Abstract
With increasing intracellular complexity, a new cell-biological problem that is the allocation of cytoplasmically synthesized proteins to their final destinations within the cell emerged. A special challenge is thereby the translocation of proteins into or across cellular membranes. The underlying mechanisms are only in parts well understood, but it can be assumed that the course of cellular evolution had a deep impact on the design of the required molecular machines. In this article, we aim to summarize the current knowledge and concepts of the evolutionary development of protein trafficking as a necessary premise and consequence of increased cellular complexity.
The progressive compartmentalization of eukaryotes required the coevolution of intracellular protein transport systems. These were most likely built on preexisting structures and mechanisms from bacterial ancestors.
The evolution of modern cells is arguably the most challenging and important problem the field of biology has ever faced …
—Carl R. Woese
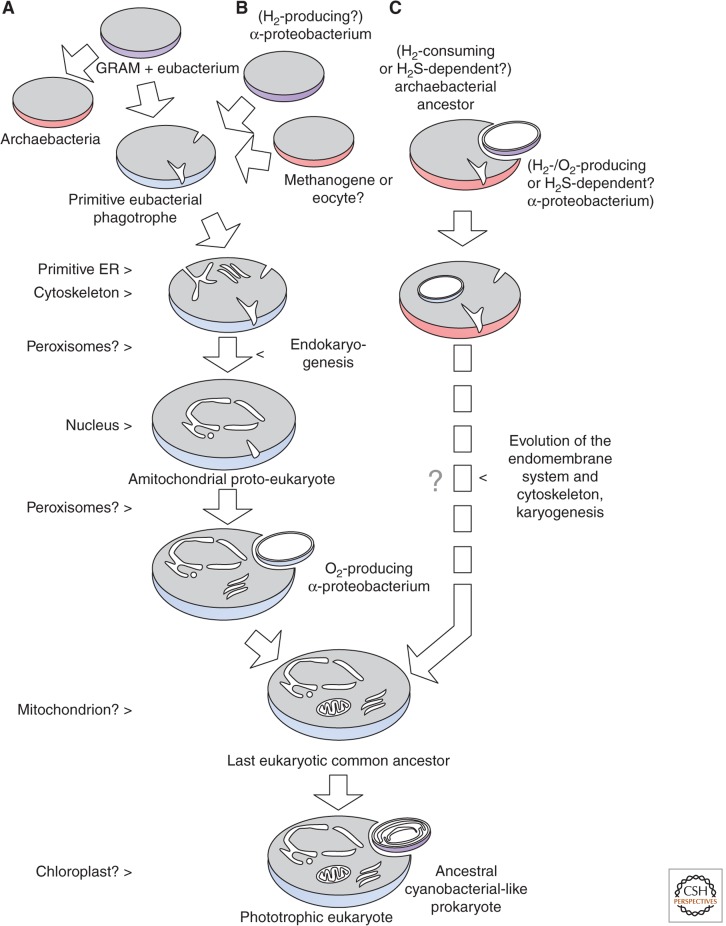
Current models may accept that all modern eukaryotic cells arose from a single common ancestor (the cenancestral eukaryote), the nature of which is—owing to the lack of direct living or fossil descendants—still highly under debate (de Duve 2007). The chimeric nature of eukaryotic genomes with eubacterial and archaebacterial shares led to a discussion about the origin of this first “proto-eukaryote.” Several models exist (see Fig. 1), which either place the evolution of the nucleus before or after the emergence of the mitochondrion (outlined in Koonin 2010; Martijn and Ettema 2013). According to the different postulated scenarios (summarized in Embley and Martin 2006), eukaryotes in the latter case might have evolved by endosymbiosis between a hydrogen-producing, oxygen-producing, or sulfur-dependent α-proteobacterium and an archaebacterial host (Fig. 1C). The resulting mitochondriate prokaryote would have evolved the nucleus subsequently. In other scenarios (Fig. 1B), the cenancestral eukaryote emerged by cellular fusion or endosymbiosis of a Gram-negative, maybe hydrogen-producing, eubacterium and a methanogenic archaebacterium or eocyte, leading to a primitive but nucleated amitochondrial (archezoan) cell (Embley and Martin 2006, and references therein). As a third alternative, Cavalier-Smith (2002) suggested a common eubacterial ancestor for eukaryotes and archaebacteria (the Neomuran hypothesis) (Fig. 1A).
Figure 1.
Evolution of the last common ancestor of all eukaryotic cells. A schematic depiction of the early eukaryogenesis. Because of the lack of living and fossil descendants, several opposing models are discussed (A–C). The anticipated order of events is shown as a flow chart. For details, see text. (Derived from Embley and Martin 2006; Koonin 2010.)
