. Residual protein binding sites in the wells were blocked for 2 h at room temperature with 2% milk in PBS. Detection of antigen-bound nanobodies was performed with a mouse anti-haemaglutinin-decapeptide-tag (clone 16B12, BAbCO) or a mouse anti-histidine-tag (Serotec), as appropriate. Subsequent detection of the mouse anti-tag antibodies was done with an alkaline phosphatase anti-mouse-IgG conjugate (Sigma), respectively. The absorption at 405 nm was measured 15 min after adding the enzyme substrate p-nitrophenyl phosphate.
2.4. EpsI:EpsJ and nanobody complex formation
The purified EpsI:EpsJ complex was incubated with the purified nanobody (Nb11) at room temperature for 30 min. The EpsI:EpsJ:Nb11 complex was then concentrated to 10 mg/ml and then separated from uncomplexed proteins via size-exclusion column chromatography, and concentrated to 8–10 mg/ml for crystallization experiments.
2.5. Crystallization
After mixing 1 μl of the EpsI:EpsJ:Nb11 protein solution with 1 μl of precipitant, plate-like crystals of EpsI:EpsJ:Nb11 grew readily in several conditions from the Index Screen (Hampton Research). While many crystals did not yield adequate diffraction, the best diffracting crystals came from 25% PEG 3350, 0.1 M ammonium sulfate, and 0.1 M Bis-Tris, pH 5.5. Crystals were cryo-protected with the mother liquor plus 1% PEG 400. Remarkably, 5% PEG 400, other PEGs, and glycerol destroyed the diffraction power of these crystals. Crystals were flash-frozen in liquid nitrogen and sent for data collection at SSRL beamline 9-2.
2.6. Structure determination
Data sets were collected at the 9-2 beamline of the Stanford Synchrotron Radiation Laboratory (SSRL) on a MarUSA MarMosaic-325 CCD detector. Data extending to 2.58 Å were processed with XDS (Kabsch, 1993).
The EpsI:EpsJ:Nb11 structure was solved using a multi-step procedure that included molecular replacement and model building. Phaser (McCoy et al., 2007) was used for molecular replacement using EpsI:EpsJ (Yanez et al., 2008b) as a search model for EpsI:EpsJ, and the antibody 2B2X (Clark et al., 2006; 72.4% amino acid sequence identity) after removal of the CDR regions as a search model for the nanobody. Although Phaser did not find a solution for the nanobody, the best EpsI:EpsJ solution from Phaser was used for further density modification (DM) in CCP4 (Collaborative Computational Project, 1994). The density maps obtained by density modification were submitted into RESOLVE (Terwilliger, 2004) for automatic model building. After RESOLVE built two substantial β-strands of the nanobody (Tyr32-Arg38 and Ala96-Arg107), superposition of 2B2X was possible using the SSM superpose command of the graphics program Coot (Emsley and Cowtan, 2004). The superimposed 2B2X molecule was merged into the same model as EpsI:EpsJ and subjected to rigid body and restrained refinement in REFMAC5 (Murshudov et al., 1997). CNS annealing was employed in the initial stages (Brünger et al., 1998), followed by rounds of REFMAC with TLS refinement with seven TLS groups for each chain of EpsI and EpsJ as determined by the TLS motion determination (TLSMD) server (Painter and Merritt, 2006). REFMAC refinement with medium main-chain and medium side-chain NCS restraints was alternated with manual model building with Coot. The stereochemical quality of the model was verified using Molprobity (Lovell et al., 2003) and PROCHECK (Laskowski et al., 1993). Final structure determination and refinement statistics are listed in Table 1.
Table 1.
Data collection and refinement statistics.
| V. vulnificus Epsl:EpsJ:Nb11 | |
|---|---|
| Data set | |
| Wavelength (Å) | 1.12709 |
| Space group | P1 |
| a (Å) | 46.8 |
| b (Å) | 67.4 |
| c (Å) | 128.6 |
| α | 96.7° |
| β | 91.6° |
| γ | 90.2° |
| Unique reflections | 49,095 |
| Resolution (Å) | 20–2.58 (2.68–2.58)b |
| Completeness (%) | 90.0 (71.0) |
| Redundancy | 2.0 (2.0) |
| Rmerge (%) | 12.9 (59.1) |
| I/σ | 14.8 (2.7) |
| Wilson B-factor (Å2) | 55.1 |
| Refinement a | |
| Resolution (Å) | 20–2.58 |
| Reflections used | 44,423 |
| Heterotrimers per unit cell | 4 |
| Residues per unit cell | 1397c |
| Atoms | 11,238 |
| Waters per unit cell | 156 |
| Chloride ion per unit cell | 4 |
| Rwork (%) | 23.2 |
| Rfree (%) | 27.9 |
| R.m.s.d bond lengths (Å2) | 0.006 |
| R.m.s.d. bond angles (°) | 0.849 |
| B-average (Å2) | 66.0 |
| EpsI | 66.6 |
| Epsj | 66.0 |
| Nb11 | 65.4 |
| B-water (Å2) | 62.9 |
| Ramachandran | |
| Most favored | 97.1% |
| Additionally allowed | 2.7% |
| Disallowed | 0.2% |
Refinement statistics are based on Refmac5 values.
