Abstract
Thirty-eight Phaeoacremonium isolates collected from pruning wounds of tropical sandalwood in Western Australia were studied with morphological and cultural characteristics as well as phylogenetic analyses of combined DNA sequences of the actin and β-tubulin genes. Three known Phaeoacremonium species were found, namely P. alvesii, P. parasiticum, and P. venezuelense. Phaeoacremonium venezuelense represents a new record for Australia. Two new species are described: P. luteum sp. nov. can be identified by the ability to produce yellow pigment on MEA, PDA, and OA, the predominance of subcylindrical to subulate type II phialides, and the mycelium showing prominent exudate droplets observed as warts; and P. santali sp. nov. which can be separated from other species producing pink colonies on MEA by the predominance of type I and II phialides, the distinct brownish olive colonies in OA, and slow growth.
Keywords: actin, β-tubulin, DNA phylogeny, Santalum, systematics, Togninia
INTRODUCTION
Tropical Sandalwood (Santalum album) is one of the world’s most valuable tropical tree species and demand has led to the overexploitation of natural sandalwood stands, in Timor from which it originates, and in India where it became naturalized over 2000 yr ago (Harbaugh & Baldwin 2007). To combat this destruction, plantations have been established in various countries including Australia, with commercial plantations established in 1999. The Ord River Irrigation Scheme near Kununurra has a Tropical sandalwood plantation estate presently occupying approximately 5 000 ha.
Sandalwood is a slow growing root hemi-parasite. The valuable sandalwood oil has been reported to start developing in the heartwood from 5 yr onwards. Plantation diseases usually only become a risk factor in plantation systems after two to three generations. However, disease risk in tropical plantation systems can become an issue in the first generation (Barry 2002, Barbour et al. 2010).
There have been few reports of fungal diseases infecting the Tropical sandalwood. Reports have been made of the presence of Phytophthora cinnamomi and in the early 1990s, an isolation of Ganoderma steyartanum was undertaken from host species, but not from Tropical sandalwood (Len Nelson pers. comm.). More recently Rural Industries Research and Development Corporation (RIRDC) supported an investigation by Barbour et al. (2010) into the identification of the heartwood rot in Tropical sandalwood and the impact on oil levels within the heartwood. Several rot fungi were isolated and sections suggested that the fungi were entering the branches and main stem via wounds made during pruning or when branches are damaged. The immediate response to this knowledge was to establish a pruning trial examining the effect of tree age and timing (season) of pruning on infection development. The pruned trees were destructively harvested after 6 and 12 mo, and over 70 endophytes, canker, and rot fungi recovered. Among these fungi were several Phaeoacremonium isolates.
The genus Phaeoacremonium was established by Crous et al. (1996), and 40 species have been described so far (Crous et al. 1996, Dupont et al. 2000, Groenewald et al. 2001, Mostert et al. 2005, 2006, Damm et al. 2008, Essakhi et al. 2008, Graham et al. 2009, Gramaje et al. 2009, 2012, Úrbez-Torres et al. 2014), including three species originally described as Togninia species with Phaeoacremonium asexual morphs: T. africana and T. griseo-olivacea (Damm et al. 2008), and T. vibratilis (Réblová & Mostert 2007). Several species of this genus have been studied intensively because of the involvement of these taxa in two complex fungal diseases of grapevine, namely Petri disease in young vines and esca disease in adult vines (Mostert et al. 2006), as well as with human infections, so-called phaeohyphomycoses (Mostert et al. 2005). However, numerous species of Phaeoacremonium have also been associated with disease symptoms of a number of woody hosts other than grapevine worldwide (Table 1).
Table 1.
The described Phaeoacremonium (and Togninia sexual morphs) species known from soil and host plants other than Vitis vinifera, and their worldwide distribution. * = epithets not yet transferred to Phaeoacremonium.
| Phaeoacremonium species | Host/Substrate | Country and Reference |
|---|---|---|
| P. aleophilum (T. minima) | Actinidia chinensis | Italy (Crous & Gams 2000) |
| Malus domestica | Iran (Arzanlou et al. 2014); South Africa (Cloete et al. 2011) | |
| Olea europea | Italy (Crous & Gams 2000); USA (Úrbez-Torres et al. 2013) | |
| Phoenix dactylifera | Iran (Mohammadi 2014) | |
| Prunus armeniaca | Iran (Arzanlou et al. 2014); South Africa (Damm et al. 2008) | |
| Prunus persica | South Africa (Damm et al. 2008) | |
| Prunus salicina | South Africa (Damm et al. 2008) | |
| Prunus pennsylvanica | USA (Hausner et al. 1992) | |
| Pyrus communis | South Africa (Cloete et al. 2011) | |
| Salix sp. | USA (Hausner et al. 1992) | |
| Soil | Spain (Agustí-Brisach et al. 2013); USA (Rooney et al. 2001) | |
| P. alvesii | Dodonaea viscosa | Australia (Mostert et al. 2005) |
| Olea europea | Italy (Nigro et al. 2013) | |
| P. amygdalinum | Prunus dulcis | Spain (Gramaje et al. 2012) |
| P. argentinense (T. argentinensis) | Soil | Argentina (Crous & Gams, 2000) |
| P. australiense | Prunus salicina | South Africa (Damm et al. 2008) |
| P. fuscum | Prunus salicina | South Africa (Damm et al. 2008) |
| P. griseorubrum | Prunus salicina | South Africa (Damm et al. 2008) |
| P. inflatipes | Hypoxylon truncatum | USA (Mostert et al. 2005) |
| Nectandra sp | Costa Rica (Groenewald et al. 2001) | |
| Quercus virginiana | USA (Groenewald et al. 2001) | |
| Soil | USA (Rooney et al. 2001) | |
| P. iranianum | Actinidia chinensis | Italy (Mostert et al. 2006) |
| Malus domestica | Iran (Arzanlou et al. 2013) | |