EARLY EUKARYOGENESIS—A BRIEF SYNOPSIS
Independent of the question of how the proto-eukaryote arose, the favored hypothesis imagines a primitive, initially noncompartmented and—most likely—a cell-wall-less “phagocyte” that was able to invaginate its cytoplasmic membrane (de Duve 2007; Cavalier-Smith 2010). These invaginations might have budded to build vesicles and (over time) convoluted membrane systems that gradually differentiated into distinct compartments such as the endoplasmic reticulum (ER), the Golgi, endo- and lysosomes, peroxisomes, and the eukaryotic plasma membrane. Arguments in favor of this concept are (1) the similar phospholipid composition of nuclear, endoplasmic, and plasma membranes (van Meer et al. 2008); (2) the striking similarities between cotranslational protein import into the ER and cotranslational transport of proteins across bacterial inner membranes (e.g., Rapoport 2007); (3) some peroxisomal membrane proteins that are inserted in the ER membranes and are subsequently rerouted to preexisting peroxisomes via vesicles (van der Zand et al. 2010, 2012); and (4) components of the peroxisomal importomer can evolutionarily be traced back to the ER-associated degradation (ERAD) machinery (Gabaldon et al. 2006; Bolte et al. 2011). However, how primitive phagocytosis evolved cannot be reconstructed today, because many different regulatory factors had to evolve in parallel, for example, mechanisms for membrane recycling (by budding and fusion) and vesicle routing along a—at least basic—cytoskeleton, involving many new factors such as SNARES, coat proteins, small G-proteins, and so on. Possible scenarios are outlined, for example, in Cavalier-Smith (2009) or Yutin et al. (2009).
The evolution of the nuclear envelope is thought to have paralleled or shortly succeeded the evolution of the endomembrane system and the cytoskeleton as suggested by its composition of cytoskeleton and endomembrane elements (Cavalier-Smith 2009). It was speculated that its de novo formation after mitosis from ER vesicles might recapitulate its evolution (Mans et al. 2004).
Independent of the origin of the nuclear envelope, comparative genomics point out that parts of the involved factors are not proto-eukaryotic innovations having evolved de novo or from eukaryotic paralogs performing other functions, but were patched together from bacterial ancestors (e.g., Cavalier-Smith 2010). For example, prototypic coat proteins seem to have provided an evolutionary basis for the nuclear pore complexes (Devos et al. 2004; Brohawn et al. 2008). Bacterial ancestors were also found for the cytoskeleton elements tubulin and actin (Nogales et al. 1998; van den Ent et al. 2001) and the HEH domain of nucleolamine-anchoring Scr1p-Man (Mans et al. 2004).
Even if not reconstructible in any detail, the proto-eukaryote most likely evolved before the change of the atmosphere from anoxic to oxic (de Duve 2007), owing to the emergence of molecular oxygen gained by oxygenic photosynthesis of phototrophic bacteria 2 to 3 billion years ago (Ertel et al. 2005; Holland 2006). This change in atmosphere is actually considered as the driving force for the acquisition of mitochondria, a hallmark in eukaryote evolution. Here, mitochondria are discussed as direct descendants of (maybe phototrophic) α-proteobacterial-like cells (e.g., Gray 1992; Andersson et al. 2003; Cavalier-Smith 2006), which were taken up by the phagotrophe and established as facultative and then obligate endosymbionts before they were gradually turned into an organelle (Poole and Penny 2007). Alternatively, it is discussed that mitochondria originated from a symbiotic relationship of a hydrogen-dependent archaeon and a facultative anaerobic eubacterium that produced hydrogen as a by-product of its anaerobic respiration (for review, see Embley and Martin 2006).
The last organelle that “joined the game” was the chloroplast, ∼2.2–1.5 billion years ago (Ertel et al. 2005). Plastids are—as mitochondria—endosymbiotic organelles that derived from the uptake and “enslavement” of a once free-living cyanobacterial-like ancestor (Sagan 1967; Martin and Kowallik 1999). Plastids—similar to mitochondria—are enveloped by a double membrane and show several characteristics that reveal their bacterial origin, for example, their remnant genome and the features/composition of their envelope membranes (Stoebe and Maier 2002). The acquisition of the ability to use energy from sunlight via photosynthesis revolutionized Earth’s life history, because it gave rise to all plants and algae we know today.
THE PROTEIN TRANSPORT “PROBLEM”
The progressive compartmentalization of the eukaryotic ancestor gave rise to complex and interdependent, but mutually affecting (e.g., aerobic and anaerobic), distinct metabolic pathways (Lopez-Garcia and Moreira 2006). This process provided enormous evolutionary benefits but necessitated the development of mechanisms to “feed” these pathways with the required enzymes and regulatory factors (Bohnsack and Schleiff 2010). Next to proteins for the exchange of metabolic intermediates, products and small solutes, and proteinaceous components for the transport of proteins were required for the proper biogenesis and preservation of the compartments (Mirus and Schleiff 2012; Sommer and Schleiff 2014). Irrespectively of whether the spatial separation of metabolic pathways is really an evolutionary advantage or simply a relic of the endosymbiotic origin of some of the compartments (as discussed by Martin 2010), it is estimated that >50% of all proteins made in the modern eukaryotic cell have to cross at least one membrane to reach their final destination within the cell (Schatz and Dobberstein 1996). Only two principles of protein traffic and transport exist within eukaryotes: (1) direct cotranslational transport across or into a membrane, as, for instance, in the case of the endoplasmic reticulum or plastid thylakoid membranes (Robinson et al. 2001; Rapoport 2007); and (2) posttranslational transport of premade proteins. Proteins for the latter mode are (a) either guided by escorting proteins to the organellar surface, where they bind to specific translocon complexes that contain pores to facilitate protein integration or translocation (e.g., in case of peroxisomes, plastids, and mitochondria, the nucleus) (e.g., Pemberton and Paschal 2005; Schleiff and Becker 2011; Platta et al. 2013), or (b) packed into vesicles that shuttle between the different compartments (as in the case of endoplasmic reticulum and downstream compartments [see, e.g., Harter and Reinhard 2000] or chloroplasts [see Villarejo et al. 2005]). In most cases, vesicular transport involves the Golgi as the central distribution center.