Values in parentheses refer to outer shell.
A total of 1397 out of 1636 residues were built into the electron density for the final EpsI:EpsJ:Nb11 model. Parts of the N- and C-termini were not present in the electron density and were omitted from the model. Poor electron density was observed for several loop regions, therefore the following residues were not included in the final structure: EpsI Pro55 and Leu86 in chain A, Lys56, Gln83, and Pro84 in chain D. Leu58, Thr81-Leu85 and Ala99 in chain G; for Epsj Glu69-Pro70 in chain B, Gly67-Pro70 and Gln102-Phe103 in chain E, Glu69-Ser71 in chain H, and Asn66-Glu69 and Pro104-Arg105 in chain K; in Nb11 only Va112-Gln13 in chain L.
3. Results
3.1. VHH library construction and selection of specific binders
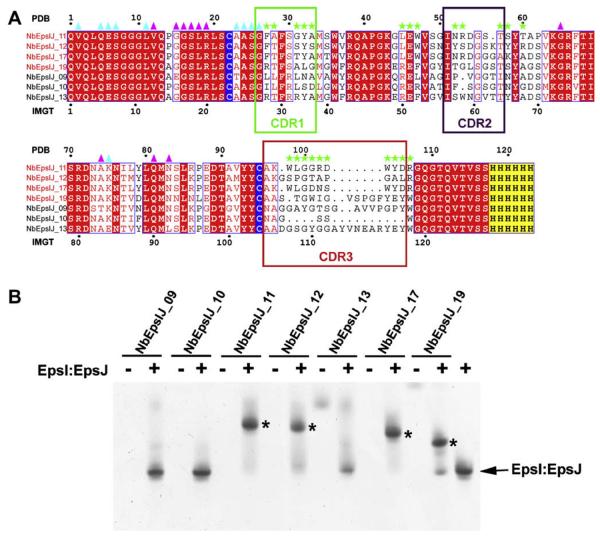
The VHH repertoire of the library resulting from the immunization was expressed on phages following procedures and selection of phage particles expressing an EpsI:EpsJ binding VHH as detailed in Section 2. A clear enrichment was observed after three consecutive rounds of selection on solid-phase coated antigen. Twice 48 randomly chosen colonies—after the second and third round—were grown for expression of their VHH as soluble protein. Of the crude periplasmic extracts tested in an ELISA, 46 were shown to be specific towards the EpsI:EpsJ complex. After HinfI/RFLP and sequence analysis on the VHH genes amplified from the clones positive in ELISA, 11 different nanobodies against the EpsI:EpsJ complex were identified. Seven of these were expressed and purified.
3.2. EpsI:EpsJ:nanobody complexes in solution
A complex of N-terminally histidine-tagged, N-terminally truncated EpsI(25–117):EpsJ(25–217) from V. vulnificus (hereafter referred to as “EpsI” and “EpsJ”, respectively) was incubated with each of the seven nanobodies in a 1:1 molar ratio to test complex formation in vitro. For four of the seven nanobodies this was successful according to native gel analysis (Fig. 1B). These four ternary EpsI:EpsJ:nanobody complexes were subsequently prepared, purified and subjected to crystallization trials. The protein–nanobody complex of EpsI:EpsJ with NbEpsIJ-11 (hereafter called Nb11) yielded well-diffracting crystals very rapidly. Throughout the remainder of the study, the residue numbering referred to in the text corresponds to the continuous numbering in the PDB file as seen at the top of the sequence alignment in Fig. 1A. To translate this residue numbering of the nanobody to the standard ImMuno-GeneTics (IMGT) numbering for immunoglobulins (Lefranc, 2005; Lefranc et al., 2003), refer to the lower line of the sequence alignment in Fig. 1A.
Fig. 1.