| Prunus armeniaca | South Africa (Damm et al. 2008) | |
| Prunus dulcis | Spain (Gramaje et al. 2012) | |
| Pyrus communis | South Africa (Cloete et al. 2011) | |
| P. mortoniae (T. fraxinopennsylvanica) | Actinidia chinensis | Italy (Prodi et al. 2008) |
| Fraxinus excelsior | Sweden (Groenewald et al. 2001) | |
| Fraxinus latifolia | USA (Eskalen et al. 2005) | |
| Fraxinus pennsylvanica | USA (Hausner et al. 1992) | |
| Prunus salicina | South Africa (Damm et al. 2008) | |
| Pyrus communis | South Africa (Cloete et al. 2011) | |
| Quercus agrifolia | USA (Lynch et al. 2013) | |
| P. novae-zealandiae (T. novae-zealandiae) | Cupressus macrocarpa | New Zealand (Hausner et al. 1992) |
| Pinus radiata | New Zealand (Hausner et al. 1992) | |
| P. parasiticum (T. parasitica) | Actinidia chinensis | Italy (Di Marco et al. 2004) |
| Aquilaria agallocha | n.d. (Mostert et al. 2006) | |
| Cupressus sp. | n.d. (Mostert et al. 2006) | |
| Nectandra sp. | Costa Rica (Hawksworth et al. 1976) | |
| Olea europea | Italy (Nigro et al. 2013) | |
| Phoenix dactylifera | Iran (Mohammadi 2014); Iraq (Hawksworth et al. 1976) | |
| Prunus armeniaca | South Africa (Damm et al. 2008); Tunisia (Hawksworth et al. 1976) | |
| Prunus avium | Greece (Rumbos 1986) | |
| Quercus virginiana | USA (Halliwell 1966) | |
| Soil | Tahiti (Dupont et al. 2002); Spain (Agustí-Brisach et al. 2013) | |
| P. pallidum | Prunus armeniaca | South Africa (Damm et al. 2008) |
| P. pruniculum | Prunus salicina | South Africa (Damm et al. 2008) |
| P. rubrigenum (T. rubrigena) | Dactylis glomerata | Spain (Sánchez-Márquez et al. 2007) |
| Olea europea | Italy (Nigro et al. 2013) | |
| P. scolyti | Prunus armeniaca | South Africa (Damm et al. 2008) |
| Prunus persica | South Africa (Damm et al. 2008) | |
| Prunus persica var. nucipersica | South Africa (Damm et al. 2008) | |
| Prunus salicina | South Africa (Damm et al. 2008) | |
| P. subulatum | Prunus armeniaca | South Africa (Damm et al. 2008) |
| P. theobromatis | Theobroma gileri | Equador (Mostert et al. 2006) |
| P. venezuelense | Prunus armeniaca | Spain (Olmo et al. 2014) |
| P. viticola (T. viticola) | Actinidia chinensis | France (Hennion et al. 2001) |
| Prunus armeniaca | South Africa (Damm et al. 2008) | |
| Prunus salicina | South Africa (Damm et al. 2008) | |
| Pyrus communis | South Africa (Cloete et al. 2011) | |
| Sorbus intermedia | Germany (Mostert et al. 2006) | |
| T. africana * | Prunus armeniaca | South Africa (Damm et al. 2008) |
| T. griseo-olivacea * | Prunus armeniaca | South Africa (Damm et al. 208) |
| T. vibratilis * | Fagus sylvatica | France and Hungary (Réblová & Mostert 2007) |
| Prunus padus | Sweden (Réblová & Mostert 2007) | |
| Sorbus sp. | Italy (Réblová & Mostert 2007) |
n.d. = no data.
The aim of the present study was to identify isolates of Phaeoacremonium collected from pruning wounds of sandalwood trees and to characterise those that appeared to be morphologically and genetically different from known species of the genus.
MATERIAL AND METHODS
Sampling and fungal isolation
Two sandalwood plantations were selected (1-yr-old and 5-yr-old). In each plantation 40 trees were selected for pruning, with the condition that more than two branches required pruning per tree. Twenty were used for the “post wet-season” pruning treatment completed on the 31 May 2011, and the remaining 20 trees were pruned for the ”pre wet-season” treatment on the 4 November 2011. Branches were pruned using secateurs, and when required pruning saws, with cuts made as close to the stem as possible without causing damage to the stem bark. Trees were pruned to a maximum of one half of tree height.
Each plantation was harvested at two time intervals, representing approximately 12 and 18 mo after pruning. Trees were cut at least 5 cm below and above the last recognizable pruning wounds and then labelled, stored in plastic polyweave bags, and transported to Perth by road freight. A bandsaw was used to cut cross sectional discs around the selected pruning wound, with cuts made approximately 1.5 cm from the edges of the wound. Samples were selected from each tree with preference from the largest to the smallest open wound. Where open wounds were not present, preference was in order of the largest to the smallest scar tissue.
Each wound sample was split longitudinally through the centre of the wound using a chisel cleaned with 70 % ethanol between samples. For each pruning wound, shavings were taken using a sterilised scalpel from the margin between stained and healthy wood. These shavings were then transferred using sterilised forceps onto two media; (1) half strength Potato Dextrose Agar (PDA, Becton Dickinson, Sparks, MD; 19.5 g/L PDA, 7.5 g/L agar) containing 133 μg/mL streptomycin; and (2) Basidiomycete selective medium (5 g/L Bacto peptone (Difco, NSW, Australia), 20 g/L agar, 0.25 g/L MgSO4•7H20, 0.5 g/L K2HPO4, 0.016 g/L benomyl, 100 μg/L streptomycin, 2 ml/L 50 % (v/v) lactic acid, and 20 ml/L 95 % ethanol). After 2 wk, representative fungal colonies were transferred onto fresh ½ strength PDA. The cultures were examined regularly and any contaminated cultures were cleaned.
Once clean, all isolates from a single harvest time were subcultured on the same day onto ½ strength PDA. This was to enable comparison of culture morphology. After 2 wk, cultures were grouped based on morphology and representative isolates from each group were selected for molecular study.
Morphological identification and characterisation
Morphological characters used in this study to distinguish Phaeoacremonium species include conidiophore morphology, phialide type and shape, size of hyphal warts, and conidial size and shape. Colony characters and pigment production on MEA, PDA and oatmeal agar (OA; 60 g oatmeal; 12.5 g agar; Difco, Madrid, Spain) (Crous et al. 2009) incubated at 25 °C were noted after 8 and 16 d.