Proteins are usually transported by virtue of distinct targeting signals that are required for the interaction with specific receptors at the organelles’ surfaces. They often are part of the amino acid sequence, share conserved amino acid sequences, or are dependent on their chemical properties. They are, in many cases, cleaved off upon successful translocation of the passenger (e.g., Martoglio and Dobberstein 1998; Gunkel et al. 2005; Schleiff and Becker 2011). Besides these peptide-based signals, posttranslational protein modifications (like, e.g., manose-6-phosphate) (Braulke and Bonifacino 2009) or motifs in the messenger RNA (mRNA) have been described as prerequisite for proper localization of proteins within the cellular context. Some mRNAs are prelocalized to the nucleus, the endoplasmic reticulum, or the mitochondria, where they are translated at distinct sites and translocated in a cotranslational manner (for review, see Weis et al. 2013). Although it was initially thought that this is an exclusively eukaryotic invention, prelocalization of mRNA was recently found in bacteria as well. This might pinpoint to a very ancient targeting mechanism that was used before proteinaceous signals and targeting factors evolved (Nevo-Dinur et al. 2011).
Compartments and their “feeding” transport mechanisms most likely coevolved. It has to be assumed that, whenever possible, the eukaryote “in-the-making” primarily recycled and adapted preexisting proteins, structures, and regulatory mechanisms from its bacterial ancestor(s) and added newly developed factors subsequently (see below). Hence, to understand which factors were adapted and which were not (and why), a reconstruction of what was present in the bacterial ancestor is necessary.
BACTERIAL SOUVENIRS
The most ancestral mode of insertion of membrane proteins into the cytoplasmic membrane of bacteria is thought to be direct insertion without proteinaceous guidance (Mirus and Schleiff 2012). Only subsequently, bacteria evolved several systems for a directed insertion of membrane proteins into their cytoplasmic membranes and the secretion of proteins across it, namely, SEC, TAT (twin-arginine translocase), and YidC. These systems most likely emerged early on in evolution, developed further, and were transmitted to modern bacteria and their eukaryotic descendants (see below) (Bohnsack and Schleiff 2010; Mirus and Schleiff 2012; Sommer and Schleiff 2014).
YidC (and their organellar homologs Oxa1 in mitochondria and Alb3 in plastids) might be the most primordial system. YidC might have evolved as an initially single-acting ribosome “docking” site to chaperone the insertion of membrane proteins (Kohler et al. 2009; Mirus and Schleiff 2012). This development must have been evolutionarily beneficial, for example, by avoiding cytosolic aggregation of membrane proteins as well as misfolding during membrane insertion. In parallel, and maybe independently to YidC, the SecYE system has evolved, which undertook the cotranslational insertion of multimeric membrane proteins using YidC to aid the lateral release of proteins from SecY (Luirink et al. 2001). It is considered that both—YidC and SecYE(G)—acted initially independently of each other and that the evolution of the amino-terminal domain of YidC (for SecF interaction) (see Xie et al. 2006) bears witness to this process (Bohnsack and Schleiff 2010). SecYE has gained SecG subsequently in evolution (Bohnsack and Schleiff 2010), which might have evolved convergently with the subunits SecB and SecA, the latter of which interacts with SecYEG through SecG (Mori et al. 1998). Therefore, SecYEG serves two functions: (1) together with the signal recognition particle (SRP), it acts as an insertase for the cotranslational insertion of membrane proteins into the cytoplasmic membrane (du Plessis et al. 2011); and (2) together with the Sec-specific chaperone SecB and the motor subunit SecA, it “pushes” secretory proteins posttranslationally across the membrane. These proteins are subsequently released via the SecDF complex, which most likely coevolved with SecYE (Lycklama and Driessen 2012).
The third system, the hetero-oligomeric TAT, facilitates the translocation of folded proteins across the cytoplasmic membrane (Fröbel et al. 2012). TatA constitutes the pore-forming subunit, whereas TatB and TatC are receptors coordinately recognizing the twin-arginine-containing signal sequence. Despite their distinct functions, the high sequence similarity of single-spanning TatA and TatB suggests a common ancestor for both. It can be imagined that TatA might have been the initial translocon with a subsequent development of TatBC (Bohnsack and Schleiff 2010).
Besides the membrane-bound translocation machineries, bacteria developed a targeting mechanism that ensures the proper feeding of the translocons with substrate proteins. It may be accepted that this mechanism evolved in the early days of self-insertion to prevent the aggregation of hydrophobic membrane proteins in the cytoplasm. Targeting and direct insertion of such proteins during their biogenesis is therefore the most logically consistent (and elegant) way. Recognition and targeting are facilitated by the SRP (signal recognition particle), which recognizes substrate proteins in their nascency at the ribosome via a hydrophobic signal sequence (the signal peptide) at the very amino terminus of the polypeptide chain. The most primitive SRP consists of the protein Ffh and the SRP-RNA (Akopian et al. 2013). Together with its receptor FtsY, Ffh might have initially acted “as an early means for RNA-based primordial ribosomes to deal with greasy polypeptide chains” (Walter et al. 2000) in that the ancestral Ffh/FtsY might have chaperoned the self-insertion of membrane proteins before the evolution of YidC and SecYE (Bohnsack and Schleiff 2010).