Anti-EpsI:EpsJ generated nanobodies and complex formation with EpsI:EpsJ. (A) Sequence alignment of nanobodies generated against V. vulnificus EpsI:EpsJ. The nanobodies that bound to EpsI:EpsJ according to gel-shift assays (Fig. 1B) are titled in red. While the numbering on top of the alignment corresponds to the continuous numbering present in the PDB file, the numbering on the bottom of the alignment corresponds to the standard IMGT numbering for antibodies and related proteins (Lefranc, 2005; Supplementary Figure S1). The conserved cysteines that form the intra-molecular disulfide bridge are highlighted in blue while the C-terminal hexahistidine tag is highlighted in yellow. Boxed segments of sequence compose the CDR regions with the green box as CDR1, purple box as CDR2, and red box as CDR3. Green stars denote residues that make contacts with EpsJ. Cyan and purple triangles reflect residues that make nanobody-nanobody contacts, with cyan as those of Chain C and purple of Chain F. (B) Ternary complex formation of N-terminal His6 EpsI:EpsJ with nanobodies. A native PAGE gel of anti-EpsI:EpsJ nanobodies alone and with V. vulnificus EpsI:EpsJ. The majority of the nanobodies do not enter the gel due to the high pI's that these protein exhibit (~9). Only NbEpsIJ-13 and NbEpsIJ-19, with pI = 8.0, enter the gel and are seen in the nanobody control lanes. Ternary complex formation between EpsI:EpsJ and the nanobody (NbEpsIJ-11, NbEpsIJ-12, NbEpsIJ-17, NbEpsIJ-19) cause a dramatic band shift in the gel and are denoted by asterisks. The EpsI:EpsJ control is seen in the far right lane. (For interpretation of color mentioned in this figure the reader is referred to the web version of the article.)
3.3. Structure solution
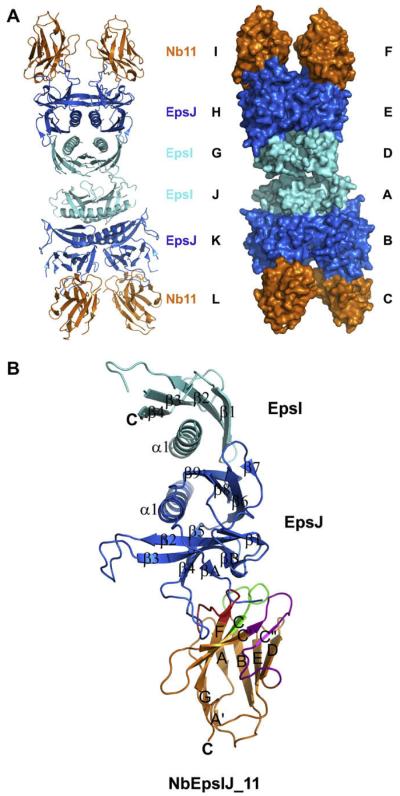
The EpsI:EpsJ:Nb11 crystals belong to space group P1, with four copies of the EpsI:EpsJ:Nb11 heterotrimer per unit cell, containing 1397 well-defined amino acids out of 1636 residues in total (Fig. 2A). The EpsI:EpsJ:Nb11 structure was refined to an Rwork = 23.2% and Rfree = 27.9% with good geometry (Table 1). The Cα carbons of the four EpsI:EpsJ:Nb11 heterotrimers per cell can be superimposed with pairwise root mean square deviations (RMSD) between 0.5 and 0.6 Å. Since these structures are so similar, one trimer (consisting of Chain A for EpsI, Chain B for EpsJ and Chain C for Nb11) will be the main ternary complex analyzed in the rest of this report.
Fig. 2.
The structure of V. vulnificus EpsI:EpsJ in complex with Nb11. (A) View of the unit cell with four EpsI:EpsJ:Nb11 ternary complexes. EpsI, light blue; EpsJ, blue; and Nb11, orange. The cartoon representation of the unit cell is seen on the left, and the surface representation is seen on the right. The chain names in the structure are next to their respective components of the unit cell. (B) General architecture and secondary structure elements of the EpsI:EpsJ:Nb11 ternary complex. EpsI, light blue; EpsJ, blue; and Nb11, orange. Nb11 is further colored by CDR, with CDR1 being green, CDR2 purple, and CDR3 red. Each α-helix and β-strand has been labeled according to its order in the protein. (For interpretation of color mentioned in this figure the reader is referred to the web version of the article.)
3.4. The structure of Nb11 and its CDRs
The four nanobody molecules per triclinic unit cell are very similar in structure with RMSD's ranging from 0.1 to 0.4 Å for 109–111 Cα atoms. Overall, the structure of Nb11 follows the classical immunoglobulin scaffold seen in other nanobody structures to date. Comparison of the Nb11 structure with the VH domain of the closest structural neighbor 1IGT, an intact IgG2a monoclonal antibody (Harris et al., 1997), shows that the major regions of difference involve CDR3 (Supplementary Figure S2). The closest nano-body DALI match among nanobodies of known structure is 1ZVH (De Genst et al., 2006) with a sequence identity of 63%, and an RMSD of 1.5 Å (Supplementary Figure S2).
The CDRs in our structure appear to display excellent electron density, allowing an evaluation of the CDR conformations to see if they adopt one of the canonical types that have been previously defined (Al-Lazikani et al., 1997; Chothia and Lesk, 1987; Chothia et al., 1992). The conformations of both of CDR1 and CDR2 from Nb11 can be classified as canonical type 1 in their respective hypervariable region repertoires.
