Summary
Autophagy is a lysosomal degradation pathway that is important in cellular homeostasis. Prior work showed a key role for the autophagy related 5 (Atg5) in resistance to Toxoplasma gondii. Here we show that the cassette of autophagy proteins involved in the conjugation of microtubule associated protein 1 light chain 3 (LC3) to phosphatidylethanolamine, including Atg7, Atg3, and the Atg12-Atg5-Atg16L1 complex play crucial roles in the control of T. gondii in vitro and in vivo. In contrast, pharmacologic modulation of the degradative autophagy pathway or genetic deletion of other essential autophagy genes had no substantial effects. Rather the conjugation system was required for targeting of LC3 and interferon-γ effectors onto the vacuolar membrane of T. gondii and its consequent disruption. These data suggest that the ubiquitin-like conjugation systems that reorganize intracellular membranes during canonical autophagy are necessary for proper targeting of immune effectors to the intracellular vacuole membranes utilized by pathogens.
Introduction
Toxoplasma gondii is a protozoan parasite that infects a broad range of animals and causes zoonotic toxoplasmosis in humans (Sibley, 2011). Toxoplasmosis is a leading cause of reportable foodborne illness in the United States and contributes to congenital disease and opportunistic disease in immunocompromised persons throughout the world (Jones et al., 2010). Although 25–30% of the world’s population are believed to carry persistent T. gondii infection for life, most people remain asymptomatic as long as the parasites are under the control of the immune system (Pappas et al., 2009). Therefore, understanding how the normal immune system controls the T. gondii infection may lead to better management of toxoplasmosis in patients, especially the immunocompromised.
The active invasion of host cells by T. gondii leads to the formation of the non-fusogenic parasitophorous vacuole (PV), a cytoplasmic membranous structure that envelops the invading T. gondii in the host cell (Sibley, 2011). Inside the PV, T. gondii replicates by endodyogeny while protected from the inimical host cytoplasm. To fight the T. gondii infection, interferon-γ (IFNγ) secretion by immune cells, its subsequent binding to host cell receptors and induction of cellular activation arms infected and naïve cells with effector molecules, such as the immunity related p47 GTPases (IRGs) and guanylate-binding proteins (GBPs) (Howard et al., 2011; Kim et al., 2012). These effectors rapidly accumulate on and around the PV membrane (PVM), leading to the disruption of PVM and subsequent death of the parasite. The proper loading of the effectors onto the PVM is known to require GTP-binding dependent oligomerization, which is controlled by regulatory interactions among the effectors (Howard et al., 2011). Moreover, we and others showed that the essential autophagy gene Atg5 (autophagy related 5) is required for the proper targeting of the effectors onto the PVM of T. gondii (Selleck et al., 2013; Zhao et al., 2009; 2008). Without Atg5, the effectors are induced by IFNγ normally, but form cytoplasmic aggregates instead of accumulating on the PVM. However, the mechanism of the Atg5-mediated targeting of the IFNγ effectors to the PVM of T. gondii is poorly understood (Howard et al., 2011).
Autophagy is an evolutionarily conserved intracellular degradation pathway that targets cellular constituents to lysosomes (Parzych and Klionsky, 2013). Among the different forms of autophagy, macroautophagy (henceforth, autophagy) is the pathway that sequesters cytoplasmic materials in double membrane bound autophagosomes and degrades the cargo through the fusion between the autophagosome and lysosome (Rubinsztein et al., 2012). Upon induction of autophagy by various signals, such as the inhibition of mammalian target of rapamycin (mTOR) kinase, the initiation complex of ULK1/2 (uncoordinated 51-like kinase 1/2)-Atg13-Atg101-FIP200 (focal adhesion kinase family-interacting protein of 200 kD) activates the phosphatidylinositol 3-kinase (PI3K) complex of Beclin1-Vps34-Vps15-Atg14L. This activation leads to the nucleation of isolation membrane formation and the elongation complex of Atg12-Atg5-Atg16L1 extends the membrane further by conjugating LC3 (microtubule associated protein 1 light chain 3) homologs to the phosphatidylethanolamine of the growing autophagosome (Rubinsztein et al., 2012).
The function of the autophagy pathway was initially identified as a starvation-induced homeostatic pathway for recycling of essential materials, but in recent years it has been expanded to include cellular remodeling, secretion, differentiation, and immune defense (Levine et al., 2011). Specifically, the role of autophagy in the selective recognition and subsequent lysosomal degradation of pathogens has been highlighted in the innate immune defense against intracellular pathogens (Levine et al., 2011). Furthermore, we showed that the autophagy elongation complex, but not the degradative autophagy pathway, plays an essential role in the control of murine norovirus (MNV) by IFNγ (Hwang et al., 2012). We also found that proximal components of the autophagy pathway including ULK1, Atg14L, and Beclin1 are required for replication of Brucella abortus in macrophages while distal components of the pathway such as Atg5, Atg7, and Atg16L1 are not (Starr et al., 2012). These observations open up the possibility that cassettes of autophagy proteins play a broad role in biology independent of the canonical degradation of cytoplasmic organelles and other constituents (Bestebroer et al., 2013; Subramani and Malhotra, 2013).
Here we show that the conjugation of LC3 through E1 Atg7, E2 Atg3, and E3 Atg12-Atg5-Atg16L1 complex is required to control T. gondii infection in vitro and in vivo. Using genetic and pharmacologic methods, we demonstrate that only the two ubiquitin-like conjugation systems of the autophagy pathway, but not the canonical degradative autophagy process nor the initiation-nucleation complex, are required for IFNγ to control T. gondii infection. These data suggest that the ubiquitin-like conjugation systems, which are normally involved in the reorganization of intracellular membranes in the canonical autophagy pathway, are also required for proper targeting of LC3 and IFNγ effectors toward the intracellular membrane structure of pathogens.
Results
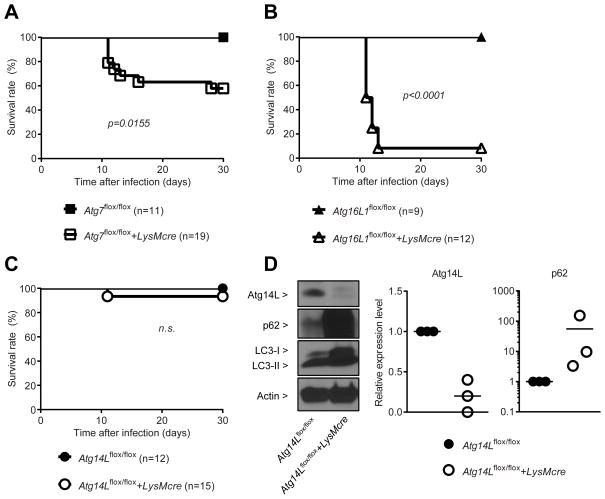
Atg5, Atg7 and Atg16L1, but not Atg14L, are required to control T. gondii infection in vivo
Previously, we showed that Atg5 in myeloid lineage cells is required for resistance of mice to infection with T. gondii (Zhao et al., 2008). Although Atg5 was required for IFNγ induced control of T. gondii in primary macrophages, autophagosomes, the hallmark of canonical autophagy, are not visualized in this process. This finding led us to hypothesize that the role of Atg5 in intracellular immunity to T. gondii infection might be independent of its role in the elongation of autophagosomal membrane required for the canonical autophagy pathway (Zhao et al., 2008). To investigate the mechanism, we examined the role of other essential autophagy genes – Atg7, Atg14L, and Atg16L1 – during the in vivo infection of T. gondii. Atg7 is the E1 activating enzyme that is required for the activation of both ubiquitin-like molecules, LC3 and Atg12. Atg16L1 binds to the Atg12-conjugated Atg5 and form the autophagosome elongation complex, which is essential for the growth of autophagosome by functioning as E3 ligase for LC3 conjugation. Atg14L functions in endoplasmic reticulum targeting of PI3K complex for the nucleation of the autophagosomal membrane (Matsunaga et al., 2010; 2009).