With the emergence of the outer membrane (OM), a new level of cellular complexity was added that required additional factors and transport machineries. Next to chaperones that assist folding or guidance of secretory proteins to the new membrane, systems for the integration into or the transport of proteins across this membrane became necessary. Secretion of proteins across the outer membrane is performed by a multitude of different mechanisms. Next to SEC-dependent mechanisms, by which proteins are secreted (e.g., autotransporters: extracellular enzymes and toxins, have been exported to the periplasm via SecYEG), Sec-independent mechanisms evolved (types I, III, and IV secretion systems) that are capable of directly secreting cytoplasmic proteins (for review, see, e.g., Mirus et al. 2010).
One of the most important things in terms of a forthcoming eukaryogenesis (see below) was the evolution of Omp85 proteins that chaperone the insertion of outer membrane proteins similarly to YidC (Löffelhardt et al. 2007). Expectedly, Omp85 is one of the oldest protein families that evolved shortly after the outer membrane (Bredemeier et al. 2007). Omp85 proteins consist of a 16-stranded β-barrel-shaped carboxy-terminal domain, as well as up to six amino-terminal polypeptide-transport-associated (POTRA) domains, the number of which differs among the different bacterial clades (Koenig et al. 2010). Like Omp85 proteins, OM proteins in general (with few exceptions) possess a β-barrel-shaped transmembrane domain, which stands in contrast to the throughout α-helical membrane domains of proteins of the cytoplasmic membrane. This entirely different buildup has most likely evolved to escape the release mechanism for α-helical membrane segments of the SecYEG complex preventing the mistargeting of these proteins to the cytoplasmic membrane. OM proteins are transferred from the cytoplasmic to the outer membrane via dedicated chaperones. Here, SurA seems to be of special interest, which might share a common ancestor with trigger factor-like proteins, the latter of which is a ribosome-associated chaperone (Bohnsack and Schleiff 2010). In addition, two further chaperones—Skp and DegP—evolved that are particularly involved in OM protein biogenesis as well (Sklar et al. 2007). Although the exact mode of targeting in the intermembrane space is not yet established, targeting of proteobacterial OM proteins relies on an invariant aromatic amino acid in the very carboxyl terminus of the proteins (Struyve et al. 1991), which is thought to be recognized by Omp85.
THE FIRST DAYS OF EUKARYOTIC PROTEIN TRAFFICKING
When discussing the evolution of protein trafficking and transport mechanisms as a necessary consequence of increased cellular complexity, it has to be considered that two different types of organelles coexist within cells: (1) those that evolved gradually de novo during early eukaryogenesis (the endomembranes, the peroxisomes and perhaps the nucleus); and (2) those of endosymbiotic origin (mitochondria and chloroplasts). The first evolved according to Darwin’s theory of “survival of the fittest” in parallel with their transport mechanisms, either by gradual transition and consequent further development of preexisting principles (e.g., Sec and SRP) (see Rapoport 2007) or by invention of totally new mechanisms (e.g., nuclear import, peroxisomal importomer, vesicle transport). One might speculate that their actual level of complexity was based on the co-occurrence of new regulatory factors. The second type of organelles evolved stepwise, starting with an autonomous coexistence of the in-cooperated endosymbiont within the host cell. In a first step of the symbiont-to-organelle transition, the host might have “evolved” mechanisms for the insertion of proteins into the symbionts’ outer membranes to benefit from their energy-rich metabolites. The most primitive base for this process might have been simple mistargeting of proteins, most likely in a cotranslational manner as typical for membrane proteins. Via gene transfer (Kleine et al. 2009), the biogenesis of symbiotic proteins might then have begun to drift to the host’s cytoplasm, being initially synthesized in- and outside of the symbiont (Mirus and Schleiff 2012). The development of a basic protein targeting and import mechanism subsequently made the loss of genes in the symbiont’s genome possible. At that early period, the most likely rather simple import mechanisms further developed by addition of sophisticated regulatory mechanisms. This allowed not only for a tight regulation of the organellar proteome by transcriptional control, but also (at the level of protein translocation) ensured rapid adaptations, for example, during plastid transitions (Bohnsack and Schleiff 2010).
As outlined above, the evolution of the endomembrane system is considered to have been crucial for the prokaryote-to-eukaryote transition. It is believed that because of the absence of bacterial ancestors and the assumed complexity, the endomembrane system evolved rapidly from a very simple trafficking system in the early eukaryote to near-modern complexity in the last common eukaryotic ancestor (Dacks and Field 2007). Thereby, the eukaryote-in-the-making frequently recycled preexisting bacterial factors for new cellular functions as, for example, Vps29, a central subunit of the retromer vesicle coat, shares similarity with bacterial phosphoesterases (Dacks and Field 2007). A second way for innovation of new factors was provided by massive gene duplication (Makarova et al. 2005). Adaptins 1, 2, and 3 of the COPI vesicle coat trace back to a single common ancestor (Schledzewski et al. 1999). All P-loop GTPases, among them the Arf/Sar family involved in vesicle coat formation and vesicle budding and the Rab/Ras family involved in vesicle fusion (Mellman and Warren 2000), were derived from a single protein ancestor (Leipe et al. 2002). Thereby, a primitive vesicle system was soon developed that diversified over time, producing different coats for distinct vesicle pathways (e.g., retrograde vs. anterograde ER–Golgi), motor proteins, GTPases, SNAREs, and so on for accurate intracellular trafficking and targeting (Nebenfuhr 2002).