Microscopic observations were made from aerial mycelium of colonies cultivated on MEA or by using the slide culture technique, as explained by Arzanlou et al. (2007) when studying the genus Mycosphaerella. Images were captured with a Nikon camera system (Digital Sight DXM 1200, Nikon, Japan). Structures were mounted in lactic acid, and 30 measurements (1000× magnification) were made. The 5th and 95th percentiles were defined for all measurements with the extremes given in parentheses. Colony colours were determined with the colour charts of Rayner (1970). Cardinal temperatures for growth were obtained by incubating MEA plates in the dark at 5–40 °C in 5 °C intervals, also including 37 °C, human body temperature. Radial growth was measured after 8 d at 25 °C.
Molecular characterization: DNA isolation and amplification
Fungal mycelium and conidia from pure cultures grown on PDA for 2 wk at 25 °C in the dark were scraped and mechanically disrupted by grinding to a fine powder under liquid nitrogen with a mortar and pestle. Total DNA was extracted with the E.Z.N.A. Plant Miniprep Kit (Omega Bio-tek, Norcross, GA) following the manufacturer’s instructions. DNA was viewed on 0.7 % agarose gels stained with ethidium bromide and stored at -20 °C.
Approximately 600 bp of the 5’ end of the β-tubulin (BT) and approximately 300 bp of the 5’ end of the actin (ACT) genes were amplified as described by Mostert et al. (2006) using primer sets T1 (O’Donnell & Cigelnik 1997) and Bt2b, and ACT-512F and ACT-783R, (Carbone & Kohn 1999), respectively. PCR products were purified with the High Pure PCR Product Purification Kit (Roche Diagnostics, Mannheim, Germany) and sequenced in both directions by Macrogen (Sequencing Center, Seoul). Sequences were edited using Sequencher software v. 4.7. (Gene Codes, Ann Arbor, MI).
Phylogenetic analyses
The new Phaeoacremonium sequences (BT and ACT), together with reference sequences (Mostert et al. 2006, Damm et al. 2008, Essakhi et al. 2008, Graham et al. 2009, Gramaje et al. 2009, 2012, Úrbez-Torres et al. 2014) and the outgroup, Pleurostomophora richardsiae (ACT = AY579271, BT = AY579334) obtained from GenBank, were aligned using MAFFT sequence alignment program v. 7 (Katoh & Standley 2013) followed by manual adjustments of the alignments in BioEdit Sequence Alignment Editor v. 7.2.3. A partition homogeneity test of the BT and ACT alignments was conducted with PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b (Swofford 2000) to test pairwise congruence between sequence data sets. Phylogenetic analyses of all aligned sequence data were performed with MEGA v. 5.05 software (Tamura et al. 2011). Alignment gaps were treated as missing data and all characters were unordered and of equal weight. Any ties were broken randomly when encountered. All characters were unordered and of equal weight.
Maximum parsimony analysis was performed for the combined Phaeoacremonium dataset using the heuristic search option with 10 random simple taxon additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm with the option of saving no more than 10 trees with a score greater than or equal to 5 (Harrison & Langdale 2006). The robustness of the trees obtained was evaluated by 1 000 bootstrap replications (Hillis & Bull 1993). Tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency index (RC), were calculated.
Sequences derived in this study were lodged at GenBank, the alignments in TreeBASE (www.treebase.org/), and taxonomic novelties in MycoBank (www.MycoBank.org; Crous et al. 2004). GenBank accession numbers of the strains collected during this study are listed in Table 2.
Table 2.
Phaeoacremonium species, accession numbers, and collection details of isolates studied.
| Species | Isolate1 | Accesion No.2 | GenBank Accesions | |
|---|---|---|---|---|
| ACT | BT | |||
| Phaeoacremonium santali (clade 1) | E2.1B | A2 | KF835395 | KF823789 |
| E9.3A | A3 | KF835396 | KF823790 | |
| E19.2C | A4 | KF835397 | KF823791 | |
| - | A5 | KF835398 | KF823792 | |
| E11.2A | A6 | KF835399 | KF823793 | |
| E15.1A | A7 | KF835400 | KF823794 | |
| F3.4 | A26 | KF835401 | KF823795 | |
| F6.7 | A27 | KF835402 | KF823796 | |
| F2.3 | A28, CBS 137498 | KF835403 | KF823797 | |
| E7.7 | A29 | KF835304 | KF823798 | |
| F2.4 | A30 | KF835405 | KF823799 | |
| E9.3 | A35 | KJ533536 | KJ533532 | |
| E9.2 | A36 | KJ533537 | KJ533533 | |
| F32.2C | A37 | KJ533538 | KJ533534 | |
| E38.3B | A38 | KJ533539 | KJ533535 | |
| Phaeoacremonium luteum (clade 2) | F3.7 | A16, CBS 137497 | KF835406 | KF823800 |
| F2.4 | A17 | KF835407 | KF823801 | |
| F3.3 | A18 | KF835408 | KF823802 | |
| F2.1 | A19 | KF835409 | KF823803 | |
| F2.5 | A20 | KF835410 | KF823804 | |
| E30.SB | A33 | KJ533542 | KJ533540 | |
| F37.3B | A34 | KJ533543 | KJ533541 | |
| Phaeoacremonium alvesii (clade 3) | F1.2A | A21 | KF790540 | KF790535 |
| F11.5B | A22 | KF790541 | KF790536 | |
| F15.3A | A23 | KF790542 | KF790537 | |
| F15.5A | A24 | KF790543 | KF790538 | |
| F5.4A | A25 | KF790544 | KF790539 | |
| Phaeoacremonium parasiticum (clade 4) | E15.3A | A8 | KF790555 | KF790545 |
| E8.3B | A9 | KF790556 | KF790546 | |
| E8.2 | A10 | KF790557 | KF790547 | |
| F11.4E | A11 | KF790558 | KF790548 | |
| E15.1B | A12 | KF790559 | KF790549 | |
| E8.3 | A13 | KF790560 | KF790550 | |
| F15.5B | A14 | KF790561 | KF790551 | |
| F11.2C | A15 | KF790562 | KF790552 | |
| F11.5C | A31 | KF790563 | KF790553 | |
| F5.5 | A32 | KF790564 | KF790554 | |
| Phaeoacremonium venezuelense (clade 5) | F32.2A | A39 | KJ496346 | KJ496345 |
1 All the isolates were collected in Kununurra (Australia) by TI Burgess.
2 CBS: Culture collection of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands.
RESULTS
Fungal identification
The fungal isolates obtained in this study were characterised by having flat slow-growing cultures on MEA. Different types of phialides that were variable in size and shape were observed in the aerial mycelium, and either discrete or integrated in conidiophores. Sporulation was abundant and conidia hyaline and aseptate. All morphological characters corresponded to the genus Phaeoacremonium (Mostert et al. 2006). Based on their appearance in culture, the isolates could be assigned to five different clades (Table 2).
