Abstract
Colletotrichum caudatum sensu lato is a widespread fungal pathogen of warm-season grasses. The fungus is easily differentiated from other Colletotrichum species through the presence of a unique filiform appendage at the apex of the conidium. Multi-locus phylogenetic analysis of four DNA sequence markers from 21 isolates of C. caudatum s.l. from six grass hosts recovered the morphospecies as a well-supported monophyletic group. Although closely related to other Colletotrichum species pathogenic to warm-season grasses (e.g. C. sublineola, C. falcatum, C. navitas, C. graminicola), the sister taxon placement of C. caudatum remained unclear. Four major subgroups and three monotypic lineages were identified from the C. caudatum s.l. isolates. Despite the presence of localized, taxon-specific incongruence between gene trees and evidence for recombination in the dataset, application of genealogical concordance species recognition criteria diagnosed the four subgroups as phylogenetic species. Traditional morphology-based species concept defines C. caudatum as one species with a broad host range; however, multi-locus phylogenetic analyses refuted this model. Instead, isolates from different hosts were mainly segregated into different lineages. In particular, isolates from the type locale and host (USA, Sorghastrum nutans) collected within a 400 km radius were divided into three distinct species that corresponded with the three sampling sites. These data established that traditional morphological and ecological features are not informative for recognition of taxa within C. caudatum s.l., although there is some evidence that some species may be host specific. To stabilize the application of the name C. caudatum, DNA sequence data from the lectotype was generated, an epitype strain consistent with the type was designated and illustrated, and an emended description of C. caudatum sensu stricto is provided. Colletotrichum alcornii, C. baltimorense, C. somersetense, and C. zoysiae are described as new morphologically cryptic species related to C. caudatum s.s.
Keywords: Andropogon gerardii, bioenergy, Colletotrichum ciliatum, Cymbopogon, Imperata cylindrica, Sorghastrum nutans, tallgrass prairie, Zoysia
INTRODUCTION
Colletotrichum caudatum sensu lato is a morphologically distinctive fungal pathogen of warm-season grasses (C4 photosynthesis, or C4) in Poaceae tribe Andropogoneae (Nag Raj 1973, Sutton 1980, 1992, Hyde et al. 2009). While many species of Colletotrichum are morphologically cryptic, and may be difficult or impossible to identify through morphologic features alone, C. caudatum s.l. is easy to recognize through visual examination of the asexual spores (Sutton 1980, Hyde et al. 2009). The fungus produces falcate-shaped conidia that bear a diagnostic filiform appendage at the apex that makes C. caudatum s.l. unique, relative to all other known Colletotrichum species (Fig. 1I–K; Nag Raj 1973). Colletotrichum caudatum was first described from the North American native grass Sorghastrum nutans (indiangrass) during the late nineteenth century (Saccardo 1880) but was not actually confirmed as a pathogen until Zeiders (1987). Based on the presence of the diagnostic conidial appendage, C. caudatum s.l. has been reported from numerous C4 grass hosts worldwide, including Bothriochloa bladhii, Cymbopogon spp., Eremochloa ophiuroides, Eragrostis spp., Imperata cylindrica, Manisuris cylindrica, and Zoysia species (Nag Raj 1973, Sutton 1980, 1992, Fuke et al. 2006, Crouch et al. 2009b, c, Farr & Rossman 2012). Colletotrichum caudatum s.l. also infects many of the perennial prairie grasses that are under investigation as candidate sources of cellulosic biomass for bioenergy production, including indiangrass, Andropogon gerardii (big bluestem), and Panicum virgatum (switchgrass; Sutton 1980, 1992, Zeiders 1987, Crouch et al. 2009a). Since these grasses are increasingly cultivated as bioenergy crops, associated fungal pathogens such as C. caudatum may become increasingly widespread. Indeed, C. caudatum was recently reported as the cause of anthracnose disease from cultivated stands of indiangrass in the USA − the first report of the disease on this host in more than 20 yr (Waxman & Bergstrom 2011).
Fig. 1.
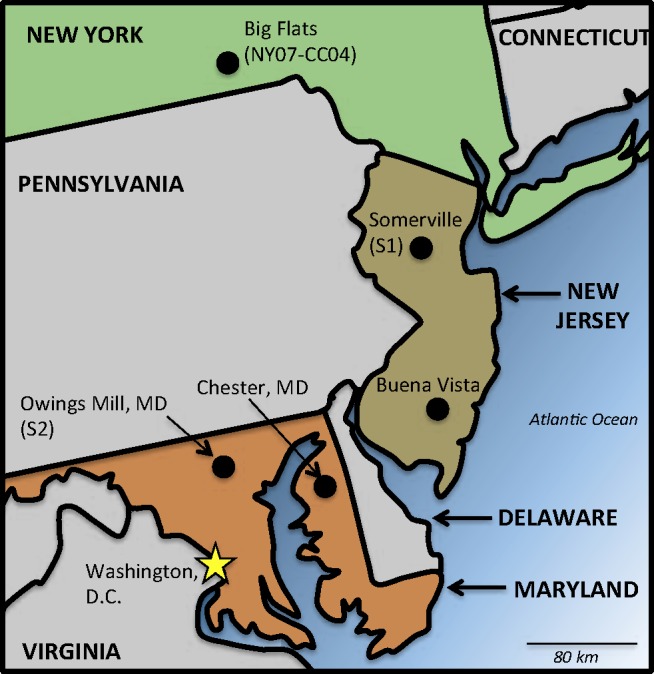
Map of Sorghastrum nutans sampling sites in the mid-Atlantic region of the United States of America. Locations are approximate.
The current ecological interpretation of C. caudatum s.l. as a widely distributed, broad-host range generalist is founded largely on the basis of the shared conidial appendage trait (Nag Raj 1973, Sutton 1980, 1992, Hyde et al. 2009); the fungus has never been systematically examined at the molecular level. Crouch (2009c) found three diverse lineages from four isolates of C. caudatum s.l. isolated from three hosts (2009c), suggesting that even in a small sample, the broad morphospecies concept does not adequately mirror the true diversity of these organisms. Furthermore, the morphological concept of C. caudatum as a broad host range pathogen is not consistent with the evolutionary history of related graminicolous Colletotrichum species, a group that is otherwise characterized by a high degree of species-level lineage diversification corresponding with host origin (Crouch et al. 2006, 2009a, b, c, Moriwaki & Tsukiboshi 2009, Crouch & Tomaso-Peterson 2012). Extended to the genus as a whole, the long standing paradigm of a few widespread, host generalist pathogens is rapidly being supplanted with discoveries of numerous morphologically cryptic species that may be limited to associations with just one or a few hosts (e.g. Damm et al. 2009, Hyde et al. 2009, Shivas & Tan 2009, Phoulivong et al. 2010, Rojas et al. 2010, Weir & Johnston 2010; Cannon et al. 2012, Weir et al. 2012, Liu et al. 2013).
Given the uncertainties surrounding the host range of C. caudatum s.l., designation of a living epitype strain consistent with the lectotype is an important prerequisite for defining this species. Once an epitype strain is circumscribed, questions of potential diversity can be addressed, including the relationship of the fungus to other grass pathogens. Recent phylogenetic studies of the Graminicola aggregate have placed C. caudatum s.l. as a taxon closely related to the grass pathogens C. falcatum and C. sublineola (Moriwaki et al. 2002, Moriwaki & Tsukiboshi 2008, Crouch et al. 2009b, c). However, none of the C. caudatum s.l. isolates included in these analyses were geographically or ecologically consistent with the lectotype of C. caudatum; as all these isolates were collected from outside North America, and on hosts other than indiangrass.
With these questions in mind, the present study was undertaken to: (1) generate fresh collections of C. caudatum s.l. from the type host and the type locale (indiangrass, USA); (2) designate and characterize an ex-epitype strain for C. caudatum; (3) confirm placement of the C. caudatum type within the Graminicola aggregate (Cannon et al. 2012); and (4) survey a collection of C. caudatum s.l. isolates to determine if the broad host range morphospecies concept corresponds with a single phylogenetic species.