The nuclear pore complex (NPC) and the nuclear import machinery partially evolved from preexisting bacterial structures (Devos et al. 2004; Mans et al. 2004; Martin and Koonin 2006; Cavalier-Smith 2010). Fully folded proteins are transported by importins, which shuttle between cyto- and nucleoplasm (through the pore complexes). The major structural motif of importin-β, the HEAT repeats (Morimoto et al. 2002), the Ras-like GTPase RAN, facilitating export of importins (Dong et al. 2007) as well as RAN-GDP import factor NTF2 (Mans et al. 2004), originated from bacterial ancestors (Bohnsack and Schleiff 2010). COPII-coat proteins and nucleoporins share structural features and arrangements (Devos et al. 2004; Sampathkumar et al. 2013) and therefore most likely a common ancestry (Brohawn et al. 2008; Debler et al. 2008; Kampmann and Blobel 2009), arguing for a coevolution of the endomembrane system and the nuclear envelope. Based on the structural conservation of the core nucleoporins, it was assumed that the nuclear pore complex emerged early on in evolution and was already established in the last common eukaryotic ancestor (Bapteste et al. 2005; DeGrasse et al. 2009).
The vast majority of membrane proteins are synthesized cotranslationally into Sec61 and are laterally released into the ER membrane. In contrast, carboxy-terminally tail-anchored (TA) proteins are inserted posttranslationally. Whereas the eukaryotic Sec61 is a systematic offspring of its bacterial predecessor SecYEG (Rapoport 2007), eukaryotes did not fall back on YidC for posttranslational membrane protein insertion. Instead, the transmembrane domain (TMD)–recognizing complex (TRC) was evolved that facilitates the insertion of TA-proteins at the ER membrane (Stefanovic and Hegde 2007). The core of this complex is Trc40/Get3, which again traces back to a bacterial ancestor, the ATPase ArsA (Rabu and High 2007).
In terms of evolution of eukaryotic protein transport systems, peroxisomes are unique. Peroxisomes import already fully folded matrix proteins. The peroxisomal importomer consists of several subunits of which Pex5 and Pex7 constitute receptor proteins for the recognition of so-called peroxisomal targeting sequences at the amino or carboxy termini of matrix proteins (Rucktäschel et al. 2011). Receptor-cargo complexes dock at the membrane via Pex13 and 14, in the case of Pex5 insert into the membrane, and release cargo into the lumen. After cargo release, Pex5 is actively extracted from the membrane via the cytoplasmic AAA-type ATPases Pex1 and 6. In line with the formation of peroxisomes as “specialized vesicles,” the peroxisomal import machinery seems to have evolved from a prototypic ERAD, as, for example, the TPR domains of Pex5, the ATPases Pex1 and 6, and so on share striking similarities to known ERAD components (Gabaldon et al. 2006; Bolte et al. 2011). In turn, ERAD, which is most likely a eukaryotic invention, must have been already evolved at an early stage of eukaryogenesis. The peroxisomal import machinery might have evolved from frequently “mistargeted” ERAD components to peroxisomes during their formation at the ER membrane, serving as the base for developing a new translocon. The apparent change in transport direction (ERAD export; peroxisomes import) and lack of components energizing this process were overcome by the loss of the assumed static pore and the evolution of an ubiquitin-dependent export-driven shuttling mechanism (Platta et al. 2013), which is reminiscent of the mechanism of dislocation of membrane-bound ERAD substrates (Bolte et al. 2011). It may be speculated that the reason for importing readily folded matrix proteins is the supposed lack of luminal chaperones (Erdmann et al. 1997). Only recently, evidence was found for small heat-shock proteins that are targeted to peroxisomes in plants (Ma et al. 2006), but whether or not this is specific for plants remains elusive to date. An alternative reason for importing folded proteins might be that enzymes for peroxismal key functions required cofactors or prosthetic groups that are synthesized and assembled outside the peroxisome only. In line with this idea, several enzymes, among them catalases or acyl-CoA-oxidases, enzymes that are involved in fatty acid oxidation and decomposition of reactive oxygen species, depend on cofactors like heme or flavin adenine dinucleotide (FAD), respectively (Kim and Miura 2004; Zamocky et al. 2008).
ENDOSYMBIOTIC ORGANELLES
As mentioned above, endosymbiotic organelles gradually evolved import machineries in their surrounding membranes by partially reusing preexisting proteinaceous factors as raw material (for review, see Mirus and Schleiff 2012; Sommer and Schleiff 2014). Despite the mechanistical differences, a convergent evolution of these machineries is particularly reflected in the very similar buildup (Schleiff and Becker 2011 and references therein). The latter is the logical consequence of both, the evolutionary development and the functional requirement. The outer membrane translocon was built by simply recycling an organellar outer membrane protein combined with eukaryotic inventions serving as receptors. The inner membrane translocons had to serve different functions, translocation across the membrane and insertion into the membrane, and thus, similar mechanistic principles evolved based on the principle “form follows function.”