Since full deletion of essential autophagy genes causes neonatal lethality, we took a conditional deletion approach (Mizushima and Levine, 2010). We infected Atg7flox/flox+LysMcre, Atg16L1flox/flox+LysMcre, Atg14Lflox/flox+LysMcre and their control littermate mice with T. gondii. Similar to Atg5flox/flox+LysMcre mice (Supplementary Figure S1A), both Atg7flox/flox+LysMcre (Figure 1A) and Atg16L1flox/flox+LysMcre (Figure 1B) mice were more susceptible to lethal infection with T. gondii than their controls. Atg7flox/flox+LysMcre mice were less susceptible to T. gondii infection than Atg5flox/flox+LysMcre and Atg16L1flox/flox+LysMcre mice, possibly due to less efficient functional deletion of Atg7 by LysMcre (Figure S1B). In contrast, both Atg14Lflox/flox and Atg14Lflox/flox+LysMcre mice showed similar resistance to the infection of T. gondii, suggesting that Atg14L is not required to control in vivo infection of T. gondii (Figure 1C). This lack of a phenotype was not due to insufficient functional deletion of Atg14L in vivo, because macrophages directly isolated from Atg14Lflox/flox+LysMcre mice have undetectable Atg14L expression and a significant accumulation of p62 protein, which is normally removed through functional autophagy, compared to control (Figure 1D) without significant induction of p62 mRNA (Figure S1C). In contrast, the conversion of LC3-I (cytosolic) to LC3-II (lipidated and membrane-bound) was largely unaffected by the deletion of Atg14L, which is consistent with previous findings (Matsunaga et al., 2009; Zhong et al., 2009). These genetic data distinguish the role of autophagy proteins in the control of in vivo infection of T. gondii from their role in canonical degradative autophagy that requires Atg14L.
Figure 1. Atg7 and Atg16L1, but not Atg14L, are required to control T. gondii infection in vivo.
Survival curves after intraperitoneal inoculation with 200 (male) or 100 (female) of T. gondii per mouse. Number of mice in parentheses. (A) Atg7flox/flox+/−LysMcre, (B) Atg16L1flox/flox+/−LysMcre, (C) Atg14Lflox/flox+/−LysMcre. Statistical analysis by Log-rank (Mantel-Cox) test. (D) A representative protein blot and quantitative analyses of Atg14L and p62 in the peritoneal exudate macrophages. Combined data as each data point with average. See also Figure S1.
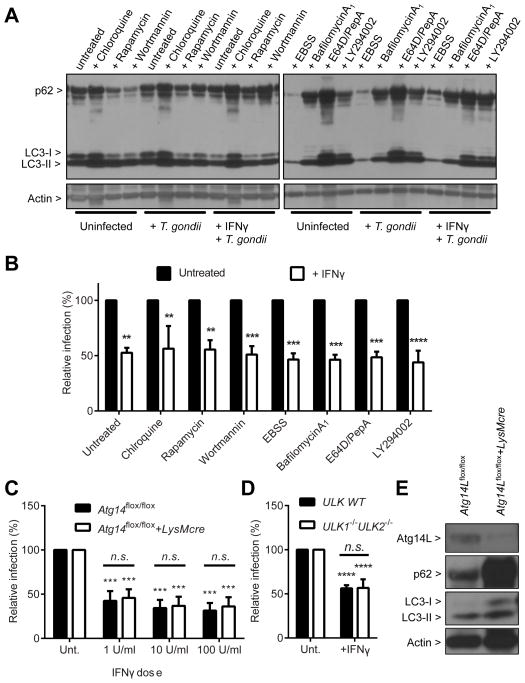
Canonical degradative autophagy pathway is not required for IFNγ to control T. gondii infection
To investigate the control of T. gondii by IFNγ in primary macrophages more efficiently than provided by previous microscopy based assays (Zhao et al., 2008), we developed a flow cytometry-based system to assess the level of T. gondii infection (Figure S2A). We compared the data of the immunofluorescence method to those of the flow cytometry method for the measurement of T. gondii infection, and found that these two assays performed comparably (as described below). Therefore, we utilized the flow cytometry based assay system for the rest of our study.
We first investigated the role of the degradative autophagy pathway in the control of T. gondii by IFNγ using pharmacologic approaches. Degradative autophagy was induced with treatment of an mTOR inhibitor (rapamycin) or starvation (Earle’s Balanced Salt Solution, EBSS) and inhibited with treatment of PI3K inhibitors (wortmannin or LY294002). Late steps of degradative autophagy, the fusion of the autophagosome with lysosome and subsequent degradation of its contents within the autolysosome, were blocked with lysosomal acidification inhibitors (chloroquine or bafilomycin A1) or lysosomal protease inhibitors (E64D and pepstatinA). Induction of autophagy is manifested as increased LC3-II and decreased p62 and its inhibition leads to decreased LC3-II and increased p62, although compensatory feedback can occur for prolonged treatment (Hwang et al., 2012; Mizushima et al., 2010). In contrast, lysosomal degradation blockers increase the level of both LC3-II and p62. The treatments with pharmacologic agents had significant and expected effects on degradative autophagy in control cells (Figure 2A), but none of the treatments significantly affected the control of T. gondii by IFNγ (Figure 2B). In fact, none of them, except starvation in EBSS medium, which caused significant death of the treated cells, significantly affected the infection level with T. gondii (Figure S2B).
Figure 2. Degradative autophagy pathway is not required for IFNγ to control T. gondii infection.
(A) Protein blot for the autophagy status of the samples as shown in (B). Samples harvested at 24 hpi. (B) Flow cytometry analysis for the relative infection (%) of T. gondii at 24 hpi (MOI=1) in BMDMs from C57BL/6 mice treated with chemicals as indicated. Cells were treated with the chemicals for 24 hrs +/− 100 U/ml (50 ng/ml) of IFNγ before infection and treated again during the infection (total 48 hrs of treatment). Autophagy inducers - rapamycin, starvation (EBSS); autophagy inhibitors - wortmannin, LY294002; degradation blockers - chloroquine, bafilomycin A1, E64D, pepstatinA. (C) Same analysis as shown in (B) for Atg14Lflox/flox+/−LysMcre BMDMs. Cells were pretreated with IFNγ for 24 hrs at the indicated doses (D) Same analysis as shown in (B) for WT and Ulk1−/−Ulk2−/− MEFs. (E) Protein blot for untreated/uninfected Atg14Lflox/flox+/−LysMcre BMDMs. Statistical analysis by one-way analysis of variance (1-ANOVA) with Tukey post test. n.s.: not significant (p>0.05), **: p<0.01, ***: p<0.001, ****: p<0.0001. Combined data as average ± SEM. See also Figure S2.