CDR3 in Nb11 consists of Ala96-Asp106 and changes its direction at the point where β-strand F begins, by ~130°, and terminates prior to the beginning of strand G (Figs. 2B, 3 and 4B). The conformations of CDR3 are more difficult to classify since CDR3's are longer and more variable than CDR1 and CDR2. In one study conformations of CDR3's were separated into β-hairpins with either a kinked base, a kinked base with an additional bulge, or an extended base (Shirai et al., 1996). A more recent CDR3 classification divides structures into three classes based on the “torso” and “head”: (1) torso with a β-bulge; (2) torso with regular β-sheet hairpin rather than β-bulge structure; (3) short regions that form short hairpins (Morea et al., 1998). The torso is formed by a two-stranded anti-parallel β-sheet composed of the first five and last six residues of the loop. All residues not in the torso make up the head of the CDR3 loop (Decanniere et al., 1999). The Nb11 structure seen here adheres to the torso with a β-bulge class in an extreme manner, with CDR3 folding over the framework and directing its β-strands perpendicular to the CDR1 and CDR2 β-strands (Fig. 4, Supplementary Figures S2B and C).
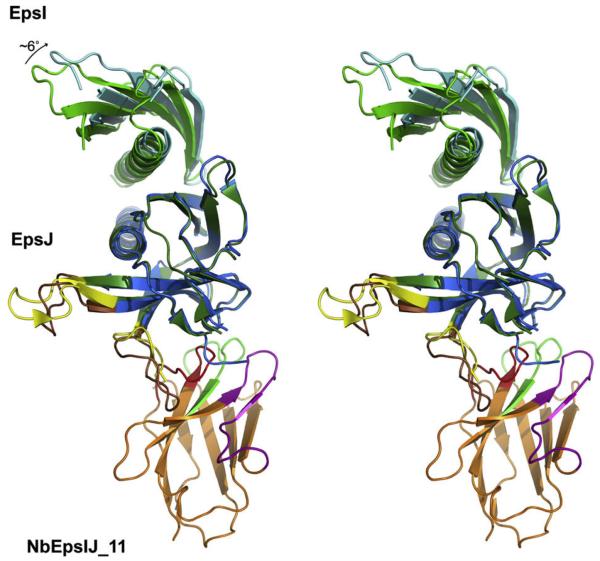
Fig. 3.
Comparison of EpsI:EpsJ heterodimers from two different structures. The EpsI:EpsJ:Nb11 structure superimposed onto the EpsI:EpsJ structure from Yanez et al. (2008b) by using only the EpsJ chains for calculating the superposition operation. The EpsI (cyan) chains differ in orientation by ~6°. Two loops differing in conformation, discussed in the text, are shown in brown in the EpsI:EpsJ:Nb11 structure and yellow in the EpsI:EpsJ structure from Yanez et al. (2008b). (For interpretation of color mentioned in this figure the reader is referred to the web version of the article.)
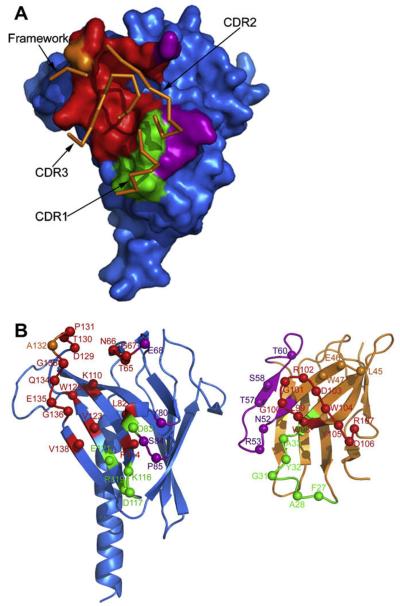
Fig. 4.
The interacting surfaces of V. vulnificus EpsJ and Nb11. (A) EpsJ is shown in blue with the residues that interact with the Nb11 CDR regions color-coded. CDR1 interactions, green; CDR2 interactions, purple; CDR3 interactions, red. The orange surface area represents residues of EpsJ interacting with the Nb11 framework. As orange ribbons are depicted the main-chain loops of the three Nb11 CDR's and framework residues interacting with residues of EpsJ. (B) “Butterfly” representation of EpsJ:Nb11 complex. Left: EpsJ: spheres denoting residues that interact with Nb11. CDR1-interacting residues, green; CDR2-interacting residues; purple; CDR3-interacting residues, red; framework-interacting residues, orange. Right: Nb11: spheres denoting residues that interact with EpsJ. CDR1, green; CDR2, purple; CDR3, red; framework, orange. “PDB entry 3CFI residue numbering” is used; see upper line of Fig. 1A. The corresponding “IMGT numbering” is shown in the lower line of Fig. 1A. (For interpretation of color mentioned in this figure the reader is referred to the web version of the article.)