Molecular identification and phylogenetic analyses
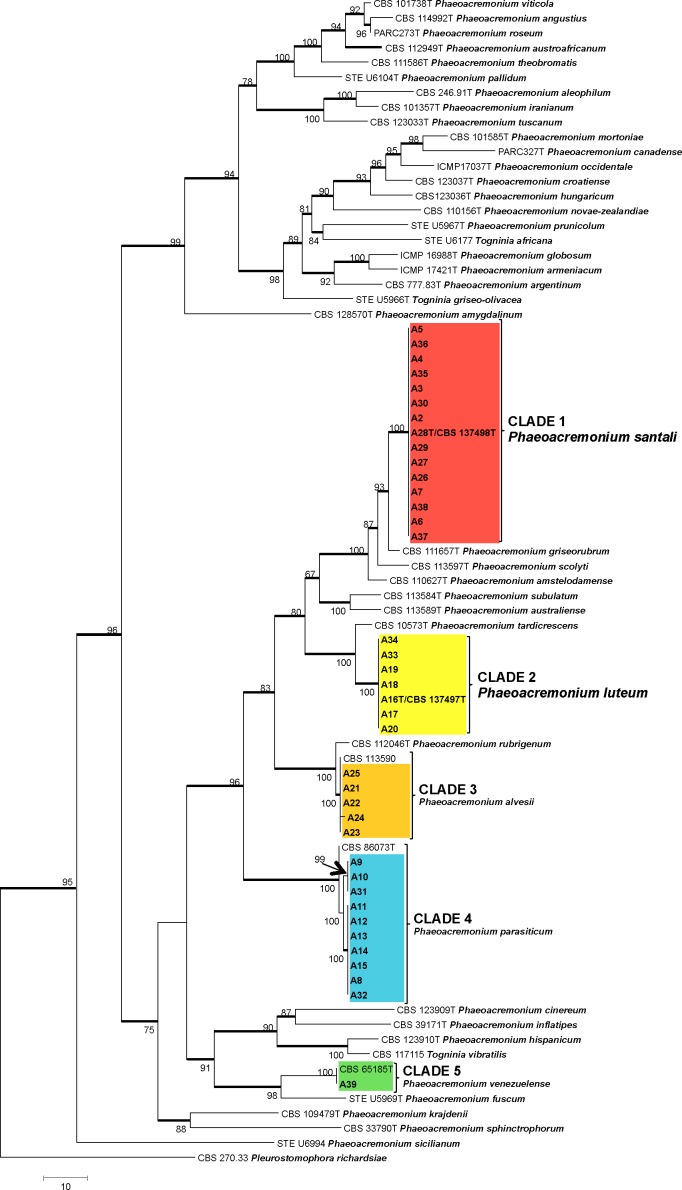
The partition homogeneity test of the BT and ACT alignments of Phaeoacremonium gave a P-value of 0.263 indicating that the datasets were congruent and could be combined. The combined sequence dataset consisted of 78 isolates including the outgroup and had 977 characters, of which 535 characters were parsimony-informative, 180 parsimony-uninformative and 262 constant. Sixty equally most parsimonious trees were retained (length = 2539 steps, CI = 0.471, RI = 0.837, RC = 0.399). A tree that closely resembled the strict consensus tree was chosen and is presented in Fig. 1. The isolates of the clade 1 grouped together in a polyphyletic clade with 100 % bootstrap support, with P. griseorubrum as closely related species. The isolates of the clade 2 grouped together in a polyphyletic clade with 100 % bootstrap support, with P. tardicrescens as closely related species. The isolates of clades 3 and 4, grouped inside the P. alvesii and P. parasiticum clades respectively, with 99 % bootstrap support.
Fig. 1.
(P. 70). One of 60 most parsimonious trees obtained from heuristic searches of a combined alignment of the TB and ACT gene sequences. Bootstrap support (1 000 replicates) above 60 % are shown at the nodes. Pleurostomophora richardsiae was used as outgroup. Ex-type strains for each species are indicated with a ‘T’ after the strain number. Thickened lines indicate branches present on strict consensus tree.
The BT and ACT sequences of the first clade of Phaeoacremonium isolates were 98 % identical to those of P. griseorubrum CBS 111657 (GenBank AY579294, AY579227). Differences were found between the first clade of Phaeoacremonium isolates and P. griseorubrum CBS 111657 sequences with five nucleotides varying in the ACT region and nine nucleotides in the BT region. The BT and ACT sequences of the second clade of Phaeoacremonium isolates were 97 % identical to those of P. tardicrescens CBS 110573 (GenBank AY579300, AY579233). Differences were found between the second clade of Phaeoacremonium isolates and P. tardicrescens CBS 110573 sequences with six nucleotides varying in the ACT region and 20 nucleotides in the BT region. The BT and ACT sequences of the third clade of isolates had 100 % identity with P. alvesii isolates CBS 113590 (GenBank AY579304) and STE-U 6988 (GenBank JQ038925), respectively. A BLASTn search showed that the BT and ACT sequences of the fourth clade of isolates had 100 % identity with isolates previously identified as P. parasiticum CBS 860.73 (GenBank AY579253). The BT and ACT sequences of the isolate corresponding to the fifth clade had 100 % identity with P. venezuelense isolate CBS 651.85 (GenBank AY579320).
TAXONOMY
Based on the DNA sequence analyses and morphological characters, two species of Phaeoacremonium proved distinct from known species, and are newly described below.
Phaeoacremonium luteum D. Gramaje, T.I. Burgess & J. Armengol, sp. nov.