We further tested the role of the canonical autophagy pathway in the control of T. gondii by IFNγ using genetic approaches. Consistent with the lack of a role for Atg14L in resistance of mice to T. gondii infection in vivo (Figure 1C), T. gondii infection in both control (Atg14Lflox/flox) and Atg14L-deficient macrophages (Atg14Lflox/flox+LysMcre) was similarly inhibited by IFNγ (Figure 2C). The ULK1 and ULK2 complex is essential in initiating autophagosome formation and deletion of both ULK1 and ULK2 causes perinatal lethality as observed for knockout of essential autophagy genes (McAlpine et al., 2013). Therefore, we utilized mouse embryonic fibroblasts (MEFs) from Ulk1−/−Ulk2−/− mice to investigate the role of the initiation complex in the control of T. gondii by IFNγ. Similar to the Atg14L-deficient cells, deletion of both ULK1 and ULK2 did not affect the control of T. gondii by IFNγ (Figure 2D). Therefore, the deletion of Atg14L in the nucleation complex (Figure 2E) and ULK1 and ULK2 in the initiation complex (Figure S2C) did not have any significant effect on the control of T. gondii by IFNγ. Collectively, pharmacologic and genetic modulation of the autophagy pathway showed that the canonical degradative autophagy pathway is not required for IFNγ to control T. gondii infection, which is consistent with in vivo data (Figure 1).
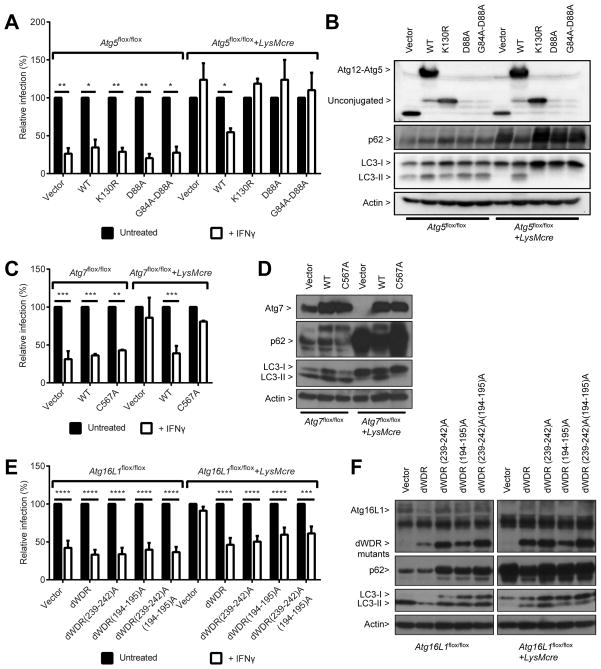
Atg12-Atg5-Atg16L1 complex is required for IFNγ to control T. gondii infection in vitro
To examine the role of Atg5 in the control of T. gondii by IFNγ, we reconstituted Atg5-deficient macrophages with wild type and mutants of Atg5 using lentiviral transduction. We first confirmed that T. gondii infection was significantly inhibited by IFNγ in control (Atg5flox/flox) but not in Atg5-deficient (Atg5flox/flox+LysMcre) macrophages transduced with control lentivirus, using both flow cytometry (Figure 3A) and immunofluorescence assays (Figure S3A). Transduction of wild type Atg5 into Atg5-deficient macrophage restored the control of T. gondii by IFNγ and basal autophagy in the transduced cells, as represented by the conversion of LC3-I to LC3-II and the decrease of p62 (Figures 3A and 3B). Atg5 functions as a complex with Atg12 and Atg16L1 in the degradative autophagy pathway. Thus, we further assessed the requirement for the Atg12-Atg5-Atg16L1 complex in the IFNγ-mediated control of T. gondii by utilizing Atg5 mutants, Atg7-deficient (Atg7flox/flox+LysMcre) and Atg16L1-deficient (Atg16L1flox/flox+LysMcre) macrophages. In contrast to wild type Atg5, the Atg12 conjugation defective mutant (Atg5-K130R) (Mizushima et al., 1998) and Atg16L1 binding defective mutants (partial defective Atg5-D88A and severely defective Atg5-G84A-D88A) of Atg5 (Hwang et al., 2012) were not able to restore the control of T. gondii by IFNγ (Figure 3A) or basal autophagy (Figure 3B) in the Atg5-deficient macrophages. These data suggest that the conjugation of Atg12 to Atg5 and the binding of Atg5 to Atg16L1 are required for IFNγ to control T. gondii infection.
Figure 3. Atg12-Atg5-Atg16L1 complex formation via Atg7 is required for IFNγ to control T. gondii infection.
(A) Flow cytometry analysis for T. gondii infection +/− 24 hr pre-treatment of 100 U/ml of IFNγ at 24 hpi (MOI=1) in Atg5flox/flox+/−LysMcre BMDMs transduced with control, WT Atg5 or mutants: K130R - defective in Atg12 conjugation, D88A and G84A/D88A – defective in Atg16L1 binding. (B) Protein blot for untreated or uninfected samples as shown in (A). (C) Same analysis as shown in (A) for Atg7flox/flox+/−LysMcre BMDMs transduced with control, WT Atg7, or enzyme-null mutant (C567A). (D) Protein blot for untreated or uninfected samples as shown in (C). (E) Same analysis as shown in (A) for Atg16L1flox/flox+/−LysMcre BMDMs transduced with control, dWDR (WD repeat deletion), dWDR(239-242)A (+FIP200 binding defective), dWDR(194–195)A (+ mutated 194th–195th), and dWDR(239-242)A(194–195)A. (F) Protein blot for untreated/uninfected samples as shown in (E). Statistical analysis by 1-ANOVA with Tukey post test. *: p < 0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001. Combined data as average ± SEM. See also Figure S3.