MATERIALS AND METHODS
Specimens and cultures
The fungal strains and specimens included in this study are listed in Table 1. The Colletotrichum caudatum lectotype designated by Nag Raj (1973) is a specimen of the fungus infecting Sorghastrum nutans (indiangrass). To obtain a potential epitype specimen with a living culture, the type host, S. nutans was sampled in September 2011 at four locations in the mid-Atlantic region of the USA; one isolate was contributed by Gary Bergstrom from New York State. Fig. 1 shows the location of sampling sites. Two locations were in the mid-Atlantic state of New Jersey: (1) a farm plot of indiangrass in Somerset (central NJ), and (2) a naturalized stand of S. nutans in Buena Vista (southern NJ, adjacent to the type locality of Newfield, NJ). Two naturalized grassland locations were sampled in the mid-Atlantic state of Maryland: (1) restored serpentine grassland trails at the Soldier’s Delight Natural Environment Area, Owings Mills (north-central MD; Fig. 2); and (2) the Grasslands Plantation and naturalized roadside stands of S. nutans near Chestertown (eastern shore, MD). Between 10–27 plants were sampled from each location. Leaf tissue was surface-sterilized through sequential immersion in 2 % (v/v) sodium hypochlorite (15 s), 70 % (v/v) ethanol (15 s), and distilled, autoclaved H2O (30 s). Sections of surface-sterilized tissue (ca. 1–2 cm) were placed on the surface of potato dextrose agar (PDA; Fisher Scientific, Hampton, NH, USA) supplemented with 40 μg/L each of penicillin, ampicillin, gentomycin, and streptomycin, followed by transfers to unamended PDA after the observation of fungal growth. Colletotrichum caudatum s.l. was isolated only from Somerset, NJ and Owings Mill, MD. Representative specimens were deposited in BPI and cultures were deposited in the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands. Additional cultures of C. caudatum were obtained from CBS; NIAS GeneBank, Ibaraki, Japan; and CABI International (IMI), Egham, Surrey.
Table 1.
Colletotrichum strains and specimens from the Graminicola aggregate analyzed in this study.
| Species or species complex | Strain or specimen no.1 | Host substrate | Location | Year |
|---|---|---|---|---|
| C. alcorni | IMI 176617 | Bothriochloa bladhii | Caboolture, Australia | 1972 |
| IMI 1766192 | Imperata cylindrica | Esk, Australia | 1973 | |
| C. baltimorense | SD11 (BPI892771)2 | Sorghastrum nutans | Maryland, USA | 2011 |
| SD2 (BPI892765) | Sorghastrum nutans | Maryland, USA | 2011 | |
| SD3 (BPI892766) | Sorghastrum nutans | Maryland, USA | 2011 | |
| SD6 (BPI892769) | Sorghastrum nutans | Maryland, USA | 2011 | |
| SD7 (BPI892768) | Sorghastrum nutans | Maryland, USA | 2011 | |
| SD9 | Sorghastrum nutans | Maryland, USA | 2011 | |
| C. caudatum | BPI4233393 | Sorghastrum nutans | New Jersey, USA | 1882 |
| NY07-CC04 (CBS 131602; BPI892767)4 | Sorghastrum nutans | New York, USA | 2007 | |
| C. sp. | BPI423330 | Andropogon sp. | Georgia, USA | 1937 |
| BPI423333 | Andropogon gerardi | North Dakota, USA | 1914 | |
| BPI423336 | Andropogon acoparius | Wisconsin, USA | 1954 | |
| BPI423338 | Sorghastrum nutans | Wisconson, USA | 1948 | |
| CBS 113172 | Cymbopogon martinii | India | 2002 | |
| MAFF 305700 | Imperata cylindrica | Tochigi, Japan | 1979 | |
| C. somersetense | JAC 11-10 | Sorghastrum nutans | New Jersey, USA | 2011 |
| JAC 11-11 (CBS 131599; BPI892770)2 | Sorghastrum nutans | New Jersey, USA | 2011 | |
| JAC 11-13 (CBS 131601; BPI892764) | Sorghastrum nutans | New Jersey, USA | 2011 | |
| JAC 11-14 | Sorghastrum nutans | New Jersey, USA | 2011 | |
| JAC 11-15 | Sorghastrum nutans | New Jersey, USA | 2011 | |
| C. zoysia | MAFF 2385732 | Zoysia tenuifolia | Yamaguchi, Japan | 1998 |
| MAFF 238574 | Zoysia tenuifolia | Hyougo, Japan | 1998 | |
| MAFF 238575 | Zoysia tenuifolia | Hiroshima, Japan | 1998 | |
| MAFF 238576 | Zoysia tenuifolia | Yamaguchi, Japan | 1998 | |
| MAFF 238577 | Zoysia tenuifolia | Hiroshima, Japan | 1998 | |
| Other species | ||||
| C. cereale | CA-FUGC11-43 | Poa annua | USA | 2003 |
| KS-20BIG (CBS 129663)4 | Bromus inermis | USA | 2004 | |
| NJ-6340 | Poa annua | USA | 2003 | |
| C. falcatum | CGMCC3.141875 | Saccharum officinarum | Indonesia | 2008 |
| MAFF 305077 | Saccharum officinarum | Japan | 1966 | |
| MAFF 306170 | Saccharum officinarum | Japan | 1991 | |
| MAFF 306299 | Saccharum officinarum | Thailand | 1989 | |
| C. graminicola | M1.001 (CBS 130836)4 | Zea mays | USA | 1978 |
| M5.001 (CBS 130839) | Zea mays | Brazil | 1990 | |
| C. hanaui | MAFF 3054042 | Digitaria ciliaris | Japan | 1975 |
| MAFF 511014 | Digitaria ciliaris | Japan | 1975 | |
| C. jacksonii | MAFF 305439 | Echinochloa esculenta | Japan | 1977 |
| MAFF 3054602 | Echinochloa esculenta | Japan | 1980 | |
| MAFF 511152 | Echinochloa esculenta | Japan | 1977 | |
| MAFF 511328 | Echinochloa esculenta | Japan | 1980 | |
| MAFF 511344 | Echinochloa esculenta | Japan | 1985 | |
| C. navitas | 9032d | Panicum virgatum | USA | 2008 |
| CBS 1250862 | Panicum virgatum | USA | 2008 | |
| C. nicholsonii | MAFF 305391 | Paspalum dilatatum | Japan | 1974 |
| MAFF 305428 | Paspalum dilatatum | Japan | 1977 | |
| MAFF 510916 | Paspalum dilatatum | Japan | 1975 | |
| MAFF 5111152 | Paspalum dilatatum | Japan | − | |
| C. paspali | MAFF 3054032 | Paspalum notatum | Japan | 1977 |
| MAFF 511000 | Paspalum notatum | Japan | 1977 | |
| C. sublineola | MAFF 305360 | Sorghum bicolor | Japan | 1957 |
| MAFF 305361 | Sorghum bicolor | Japan | 1957 | |
| MAFF 510020 | Sorghum bicolor | Japan | 1957 | |
| MAFF 510021 | Sorghum bicolor | Japan | 1957 | |
| S3.001 (CBS 131301)4 | Sorghum bicolor | Burkina Fasso | − | |
1 All herbarium specimens are indicated by the prefix BPI in their name. DNA was extracted from living strains except for case where only an herbarium specimen was available;
2 Ex-holotype strain;
3 Ex-lectotype strain;
4 Ex-epitype strain;
5 Ex-neotype strain.
Fig. 2.