3.5. EpsJ:Nb11 interactions
The EpsJ:Nb11 interface buries 1461 Å2 solvent accessible surface, close to the average of 1554 Å2 buried surface for antibody-protein complexes (Jones and Thornton, 1996). The tightness of the interface can be judged with the gap volume index which is a measure of the complementarity of surfaces (Jones and Thornton, 1996). In the case of the of the EpsJ:Nb11 interface, the gap volume index is 2.6 Å, which is below the average gap volume index of 3.0 Å for antibody-protein complexes (Jones and Thornton, 1996). EpsJ buries 731 Å2 in the EpsJ:nanobody interface which is ~8% of its total accessible protein surface involving 26 interface residues in EpsJ (interactions are described in the Supplementary Text). The interacting residues on EpsJ are all from the first β-sheet of the so-called “variable region” that includes strands β1–β5.
A total of 22 residues from Nb11 are engaged in contacts with EpsJ (Figs. 1A and 4B; Supplementary Table 1). CDR1 and CDR2 make rather minor contacts, burying 121 Å2 and 87 Å2, respectively. CDR3 buries 478 Å2 surface area, providing the majority of the interface. CDR3 serves as the primary binding loop to EpsJ by burying itself into an extensive pocket formed by the βA–βB, β4–bβ and β1–β5 loops of EpsJ (Fig. 4). This EpsJ pocket contains several charged residues that are complementary with the charged residues on CDR3, as described in the Supplementary Text.
4. Discussion
4.1. The EpsI:EpsJ:Nb11 complex reveals structural changes that may be relevant to pseudopilus function
It is of interest to compare EpsI, EpsJ as well as the EpsI:EpsJ heterodimer in our current structure (Fig. 2B) with those of the V. vulnificus EpsI:EpsJ structure described by Yanez et al. (2008b) since this is the first time the effect of nanobody binding on a heterodimer can be evaluated.
The two structures of EpsI from these two studies superimpose with an RMSD of 0.7 Å for 75 equivalent Cα atoms. The major differences reside in flexible loops with the largest divergence at Asn57 with a shift of 3.8 Å for the Ca atoms between the two structures.
The EpsJ molecule in the current structure superimposes with EpsJ in the previous structure of Yanez et al. (2008b), with an RMSD of 0.9 Å for 152 equivalent Cα atoms, but this requires ignoring two loops which are very different in conformation:
-
(i)
In the heterodimer without nanobody, the β2–β3 loop is in an extended conformation in all four EpsJ molecules, whereas this loop in is bent by almost 90° in all four EpsJ molecules of the current EpsI:EpsJ:nanobody crystals (Fig. 3). At the Cα of Gln100, the loop deviates by more than 16 Å if we compare both complexes. The functional implications of this dramatic change in conformation of the β2–β3 loop in the two crystal forms remain unclear but might indicate structural changes pseudopilins are likely to undergo during assembly into a pseudopilus or during transport of secreted proteins out of the cell by the T2SS.
-
(ii)
The β4–β5 loop (Trp125-Val137) with a largest difference of 4.8 Å at the position of Pro128 (Fig. 3). This β4–β5 loop difference is probably due to interactions of the loop with the nanobody in the EpsI:EpsJ:Nb11 structure, with the nanobody causing these residues to adopt an altered and well-defined conformation in the ternary complex (Figs. 3 and 4; Supplementary Text). In contrast, several residues (Ala132-Gly133) from this loop are highly flexible in the previous EpsI:EpsJ structure (Yanez et al., 2008b).
Our new structure allows a comparison of the entire EpsI:EpsJ heterodimer in the current EpsI:EpsJ:Nb11 ternary complex (Fig. 2B) with the V. vulnificus EpsI:EpsJ heterodimer described by Yanez et al. (2008b). When excluding the two major deviating loops in EpsJ discussed above, the two heterodimers can be superimposed with an RMSD of 1.1 Å for 218 equivalent Cα atoms. In both structures, the arrangement of EpsI and EpsJ corresponds with a right-handed nature of the T2SS pseudopilus (Korotkov and Hol, 2008; Yanez et al., 2008b), a key element of the T2SS for which right-handed and left-handed arrangements have been proposed (Craig et al., 2006; Köhler et al., 2004). The orientation of EpsI versus EpsJ differs by approximately 6° in the two heterodimers (Fig. 3). This flexibility might be important during the assembly and/or functioning of the dynamic piston-like pseudopilus in the central periplasmic space of the T2SS.
4.2. Nanobodies can promote crystal growth in multiple ways
Yanez et al. (2008b) recently described the difficulties which had to be overcome to obtain suitable crystals for the structure determination of the EpsI:EpsJ heterodimer. Compared to the approximately 10 months and 17 protein variants it had taken to start from EpsI:EpsJ and to arrive at well-diffracting crystals of the mutated and truncated form, crystallization of wild-type N-terminally truncated EpsI:EpsJ in complex with nanobody Nb11 merely took 15 days. A total of 192 crystallization conditions were tested. Only optimization of the cryoprotectant was required.