T. gondii infection was not inhibited by IFNγ in Atg7-deficient macrophages, but transduction with wild type Atg7 restored the control of T. gondii by IFNγ as well as basal autophagy in Atg7-deficient macrophages (Figures 3C and 3D). However, an enzyme-null mutant of Atg7 (Atg7/C567A) (Fujita et al., 2008) was not able to restore either the control of T. gondii by IFNγ or basal autophagy (Figures 3C and 3D). It was recently shown that autophagic sequestration of invading bacteria and the subsequent degradation of the cargo (called xenophagy) can be targeted through the ubiquitination of the endosomes that contain the invading bacteria (Fujita et al., 2013; Levine et al., 2011). The elongation complex localizes and directs LC3 to the ubiquitinated target because Atg16L1 recognizes the ubiquitinated substrates via three independent mechanisms. First, Atg16L1 directly binds to ubiquitin via the WD repeat domain at its C-terminus. Second, Atg16L1 binds to FIP200 in the initiation complex, which is recruited to the ubiquitinated target independently. This interaction is also required for the proper targeting of Atg16L1 to the site of autophagosome initiation for canonical autophagy (e.g. starvation-induced) (Gammoh et al., 2013; Nishimura et al., 2013). Third, amino acids 194–195 of Atg16L1 play a role through an unknown mechanism (Fujita et al., 2013). We therefore reconstituted Atg16L1-deficient macrophages with various mutants of Atg16L1 and investigated the IFNγ-mediated control of T. gondii. As observed for Atg5- and Atg7-deficient macrophages, T. gondii infection was not controlled by IFNγ in the absence of Atg16L1 (Figure 3E). Upon transduction of wild type Atg16L1 and its WD repeat deletion mutant (dWDR), the control of T. gondii by IFNγ and basal autophagy were restored (Figures S3B, 3E and 3F). Further, transduction of FIP200-binding defective derivative, dWDR(239–242)A, and a derivative carrying mutations in amino acids 194–195, dWDR(194–195)A, of WD repeat deletion mutant as well as triple mutant, dWDR(239–242)A(194–195)A (Fujita et al., 2013), restored the control of T. gondii by IFNγ significantly (Figure 3E). However, those mutants did not fully restore basal autophagy (no significant reduction in p62 level) while LC3-II conversion was restored (Figure 3F). These data suggest that the C-terminal WD repeat domain, binding to FIP200 and concomitant ubiquitin-binding activity of Atg16L1 are not required for IFNγ-mediated control of T. gondii and that the N-terminal conserved domain of Atg16L1, which includes a domain for Atg5-binding, is sufficient for its function. Taken together, all these data suggest that the whole elongation complex of Atg12-Atg5-Atg16L1 is required for IFNγ to control T. gondii infection in vitro and further indicated that the function of Atg12-Atg5-Atg16L1 complex in the control of T. gondii by IFNγ is different from its role in canonical autophagy and ubiquitin-dependent xenophagy.
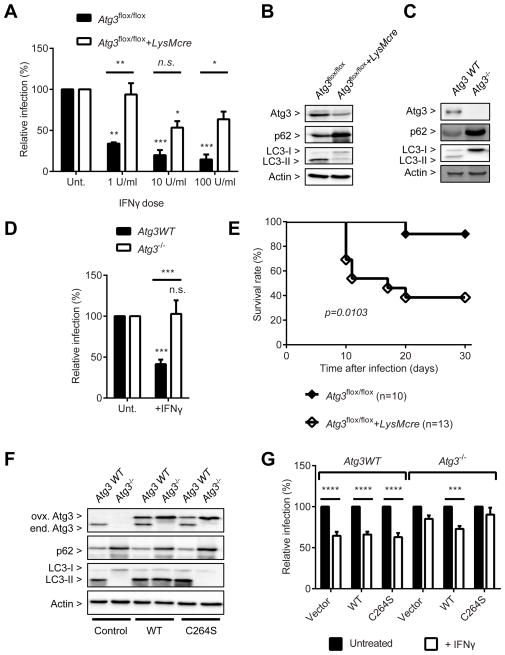
Atg3 is required for IFNγ to control T. gondii infection
The only known function of the Atg12-Atg5-Atg16L1 complex so far is to provide an E3 ligase activity for the targeted conjugation of LC3 homologs to phosphatidylethanolamine on the growing autophagosome (Rubinsztein et al., 2012). Although the canonical autophagy pathway is not required for IFNγ to control T. gondii infection, it was still possible that the conjugation of LC3 homologs to membranes through the E3 ligase activity of the complex is required. To investigate the requirement for LC3 conjugation in the control of T. gondii by IFNγ, we utilized Atg4B- and Atg3-deficient macrophages because of the redundancy of LC3 family members in mammalian system (Shpilka et al., 2011). Atg4B is the dominant isoform of Atg4 in macrophages that is required for the proteolytic processing of LC3 homologs for both conjugation to and deconjugation from autophagosomal membranes (Hwang et al., 2012; Mariño et al., 2010). Atg3 is the essential E2 enzyme only for the conjugation of LC3 homologs (Sou et al., 2008), although Atg7 is required for conjugation of both Atg12 and LC3 homologs. In Atg4B-deficient macrophages, T. gondii infection was controlled by IFNγ significantly better than its control macrophages (Figure S3C). In contrast, in Atg3-deficient macrophages T. gondii infection was controlled by IFNγ significantly less than in control macrophages (Figure 4A). Since the deletion of Atg3 by Cre recombinase in the macrophages was not complete (Figure 4B), we obtained MEFs with complete functional deficiency of Atg3 (Atg3−/−, Figure 4C) and confirmed that T. gondii infection was not controlled by IFNγ (Figure 4D), which is consistent with the recent findings of others (Haldar et al., 2014). We further examined the physiological significance of Atg3 during the in vivo T. gondii infection by infecting Atg3flox/flox+LysMcre and littermate control mice with T. gondii. Consistent with the in vitro data, mice with myeloid lineage specific deletion of Atg3 were significantly more susceptible to the lethal infection of T. gondii than control mice (Figure 4E). We further examined the importance of E2 enzyme activity of Atg3 in the IFNγ-mediated control of T. gondii by transducing Atg3−/− MEF with wild type and enzyme-null mutant (C264S) of Atg3 (Figure 4F) (Sou et al., 2008). T. gondii infection was significantly controlled by IFNγ in Atg3−/− MEF transduced with wild type Atg3 but not with the enzyme-null mutant, demonstrating that the E2 enzyme activity of Atg3 is required for the control of T. gondii by IFNγ (Figure 4G). Thus, E2 conjugating enzyme Atg3, in addition to E1 activating enzyme Atg7 and E3 ligase Atg12-Atg5-Atg16L1 complex, is required for the IFNγ-mediated control of T. gondii infection in vitro and in vivo.
Figure 4. Atg3 is required for the control of T. gondii infection.
(A) Flow cytometry analysis for T. gondii infection at 24 hpi (MOI=1) in Atg3flox/flox+/−LysMcre BMDMs +/− 24 hr pre-treatment of IFNγ at the indicated doses. (B) Protein blot for untreated or uninfected Atg3flox/flox+/−LysMcre BMDMs. (C) Protein blot for WT and Atg3−/− MEF. (D) Same analysis as shown in (A) for the WT and Atg3−/− MEF. (E) Survival curves of Atg3flox/flox+/−LysMcre mice after intraperitoneal inoculation with T. gondii. (F) Protein blot for untreated or uninfected samples as shown in (G). (G) Same analysis as shown in (A) for WT and Atg3−/− MEFs transduced with control, WT Atg3, or its enzyme-null mutant (C264S) +/− 24 hr pre-treatment of 100 U/ml of IFNγ. Statistical analysis by 1-ANOVA with Tukey post test or Log-rank (Mantel-Cox) test. n.s.: not significant (p>0.05), **: p<0.01, ***: p<0.001, ****: p<0.0001. Combined data as average ± SEM. See also Figure S3.