Morphological features of Colletotrichum caudatum and related taxa in the Caudatum sub-aggregate. A–G. C. caudatum ex-epitype strain NY07-CC04 (CBS131602). A. On potato dextrose agar (PDA). B–C. conidia. D. Setae. E–F. Hyphal appressoria. G. Anthracnose lesions on Sorghastrum nutans infected with NY07-CC04; photograph courtesy of Gary Bergstrom. H. C. somersetense JAC 11-10 on PDA. I. C. somersetense JAC 11-13 on PDA. J–P. C. somersetense ex-holotype strain JAC 11-11 (CBS131599). J. On PDA. K. Setae. L–P. Hyphal appressoria. Q. C. alcornii IMI 176617 on PDA. R–U. C. alcornii ex-holotype strain. R. On PDA. S. Conidia. T. Hyphal appressoria. U. Setae. V–W. C. baltimorense ex-holotype strain SD2. V. On PDA. W. Conidia. X. Underside of PDA plates. Left to right: C. somersetense JAC 11-10, C. caudatum NY07-CC04, C. baltimorense SD2. Y–Z. C. baltimorense ex-holotype strain SD2. Y. Hyphal appressoria. Z. Setae. AA–AB. C. baltimorense type locale. AC–AF. C. zoysiae ex-holotype strain MAFF 238573. AC. On PDA. AD. Conidia. AE–AF. Hyphal appressoria. AG–AF. Setae. Bars = 30 μm.
Morphology
Morphological descriptions are based on material mounted in lactic acid, with or without bromophenol blue, and visualized using a Zeiss Axioplan 2 microscope (Carl Zeiss Microimaging, Oberkochen, Germany) using differential interference contrast illumination. Morphological observations of conidia, hyphae and setae from living materials were completed at 5–7 d from PDA cultures grown at 25 °C with a 12 h cool white fluorescent light photoperiod. Appressorium development from vegetative hyphae was inititated as described by Sutton (1968), with the substitution of malt dextrose agar (MP Biomedicals, Solon, OH; 15 % w/v) for potato-carrot agar. Photomicrographs and measurements were generated using Zeiss AxioVision v. 4. 8; all measurements are based on 30 observations. Colony characters were noted after 7 d growth on unsealed plates of PDA incubated at 25 °C under continuous fluorescent light illumination as above. Colony colours were designated according to Rayner (1970).
Genomic DNA isolation, amplification and sequencing
Genomic DNA was extracted from fungal hypae scraped from the surface of PDA cultures using a sterile razor blade. For fungarium materials used for DNA extractions, approximately 5 cm segments of diseased host tissue showing visible signs of Colletotrichum fungus (e.g. setae) were excised and sliced into small fragments using a sterile blade. Cell lysis was performed through homogenization of harvested tissue in an MP FastPrep 24 (MP BioMedicals) in the presence of Q-BioGene Lysing Matrix C (MP Biomedicals) for two cycles of 20s each. DNA was extracted from the homogenate using the Omni Prep DNA Extraction Kit (G-Biosciences, Maryland Heights, MO). Prior to the final precipitation step, DNA extractions from fungarium specimens were treated with the Nucleon resin component from the Nucleon PhytoPure Genomic DNA kit (GE Biosciences, Piscataway, NJ) to bind and remove excess polysaccharides from solution. Final nucleic acid concentrations were assessed using a Nano-Drop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Four DNA sequence markers from three loci were amplified from the DNA of living fungal samples as previously described: (1) the internal transcribed spacer region (ITS; White et al. 1990); (2) portions of the single copy manganese superoxide dismutase (Sod2; Crouch et al. 2006); (3) the 3’ end of the apurinic DNA lyase 2 (Apn2; Crouch et al. 2009c); and (4) the combined 5’ end of the mating type idiomorph MAT1, intergenic DNA and the 5’ end of the Apn2 gene (Mat/Apn2; Crouch et al. 2009c). After agarose gel visualization, amplicon DNA was prepared for Sanger sequencing using ExoSap-It (Affymetrix, Cleveland, OH). From fungarium DNA, a sequencing marker from the Sod2 gene was PCR amplified as previously described using the Sod220F/Sod2226R primers (Crouch & Tomaso-Peterson 2012). Labeled sequence reactions were prepared from amplicons using BigDye 3 Terminator cycle sequencing chemistry (Applied Biosystems, Inc., Carlsbad, CA) and read using an ABI 3130 capillary sequencer (Applied Biosystems Inc.). Sequence reactions were primed from amplicons using the corresponding amplification primers to generate reads in both directions; the resultant sequence reads were edited and assembled using Sequencher v. 4.1 (Gene Codes, Madison, WI). New sequence data was submitted to GenBank under accession numbers JX076857-JX076932.
Phylogenetic analysis
Sequence data from representative taxa in the Graminicola aggregate of Colletotrichum (e.g., those associated with grass host plants; Crouch & Beirn 2009) were included in the phylogenetic analysis, with data from ex-type strains included wherever possible. Sequence data from three representative C. cereale isolates were included as the outgroup taxa to root the tree (Crouch et al. 2009c, O’Connell et al. 2012). Multiple sequence alignments were constructed using the CLC Workbench v. 6.1.1 (CLC Bio, Germantown, MD). Alignments were manually adjusted, with gaps and ambiguously aligned bases eliminated from the datasets using a text editor, then coded as single multi-state characters where positional homology could be assessed. Phylogenetic analysis was performed using PAUP v. 3.0b4 (Swofford 2000) under the maximum parsimony (MP) optimality criterion using heuristic search strategies, 100 random addition sequence replicates and tree bisection reconnection branch swapping. Trees produced from each analysis were used to construct 50 % majority rule trees for each of the single-locus and combined-locus datasets. Bootstrap analyses were run using MP (1000 replicates, 10 random addition sequence replicates). Trees were imported into FigTree v. 1.3.1 (Rambaut 2009) for visualization. Nucleotide identity between taxa was calculated in SplitsTree v. 4.12.3 (www.splitstree.org) using uncorrected P distances.
Network and recombination analysis
Reticulate networks were constructed in SplitsTree v. 4.12.3 (www.splitstree.org) using the split decomposition method to visualize homoplasies, where datasets were partitioned into “splits” using parsimony, then combined consecutively to generate genealogies. Branch support was estimated through 10 000 bootstrap replicates, then used to construct 95 % frequency confidence trees, with the splits transformations generated as cluster networks, and topologies drawn as rectangular reticulate phylograms. Incompatible, contradictory relationships indicative of recombination or homoplasy were plotted in the network as loops, with multiple connections between taxa. Network topologies were rooted with two isolates of the outgroup taxa C. navitas.
The dataset was evaluated for the presence of recombination using the pairwise homoplasy index (PHI) test (Bruen et al. 2006) implemented by SplitsTree, where the minimum number of homoplasies required to describe the genealogical history between sites was calculated. If recombination, rather than recurrent mutation, was responsible for the observed homoplasy, the PHI scores would be lower than expected for recurrent mutation. Statistical significance of PHI scores was estimated by 1 000 random permutation of sites in the dataset while simulating the absence of recombination, then calculating whether the frequency of the non-recombinant permutated PHI score is less than the observed score.
Conflicting signal was assessed across taxa through calculation of delta scores and Q-residual scores (Holland et al. 2002) in SplitsTree. Both are quartet-based analytical distance methods, where data additively is tested, and each taxon is evaluated to determine the extent that it is involved in conflicting signal. Scores are assigned from 0 and 1, where 0 reflects an exact fit to a bifurcating tree (strict additivity, no conflict) and 1 reflects a complete departure from a tree-like dataset (Holland et al. 2002, Gray et al. 2010).
Species recognition
Geneaological concordance phylogenetic species recognition (GCPSR) criteria (Avise & Ball 1990, Taylor et al. 2000, de Queiroz 2007) was used as the null hypothesis for species diagnosis. The following minimum attributes were required to recognize any lineage within the C. caudatum morphospecies as a phylogenetic species: (1) monophyly in the phylogenetic tree inferred from the combined dataset; (2) genealogical concordance; and (3) bootstrap support in the combined dataset phylogeny. Genealogical concordance was assessed by constructing a strict consensus tree topology from the four single-locus 50 % consensus MP trees; clades recovered in the strict consensus tree were recognized as phylogenetic species.