The vast majority of nuclear-encoded mitochondrial or plastid proteins are equipped with nonhydrophobic, cleavable amino acid sequences serving as targeting or transit peptides, respectively. These signals facilitate the interaction with guiding factors and chaperones, which target the proteins to the organellar surface, where they are bound by specific receptors as part of TOM/TOC (translocons of the outer membranes of mitochondria or chloroplasts). These oligomeric complexes have a central pore-forming subunit, which in the case of mitochondria (Tom40) is closely related to the mitochondrial porin VDAC (Pusnik et al. 2009; Gessmann et al. 2011), although it appears obvious that Tom40 traces back to a proteobacterial β-barrel protein, the nature of which is unclear. Several hypotheses have been formulated with different plausibilities. It was recently claimed that Tom40 might be an Omp85 descent (Pusnik et al. 2011; Harsman et al. 2012), which appears rather unlikely based on the observed structures of Omp85-like proteins and VDAC (Clantin et al. 2007; Hiller et al. 2008; Ujwal et al. 2008) and its phylogenetic relation to VDAC (Flinner et al. 2012). It was proposed earlier that a proteobacterial porin served as the ancestor of Tom40 (Cavalier-Smith 2006). Lately, a third mechanism of VDAC/Tom40 evolution has been proposed, namely, the pentamerization of a 4-β-sheet protein segment with a subsequent loss of one strand (Zeth and Thein 2010). Hence, at present, it cannot be stated with certainty whether or not this protein family is indeed of bacterial origin, although it appears plausible, because host membrane proteins have most likely been α-helical.
The pore-forming subunit of the TOC complex, Toc75, is derived from a cyanobacterial Omp85 protein (Bredemeier et al. 2007), having not only changed its function, but also its topology in evolution to turn from an insertase (see above) to a translocase for plastid proteins (Sommer et al. 2011). Considering the different pore properties of modern α-proteobacterial and cyanobacterial Omp85 proteins (Bredemeier et al. 2007; Wunder et al. 2009), an Omp85 descent of Tom40 is unlikely, because only the latter possesses an adequate pore size that allows for the passage of an unfolded polypeptide chain. Toc75 possesses both, primitive receptor and channel function (Schleiff et al. 2011; Sommer et al. 2011), which is why it was speculated that the protein displays the most primitive TOC with all other subunits having evolved subsequently (Sommer and Schleiff 2014). This indicates that initial, rather simple transport mechanisms might have been suited to account for a successful symbiont-to-organelle transition.
TOM and TOC serve as major import gates for proteins into the organelles. Subsequently, after their passage, the proteins are sorted and distributed into the various suborganellar compartments (e.g., outer and inner membrane, intermembrane space, matrix/stroma, and in case of plastids, additionally, thylakoid membrane and lumen) supported by a multitude of different chaperones and translocon subcomplexes (Schleiff and Becker 2011). For instance, the sorting and assembly machinery (SAM) in the mitochondrial outer membrane facilitates the insertion of barrel-shaped outer membrane proteins. The core of this complex is the Omp85 descendant Sam50, which acts in a similar way as its bacterial counterpart (Schleiff and Soll 2005). Passage of the inner membrane is facilitated by the TIM23 complex, whose eponymous core component Tim23 shares homology to the bacterial permeases LivH (Rassow et al. 1999). Such other TIM23 subunits as Tim44 and the motor subunits Pam18 and Hsp70 trace back to bacterial ancestors as well (Hewitt et al. 2011). Inner membrane proteins that are inserted from the plasmic phase (matrix) use Oxa1, a clear YidC homolog (Luirink et al. 2001).
The situation is comparable in chloroplasts. Only a few core components of the modern TOC/TIC complexes can be traced back to cyanobacteria or the most prototypic plastids known today (Mirus and Schleiff 2012; Sommer and Schleiff 2014). These are next to Toc75 only Tic20/21, Tic55, Tic62, and Tic32 with different functions in cyanobacteria and Tic110—the supposed pore-forming subunit of TIC—as early eukaryotic inventions. Only the intermembrane space chaperone Tic22 has a cyanobacterial ancestor with a similar function in plants (Tripp et al. 2012). The functional homolog of Oxa1 in plants, Alb3, is also a YidC descendant (Luirink et al. 2001). Whereas the bacterial Sec and TAT systems were abandoned in mitochondria, they experienced a renaissance in plastids. Both systems facilitate the insertion and import of thylakoid proteins into thylakoid membrane and lumen in a very similar way to the bacterial counterparts, respectively (e.g., Albiniak et al. 2012). Altogether it can be stated that proteins of bacterial origins are predominantly reused in the inner membranes of both organelles. That might make sense if one considers that especially the TOM/TOC complexes rather than TIM/TIC serve as regulatory interfaces for a harmonized coordination of organellar biogenesis and function and the host.