Colocalization of LC3 with the parasitophorous vacuole membrane of T. gondii
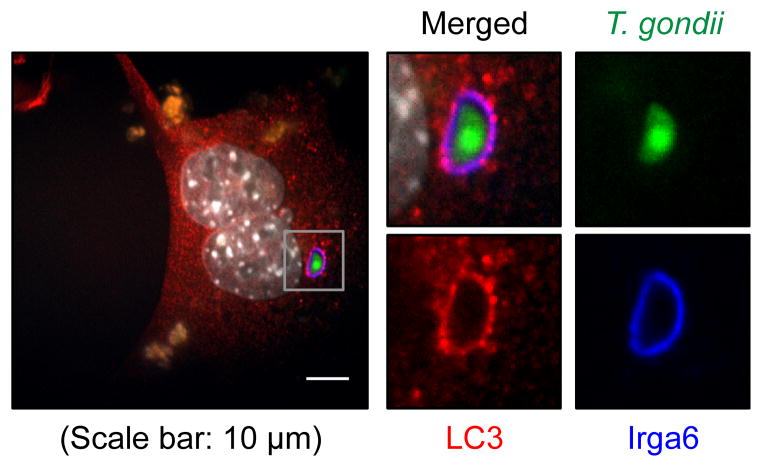
We and others have showed that Atg5 is required for the proper targeting of the IFNγ-induced effectors to the PVM of T. gondii without the involvement of autophagosome (Selleck et al., 2013; Zhao et al., 2009; 2008). One plausible mechanism based on our data was that the conjugation of LC3 homologs plays a crucial role in the recruitment of the IFNγ effectors onto the PVM of T. gondii without the involvement of functional modules for autophagosome initiation or nucleation and lysosomal degradation. Therefore, we reasoned that LC3 homologs might be directly conjugated onto the PVM of T. gondii. However, it was previously shown that GFP-LC3, conventional marker for autophagosomal membrane, does not co-localize with the PVM of T. gondii (Martens et al., 2005). Since a discrepancy in cellular localization and function between the N-terminal GFP-tagged LC3 and endogenous LC3 has been reported (Reggiori et al., 2010), we examined the localization of endogenous LC3 with regard to invading T. gondii and IFNγ-induced effectors, such as immunity-related GTPase 6 (Irga6, a.k.a. interferon inducible GTPase 1, IIGP1). We first investigated the localization of LC3 in non-phagocytic cells, MEFs, because LC3 can be recruited to the phagosomal single membrane (Sanjuan et al., 2007). Live T. gondii enters cells by an active parasite-driven process that is independent of phagocytosis (Sibley, 2011). At 2 hour-post-infection (hpi) of IFNγ-treated MEFs, Irga6 localized on the PVM of T. gondii and the colocalization of endogenous LC3 with Irga6 was detected on the PVM (Figure 5). Therefore, endogenous LC3 can localize on or near the PVM of T. gondii and, considering the absence of double membrane bound autophagosome near the PVM (Zhao et al., 2008) and the necessity of LC3 conjugation machinery for the control of T. gondii by IFNγ, it is highly likely that LC3 is conjugated to the PVM of T. gondii.
Figure 5. LC3 and Irga6 localize on the PVM of T. gondii.
(A) A representative image of immunofluorescence for WT MEF at 2 hpi (MOI=1) of T. gondii infection with 24 hr pre-treatment of 100 U/ml of IFNγ. Experiments were performed more than thrice.
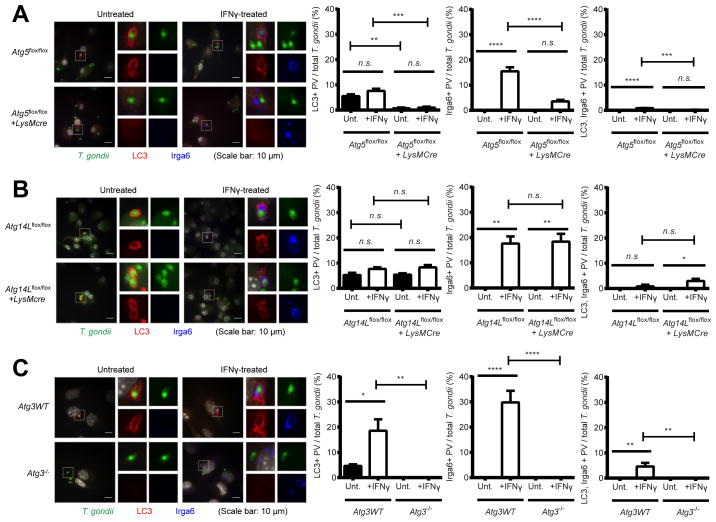
Atg5 and Atg3, but not Atg14L, are required for the localization of LC3 and Irga6 on the PVM
To examine the genetic requirement of LC3 and Irga6 localization on the PVM of T. gondii, we further investigated their localization with regard to invading T. gondii in Atg5- and Atg14L-deficient macrophages, as well as in Atg3−/− MEFs. In IFNγ-treated control macrophages, Irga6 and LC3 localized on the PVMs of T. gondii both individually and concurrently (Figure 6A). Even in untreated macrophages, LC3 alone localized on the PVM of a small percentage of T. gondii in the absence of Irga6, suggesting that the localization of LC3 on the PVM of T. gondii is not dependent on induction or recruitment of IFNγ effectors. In contrast, in Atg5-deficient macrophages the localizations of LC3 and Irga6 on the PVM of T. gondii were significantly reduced and Irga6 formed cytoplasmic aggregates as shown previously (Zhao et al., 2008). These data demonstrate that Atg5 is required for the proper targeting of both LC3 and Irga6 to the PVM of T. gondii as well as for the control of T. gondii by IFNγ.
Figure 6. Atg5 and Atg3, but not Atg14L, are required for the localization of LC3 and Irga6 on the PVM of T. gondii.
Representative images (left) and quantitation (right) of immunofluorescence for T. gondii, LC3, and Irga6 in (A) Atg5flox/flox+/−LysMcre and (B) Atg14Lflox/flox+/−LysMcre BMDMs and (C) Atg3 WT and Atg3−/− MEFs at 2 hpi (MOI=1) of T. gondii infection +/− 24 hr pre-treatment of 100 U/ml of IFNγ. At least 100 cells infected T. gondii were analyzed for quantitation. Statistical analysis by 1-ANOVA with Tukey post test. n.s.: not significant (p>0.05), **: p<0.01, ***: p<0.001, ****: p<0.0001. Combined data as average ± SEM.
In contrast to Atg5-deficient macrophages, in Atg14L-deficient macrophages there was no reduction in the localization of LC3 and Irga6 to the PVM of T. gondii (Figure 6B) and Irga6 did not form aggregates, indicating that localization of LC3 and Irga6 onto the PVM as well as the control of T. gondii by IFNγ do not require a gene essential for canonical degradative autophagy. However, in Atg3−/− MEFs, LC3 did not localize on the PVM of T. gondii and Irga6 formed cytoplasmic aggregates (Figure 6C). Again, LC3 was detected on the PVM of a small percentage of T. gondii even in the absence of IFNγ treatment, confirming that the localization of LC3 on the PVM requires functional Atg3 but not IFNγ. Collectively, our data demonstrated that Atg5 and Atg3, but not Atg14L, are required for the localization of LC3 and Irga6 on the PVM of T. gondii and that LC3 localizes on the PVM of T. gondii independent of IFNγ.