RESULTS
Placement relative to other graminicolous Colletotrichum species
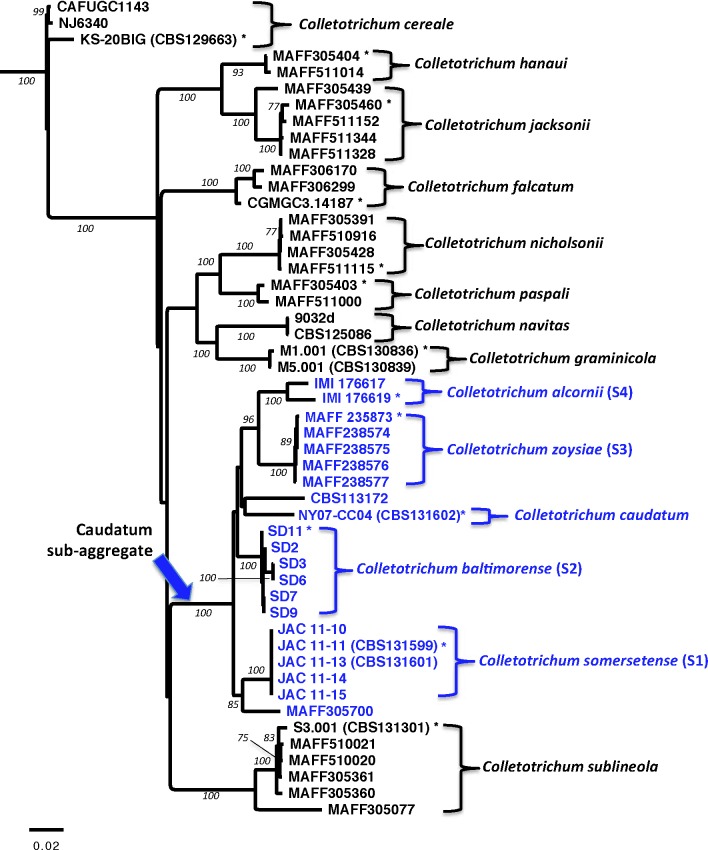
Nucleotide sequences generated using four markers (Apn2, Sod2, ITS and Mat/Apn2) were aligned to produce a 3098 character dataset, which included 3015 nucleotide bases and 83 standard characters manually coded from indels. The dataset included 21 isolates of C. caudatum s.l. and 31 representative strains of other graminicolous Colletotrichum taxa. A summary of the dataset is presented in Table 2. All C. caudatum s.l. isolates clustered together as a monophyletic group in the phylogenetic trees constructed from single- and multi-locus datasets (bootstrap [BS] value of 100). Isolates of C. caudatum s.l. shared between 94.1−100 % nucleotide identity. The 50 % consensus phylogeny generated using the multi-locus dataset is shown in Fig. 3.
Table 2.
Summary of characteristics of the four marker datasets used to conduct molecular phylogenetic analyses. All analyses were run from datasets consisting of the living cultures of Colletotrichum strains, as listed in Table 1.
| General dataset parameters | Apn2 | ITS | Mat1 | Sod2 | All data |
|---|---|---|---|---|---|
| DNA characters | 798 | 401 | 1297 | 519 | 3015 |
| Coded gap characters | 2 | 7 | 61 | 13 | 84 |
| Total aligned characters | 800 | 408 | 1358 | 532 | 3098 |
| Constant characters | 494 | 276 | 533 | 306 | 1609 |
| Parsimony informative characters | 275 | 54 | 691 | 189 | 1209 |
| PHI tests for recombination1 | |||||
| All C. caudatum s.l. isolates | YES | NO | YES | YES | |
| p = 0.0086 | p = 0.2487 | p = 0 | p = 0 | ||
| C. caudatum s.l. isolates, excluding incongruent taxa2 | NO | NO | NO | ||
| YES | p = 1.0 | p = 1.0 | p = 1.0 | ||
| p = 0.0055 | |||||
| Delta score3 | 0.164 | 0.067 | 0.188 | 0.256 | |
| Q-residual score3 | 0.022 | 0.005 | 0.08 | 0.122 | |
1Pairwise homoplasy index (PHI) test performed using a dataset composed of Colletotrichum in the Caudatum subaggregate: C. alcornii, C. baltimorense, C. caudatum, C. somersetense, C. zoysiae, and isolates CBS112172 and MAFF 305700.
2Incongruent taxa = CBS113172 and NY07-CC04.
3Where delta score and Q-residual score=0 when distances between taxa exactly fit a tree. Scores ranging from 0 (tree-like) to 1 (non-tree-like).
Fig. 3.
Phylogenetic tree showing the relationship between Colletotrichum caudatum and other grass-associated species of Colletotrichum estimated through maximum parsimony analysis of a four gene combined nucleotide sequence alignment. Isolates within the Caudatum subaggregate are shown in blue. The tree is outgroup rooted with C. cereale. Bootstrap support values >75 are shown at nodes. * = Ex-type strain.
Identification of phylogenetic species
Phylogenetic analysis of the molecular datasets showed that the C. caudatum s.l. clade was subdivided into four major subgroups that were supported by BS values >75; these groups were designated S1−S4. Single- and multi-locus phylogenetic trees are shown in Fig. 3–4.
Fig. 4.
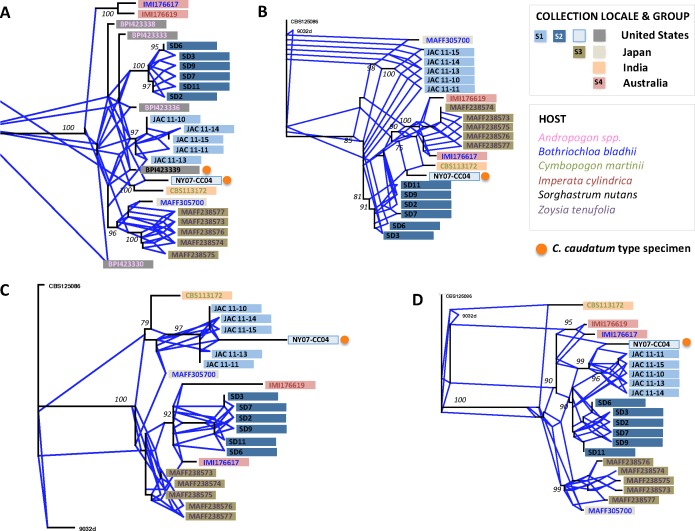
Confidence split decomposition networks (95 %) constructed from Colletotrichum isolates with caudate conidia, and illustrated as rectangular reticulate phylograms. Trees are outgroup rooted using C. navitas isolates CBS125086 and 9032d (not illustrated in all trees). Bootstrap support values >75 are shown at nodes. Reticulating angular branches coloured blue represent alternative, conflicting signal in the dataset. Group designations S1–S4 are given based on clades determined to be genealogically concordant across the four single-locus phylogenies. A. Sod2 dataset, including fungarium specimen sequence data. B. Mat/Apn2 dataset. C. ITS dataset. D. Apn2 dataset.
Groups S1, S2 and S3 were recovered as distinct monophyletic groups in the single- and multi-locus analyses (BS = 90−100). Groups S1 and S2 were populated by indiangrass isolates of C. caudatum s.l. collected at two sampling sites in the mid-Atlantic region of the USA; the S1 and S2 sampling sites were separated by ~270 km (Fig. 1). All five S1 isolates shared 100 % nucleotide identity. S2 isolates shared between 99.5−100 % nucleotide identity, with isolates SD3 and SD6 clustering together within the S2 group. Group S3 was populated by isolates from Z. tenuifolia hosts collected from three Japanese prefectures. S3 isolates shared between 99.5−100 % nucleotide identity; no clustering of the individual S3 group isolates was observed. Groups S1, S2 and S3 were all recovered in the strict consensus tree (Fig. 3). Thus, groups S1, S2 and S3 were genealogically concordant and met the criteria for recognition as phylogenetic species; they are each referred to hereafter as species S1, S2 and S3.