MycoBank MB808419
(Fig. 2)
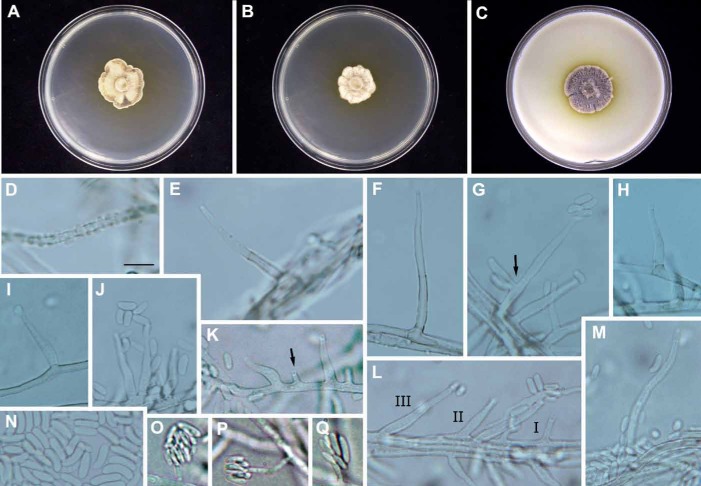
Fig. 2.
Phaeoacremonium luteum (CBS 137497 – ex-type culture A 16). A–C. Sixteen-day-old colonies incubated at 25 °C on MEA (A), PDA (B) and OA (C). D–N. Aerial structures on MEA; D. Mycelium showing prominent exudate droplets observed as warts; E–F. Conidiophores; G. Conidiophore (indicated by arrow) with terminal and 1 lateral phialide; H–J. Type II phialides; K. Type I phialide (indicated by arrow); L. Type III, type II and type I phialides. M. Type III phialide; N. Conidia. O–Q. Structures on the surface of and in MEA: Phialides with conidia. Bar: D = 10 μm (applies also to E–Q).
Etymology: Named after the yellow pigment produced that diffused into the agar ahead of the leading edge of the colony in all culture media.
Diagnosis: Phaeoacremonium luteum can be distinguished from the other species producing yellow pigment on MEA, PDA and OA, namely P. alvesii, P. subulatum, P. globosum, and the asexual morph of Toginia africana, by the predominance of type II phialides, the mycelium having prominent exudate droplets evident as warts, and slow growth.
Type: Australia: Western Australia: Kununurra, isolated from Santalum album trees, Dec. 2012, T. I. Burgess (CBS H-21622 – holotype; CBS 137497 – ex-type culture A16).
Description: Aerial structures in vitro on MEA: Mycelium consisting of branched, septate hyphae that occurs singly or in bundles of up to 8; hyphae tuberculate with warts to 2 μm diam, verruculose, medium to pale brown and 1.5–2.5 μm wide. Conidiophores mostly short, usually unbranched, arising from aerial or submerged hyphae, erect to flexuous, to 5-septate, sometimes bearing next to the terminal phialide 1–2 lateral ones, medium brown to pale brown, verrucose on the lower part, (15–)15.5–30–40(–59) μm long and 1.5–2.2–3 μm wide. Conidiogenous cells phialides, terminal or lateral, mostly monophialidic, smooth to verruculose, hyaline, collarettes 1.5–2.5 μm long, 1–1.5 μm wide; type I phialides mostly cylindrical, (3–)4–5–6(–6.5) × 1.5–2(–2.5) μm; type II phialides predominant, subcylindrical to subulate, (10–)10.5–13.5–17 × 2–3.5 μm; type III phialides cylindrical to subcylindrical, 20–24.5–30(–32) × 2–2.5–3 μm. Conidia hyaline, oblong or obovate, some reniform, 4–5–6(–7.5) × (1.5–)2–2.5–3 (av. = 5 × 2.5) μm, L/W ratio = 2.2. On surface or submerged in the agar: Phialides hyaline, mostly cylindrical, 3–6–9(–10) × 1–1.5–2 μm. Conidia hyaline, mostly allantoid, few reniform, 4.5–6(–7) × 1–1.5–2, L/W ratio = 4.3.
Culture characteristics: Colonies reaching a radius of 4.5–5 mm after 8 d at 25 °C. Minimum temperature for growth 15 °C, optimum 30–35 °C, maximum 37 °C. Colonies on MEA flat, felty to powdery, with crenate margin; after 8 d and 16 d, brownish olive towards the edge above, buffy brown to buff-yellow in reverse. Colonies on PDA flat, felty or woolly textured, with crenate margins; after 8 d and 16 d, buff-yellow and dark greyish brown above, buff-yellow to buffy brown in reverse. Colonies on OA flat, with woolly tufts, with entire margin; after 8 d and 16 d olive-grey and brownish vinaceous above, buffy brown to greyish brown in reverse. Yellow pigment produced on MEA, PDA and OA.
Additional cultures examined: Australia: Western Australia: Kununurra, isolated from Santalum album trees, Dec. 2012, T.I. Burgess A17, A18, A19, and A20.
Phaeoacremonium santali D. Gramaje, T.I. Burgess, J. Armengol, sp. nov.
MycoBank MB808420
(Fig. 3)
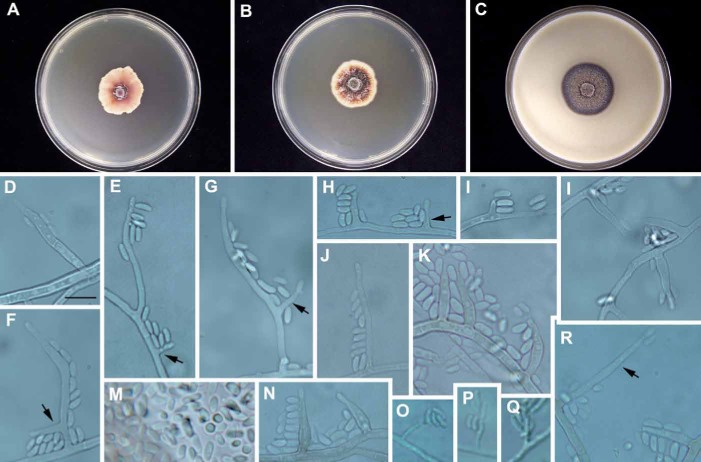
Fig. 3.
Phaeoacremonium santali (CBS 137498 – ex-type culture A 28). A–C. Sixteen-day-old colonies incubated at 25 °C on MEA (A), PDA (B) and OA (C). D–N. Aerial structures on MEA; D–E. Conidiophores and type I phialide (indicated by arrow); F–G. Conidiophores (indicated by arrows) with terminal and 1 lateral phialide; H. Type II and Type I phialides (indicated by arrow); I. Type I phialide; J. Type III phialide. K–L. Type II phialides; M. Conidia; N. Type II phialides; O–Q. Structures on the surface of and in MEA: phialides with conidia; R. Type III phialide (indicated by arrow). Bar: D = 10 μm (applies also to E–R).