As mentioned above, most, but not all plastid proteins possess canonical targeting signals (Schleiff and Becker 2011). In higher plants, a small group of chloroplast proteins is targeted to the organelle via the endomembrane system, such as the α-carbonic anhydrase, the nucleotide pyrophosphatase/phosphodiesterase, or the α-amylase (Villarejo et al. 2005; Nanjo et al. 2006; Kitajima et al. 2009). These proteins are glycosylated and possess cleavable signal peptides similar to secretory proteins. This discovery led to the speculation that all nuclear-encoded plastid proteins might have been initially targeted to the chloroplasts via the endomembrane system and that TOC and TIC evolved only subsequently (Bhattacharya et al. 2007). According to this scenario, the TOC/TIC pathway became more efficient over time, and consequently, signal peptides would have changed into transit peptides (in the course of evolution), which would have gradually shifted hundreds of nuclear-encoded plastid proteins from the ER/Golgi transport route to the TOC/TIC system (Bhattacharya et al. 2007). Very recently, phylogenetic analyses refuted this scenario, because it was shown that all described endomembrane system-targeted proteins were subsequent eukaryotic innovations and not targeted to chloroplasts in the early days of organellar evolution (Gagat et al. 2013). Accordingly, it was concluded that these proteins constitute only a specific class of proteins with a peculiar evolutionary history and that TOC/TIC was the initial system for protein uptake (Gagat et al. 2013).
SIGNAL EVOLUTION—PREREQUISITE OR CONSEQUENCE FOR TRANSLOCON EVOLUTION IN ENDOSYMBIOTIC ORGANELLES?
Given the molecular crowding of the cytoplasm (Ellis 2001), the question arises, why—despite the endomembrane system—most organelles are fed posttranslationally with proteins? It is assumed that diffusion as the driving force for protein trafficking to organelles is energetically favorable, but too inefficient, because diffusion rates are strongly dependent on the molecular weight (Ellis 2001). Prepositioning of ribosomes mise-en-place on first sight might enhance efficiency but would require sophisticated mechanisms for mRNA targeting and transport to the organellar surface as well. The question how posttranslational targeting mechanisms evolved may help to answer why.
Reconstructing the origins of this development, it can be assumed that the first cytoplasmically synthesized soluble mitochondrial and plastid proteins lacked suitable targeting information and consequently folded and accumulated in the cytoplasm. Import of those proteins into the organelles would occur only accidentally after diffusion of the proteins to the organellar surface, provided that primitive translocons already had evolved (Fig. 2, step 1). Basic targeting signals might have evolved already shortly after by different mechanisms, for example, “including” the 5′ UTR into the open reading frame or “occurrence” of a new exon (Mirus and Schleiff 2012). However, certainly their evolution was funneled by certain constraints: a targeting signal (1) would only make sense if there is a receptor (at least as part of the primitive translocon) for its recognition; (2) it needs to be exposed in that it can serve as the “seed” for interactions; and (3) it must not share overlapping properties with other targeting signals, for example, for nuclear import or for the endoplasmic reticulum (Fig. 2, step 2). Accordingly, it may be accepted that mitochondrial and plastid targeting signals evolved “purposely” de novo with unique biochemical properties.
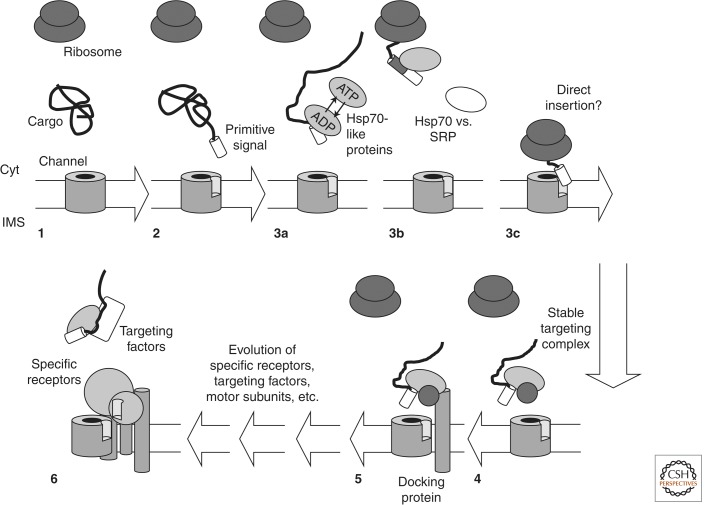
Figure 2.
Evolution of posttranslational targeting of proteins to the surfaces of the endosymbiontic organelles. A model showing the evolution of posttranslational targeting of proteins to chloroplast and/or mitochondria is depicted. (1) Nuclear-encoded organellar proteins (as black line), initially lacking appropriate targeting signals, are synthesized and most likely accumulated in the cytoplasm. Import of those proteins into the organelles would occur only accidentally after diffusion of the proteins to the organellar surface (outer membrane as white bar), provided that a primitive translocon (dark gray) already had evolved. (2–6) The anticipated order of events for the evolution of appropriate targeting and translocon complexes. For details, see text. Cyt, cytosol; IMS, intermembrane space.