Group S4, composed of two C. caudatum s.l. isolates from Australia that shared 98.1 % nucleotide identity, was recovered as a monophyletic group in the multi-locus analysis (BS = 100), and in two of the four single-locus analyses (Sod2 and Apn2; BS = 95, 100, respectively; Fig. 3). In the ITS tree, the Australian S4 isolates positioned with with S2 isolates from the USA, and in the Mat/Apn2 tree the S4 isolates grouped with the S3 isolates from Japan (Fig. 4). Although the S4 isolates were placed in the context of larger clades in the ITS and Mat/Apn2, they were still members of the same clade, reflecting a lack of signal rather than incongruence. Overall, the strict consensus tree topology supported the grouping of the two S4 isolates as genealogically concordant, and S4 was monophyletic and supported by bootstrap estimates in the multi-locus phylogeny (Fig. 3). Therefore, group S4 met the criteria for recognition as a phylogenetic species and is referred to hereafter as species S4.
In addition to phylogenetic species S1−S4, three single isolate lineages were identified from the multi-locus dataset: NY07-CC04, MAFF305700, CBS112172 (Fig. 4). The placement of these isolates varied across each of the four single-locus phylogenies. Isolate NY07-CC04 collected from indiangrass in the USA was placed as the sister taxa to species S1 (also collected from indiangrass in the USA) in all single-locus phylogenies except Mat/Apn2, where it was positioned closely to species S3 from Japan. Japanese isolate MAFF30570 grouped most closely to species S3 in two of the single-locus trees (Sod2, Apn2), but was positioned near species S1 from the USA in the ITS and Mat/Apn2 trees. Isolate CBS113172 from India was positioned at a basal position in the Apn2 tree, grouped closest to species S3 in the Mat/Apn2 tree, and closest to species S1 in the ITS and Sod2 trees. For NY07-CC04, MAFF305700, and CBS112172, the conflict between individual genealogies was not resolved in the strict consensus tree topology. Each of these isolates was distinct from phylogenetic species S1−S4, and each was distinct from one another (not shown).
Reticulate evolution and recombination
Split decomposition analysis, pairwise homoplasy indices (PHI’s), delta scores and Q-residual scores all provided evidence of conflicting data from the single- and multi-locus datasets. Fig. 4 shows the 95 % confidence networks constructed from the individual loci (Fig. 4) and multi-locus datasets using split decomposition. Multiple branching connections were identified between individual taxa and between species groups in the splits tree topologies. PHI tests substantiated the conflicting data observed in the splits trees for all datasets except the ITS (Table 2, 3), with statistically significant levels of recombination present.
Table 3.
Comparison of delta and Q-residual scores for Colletotrichum taxa in the Caudatum subaggregate across four single-locus and combined nucleotide sequence datasets. Scores range from 0 to 1, where 0=tree-like data (additive) and 1=non-tree-like. Scores that depart from the mean score by >1 standard deviation are highlighted in grey.
| Strain no. | Apn2 | ITS | Mat1/Apn2 | Sod2 | Combined | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Delta score | Q-residual score | Delta score | Q-residual score | Delta score | Q-residual score | Delta score | Q-residual score | Delta score | Q-residual score | |
| IMI 176617 | 0.16 | 0.014 | 0.126 | 0.006 | 0.159 | 0.056 | 0.09 | 0.005 | 0.175 | 0.026 |
| IMI 176619 | 0.178 | 0.016 | 0.146 | 0.01 | 0.202 | 0.057 | 0.112 | 0.007 | 0.213 | 0.029 |
| CBS113172 | 0.251 | 0.045 | 0.152 | 0.017 | 0.349 | 0.149 | 0.139 | 0.004 | 0.355 | 0.071 |
| NY07-CC04 | 0.222 | 0.054 | 0.062 | 0.004 | 0.269 | 0.099 | 0.221 | 0.004 | 0.274 | 0.041 |
| JAC 11-10 | 0.118 | 0.017 | 0.053 | 0.004 | 0.143 | 0.067 | 0.072 | 0.002 | 0.142 | 0.028 |
| JAC 11-11 | 0.118 | 0.017 | 0.053 | 0.004 | 0.143 | 0.067 | 0.072 | 0.002 | 0.142 | 0.028 |
| JAC 11-13 | 0.118 | 0.017 | 0.053 | 0.004 | 0.143 | 0.067 | 0.072 | 0.002 | 0.142 | 0.028 |
| JAC 11-14 | 0.118 | 0.017 | 0.053 | 0.004 | 0.143 | 0.067 | 0.072 | 0.002 | 0.142 | 0.028 |
| JAC 11-15 | 0.118 | 0.017 | 0.053 | 0.004 | 0.143 | 0.067 | 0.072 | 0.002 | 0.142 | 0.028 |
| MAFF238573 | 0.197 | 0.027 | 0.056 | 0.004 | 0.127 | 0.059 | 0.07 | 0.002 | 0.167 | 0.024 |
| MAFF238574 | 0.115 | 0.017 | 0.056 | 0.004 | 0.127 | 0.059 | 0.065 | 0.002 | 0.152 | 0.025 |
| MAFF238575 | 0.115 | 0.017 | 0.056 | 0.004 | 0.127 | 0.059 | 0.065 | 0.002 | 0.152 | 0.025 |
| MAFF238576 | 0.115 | 0.017 | 0.056 | 0.004 | 0.127 | 0.059 | 0.065 | 0.002 | 0.154 | 0.025 |
| MAFF238577 | 0.123 | 0.02 | 0.056 | 0.004 | 0.127 | 0.059 | 0.07 | 0.002 | 0.149 | 0.025 |
| MAFF305700 | 0.131 | 0.016 | 0.088 | 0.009 | 0.2 | 0.075 | 0.072 | 0.003 | 0.271 | 0.053 |
| SD11 | 0.288 | 0.04 | 0.05 | 0.004 | 0.215 | 0.082 | 0.062 | 0.001 | 0.237 | 0.038 |
| SD2 | 0.184 | 0.017 | 0.05 | 0.004 | 0.215 | 0.082 | 0.062 | 0.001 | 0.227 | 0.038 |
| SD3 | 0.184 | 0.017 | 0.05 | 0.004 | 0.283 | 0.147 | 0.062 | 0.001 | 0.231 | 0.047 |
| SD6 | 0.249 | 0.032 | 0.05 | 0.004 | 0.283 | 0.147 | 0.062 | 0.001 | 0.238 | 0.051 |
| SD7 | 0.166 | 0.019 | 0.05 | 0.004 | 0.215 | 0.082 | 0.062 | 0.001 | 0.217 | 0.036 |
| SD9 | 0.166 | 0.019 | 0.05 | 0.004 | 0.215 | 0.082 | 0.062 | 0.001 | 0.217 | 0.036 |
| Mean | 0.164 | 0.022 | 0.067 | 0.005 | 0.188 | 0.08 | 0.081 | 0.002 | 0.197 | 0.035 |
| Standard deviation | 0.053 | 0.011 | 0.032 | 0.003 | 0.065 | 0.03 | 0.037 | 0.001 | 0.058 | 0.012 |
Delta scores and Q-residuals, which provide an assessment of data additivity and tree-like (bifurcating) evolutionary patterns, also showed the ITS and Sod2 datasets to be free of conflicting signal, with scores close to 0 (where zero = additive and 1 = non-tree-like; Table 3). In contrast, delta scores and Q-residual estimates for the Apn2, Sod2 and Mat/Apn2 datasets each exhibited a moderate level of noise, consistent with the PHI evidence for recombination in these datasets.
Delta scores and Q-residuals for C. caudatum s.l. isolates were compared against dataset means to identify individual isolates with disproportionate levels of conflicting signal (Table 3). In all of four of the recombinant datasets, isolate NY07-CC04 had scores that were >1 standard deviation from the mean; in the three of the four recombinant datasets, CBS113172 had scores that were >1 standard deviation from the mean. By removing CBS113172 and NY07-CC04 from the dataset, the recombinant signal was eliminated from all but the Apn2 dataset, indicating that the majority of conflicting data came from these two isolates.
Which of the phylogenetic species is Colletotrichum caudatum sensu stricto?