Several of the four nanobodies shown to bind to EpsI:EpsJ (Fig. 1B) gave crystals with both N- and C-terminal His6-tagged EpsI:EpsJ; however the crystals with Nb11 that lead to structure determination were the best diffracting and required no optimization. Interestingly, the most robust crystal producing complex contained an EpsI variant with a cleaved N-terminal His6-tag. This variant of EpsI:EpsJ had formerly, without Nb11, given no crystal hits whatsoever. Clearly, the addition of a nanobody converted a protein that was very difficult to crystallize into a complex which could be crystallized to diffraction quality within days.
Crystal contacts between nanobodies from neighboring EpsI:EpsJ:Nb11 heterotetramers bury 464 Å2 solvent accessible surface, involving 10 residues from each nanobody (Figs. 1A and 4B). Essentially all these contacts are provided by framework side chains. Fig. 5 shows how Nb11 assists in crystal packing: layers of nanobodies contacting each other traverse the crystal linking layers of EpsI:EpsJ heterodimers.
Fig. 5.
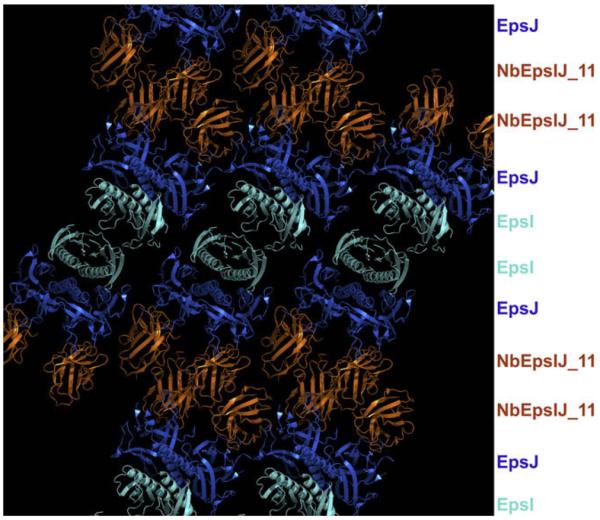
View of the “layers” of the heterotrimer in the crystal lattice. Crystal contacts between the nanobodies within the crystal allow the lattice to be composed of layers of EpsI, EpsJ, and Nb11 molecules. Nb11 in gold; EpsI in light blue; EpsJ dark blue. (For interpretation of color mentioned in this figure the reader is referred to the web version of the article.)
It is of interest to compare the way by which nanobody Nb11 promotes crystallization of the V. vulnificus EpsI:EpsJ heterodimer with the way by which another nanobody, NbGspD-7, facilitated crystal formation of the flexible periplasmic domain of the T2SS secretin GspD (“peri-GspD”) as described by Korotkov et al., in press. In the latter case, NbGspD-7 generates a compact heterotetramer formed by two nanobodies and two peri-GspD chains. Even though NbGspD-7 also engages in crystal contacts with adjacent tetramers, the main effect of this nanobody on the crystal growth in this case is most likely a decrease of the conformational freedom of its target protein with a concomitant increase of the crystallization probability.
These two cases of substantial acceleration of crystal growth of two recalcitrant proteins are therefore achieved mainly by “gluing” in the case of Nb11 and by “freezing” in the case of NbGspD-7. Future studies are needed to evaluate which of these modes of operation will be predominant in general. Both nanobodies accelerated crystal growth dramatically and this bodes well for future applications of nanobodies as “crystallization chaperones”.
5. Protein Data Bank accession codes
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank and are available under accession code 3CFI.
Supplementary Material
Acknowledgments
We thank Jürgen Bosch, Stewart Turley, and Jan Abendroth for help and valuable discussions. We thank Nele Buys for the selection, expression and purification of the nanobodies. We are indebted to the support staff of beamline 9-2 of the SSRL for assistance during data collection. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), supported by the Department of Energy and by NIH. This work was supported by Grant AI34501 from the NIH, by the Howard Hughes Medical Institute (HHMI) to W.G.J.H., and University of Washington's Molecular Biophysics Training Grant (5 T32 GM008268-19) from the NIH. This work was also supported by the Belgian Government under the framework of the Interuniversity Attraction Poles (I.A.P. P6/19).
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.jsb.2008.11.008.