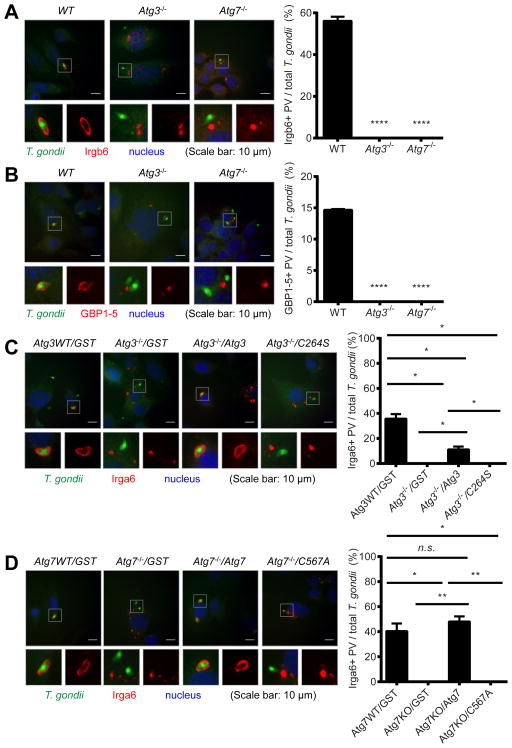
The ubiquitin-like conjugation systems are required for the localization of IFNγ effectors onto PVM
We confirmed that another member of the IRG family (Irgb6, Figure 7A) and GBPs (GBP1-5, Figure 7B) (Howard et al., 2011; Kim et al., 2012) were also recruited onto the PVM of T. gondii only in the presence of Atg3 and Atg7. Further, when we tested the control of endodyogenic replication of T. gondii within a parasitophorous vacuole by IFNγ, IFNγ also significantly reduced the replication of T. gondii in the vacuole, in an Atg3- and Atg7-dependent manner (Figure S4A). Lastly, to examine the necessity of the enzyme activity of the Atg3 and Atg7 in the recruitment of IFNγ effectors onto the PVM of T. gondii, we reconstituted Atg3−/− and Atg7−/− MEFs with wild type and catalytically inactive mutants of Atg3 (C264S) and Atg7 (C567A), respectively (Figures 4F and S4B). Consistent with the genetic data (Figures 3C and 4G), wild type Atg3 and Atg7 were able to restore the recruitment of Irga6 onto the PVM of T. gondii in Atg3−/− and Atg7−/− MEF, respectively (Figures 7C and 7D). However, catalytically inactive mutants of Atg3 and Atg7 were incapable of restoring the recruitment of the IFNγ effector onto the PVM. Collectively, these data demonstrate the essential role of the ubiquitin-like conjugation systems of the autophagy pathway in the recruitment of IFNγ effectors onto the PVM of T. gondii and subsequent control of T. gondii infection and replication.
Figure 7. The ubiquitin-like conjugation systems of the autophagy pathway are required for the localization of IFNγ effectors onto the PVM of T. gondii infection.
Immunofluorescence of MEFs at 2 hpi (MOI=1) of T. gondii infection with 24 hr pre-treatment of 100 U/ml of IFNγ. Representative images (left) and quantitation (right) of immunofluorescence. Irgb6 (A) and GBP1-5 (B) in WT, Atg3−/− and Atg7−/− MEFs. At least 50 of infected T. gondii were analyzed for quantitation. Statistical analysis by 1-ANOVA with Tukey post test. ****: p < 0.0001. Experiments were performed twice. Combined data as average ± SEM. (C) T. gondii and Irga6 in Atg3 WT and Atg3−/− MEFs transduced with control (GST, Glutathione-S-transferase), WT Atg3, or enzyme-null mutant (C264S). (D) T. gondii and Irga6 in Atg7 WT and Atg7−/− MEFs transduced with control (GST), WT Atg7, or enzyme-null mutant (C567A). At least 100 of infected T. gondii were analyzed for quantitation. Statistical analysis by unpaired t-test. n.s.: not significant. *: p < 0.05, **: p < 0.01. Experiments were performed twice. Combined data as average ± SEM. See also Figure S4.
Discussion
Degradative autophagy has been shown to play crucial roles in immune defense against intracellular pathogens as well as its conventional roles in recycling and remodeling (Levine et al., 2011). In addition, the evolutionarily conserved autophagy proteins have evolved and acquired unique functions in the immune system and myeloid cells, independent of their roles in the degradation pathway (Bestebroer et al., 2013; DeSelm et al., 2011; Hwang et al., 2012; Reggiori et al., 2010; Starr et al., 2012; Subramani and Malhotra, 2013). In this report, we extended these observations by demonstrating that the ubiquitin-like conjugation machinery of the autophagy pathway, E1 Atg7, E2 Atg3, and E3 Atg12-Atg5-Atg16L1, are required for the proper targeting of IFNγ effectors onto the PVM of T. gondii and subsequent control of in vitro and in vivo T. gondii infection. Although degradative autophagy has been proposed to play a role in the CD40 ligation-mediated defense against T. gondii in non-hematopoietic cells (Van Grol et al., 2013), the multiple aspects of the overall autophagy pathway including the ULK1and ULK2 initiation complex, the Atg14L nucleation complex, and lysosomal degradation were not required for the control of T. gondii by IFNγ in MEFs or macrophages. We further found that LC3 localized on, and possibly was conjugated to, the PVM of T. gondii, and confirmed that the recruitment of IFNγ effectors to the PVM was dependent on the functional ubiquitin-like conjugation systems of the autophagy pathway. These data suggest that the ubiquitin-like conjugation systems utilized for the reorganization of intracellular membranes in the canonical autophagy pathway have evolved to target immune effectors toward the intracellular vacuole membrane structure of pathogens.
Although IRGs and GBPs are thought to participate in disrupting the PVM, the molecular mechanism of their recruitment to this interface through Atg5 is incompletely understood (Howard et al., 2011). The autophagosome itself is not directly involved in the control of T. gondii per se (Zhao et al., 2009; 2008). Nevertheless, it was possible that a functional degradative autophagy pathway may be required to remove prematurely activated IFNγ effectors, which tend to aggregate and subsequently inhibit the proper loading of the effectors on the target membrane (Selleck et al., 2013). In this setting, one might predict that the entire degradative autophagy pathway and all essential autophagy proteins are required for proper effector function. However, our data clearly demonstrated that the degradative autophagy pathway and the other functional cassettes of autophagy proteins were not required for efficient control of T. gondii by IFNγ. In fact, unlike the situation for Atg5-deficient cells, we did not detect any aggregation of the activated IFNγ effectors in Atg14L-deficient macrophages, even though canonical autophagy was significantly inhibited. Thus, our current data do not support the role of degradative autophagy for the proper maintenance of activated IFNγ effectors.
Independent of IFNγ effector targeting to T. gondii, LC3 localized on a small percentage of PVMs in a ubiquitin-like conjugation system dependent process. This finding is reminiscent of Atg16L1 localization on the membranous replication complex of MNV without IFNγ-treatment (Hwang et al., 2012). The Atg12-Atg5-Atg16L1 complex, particularly Atg16L1, can specify the lipidation site of LC3 for the membrane biogenesis in both nonselective autophagy (Fujita et al., 2008) and selective xenophagy (Fujita et al., 2013). Thus, it is tempting to speculate that the Atg12-Atg5-Atg16L1 complex may function as a module to recognize and/or mark the “non-self” intracellular membrane structure of pathogens through the conjugation of LC3. The conjugated LC3 may recruit the effectors through direct interaction or indirectly through adaptor proteins in a similar way to its function in canonical autophagy (Shpilka et al., 2011). Alternatively, although the localization of LC3 on the PVM of T. gondii happened even in non-phagocytic cells, it is still possible that these events may represent parasites that were engulfed by cellular processes other than pathogen-driven invasion. Resolving these respective roles will take further studies on the kinetics of LC3 recruitment to PVMs in non-IFNγ-treated cells.