The identification of four phylogenetic species and three divergent isolates within the broad circumscription of the Colletotrichum caudatum s.l. morphospecies added an unexpected complication to the process of identifying an appropriate sequenced epitype for the species. Twelve candidate epitype strains were collected from three separate sites within 450 km of Newfield, NJ, where the lectotype originated. Each of the three sampling sites yielded C. caudatum s.l. isolates from indiangrass bearing a unique multi-locus molecular signature. Colletotrichum isolates collected from Somerset, NJ were members of phylogenetic species group S1; isolates collected from Owings Mills, MD were members of phylogenetic species group S2; and isolate NY07-CC04 collected from Big Flats, NY was recovered as a single taxon lineage with connections to species group S1 (Fig. 4).
The twelve candidate C. caudatum epitype strains were morphologically indistinguishable from one another, but did exhibit variation in the production of pigment on PDA media. Species S1 did not produce a visually discernable pigment, species S2 produced a sienna orange pigment, and isolate NY07-CC04 produced a luteous yellow pigment. However, since the C. caudatum protologue did not provide cultural details, this character was not useful for determination of a suitable epitype strain from among the twelve epitype candidates.
Molecular phylogenetic analysis of the 246-bp Sod2 marker sequenced from the C. caudatum lectotype (BPI423339) and four additional fungarium specimens was conducted to determine if any of the candidate epitype isolates were consistent with the type on the molecular level. Attempts were made to analyze two additional short marker regions nested within the Apn2 gene as described for the C. sublineola lectotype (Crouch & Tomaso-Peterson 2012), but PCR amplifications were unsuccessful even after several attempts to optimize the reaction conditions (data not shown). Sod2 sequences of the C. caudatum lectotype shared 98.8 % identity with isolate NY07-CC04 and members of phylogenetic species S1. Within the Sod2 gene tree, BPI42339 was positioned in the tree topology between species S1 and NY07-CC04, clustering most closely with NY007-CC04. NY07-CC04 has been previously confirmed as a pathogen of indiangrass, consistent with the type diagnosis (Waxman & Bergstrom 2011). In contrast, Koch’s postulates have not been fulfilled for the members of phylogenetic species S1; these isolates may not be pathogens of the host plant. Based on nucleotide similarities, sister taxon relationship, and shared host and geographic origin, NY07-CC04 is designated as the epitype strain for C. caudatum s. str. here.
TAXONOMY
Colletotrichum caudatum (Peck ex Sacc.) Peck, Bull. N.Y. State Mus. 131: 81 (1909).
MycoBank MB152590
Etymology: Named for the caudate conidial appendage.
Basionym: Ellisiella caudata Sacc., Michelia 2: 147 (1880); as “Ellisiella caudata (Peck) Sacc.”
Synonym: Ellisiellina caudata (Sacc.) Camara, Agron. Lusitana 11: 72 (1949); as “Ellisiellina caudata (Peck) Sacc.”
Type: USA: New Jersey: Newfield, on the leaves of Sorghastrum nutans, 1882, J. B. Ellis [S. M. Tracy Herb.] (BPI 423339 – lectotype designated by Nag Raj 1973); New York: Big Flats, Chemung Co., on the leaves of Sorghastrum nutans ‘Rumsey’, 2007, G. C. Bergstom (BPI 892767– epitype designated here, MBT177387; CBS 131602–ex-epitype culture, = NY07-CC04 (Waxman & Bergstrom 2011)).
Description: Setae septate with 4–5(–6) smooth walled cells, dark chestnut to black, tapering to an acute apex, septate, straight (–bent), usually emerging from darkened hyphae, 70–145(–285) μm long, 6–10.5 μm wide at the base, 1.0–3.5 μm wide at the apex. Conidia enteroblastic, falcate, unicellular, hyaline, smooth walled, apex reduced into a filiform appendage 7.0–15.0 μm long, curved (–straight), 30.0–56.0 μm long (including appendages), 4.0–5.5 μm wide. Hyphal appressoria fuscous black to black, unicellular, globose, ovoid or obovoid or clavate, mostly smooth but sometimes or lobate, apice cylindrical or obtuse, edges entire, (7.0–)12.5–24.0 × 6.0–14.5 μm.
Cultures: Colonies on PDA at 25 °C with sparse mycelium, umber to ochreous surrounded with a luteous yellow pigment diffusing into agar, reverse umber to ochreous surrounded with luteous yellow pigment.
Habitat & Distribution: On Sorghastrum nutans and known only from the eastern United States.
Notes: Morphologically similar to Colletotrichum alcornii, C. baltimorense, C. somersetense, and C. zoysiae, but differs in nucleotide polymorphisms at Apn2, ITS, Sod2, Mat/Apn2 DNA sequences. This amended description of C. caudatum marks a return to the original concept of the species given by Saccardo (1880), with the known distribution of the fungus limited to Sorghastrum nutans in the mid-Atlantic states of the USA. Several authors have treated any Colletotrichum bearing a conidial appendage as C. caudatum s.l., regardless of host (e.g. Nag Raj 1973, Sutton 1980, 1992, Moriwaki et al. 2002, Fuke et al. 2006, Moriwaki & Tsukiboshi 2008, Crouch & Beirn 2009, Crouch et al. 2009b, Hyde et al. 2009), but phylogenetic data do not support such a classification.
Ramakrishnan & Ramakrishnan (1947) described C. ciliatum from Cymbopogon polyneuros in India as a distinct species because of the filiform conidial appendage. Subsequent authors concluded that C. ciliatum was a synonym of C. caudatum as both taxa produced caudate conidia (Arx 1957, Agnihothrudu 1966, Nag Raj 1973, Sutton 1980, 1992). Since the conidial appendage was shown to be an unreliable character in this work, the status of C. ciliatum as a unique taxon is currently unresolved.
Colletotrichum alcornii J. A. Crouch, sp. nov.
MycoBank MB800276
Etymology: Named for the mycologist who first collected the species, J. L. Alcorn.
Diagnosis: Morphologically similar to C. caudatum, but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Type: Australia: Queensland: Caboolture, on leaves of Imperata cylindrica var. major, 26 March 1973, J. L. Alcorn (IMI 176619 – holotype).
Description: Setae septate, with 4–5(–7) smooth walled cells, dark chestnut to black, tapering to an acute apex, septate, straight (–bent), usually emerging from darkened hyphae, 67–150 μm long, 6.0–10.5 μm wide at the base, 1.0–3.0 μm wide at the apex. Conidia formed enteroblastically, falcate, unicellular, hyaline, smooth walled, apex reduced into a filiform appendage 2.0–6.0 μm long, curved (–straight), (–16.5)29.5–39.5 μm long (including appendages), 4.0–5.5 μm wide. Hyphal appressoria fuscous black to black, unicellular, globose, ovoid or obovoid or clavate, mostly smooth but sometimes or lobate, apice cylindrical or obtuse, edges entire, 15.0–18.0 × 14.5–18.0 μm.
Cultures: Colonies on PDA grown at 25 °C with sparse mycelium, light orange to salmon.
Habitat & Distribution: Known from Imperata cylindrica and Bothriochloa bladhii in Australia.
Notes: Morphologically similar to Colletotricum caudatum, C. baltimorense, C. somersetense, and C. zoysiae but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Additional specimen examined: Australia: Queensland: Esk, on leaves of Bothriochloa bladhii, 13 December 1972, J. L. Alcorn (IMI 176617).
Colletotrichum baltimorense J. A. Crouch, sp. nov.
MycoBank MB807151
Etymology: Named for the location where the fungus was collected, Baltimore County, Maryland, USA.
Diagnosis: Morphologically similar to C. alcornii, C. caudatum, C. somersetense, and C. zoysiae but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Type: USA: Maryland: Baltimore County, Owings Mills, Natural Environment Area, on leaves of Sorghastrum nutans, 12 September 2011, J. A. Crouch (BPI I1892771 – holotype; SD-11 – ex-holotype culture).
Description: Setae septate with 4–5(–6) smooth walled cells, dark chestnut to black, tapering to an acute apex, septate, straight (–bent), usually emerging from darkened hyphae, 78.5–155.0(–202.0) μm long, 6.0–10.5 μm wide at the base, 0.9–3.0 μm wide at the apex. Conidia enteroblastic, falcate, unicellular, hyaline, smooth walled, apex reduced into a filiform appendage 7.5–10.0 μm long, curved (–straight), 32.5-57.5 μm long (including appendages), 4.0–5.5 μm wide. Hyphal appressoria fuscous black to black, unicellular, globose, ovoid or obovoid or clavate, mostly smooth but sometimes or lobate, apice cylindrical or obtuse, edges entire, 15.0 – 20.0 × 14.5–17.5 μm.