References
- Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- Bortoli-German I, Brun E, Py B, Chippaux M, Barras F. Periplasmic disulphide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol. Microbiol. 1994;11:545–553. doi: 10.1111/j.1365-2958.1994.tb00335.x. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM, Gherardi E, Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. Structural repertoire of the human VH segments. J. Mol. Biol. 1992;227:799–817. doi: 10.1016/0022-2836(92)90224-8. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, Hanf KJ, Jarpe M, Liparoto SF, Li Y, Lugovskoy A, Miller S, Rushe M, Sherman W, Simon K, Van Vlijmen H. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Sci. 2006;15:949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Connell TD, Metzger DJ, Wang M, Jobling MG, Holmes RK. Initial studies of the structural signal for extracellular transport of cholera toxin and other proteins recognized by Vibrio cholerae. Infect. Immun. 1995;63:4091–4098. doi: 10.1128/iai.63.10.4091-4098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Li J. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- De Genst E, Handelberg F, Van Meirhaeghe A, Vynck S, Loris R, Wyns L, Muyldermans S. Chemical basis for the affinity maturation of a camel single domain antibody. J. Biol. Chem. 2004;279:53593–53601. doi: 10.1074/jbc.M407843200. [DOI] [PubMed] [Google Scholar]
- De Genst E, Silence K, Ghahroudi MA, Decanniere K, Loris R, Kinne J, Wyns L, Muyldermans S. Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires. J. Biol. Chem. 2005;280:14114–14121. doi: 10.1074/jbc.M413011200. [DOI] [PubMed] [Google Scholar]
- De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decanniere K, Muyldermans S, Wyns L. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes? J. Mol. Biol. 2000;300:83–91. doi: 10.1006/jmbi.2000.3839. [DOI] [PubMed] [Google Scholar]
- Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure. 1999;7:361–370. doi: 10.1016/s0969-2126(99)80049-5. [DOI] [PubMed] [Google Scholar]
- Decanniere K, Transue TR, Desmyter A, Maes D, Muyldermans S, Wyns L. Degenerate interfaces in antigen–antibody complexes. J. Mol. Biol. 2001;313:473–478. doi: 10.1006/jmbi.2001.5075. [DOI] [PubMed] [Google Scholar]
- Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J. Biol. Chem. 2001;276:26285–26290. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- Desmyter A, Spinelli S, Payan F, Lauwereys M, Wyns L, Muyldermans S, Cambillau C. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J. Biol. Chem. 2002;277:23645–23650. doi: 10.1074/jbc.M202327200. [DOI] [PubMed] [Google Scholar]
- Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- Dolk E, van der Vaart M, Lutje Hulsik D, Vriend G, de Haard H, Spinelli S, Cambillau C, Frenken L, Verrips T. Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo. Appl. Environ. Microbiol. 2005;71:442–450. doi: 10.1128/AEM.71.1.442-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoulin M, Last AM, Desmyter A, Decanniere K, Canet D, Larsson G, Spencer A, Archer DB, Sasse J, Muyldermans S, Wyns L, Redfield C, Matagne A, Robinson CV, Dobson CM. A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme. Nature. 2003;424:783–788. doi: 10.1038/nature01870. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Essen LO, Harrenga A, Ostermeier C, Michel H. 1.3 A X-ray structure of an antibody Fv fragment used for induced membrane-protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2003;59:677–687. doi: 10.1107/s0907444903002233. [DOI] [PubMed] [Google Scholar]
- Filloux A. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–468. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hardie KR, Schulze A, Parker MW, Buckley JT. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bond formation. Mol. Microbiol. 1995;17:1035–1044. doi: 10.1111/j.1365-2958.1995.mmi_17061035.x. [DOI] [PubMed] [Google Scholar]
- Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- Hirst TR, Holmgren J. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 1987;84:7418–7422. doi: 10.1073/pnas.84.21.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M, Mattick JS. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- Huber T, Steiner D, Rothlisberger D, Pluckthun A. In vitro selection and characterization of DARPins and Fab fragments for the co-crystallization of membrane proteins: the Na(+)-citrate symporter CitS as an example. J. Struct. Biol. 2007;159:206–221. doi: 10.1016/j.jsb.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Hunte C, Michel H. Crystallisation of membrane proteins mediated by antibody fragments. Curr. Opin. Struct. Biol. 2002;12:503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- Johnson TL, Abendroth J, Hol WGJ, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol. Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- Köhler R, Schafer K, Muller S, Vignon G, Diederichs K, Philippsen A, Ringler P, Pugsley AP, Engel A, Welte W. Structure and assembly of the pseudopilin PulG. Mol. Microbiol. 2004;54:647–664. doi: 10.1111/j.1365-2958.2004.04307.x. [DOI] [PubMed] [Google Scholar]