Interestingly, T. gondii infection was controlled by IFNγ more efficiently in Atg4B-deficient cells than in control cells, which also happened in the control of MNV by IFNγ (Hwang et al., 2012). A major function of Atg4 family proteins is to cleave the membrane bound LC3 as well as to proteolytically process LC3 for membrane conjugation. Thus, in the absence of Atg4B, the deconjugation or delipidation of LC3 from the membrane is slowed down, which may lead to the more recruitment of the IFNγ effectors and thus more efficient control of pathogens by IFNγ. In this regard, it is interesting to note that Atg4B can be recruited onto the membrane structure of pathogens by one of the IFNγ effectors, GBP7 (Kim et al., 2011). The recruitment of Atg4B to the membrane by the IFNγ effectors and subsequent cleavage of LC3 from the membrane might be a negative feedback mechanism for proper activity of IFNγ effectors.
Although both LC3 and the IFNγ effector Irga6 localized on the PVM of T. gondii, there was relatively little concurrent colocalization between LC3 and Irga6 on the PVM. These data suggest that LC3 and the IFNγ effectors may not work simultaneously and their recruitment to the PVM of T. gondii may be spatially and temporally regulated for efficient targeting and disruption of the membrane structure of the pathogens. Alternatively, the ubiquitin-like conjugation system of the autophagy pathway might be required for some yet undefined role that is necessary for proper homeostasis of the IFNγ effectors, since they form aggregates when these autophagy proteins are absent. Indeed, it was previously proposed that Atg5 may be required for the normal function of regulatory IRGs (GMS subfamily, e.g. Irgm1, Irgm3) to demarcate endosomal membranes and special pathogen-oriented membranes and/or to regulate nucleotide exchange and activation of the effector IRGs (GKS subfamily, e.g. Irga6, Irgb6) (Haldar et al., 2013; Hunn et al., 2008). This explanation is not mutually exclusive with the mechanisms proposed here. Further studies are required to elucidate how the conjugation of LC3 homologs by the ubiquitin-like conjugation systems determines the recruitment of the IFNγ effectors onto the PVM of T. gondii.
It is interesting to note that a similar dependence on Atg5 and Atg7 and targeting of LC3 to single membranes has been observed for the secretion of lysosomal proteins by osteoclasts and mucin by colonic goblet cells, LC3-associated phagocytosis (LAP) and entotic cell engulfment (DeSelm et al., 2011; Florey et al., 2011; Martinez et al., 2011; Patel et al., 2013). Particularly, considering the enhanced killing of phagocytosed microbes through LAP (Sanjuan et al., 2007), it will be important to further study the shared and differentiated mechanism between LAP and the process described here that targets intracellular membrane structures in host immune defense. Notably, the parasitophorous vacuole of T. gondii is actively made by the pathogen itself even in non-phagocytic cells, suggesting that, even if the same machinery is required by both LAP and destruction of intracellular T. gondii, the membrane events involved may differ topologically.
Our findings confirm and extend recently published findings showing the critical role of Atg3, Atg7, and Atg16L1, but not Atg9a and Atg14, for the control of T. gondii by IFNγ effectors (Haldar et al., 2014; Ohshima et al., 2014). The data we provide here show in vivo relevance and molecular proof of the specific functions of the required autophagy proteins (e.g. enzymatic activity of Atg3 and Atg7, binding of Atg5 to Atg16L1) in intracellular killing of T. gondii. Importantly, we also show here the functions of the autophagy proteins in macrophages, primary cells that are involved in resistance to T. gondii. Thus, our study firmly demonstrated the nondegradative function of ubiquitin-like conjugation systems of the autophagy pathway in cell-autonomous immune defense system. Together, these studies establish the paradigm that autophagy proteins function in intracellular immunity in ways that do not require the entire degradative autophagy pathway.
In summary, we found that the ubiquitin-like conjugation machinery of the autophagy pathway, but not the degradative autophagy pathway itself, plays crucial roles in the anti-parasitic activity of IFNγ in that it is required for the proper targeting of the IFNγ effectors to the vacuole membrane structure of the parasites. Elucidating such nondegradative roles of autophagy proteins in sensing and inducing the destruction of pathogenic membrane structures in the cytoplasm may lead to novel therapeutic and/or prophylactic treatments for the infectious diseases caused by these pathogens as well as to a greater appreciation and understanding of fundamental biological processes of the genes in the autophagy pathway. The evolutionary conservation of autophagy from the earliest stages of metazoan evolution may have favored the diversification of autophagy proteins to contribute differentially to host defense and thereby to play key roles both through degradative functions of autophagy and through other, perhaps as yet undiscovered, mechanisms of intracellular immunity.
Experimental Procedures
Mice and Cells
Atg5flox/flox+/−LysMcre, Atg7flox/flox+/−LysMcre, Atg16L1flox/flox+/−LysMcre mice were previously described (Hwang et al., 2012). Atg14Lflox/flox+/−LysMcre mice were kindly provided by Dr. Shizuo Akira, Osaka University, Japan. Atg3flox/flox+/−LysMcre mice were derived from Atg3flox/flox mice kindly provided by Dr. You-Wen He, Duke University, U.S.A. (Jia and He, 2011). All mice were housed and bred at Washington University in St. Louis, St. Louis MO and University of Chicago, Chicago IL under specific-pathogen-free conditions in accordance with federal and university guidelines. Bone marrow derived macrophages (BMDMs), MEFs, and 293T cells were used for T. gondii infection and lentiviral transduction. See the Supplemental Experimental Procedures for the details.
Infection with T. gondii
T. gondii (type-II strains) expressing luciferase (ME49) and green fluorescent protein (PTG) were maintained in HFF cells (Zhao et al., 2008). At the time of infection, an inoculum of infectious T. gondii tachyzoites was prepared by disrupting the infected HFF cells using 26G needle and syringe, centrifuging the lysate at 400 x g for 10 minutes, and resuspending the precipitate in culture media (in vitro) or serum free DMEM (in vivo). For in vitro experiments, cells were pretreated with recombinant murine IFNγ at the indicated doses (e.g. 100 U/ml = 50 ng/ml) for 24 hours, infected with T. gondii tachyzoites at the multiplicity of infection (MOI) of 1. At 24 hr-post-infection (hpi), the infected cells were fixed with 2% formaldehyde (Ted Pella; 18505) in PBS for 10–20 minutes at room temperature. The fixed cells were permeabilized with PBS + 0.1% Triton X-100 (PBS-T) overnight at 4°C and T. gondii infection was assessed by indirect immunofluorescence or flow cytometry. For in vivo experiments of Atg5flox/flox+/−LysMcre, Atg7flox/flox+/−LysMcre, Atg14Lflox/flox+/−LysMcre, and Atg16L1flox/flox+/−LysMcre mice, 9~11 week old mice were inoculated intraperitoneally with 200 (male) or 100 (female) of T. gondii tachyzoites per mouse (Zhao et al., 2008) and the survival of the infected mice was monitored for 30 days in accordance with the guideline of Washington University in St. Louis. No significant phenotypic difference was observed between male and female mice. The T. gondii infection study of Atg3flox/flox+/−LysMcre mice were performed in the presence of 0.6 ug/ml buprenorphine under the guideline of University of Chicago. Two or three independent experiments were done for each in vivo infection study.