Cultures: Colonies on PDA at 25 °C with sparse white mycelium towards the centre, light orange to salmon, reverse orange to salmon.
Habitat & Distribution: Known only from Sorghastrum nutans in Maryland, USA.
Notes: Morphologically similar to Colletotrichum alcornii, C. caudatum, C. somersetense, and C. zoysiae, but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Additional specimens examined: USA: Maryland: on leaves of Sorghastrum nutans, 12 Sept. 2011, J. A. Crouch (BPI 892765); on leaves of S. nutans, 12 Sept. 2011, J. A. Crouch (BPI 892766); on leaves of S. nutans, 12 Sept 2011, J. A. Crouch (BPI 892768); on leaves of S. nutans, 12 Sept. 2011, J. A. Crouch (BPI 892769).
Colletotrichum somersetense J. A. Crouch, sp nov.
MycoBank MB807152
Etymology: Named for the location where the fungus was collected, Somerset County, New Jersey.
Diagnosis: Morphologically similar to C. alcornii, but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Type: USA: New Jersey: Somerset County, Somerset, on the leaves of Sorghastrum nutans, 23 Sept. 2011, J. A. Crouch (BPI 892770 – holotype; CBS131599 – ex-holotype culture, also known as JAC 11-11).
Description: Setae septate with 4–5(–8) smooth walled cells, dark chestnut to black, tapering to an acute apex, septate, straight (–bent), usually emerging from darkened hyphae, 70–142 μm long, 6.0–11.0 μm wide at the base, 1.0–2.5 μm wide at the apex. Conidia enteroblastic, falcate, unicellular, hyaline, smooth walled, apex reduced into a filiform appendage 8.0–12.5 μm long, curved (–straight), 30.5–54.5(–60.5) μm long (including appendages), 4.0–5.0 μm wide. Hyphal appressoria fuscous black to black, unicellular, globose, ovoid or obovoid or clavate, mostly smooth but sometimes or lobate, apice cylindrical or obtuse, edges entire, (6.0–)13.0–23.0 × 7.5–17.0 μm.
Culture characteristics: Colonies on PDA grown under full light at 25 °C with sparse mycelium, with salmon pigmentation overlaid with parallel rows of clustered black setae.
Habitat & Distribution: On leaves of Sorghastrum nutans. Known only from Maryland, USA.
Notes: Morphologically similar to Colletotrichum alcornii, C. caudatum, C. baltimorense, and C. zoysiae but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Additional specimen examined: USA: New Jersey: Somerset County, Somerset, on the leaves of Sorghastrum nutans, 23 Sept. 2011, J. A. Crouch (BPI 892764; CBS131601; also known as JAC 11-13).
Colletotrichum zoysiae J. A. Crouch, sp. nov.
MycoBank MB800278
Etymology: Named for the host plant genus.
Diagnosis: Morphologically similar to C. alcornii, C. baltimorense, C. caudatum, and C. somersetense, but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Type: Japan: Yamaguchi: on leaves of Zoysia tenuifolia, May 1998, A. Tanaka (BPI 884090–holotype; NIAS Genebank, = MAFF238573 – ex-holotype culture).
Description: Setae septate with 4–6(–7) smooth walled cells, dark brown, tapering to an acute apex, septate, straight (–bent), usually emerging from darkened hyphae, (52–)77–135 μm long, 6.0–9.5 μm wide at the base, 1.0–3.0 μm wide at the apex. Conidia enteroblastic, falcate, unicellular, hyaline, smooth walled, apex reduced into a filiform appendage (1.0–)6.0–8.0 μm long, curved (–straight), 25.0–47.5 μm long (including appendages), 4.0–6.5 μm wide. Hyphal appressoria fuscous black to black, unicellular, elongate or rounded or irregular or lobate or multi-lobate, apice cylindrical or obtuse, edges entire, 17.0–22.5(–26.0) × 10.0–14.5 μm.
Culture characteristics: Colonies on PDA grown under full light at 25 °C with raised mycelium without visible conidia, mycelium mouse grey to pale mouse grey to pale olivaceous grey at the colony edge.
Habitat & Distribution: On leaves of Zoysia tenuifolia. Known only from Japan.
Notes: Morphologically similar to Colletotrichum alcornii, C. baltimorense, C. caudatum, and C. somersetense, but differing in nucleotide polymorphisms at Apn2, ITS, Sod2, and Mat/Apn2.
Additional specimens examined: Japan: Yamaguchi: on leaves of Zoysia tenuifolia, May 1998, A. Tanaka (MAFF238576). Hyougo: on leaves of Zoysia tenuifolia, May 1998, A. Tanaka (MAFF238574). Hiroshima: on leaves of Zoysia tenuifolia, May 1998, A. Tanaka (MAFF238575); on leaves of Zoysia tenuifolia, May 1998, A. Tanaka (MAFF238577).
DISCUSSION
Despite the morphological similarity, Colletotrichum caudatum s.l. was found to encompass several distinct molecular phylogenetic lineages. Four of these lineages were diagnosable through strict GCPSR criteria as phylogenetic species. Although the relatively opportunistic sampling of C. caudatum in this study limits an overly broad interpretation of the results, some longstanding assumptions about these organisms are brought into question by these data. In particular, the concept of C. caudatum s.l. as a morphologically definable fungus that inhabits numerous warm-season grasses is not supported by the observed patterns of lineage diversification.
Following the informal Colletotrichum aggregate naming convention introduced by Cannon et al. (2012) to facilitate communication about new taxonomy in the genus, whereby monophyletic groups of closely related species are referred to by the name of the most well-known member (e.g. Gloeosporioides aggregate, Graminicola aggregate, Acutatum aggregate), the Caudatum subaggregate is proposed to collectively refer to C. alcornii, C. baltimorense, C. caudatum s. str., C. somersetense, C. zoysiae, and other closely related lineages. The Caudatum subaggregate is a member of the Graminicola aggregate, a monophyletic group of species associated with grass hosts and possessing falcate conidia. The Caudatum subaggregate is united through shared ancestry, and the posession of caudate conidia – a morphological novelty in the genus Colletotrichum.
Colletotrichum strains in the Caudatum subaggregate from indiangrass, Zoysia tenuifolia, Imperata cylindrica, Bothriochloa bladhii, Cymbopogon martinii, and Andropogon sp. all clustered into separate lineages. None of the isolates from hosts other than indiangrass clustered with the lectotype in the Sod2 genealogy. Clustering of isolates also corresponded with the sampling locale, both on the continental (North America, Japan, Australia, India) and the regional (Maryland, New Jersey, New York) scale. One of the most striking observations from this study was the differentiation of indiangrass isolates within the C. caudatum complex into three lineages corresponding to three separate sampling sites located within a 400 km radius. All three sites were maintained monocultured stands of indiangrass: the New York and New Jersey sites were established for biomass agricultural production on farmland, and the Maryland site was established as a serpentine grassland restoration on dry, nutrient poor soil (Fig. 1). Given the sample size limitations and the overlapping patterns of lineage differentiation that corresponded with host and geographic origin, it is premature to draw any conclusions about the correlation of such factors with the fungal genotype. Additional study is required to address the issue of ecological or geographic specialization within this complex.
