Abstract
Numerous studies over the past decade have identified increasing numbers of long noncoding RNAs (lncRNAs) across many organisms. Research since has shown that lncRNAs constitute an important layer of genome regulation in diverse biological processes and disease. Here, we discuss the common emerging theme of lncRNAs interfacing with epigenetic machinery. This, in turn, modulates the activity and localization of the epigenetic machinery during cell fate specification.
Long noncoding RNAs (lncRNAs) physically associate with chromatin-modifying complexes and guide them to specific genomic loci. For example, HOTAIR interacts with PRC2, resulting in the trans repression of HOX genes.
Cellular identity is achieved through a complex choreography of DNA regulatory elements interacting with protein regulatory complexes to shape a myriad of unique epigenetic landscapes. The vast arrays of epigenetic landscapes are established by using ubiquitously expressed chromatin-modifying and -remodeling complexes, to give rise to countless unique combinations of histone posttranslational modifications and DNA methylation patterns. This raises the age-old question: How are the enzymatic complexes being guided to place these marks at a specific combination of sites under different cellular contexts? It has long been suspected that noncoding RNA molecules may provide some specificity to target these complexes to their sites of action. Indeed, it is becoming increasingly clear that a contingent of thousands of long noncoding RNAs (lncRNAs) represent a key layer of epigenetic control (see Fig. 24 of Allis et al. 2014).
A dramatic example of RNA-based epigenetic regulation that operates in mammalian dosage compensation has been known for more than 20 years (Brockdorff and Turner 2014). Specifically, a long intergenic noncoding RNA (lincRNA) termed XIST (X inactive-specific transcript) is expressed from one female X chromosome resulting in the recruitment of Polycomb group complexes (PcG), such as PRC2, to this chromosome with the concomitant transcriptional silencing across a majority of the X chromosome. In other words, a single lncRNA gene is able to target and silence the majority of a chromosome in cis. This is a powerful precedent for RNA-mediated epigenetic regulation. However, the link between the RNA and PRC2 recruitment was, for a long time, elusive. A clue came from studying the epigenetic dynamics of a different lincRNA termed HOTAIR (Hox transcript antisense intergenic RNA; Rinn et al. 2007).
HOTAIR is expressed from one of the four clusters of HOX transcription factors (i.e., HOXC). The HOX family of proteins is a key regulator of body plan during development. HOTAIR was identified because of its distinctive pattern of expression in posterior and distal mouse tissues during development and adult human fibroblasts (Rinn et al. 2007). Intriguingly, HOTAIR expression demarcated a distinct epigenetic boundary between euchromatic and heterochromatic regions within the HOXC cluster. Moreover, the euchromatic-heterochromatic regions were inverted between anterior and posterior cell types; we termed these “diametric” chromatin domains. Collectively, the data led to the initial hypothesis that HOTAIR serves as an epigenetic boundary in cis within the HOXC cluster. To our surprise the HOXC chromatin boundary remained unchanged when HOTAIR RNA function was lost; however, the HOXD cluster, located on a separate chromosome, became active. Thus, similar to XIST, the expression of HOTAIR from the HOXC cluster results in the epigenetic silencing, yet the HOX cluster that it regulates (HOXD) is located on a different chromosome, i.e., is regulated in trans. This raised the question: How?
The answer was determined by a few critical experiments, which indicated that the HOTAIR lincRNA interacted with PRC2 and that this was required for proper localization of PRC2. The first experiment, using immunoprecipitation of the PRC2 complex, identified HOTAIR as being physically associated or coprecipitated with PRC2 (Rinn et al. 2007). The reciprocal experiment, which isolated proteins bound to the HOTAIR transcript, also revealed an association between HOTAIR and components of the PRC2 complex but not other chromatin regulatory factors. The final missing link was the demonstration that an RNA–protein interaction between HOTAIR and PRC2 is required for proper localization of PRC2 across the HOXD cluster. Indeed, depletion of HOTAIR resulted in the mislocalization of PRC2 from the HOXD cluster, in trans, and concomitant activation of HOXD genes. Together, these findings showed that the physical association between HOTAIR and PRC2 was required for the proper targeting of the chromatin regulatory machinery to their target sites. Thus, a novel layer of RNA-based modulation of epigenetic landscapes was unequivocally revealed.