Etymology: Named after the host it was isolated from, Tropical sandalwood (Santalum album).
Diagnosis: Phaeoacremonium santali can be distinguished from the other species producing pink colonies on MEA, namely P. alvesii, P. armeniacum, P. griseorubrum, P. rubrigenum, P. scolyti, and P. viticola, by the predominance of type I and II phialides, the brownish olive colour in OA, and slow growth. Colonies reached a radius of only 6.6–7.5 mm in 8 d at 25 °C on MEA. Phaeoacremonium griseorubrum overlaps with P. santali in growth rate, but has a temperature maximum for growth of 40 °C, compared with 37 °C in the latter species.
Type: Australia: Western Australia: Kununurra, isolated from Santalum album trees, Dec. 2012, T.I. Burgess (CBS H-21621 – holotype; CBS 137498 – ex-type culture A 28).
Description: Aerial structures in vitro on MEA: Mycelium consisting of branched, septate hyphae that occurs singly or in bundles of up to six; smooth or rarely with warts, verruculose, yellow-brown to hyaline, 2–3.5 μm wide. Conidiophores mostly short, usually unbranched, arising from aerial or submerged hyphae, erect to flexuous, to 3-septate, often bearing besides the terminal phialide 1–2 lateral ones, pale brown, smooth to verruculose, (10–)10.5–15–27(–31) μm long and 1.5–2(–2.5) μm wide. Conidigoenous cells phialides, terminal or lateral, mostly polyphialidic, smooth to verruculose, hyaline, collarettes 1.5–2.5 μm long, 1–1.5 μm wide; type I and II phialides predominant, type I phialides cylindrical, occasionally widened at the base, tapering towards the apex, (2–)2.5–6–7.5(–8) × 1–1.5–2 μm; type II phialides elongate-ampulliform and attenuated at the base, or navicular, tapering towards the apex, (5.5–)6–7–9 × 2–3–3 μm; type III phialides subcylindrical to navicular, 12–14–19(–20) × 1.5–2–2.5 μm. Conidia hyaline, oblong ellipsoidal, some obovoid or reniform, (3–)4–4.5–5(–6) × 1.5–2–3 μm, L/W ratio = 2.1. On surface or submerged in the agar: Phialides hyaline, mostly cylindrical, 5–7.5–10(–12) × 1–1.5–2 μm. Conidia hyaline, mostly allantoid, 5–6–7(–11) × 1–1.5–2, L/W ratio = 4.1.
Culture characteristics: Colonies reaching a radius of 6.6–7.5 mm after 8 d at 25 °C. Minimum temperature for growth 15 °C, optimum 30 °C, maximum 37 °C. Colonies on MEA flat, erose or dentate; after 8 d pale rose to pinkish vinaceous towards the edge above, pale rose towards the edge in reverse, after 16 d rosy vinaceous to pinkish buff towards the edge above, vinaceous pink near the centre and pinkish buff towards the edge in reverse. Colonies on PDA flat, felt-like with few woolly tufts near the centre, with entire margin; after 8 d pale pinkish buff towards the edge above and in reverse, after 16 d isabelline to oliveacous towards the edge above, violet-brown towards the centre and pale brown to orange-grey towards the edge in reverse. Colonies on OA flat, felty to powdery, with entire margin; after 8 d and 16 d dull green to olive green above, brownish olive to dark vinaceous-brown towards the edge in reverse.
Additional cultures examined: Australia: Western Australia: Kununurra, isolated from Santalum album trees, Dec. 2012, T.I. Burgess A2, A3, A4, A5, A6, A7, A26, A27, A29, and A30.
DISCUSSION
In this study, the integration of morphology, cultural characters, and DNA sequence data revealed the presence of five Phaeoacremonium species within pruning wounds of Santalum album in tropical Western Australia. Phaeoacremonium species were commonly isolated from both 1- and 5-year-old trees and in both harvests, approximately 12 and 18 mo after pruning. Two novel species of Phaeoacremonium, P. luteum and P. santali, were obtained from sandalwood, bringing the total number of known species of the genus to 42.
Micromorphological traits, such as conidiophore morphology, phialide type and shape, size of hyphal warts, and cultural characters are useful in distinguishing Phaeoacremonium species (Mostert et al. 2005). In addition, molecular analyses of part of the β-tubulin and actin gene regions have been shown to give high phylogenetic resolution within Phaeoacremonium in previous studies (Mostert et al. 2005, 2006, Réblová & Mostert 2007, Essakhi et al. 2008, Graham et al. 2009, Gramaje et al. 2012). Distinct features of P. luteum include its ability to produce yellow pigment on MEA, PDA and OA, the predominance of subcylindrical to subulate type II phialides and the mycelium showing prominent exudate droplets observed as warts. Yellow pigment production on PDA and OA is a common culture characteristic of some Phaeoacremonium species, and is considered useful in distinguishing species in the genus, especially on OA, which is an excellent medium to test pigment production (Mostert et al. 2006). It is also important to note the slow growth of this species on malt extract agar, with colonies reaching a radius of only 4.5–5 mm after 8 d. Phaeoacremonium santali could be distinguished from the other species producing pink colonies on MEA by the predominance of type I and II phialides, its distinct brownish olive colonies in oatmeal agar, and slow growth.
Growth temperature studies showed that all the isolates of P. luteum and P. santali had a maximum growth temperature of 37 °C, suggesting that they have the potential to survive at human body temperature. Several thermotolerant Phaeoacremonium species, such as P. alvesii, P. griseorubrum, P. krajdenii, P. parasiticum, and P. venezuelense, are associated with phaeohyphomycosis in humans and also have been isolated from woody hosts (Mostert et al. 2005, Essakhi et al. 2008).
In addition to the two new taxa, three previously known species were also found on sandalwood. Phaeoacremonium alvesii and P. parasiticum had been previously reported from Dodonaea viscosa (Mostert et al. 2005) and Vitis vinifera (Pascoe & Cottral 2000) in Australia, respectively. Phaeoacremonium venezuelense represents a new record for Australia and has previously been reported from humans in Brazil (Guarro et al. 2006), Canada (Mostert et al. 2005), France (Mostofi et al. 2012), and Venezuela (de Albornoz 1974), from grapevine in Algeria (Berraf-Tebbal et al. 2011) and South Africa (Mostert et al. 2005), and more recently from wood decay of apricot trees in Spain (Olmo et al. 2014).