Acknowledgments
Ed Ismael provided technical support that is greatly appreciated. We thank the U.S. National Fungus Collections (BPI) for the loan of fungarium specimens, Amy Y. Rossman for comments on the manuscript, and Gary Bergstrom and Katie Waxman for sharing isolates of C. caudatum. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
REFERENCES
- Agnihothrudu V. (1966) A note on the occurence of Ellisiella caudata (Peck) da Camara and its synonmy. Mycopathologia et Mycologia Applicata 28: 193–195 [Google Scholar]
- Avise JC, Ball RM. (1990) Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Survey of Evolutionary Biology 7: 45–67 [Google Scholar]
- Bruen TC, Philippe H, Bryant D. (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon PF, Damm U, Johnston PR, Weir BS. (2012) Colletotrichum – current status and future directions. Studies in Mycology 15: 181–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch JA, Beirn LA. (2009) Anthracnose of cereals and grasses. Fungal Diversity 39: 19–44 [Google Scholar]
- Crouch JA, Beirn LA, Cortese LA, Bonos SB, Clarke BB. (2009a) Anthracnose disease of switchgrass caused by the novel fungal species Colletotrichum navitas. Mycological Research 113: 1411–1421 [DOI] [PubMed] [Google Scholar]
- Crouch JA, Clarke BB, Hillman BI. (2006) Unraveling evolutionary relationships among the divergent lineages of Colletotrichum causing anthracnose disease in turfgrass and corn. Phytopathology 96: 46–60 [DOI] [PubMed] [Google Scholar]
- Crouch JA, Clarke BB, White JFJ, Hillman BI. (2009b) Systematic analysis of the falcate-spored graminicolous Colletotrichum and a description of six new species of the fungus from warm season grasses. Mycologia 101: 717–732 [DOI] [PubMed] [Google Scholar]
- Crouch JA, Tomaso-Peterson M. (2012) Anthracnose disease of centipedegrass turf caused by Colletotrichum eremochloae, a new fungal species closely related to Colletotrichum sublineola. Mycologia 104:1085–1096 [DOI] [PubMed] [Google Scholar]
- Crouch JA, Tredway LP, Clarke BB, Hillman BI. (2009c) Phylogenetic and population genetic divergence correspond with habitat for the pathogen Colletotrichum cereale and allied taxa across diverse grass communities. Molecular Ecology 18: 123–135 [DOI] [PubMed] [Google Scholar]
- Damm U, Woudenberg JHC, Cannon PF, Crous PW. (2009) Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 39: 45–87 [Google Scholar]
- de Queiroz K. (2007) Species concepts and species delimitation. Systematic Biology 56: 879–886 [DOI] [PubMed] [Google Scholar]
- Farr DL, Rossman AY. (2012) Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. http://nt.ars-grin.gov/fungaldatabases/fungushost/fungushost.cfm. [Google Scholar]
- Fuke K, Hozumi N, Enami Y, Matsuura K, Tajimi A, Fuke K, Hozumi N, Enami Y, Matsuura K, Tajimi A. (2006) Anthracnose of centipede grass caused by Colletotrichum caudatum. Journal of General Plant Pathology 72: 74–75 [Google Scholar]
- Gray RD, Bryant D, Greenhill SJ. (2010) On the shape and fabric of human history. Philosophical Transactions of the Royal Society of London, Series B-Biological Sciences 365: 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland BR, Huber KT, Dress A, Moulton V. (2002) δ Plots: a tool for analyzing phylogenetic distance data. Molecular Biology and Evolution 19: 2051–2059 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Cai L, Cannon PF, Crouch JA, Crous PW, Damm U, Goodwin PH, Chen H, Johnston PR, Jones EBG, Liu ZY, McKenzie EHC, Moriwaki J, Noireung P, Pennycook SR, Pfenning LH, Prihastuti H, Sato T, Shivas RG, Tan YP, Taylor PWJ, Weir BS, Yang YL, Zhang JZ. (2009) Colletotrichum – names in current use. Fungal Diversity 39: 147–183 [Google Scholar]
- Liu F, Cai L, Crous PW, Damm U. (2013) Circumscription of the anthracnose pathogens Colletotrichum lindemuthianum and C. nigrum. Mycologia 105: 844–860 [DOI] [PubMed] [Google Scholar]
- Moriwaki J, Tsukiboshi T. (2008) Colletotrichum echinichloae, a new species on Japanese barnyard millet (Echinochloa utilis). Mycoscience 50: 273–280 [Google Scholar]
- Moriwaki J, Tsukiboshi T, Sato T. (2002) Grouping of Colletotrichum species in Japan based on rDNA sequences. Journal of General Plant Pathology 68: 307–320 [Google Scholar]
- Nag Raj TR. (1973) Genera coelomycetum. X. Ellisiella, Samukuta and Sakireeta. Canadian Journal of Botany 51: 2463–2472 [Google Scholar]
- O’Connell R, Thon M, Hacquard S, van Themaat E, Amyotte S, Kleeman J, Torres-Quintero M, Damm U, Buiate E, Epstein L, Alkan N, Altmuller J, Alvarado-Balderrama L, Bauser C, Becker C, Birren B, Chen Z, Crouch JA, Duvick J, Farman M, Gan P, Heiman D, Henrissat B, Howard R, Kabbage M, Koch C, Kubo Y, Law A, Lebrun M-H, Lee Y, Miyara I, Moore N, Neumann U, Pannaccione D, Panstruga R, Place M, Proctor R, Prusky D, Rech G, Reinhardt R, Rollins J, Rounsley S, Schardl C, Schwartz D, Shenoy N, Shirasu K, Stuber K, Sukno S, Sweigard J, Takano Y, Takahara H, van der Does H, Voll L, Will I, Young S, Zeng Q, Zhang J, Zhou S, Dickman M, Schulze-Lefert P, Ma L, Vaillancourt L. (2012) Life-style transitions in plant pathogenic Colletotrichum fungi defined by genome and transcriptome analyses. Nature Genetics 44: 1060-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoulivong S, Cai L, Chen H, McKenzie EHC, Abdelsalam K, Chukeatirote E, Hyde KD. (2010) Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity 44: 33–43 [Google Scholar]
- Ramakrishnan TS, Ramakrishnan K. (1947) Additions to the fungi of Madras-II. Proceedings of the Indian Academy of Sciences, Section B 25: 178–187 [Google Scholar]
- Rambaut A. (2009) FigTree v. 1.3.1. Computer program and documentation provided by the author at http://tree.bio.ed.ac.uk/software/ [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Kew: Commonwealth Mycological Institute; [Google Scholar]
- Rojas EI, Rehner SA, Samuels GJ, VanBael SA, Herre EA, Cannon PF, Chen R, Pang J, Wang R, Zhang Y, Peng Y-Q, Sha T. (2010) Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: Multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102: 1318–1338 [DOI] [PubMed] [Google Scholar]
- Saccardo PA. (1880) Fungorum extra-europaerum pugillus. Michelia 2: 147 [Google Scholar]
- Shivas RG, Tan YP. (2009) A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Diversity 39: 111–122 [Google Scholar]
- Sutton BC. (1968) The appressoria of Colletotrichum graminicola and C. falcatum. Canadian Journal of Botany 46: 873–876 [Google Scholar]
- Sutton BC. (1980) The Coelomycetes. Kew: Commonwealth Mycological Institute; [Google Scholar]
- Sutton BC. (1992) The genus Glomerella and its anamorph Colletotrichum. In: Bailey JA, Jeger MJ. (eds), Colletotrichum: Biology, pathology and control: 1–26 CAB International, Wallingford, England: [Google Scholar]
- Swofford DL. (2000) PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA, USA: [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32 [DOI] [PubMed] [Google Scholar]
- von Arx JA. (1957) Die Arten der Gattung Colletotrichum Cda. Phytopathologische Zeitschrift 29: 413–468 [Google Scholar]
- Waxman K, Bergstrom GC. (2011) First report of anthracnose caused by Colletotrichum caudatum on indiangrass in New York. Plant Disease 95: 1189 [DOI] [PubMed] [Google Scholar]
- Weir BS, Johnston PR. (2010) Characterisation and neotypification of Gloeosporium kaki Hori as Colletotrichum horii nom. nov. Mycotaxon 111: 209–219 [Google Scholar]
- Weir BS, Johnston PR, Damm U. (2012) The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: A guide to methods and applications 315–322 Academic Press, San Diego, California, USA: [Google Scholar]
- Zeiders KE. (1987) Leaf spot of indiangrass caused by Colletotrichum caudatum. Plant Disease 71: 348–350 [Google Scholar]