- Koide A, Tereshko V, Uysal S, Margalef K, Kossiakoff AA, Koide S. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J. Mol. Biol. 2007;373:941–953. doi: 10.1016/j.jmb.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Hol WG. Structure of the GspK–GspI–GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat. Struct. Mol. Biol. 2008;15:462–468. doi: 10.1038/nsmb.1426. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Krumm B, Bagdasarian M, Hol WG. Structural and functional studies of EpsC, a crucial component of the type 2 secretion system from Vibrio cholerae. J. Mol. Biol. 2006;363:311–321. doi: 10.1016/j.jmb.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Kovari LC, Momany C, Rossmann MG. The use of antibody fragments for crystallization and structure determinations. Structure. 1995;3:1291–1293. doi: 10.1016/s0969-2126(01)00266-0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. IMGT, the international ImMunoGeneTics information system: a standardized approach for immunogenetics and immunoinformatics. Immunome Res. 2005;1:3. doi: 10.1186/1745-7580-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Loris R, Marianovsky I, Lah J, Laeremans T, Engelberg-Kulka H, Glaser G, Muyldermans S, Wyns L. Crystal structure of the intrinsically flexible addiction antidote MazE. J. Biol. Chem. 2003;278:28252–28257. doi: 10.1074/jbc.M302336200. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Alm RA. Response from Mattick and Alm: common architecture of type 4 fimbriae and complexes involved in macromolecular traffic. Trends Microbiol. 1995;3:411–413. [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morea V, Tramontano A, Rustici M, Chothia C, Lesk AM. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 1998;275:269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- Overbye LJ, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Perez JM, Renisio JG, Prompers JJ, van Platerink CJ, Cambillau C, Darbon H, Frenken LG. Thermal unfolding of a llama antibody fragment: a two-state reversible process. Biochemistry. 2001;40:74–83. doi: 10.1021/bi0009082. [DOI] [PubMed] [Google Scholar]
- Pugsley AP. Translocation of a folded protein across the outer membrane in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1992;89:12058–12062. doi: 10.1073/pnas.89.24.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revets H, De Baetselier P, Muyldermans S. Nanobodies as novel agents for cancer therapy. Expert Opin. Biol. Ther. 2005;5:111–124. doi: 10.1517/14712598.5.1.111. [DOI] [PubMed] [Google Scholar]
- Sandkvist M. Biology of type II secretion. Mol. Microbiol. 2001a;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- Sandkvist M. Type II secretion and pathogenesis. Infect. Immun. 2001b;69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M, Bagdasarian M, Howard SP. Characterization of the multimeric Eps complex required for cholera toxin secretion. Int. J. Med. Microbiol. 2000;290:345–350. doi: 10.1016/S1438-4221(00)80038-7. [DOI] [PubMed] [Google Scholar]
- Sandkvist M, Michel LO, Hough LP, Morales VM, Bagdasarian M, Koomey M, DiRita VJ, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N, Vignon G, Pugsley AP, Gounon P. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 2000;19:2221–2228. doi: 10.1093/emboj/19.10.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H, Kidera A, Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996;399:1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- Spinelli S, Desmyter A, Verrips CT, de Haard HJ, Moineau S, Cambillau C. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat. Struct. Mol. Biol. 2006;13:85–89. doi: 10.1038/nsmb1029. [DOI] [PubMed] [Google Scholar]
- Stumpp MT, Amstutz P. DARPins: a true alternative to antibodies. Curr. Opin. Drug. Discov. Devel. 2007;10:153–159. [PubMed] [Google Scholar]
- Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegoni M, Spinelli S, Verhoeyen M, Davis P, Cambillau C. Crystal structure of a ternary complex between human chorionic gonadotropin (hCG) and two Fv fragments specific for the alpha and beta-subunits. J. Mol. Biol. 1999;289:1375–1385. doi: 10.1006/jmbi.1999.2845. [DOI] [PubMed] [Google Scholar]
- Tereshko V, Uysal S, Koide A, Margalef K, Koide S, Kossiakoff AA. Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci. 2008;17:1175–1187. doi: 10.1110/ps.034892.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- Transue TR, De Genst E, Ghahroudi MA, Wyns L, Muyldermans S. Camel single-domain antibody inhibits enzyme by mimicking carbohydrate substrate. Proteins. 1998;32:515–522. doi: 10.1002/(sici)1097-0134(19980901)32:4<515::aid-prot9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- van der Linden RH, Frenken LG, de Geus B, Harmsen MM, Ruuls RC, Stok W, de Ron L, Wilson S, Davis P, Verrips CT. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- Whitlow M, Bell BA, Feng SL, Filpula D, Hardman KD, Hubert SL, Rollence ML, Wood JF, Schott ME, Milenic DE, et al. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng. 1993;6:989–995. doi: 10.1093/protein/6.8.989. [DOI] [PubMed] [Google Scholar]
- Wolfson W. Ablynx makes nanobodies from llama bodies. Chem. Biol. 2006;13:1243–1244. doi: 10.1016/j.chembiol.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Yanez M, Korotkov K, Abendroth J, Hol WG. Structure of the minor pseudopilin EpsH from the type 2 secretion system of Vibrio cholerae. J. Mol. Biol. 2008a;377:91–103. doi: 10.1016/j.jmb.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez ME, Korotkov KV, Abendroth J, Hol WG. The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus. J. Mol. Biol. 2008b;375:471–486. doi: 10.1016/j.jmb.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.