Species of Phaeoacreomonium obtained in this study were all collected from pruning wounds. Some Phaeoacremonium spp., such as P. aleophilum and P. mortoniae, produce perithecia (i.e. a Togninia sexual morph) in old, rotted, vascular tissue of pruning wounds and in deep cracks in cordons, trunks, and spurs of grapevine (Eskalen et al. 2005, Rooney-Latham et al. 2005, Baloyi et al. 2013). Ascospores are released from these overwintering structures by rain and can infect the grapevine through fresh pruning wounds, which are recognized as the main point of entry for Phaeoacremonium species into grapevines (Eskalen et al. 2005). Of the species found in this study, only P. parasiticum has a known sexual morph and could possibly be present as perithecia on sandalwood trees. Aerial inoculum could be released by these ascomata on infected tress, thus becoming a major source of fungal infection. Insect transmission of sexual spores or conidia may also occur (Moyo et al. 2014).
Acknowledgments
We acknowledge Pablo Castillo (IAS-CSIC) for sharing the equipment to perform microscopic observations. Isolations were made under project PRJ-004677 “Heartwood Rot Identification and Impact in Sandalwood (Santalum album)” funded by the Rural Industries Research and Development Corporation of Australia. We thank Len Norris and Diane White for assistance with collection and isolation.
REFERENCES
- Agustí-Brisach C, Gramaje D, García-Jiménez J, Armengol J. (2013) Detection of black-foot and Petri disease pathogens in natural soils of grapevine nurseries and vineyards using bait plants. Plant and Soil 364: 5–13 [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW. (2007) Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzanlou M, Narmani A, Khodaei S, Moshari S. (2014) Pome and stone fruit trees as possible reservoir hosts for Phaeoacremonium spp., the causal agents of grapevine esca disease in Iran. Archives of Phytopathology and Plant Protection 47: 717–727 [Google Scholar]
- Baloyi MA, Halleen F, Mostert L, Eskalen A. (2013) First report of Togninia minima perithecia on esca- and Petri-diseased grapevines in South Africa. Plant Disease 97: 1247. [DOI] [PubMed] [Google Scholar]
- Barbour L, Norris L, Burgess TI. (2010) Heartwood rot identification and impact in sandalwood (Santalum album). Barton, ACT: Rural Industries Research and Development Corporation [Google Scholar]
- Barry K. (2002) Heartrots in Plantation Hardwoods in Indonesia and Australia. [Technical Report no. 51.] Canberra, ACT: Australian Center for International Agricultural Research [Google Scholar]
- Berraf-Tebbal A, Bouznad Z, Santos JM, Coelho MA, Péros JP, Phillips AJL. (2011) Phaeoacremonium species associated with Eutypa dieback and esca of grapevines in Algeria. Phytopathologia Mediterranea 50: S86–S97 [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556 [Google Scholar]
- Cloete M, Fourie PH, Damm U, Crous PW, Mostert L. (2011) Fungi associated with die-back symptoms of apple and pear trees, a possible inoculum source of grapevine trunk disease pathogens. Phytopathologia Mediterranea 50: S176–S190 [Google Scholar]
- Crous PW, Gams W. (2000) Phaeomoniella chlamydospora gen. et comb. nov., a causal organism of Petri grapevine decline and esca. Phytopathologia Mediterranea 39: 112–188 [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Crous PW, Gams W, Wingfield MJ, Van Wyk PS. (1996) Phaeoacremonium gen. nov. associated with wilt and decline diseases of woody hosts and human infections. Mycologia 88: 786–796 [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, Samson RA. (eds) (2009) Fungal Biodiversity. [CBS Laboratory Manual Series no. 1.] Utrecht: CBS-KNAW Fungal Biodiversity Centre [Google Scholar]
- Damm U, Mostert L, Crous PW, Fourie PH. (2008) Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 20: 87–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albornoz MB. (1974) Cephalosporium serrae, agente etiológico de micetomas. Mycopathologia et Mycologia Applicata: 54: 485–498 [DOI] [PubMed] [Google Scholar]
- Di Marco S, Calzarano F, Osti F, Mazzullo A. (2004) Pathogenicity of fungi associated with a decay of kiwifruit. Australasian Plant Pathology 33: 337–342 [Google Scholar]
- Dupont J, Laloui W, Magnin S, Larignon P, Roquebert MF. (2000) Phaeoacremonium viticola, a new species associated with esca disease of grapevine in France. Mycologia 92: 499–504 [Google Scholar]
- Dupont J, Magnin S, Césari C, Gatica M. (2002) ITS and β-tubulin markers help delineate Phaeoacremonium species, and the occurrence of P. parasiticum in grapevine disease in Argentina. Mycological Research 106: 1143–1150 [Google Scholar]
- Eskalen A, Rooney-Latham S, Gubler WD. (2005) Occurrence of Togninia fraxinopennsylvanica on esca-diseased grapevines (Vitis vinifera) and declining ash trees (Fraxinus latifolia) in California. Plant Disease 89: 528. [DOI] [PubMed] [Google Scholar]
- Essakhi S, Mugnai L, Crous PW, Groenewald JZ, Surico G. (2008) Molecular and phenotypic characterization of novel Phaeoacremonium species associated with Petri disease and esca of grapevine. Persoonia 21: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramaje D, Agustí-Brisach C, Pérez-Sierra A, Moralejo E, Olmo D, Mostert L, Damm U, Armengol J. (2012) Fungal trunk pathogens associated with wood decay of almond trees on Mallorca (Spain). Persoonia 28: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramaje D, Armengol J, Mohammadi H, Banihashemi Z, Mostert L. (2009) Novel Phaeoacremonium species associated with Petri disease and esca of grapevines in Iran and Spain. Mycologia 101: 920–929 [DOI] [PubMed] [Google Scholar]
- Graham AB, Johnston PR, Weir BS. (2009) Three new Phaeoacremonium species on grapevines in New Zealand. Australasian Plant Pathology 38: 505–513 [Google Scholar]