Chemicals
Rapamycin (sc-3504, 5 mM in DMSO), wortmannin (sc-3505, 5 mM in DMSO), bafilomycin A1 (sc-201550, 100 uM in DMSO), E64D (sc-201280, 5 mg/ml in DMSO), pepstatin A (sc-45036, 5 mg/ml in DMSO) from Santa Cruz Biotechnology; chloroquine (C6628, 20 mM in water) from Sigma; LY294002 (#9901, 10 mM in DMSO) from Cell Signaling; mouse IFNγ (485MI/CF, 200 U/ul (100 ng/ul) in PBS/0.1%BSA) from R&D Systems.
Protein analysis by western blot
Total cellular proteins were harvested and proteins were analyzed as previously described (Hwang et al., 2012). Peritoneal exudate cells (PECs) were obtained by lavage, incubated at 37°C for 4 hours on tissue culture treated plates, and washed to purify adherent macrophages. Cell lysates were then harvested and analyzed by western blot. Commercial antibodies used in this study are as follows: 2A (Millipore, ABS31), Atg3 (MBL, M133-3), Atg7 (Sigma, A2856), Atg14L (Sigma, A6358), Atg16L1 (Sigma, A7356), Beta-actin (Sigma, A5316), LC3B (Sigma, L7543), and p62 (Sigma, P0067).
Immunofluorescence and Flow cytometry
Fixed and permeabilized cells were analyzed for T. gondii infection by immunofluorescence and flow cytometry using rabbit polyclonal anti-GRA7 (dense granule protein 7) (Selleck et al., 2013) or GFP-expressing reporter stain (PTG). See the Supplemental Experimental Procedures for the details.
Statistical analysis
All data were analyzed with Prism software (GraphPad). Unless otherwise stated, all experiments were performed at least three times and the data were combined for presentation as average ± SEM. All differences not specifically indicated to be significant were not significant (n.s., p > 0.05). *: p < 0.05, **: p < 0.01, *** p < 0.001, ****: p < 0.0001.
Supplementary Material
Highlights.
A cassette of Atg7, Atg3, Atg12-Atg5-Atg16L1 mediates IFNγ control of T. gondii.
The degradative pathway of autophagy is not required for IFNγ control of T. gondii.
Autophagy proteins mark membrane structures by pathogens for intracellular immunity.
Autophagy proteins are required to direct IFNγ effectors to membranes of pathogens.
Acknowledgments
This work was supported by U19 AI109725, RO1 AI054483, RO1 AI084887, and CCFA grant #274415 to HWV, start-up funds to SH and RO1 AI03669 to LDS. We thank Virgin lab and Hwang lab members for their comments on the manuscript and D. Kreamalmeyer and M. White for managing mouse colonies. Experimental support was provided by the Speed Congenics Facility of the Rheumatic Diseases Core Center. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the National Institutes of Health, under Award Number P30AR048335.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bestebroer J, V’kovski P, Mauthe M, Reggiori F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic. 2013;14:1029–41. doi: 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21:966–74. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–43. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–28. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Str Mol Biol. 2013;20:144–9. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-Like Conjugation Enzyme Atg3 Promotes Binding of IRG and Gbp Proteins to Chlamydia- and Toxoplasma-Containing Vacuoles and Host Resistance. PLoS ONE. 2014;9:e86684. doi: 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J. IRG and GBP Host Resistance Factors Target Aberrant, “Non-self” Vacuoles Characterized by the Missing of ‘Self’ IRGM Proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JC, Hunn JP, Steinfeldt T. The IRG protein-based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii. Curr Opin Micro. 2011;14:414–21. doi: 10.1016/j.mib.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO. 2008;27:2495–509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Micro. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, He Y-W. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immun. 2011;186:5313–22. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- Jones JL, Krueger A, Schulkin J, Schantz PM. Toxoplasmosis prevention and testing in pregnancy, survey of obstetrician-gynaecologists. Zoon Pub Health. 2010;57:27–33. doi: 10.1111/j.1863-2378.2009.01277.x. [DOI] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A Family of IFN-γ-Inducible 65-kD GTPases Protects Against Bacterial Infection. Science. 2011;332:717–21. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- Kim B-H, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-γ-Inducible GTPases in Host Cell Defense. Cell Host Micro. 2012;12:432–44. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, Salvador-Montoliu N, Vega JA, Germanà A, Fueyo A, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC. Disruption of Toxoplasma gondii Parasitophorous Vacuoles by the Mouse p47-Resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA. 2011;108:17396–401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, AKIRA S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McAlpine F, Williamson LE, Tooze SA, Chan EYW. Regulation of nutrient- sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013;9:361–73. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–92. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in Mammalian Autophagy Research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, Mizushima N. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 2013;14:284–91. doi: 10.1038/embor.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, Matsuura Y, Pann-Ghill S, Hayashi M, Ebisu S, et al. Role of Mouse and Human Autophagy Proteins in IFN-γ-Induced Cell-Autonomous Responses against Toxoplasma gondii. J Immun. 2014;192:3328–35. doi: 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- Pappas G, Roussos N, Falagas ME. Toxoplasmosis snapshots: global status of Toxoplasma gondii seroprevalence and implications for pregnancy and congenital toxoplasmosis. Inter J Parasit. 2009;39:1385–94. doi: 10.1016/j.ijpara.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Parzych KR, Klionsky DJ. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid Redox Sig. 2013;20:460–73. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO. 2013;32:3130–44. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, Bernasconi R, de Haan CAM, Molinari M. Coronaviruses Hijack the LC3-I-Positive EDEMosomes, ER-Derived Vesicles Exporting Short-Lived ERAD Regulators, for Replication. Cell Host Micro. 2010;7:500–8. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of Autophagosome Biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–57. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HW, MacMicking JD, Sibley LD. Guanylate-binding Protein 1 (Gbp1) Contributes to Cell-autonomous Immunity against Toxoplasma gondii. PLoS Pathog. 2013;9:e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Gen Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD. Invasion and intracellular survival by protozoan parasites. Immunol Rev. 2011;240:72–91. doi: 10.1111/j.1600-065X.2010.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata JI, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Micro. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–51. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Grol J, Muniz-Feliciano L, Portillo JAC, Bonilha VL, Subauste CS. CD40 Induces Anti-Toxoplasma gondii Activity in Nonhematopoietic Cells Dependent on Autophagy Proteins. Infect Immun. 2013;81:2002–11. doi: 10.1128/IAI.01145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YO, Khaminets A, Hunn JP, Howard JC. Disruption of the Toxoplasma gondii Parasitophorous Vacuole by IFNγ-Inducible Immunity-Related GTPases (IRG Proteins) Triggers Necrotic Cell Death. PLoS Pathog. 2009;5:e1000288. doi: 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-Independent Essential Function for the Autophagy Protein Atg5 in Cellular Immunity to Intracellular Pathogens. Cell Host Micro. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1 phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.