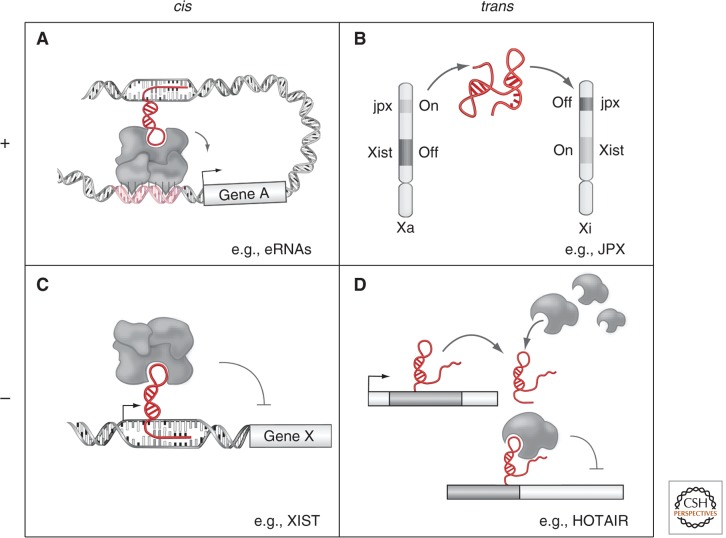
Since unraveling the fact that PRC2 physically associates with HOTAIR or XIST, so targeting the complex to specific genomic regions, this mechanism has been shown to apply to numerous lncRNAs that physically associate with PRC2. Many of these associations are, in fact, required for the proper localization of the epigenetic regulatory machinery in cis and trans (see Fig. 1). Two independent studies, in particular, determined that in both human and mouse cells, hundreds of lncRNAs, accounting for up to 30% of the transcriptome, coprecipitate with PRC2. Moreover, many of the tested PRC2-bound lncRNAs were required for proper epigenetic and transcriptional regulation of PRC2 targets (Khalil et al. 2009; Zhao et al. 2010). It was further noted from these studies that one RNA could be bound to many different chromatin regulatory proteins, suggesting that it could function as an RNA bridge across multiple complexes (Khalil et al. 2009). Indeed, a detailed biochemical analysis of HOTAIR showed that in addition to binding PRC2, it also bound the histone demethylase LSD1 and NCOR. This pointed to a novel model for epigenetic regulation whereby a single lncRNA recruits several synergistic chromatin regulatory complexes to help guide, dock (NCOR), and facilitate heterochromatin formation (LSD1 and PRC2). Collectively, these studies led to the idea that an RNA scaffold can bridge numerous chromatin and additional regulatory complexes to impart genome target specificity (Fig. 1D). The recent finding that HOTAIR overexpression is a hallmark in metastatic breast cancer (Gupta et al. 2010) has underscored the importance of HOTAIRs role in epigenetic regulation. In fact, HOTAIR serves as an “onco-lncRNA,” inducing metastasis in breast cancer when overexpressed by remodeling the epithelial epigenome to resemble that of stromal cells. Cumulatively, the lessons learned from HOTAIR over the past five years have shown that lncRNAs play a critical role in interfacing with and modulating chromatin complexes during development and disease.
Figure 1.
Models of how lncRNAs may function in the epigenetic control of gene expression, both activating and repressing transcription in cis and in trans. (A) Enhancer RNAs have been shown to play an RNA-mediated role in forming enhancer interactions resulting in long-range cis gene regulation by lncRNAs. (B) Activation by the JPX lncRNA as an example of a trans-acting lncRNA, which in this case facilitates Xist activation. (C) Xist as an example of an lncRNA that facilitates gene repression in cis across the majority of the X chromosome. (D) HOTAIR expression results in trans repression of HOX genes.
In the 50 years since RNA was identified as a central component in the flow of genetic information, it has become increasingly clear that RNA is more than a mere messenger and instead performs vast and diverse functions (Amaral et al. 2008). lncRNAs are, in fact, emerging as a critical layer of epigenetic regulation in which different lncRNAs are associated with distinctive epigenetic states, yet share a common mechanism; they physically associate with chromatin-modifying and -remodeling complexes, and guide them to specific genomic loci that are crucial for proper cellular function. However, this is only one facet of lncRNA biology; there is a diversity of other functional roles they play across numerous biological processes alluded to in Amaral et al. (2008).
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- *.Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 [DOI] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS 2008. The eukaryotic genome as an RNA machine. Science 319: 1787–1789 [DOI] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner BM 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]