- Groenewald M, Kang J-C, Crous PW, Gams W. (2001) ITS and beta-tubulin phylogeny of Phaeoacremonium and Phaeomoniella species. Mycological Research 105: 651–657 [Google Scholar]
- Guarro J, Silvestre AM, Jr, Verkley G, Cano J, Gompertz OF, Gené J, Ogawa MM, Tomimori-Yamashita J, Teixeira SP, de Almeida FA. (2006) Limitations of DNA sequencing for diagnosis of a mixed infection by two fungi, Phaeoacremonium venezuelense and a Plectophomella sp., in a transplant recipient. Journal of Clinical Microbiology 44: 4279–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell RS. (1966) Association of Cephalosporium with a decline of oak in Texas. Plant Disease Reporter 50: 75–78 [Google Scholar]
- Harrison CJ, Langdale JA. (2006) A step by step guide to phylogeny reconstruction. Plant Journal 45: 561–572 [DOI] [PubMed] [Google Scholar]
- Harbaugh DT, Baldwin BG. (2007) Phylogeny and biogeography of the sandalwoods (Santalum, Santalaceae): repeated dispersals throughout the Pacific. American Journal of Botany 94: 1028–1040 [DOI] [PubMed] [Google Scholar]
- Hausner G, Eyjólfsdóttir G, Reid J. (1992) Two additional species of the genus Togninia. Canadian Journal of Botany 70: 724–734 [Google Scholar]
- Hawksworth DL, Gibson IAS, Gams W. (1976) Phialophora parasitica associated with disease conditions in various trees. Transactions of British Mycological Society 66: 427–431 [Google Scholar]
- Hennion B, Baudry A, Lecompte P, Durpaire MP, Mouyon M, Tailleur JL, Larignon P. (2001) Dépérissement du kiwi par maladie du bois. Infos-Ctifl 176: 25–27 [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192 [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability (outlines version 7). Molecular Biology and Evolution 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SC, Zambino PJ, Mayorquin JS, Wang DH, Eskalen A. (2013) Identification of new fungal pathogens of coast live oak in California. Plant Disease 97: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Mohammadi H. (2014) Phaeoacremonium spp. and Botryosphaeriaceae spp. associated with date palm (Phoenix dactylifera L.) decline in Iran. Journal of Phytopathology DOI: 10.1111/jph.12229 [Google Scholar]
- Mostert L, Groenewald JZ, Summerbell RC, Gams W, Crous PW. (2006) Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Studies in Mycology 54: 1–115 [Google Scholar]
- Mostert L, Groenewald JZ, Summerbell RC, Robert V, Sutton DA, Padhye AA, Crous PW. (2005) Species of Phaeoacremonium associated with human infections and environmental reservoirs in infected woody plants. Journal of Clinical Microbiology 43: 1752–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostofi K, Blanchet D, Marnet D, Abboud P, Jeanbourquin D, Charles JI, Belzunc C. (2012) Cervical spondylitis due to Phaeoacremonium venezuelense in an immunocompetent patient. A first case report. Journal de Mycologie Médicale 22: 197–200 [DOI] [PubMed] [Google Scholar]
- Moyo P, Allsopp E, Roets F, Mostert L, Halleen F. (2014) Arthropods vector grapevine trunk disease pathogens. Phytopathology doi: http://dx.doi.org/10.1094/PHYTO-11-13-0303-R [DOI] [PubMed] [Google Scholar]
- Nigro F, Boscia D, Antelmi I, Ippolito A. (2013) Fungal species associated with a severe decline of olive in southern Italy. Journal of Plant Pathology 95: 668 [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116 [DOI] [PubMed] [Google Scholar]
- Olmo D, Gramaje D, Agustí-Brisach C, Leon M, Armengol J. (2014) First report of Phaeoacremonium venezuelense associated with decay of apricot trees in Spain. Plant Disease DOI:http://dx.doi.org/10.1094/PDIS-12-13-1198-PDN [DOI] [PubMed] [Google Scholar]
- Pascoe I, Cottral E. (2000) Developments in grapevine trunk diseases research in Australia. Phytopathologia Mediterranea 39: 68–75 [Google Scholar]
- Prodi A, Sandalo S, Tonti S, Nipoti P, Pisi A. (2008) Phialophora-like fungi associated with kiwifruit elephantiasis. Journal of Plant Pathology 90: 487–494 [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Kew: Commonwealth Mycological [Google Scholar]
- Réblová M, Mostert L. (2007) Romellia is congeneric with Togninia, and description of Conidiotheca gen. nov. for one species of this genus with polysporous asci. Mycological Research 111: 299–307 [DOI] [PubMed] [Google Scholar]
- Richter H, Gindro K, Pezet R, Viret O. (2007) Localization and quantification of fungi in esca diseased grapevine trunks. Phytopathologia Mediterranea 46: 105–106 [Google Scholar]
- Rooney S, Eskalen A, Gubler WD. (2001) Recovery of Phaeomoniella chlamydospora and Phaeoacremonium inflatipes from soil and grapevine tissues. Phytopathologia Mediterranea 40: S351–356 [Google Scholar]
- Rooney-Latham S, Eskalen A, Gubler WD. (2005) Teleomorph formation of Phaeoacremonium aleophilum, cause of esca and grapevine decline in California. Plant Disease 89: 177–184 [DOI] [PubMed] [Google Scholar]
- Rumbos I. (1986) Phialophora parasitica, causal agent of cherry dieback. Journal of Phytopathology 117: 283–287 [Google Scholar]
- Sánchez-Márquez S, Bills GF, Zabalgogeazcoa I. (2007) The endophytic mycobiota of the grass Dactylis glomerata. Fungal Diversity 27: 171–195 [Google Scholar]
- Swofford DL. (2000) PAUP* 4.0: Phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úrbez-Torres JR, Haag P, Bowen P, O’Gorman DT. (2014) Grapevine trunk diseases in British Columbia: incidence and characterization of the fungal pathogens associated with esca and Petri diseases of grapevine. Plant Disease 98: 469–482 [DOI] [PubMed] [Google Scholar]
- Úrbez-Torres JR, Peduto F, Vossen PM, Krueger WH, Gubler WD. (2013) Olive twig and branch dieback: etiology, incidence, and distribution in California. Plant Disease 97: 231–244 [DOI] [PubMed] [Google Scholar]