The arrival of the protein in an unfolded state at the organellar surface might have been evolutionarily beneficial, circumventing energetically expensive unfolding of the polypeptide for translocation. Consequently, Hsp70-like chaperones undergoing repeated ATP-dependent binding and release cycles (Mayer and Bukau 2005) might have served as initial “targeting” complexes (Fig. 2, steps 3a–c). To this end, it would have been of evolutionary “advantage” if otherwise soluble proteins would possess an amino acid extension that results in protein misfolding, necessitating the sequestering chaperones. This is in line with the observed insolubility of the precursor form of organellar proteins, but solubility of the mature form when heterologously expressed (e.g., for FNR) (Ceccarelli et al. 1991). Alternatively, amino-terminal extensions might have evolved to form exposed parts of the proteins to ensure sufficient insertion already during translation (Fig. 2, step 3c). In this case, the association with the chaperones would have been a subsequent event. A third scenario that can be envisioned is the evolution of an amino-terminal segment in front of the coding region of a membrane protein. The hydrophobicity of the transmembrane segment would have been automatically recognized by SRP leading to its mistargeting to the ER. A nonhydrophobic segment, however, would have circumvented such targeting but would have led to a cytoplasmically localized hydrophobic protein with a tendency to aggregate, again enforcing Hsp70 interaction (Fig. 2, step 3b). All three scenarios would explain why for both, mitochondrial and chloroplast preproteins, a major component of every so-far-described “guiding complex” is Hsp70. And all the more, it might be also not surprising that 14–3–3 proteins and Hsp90 are part of these guiding complexes as well, because they are generally known to act in concert with Hsp70 (e.g., Wegele et al. 2004; Yano et al. 2006). Subsequently, chaperone binding might have been stabilized by the evolution of special cofactors that impede nucleotide exchange to prevent elevated energy consumption (Fig. 2, step 4). The existence of such chaperone-containing complexes might have then enforced the evolutionary occurrence of chaperone docking proteins like Tom70 or Toc64 at the organellar surfaces (Schlegel et al. 2007; Mirus et al. 2009), which would omit random diffusion by producing a chemical gradient (Fig. 2, step 5). However, at this stage, specificity of targeting would not have been provoked. It is known that at least 20% of all cytosolic proteins require guidance of Hsp70 in folding (Hartl and Hayer-Hartl 2002), which would then be—according to this scenario—targeted to the organellar surface. Moreover, protein targeting into the nucleus depends on positively charged regions within the proteins as well and amino-terminal hydrophobic regions serve as signals for cotranslational targeting to the SEC complex. Hence, to ensure the required specificity of targeting, further guiding factors and receptors as well as more specific signals would have to evolve, especially, for example, to discriminate between mitochondria and plastids in plant cells (Fig. 2, transition 5 to 6). Accordingly, evolution had to be directed toward alternative signal sequences still serving their function but avoiding mistargeting. In line, the substrate specificity of Hsp70 is usually mediated by cochaperones (Mayer and Bukau 2005).
In summary, the mentioned scenario might explain why a posttranslational import mode was established during evolution. Cotranslational import, based on a hydrophobic signal, would have mistargeted the proteins to the endoplasmic reticulum, and a complex vesicle transport and fusion system to the organelles would have been required, which at the early period of eukaryogenesis might not have been present in the required differentiation. Nevertheless, the freedom of evolution was early recognized, because 25% of all randomly synthesized peptides are suited as basic import signals for mitochondria (Baker and Schatz 1987). That shows that the specificity of the receptor units at the surface is rather broad, and, therefore, receptors most likely evolved subsequently to the topogenic signals.
EVOLUTIONARY VARIATIONS OF COMPLEXITY
For many years, the finding of eukaryotes that lack mitochondria was seen as proof for the existence of a premitochondrial phagotrophic proto-eukaryote, but it turned out that these were only secondarily “amitochondriates” still possessing remnants of the original mitochondrion (i.e., mitosomes and maybe hydrogenosomes) (Hackstein et al. 2006). These organelles show a highly reduced complexity in comparison to real mitochondria and teach us the possible simplicity of translocation pathways. In fact, the most simplistic TOM/TIM is constituted of the channel proteins Tom40 and Tim23 as well as of Sam50 only (Lithgow and Schneider 2010).
At the further end of this development are plastids, which are surrounded by more than two envelope membranes (complex plastids). These plastids arose by secondary endosymbiosis and are found in many phototrophic protists and their nonphototrophic relatives (e.g., Agrawal and Striepen 2010; Keeling 2010) In here, eukaryotes have gained plastids in lateral transfer by taking up already “simple” plastid-bearing eukaryotic cells (an ancestral red or green algae, respectively). This new level of complexity is the ultimate challenge in terms of protein trafficking because plastid proteins have to be transported across up to five membranes (e.g., Gould et al. 2007). Reutilization and adaptation of preexisting transport machineries were carried to the extreme (Hempel et al. 2007; Bolte et al. 2009).
CONCLUDING REMARKS
As mentioned above, the evolution of the eukaryotic cell was and will be further on a challenging task. Despite all efforts in understanding the evolutionary coherencies, we will probably never be able to fully reconstruct all antecedents of early eukaryogenesis in all details, simply because we do not know how the last common eukaryotic ancestor really looked and which differentiation grade the individual organelles had at that time compared with modern cells. All the more, it is exciting that the finding of a plastid in-birth (Nakayama and Ishida 2009) affords the opportunity to study evolution “in real time.” The cercozoan testate amoeba Paulinella chromatophora possesses a cyanobacterial symbiont that traces back to a recent endosymbiotic event that took place a billion years after the primary endosymbiosis of all other plastids (Yoon et al. 2009). At that point, eukaryotic cells were already differentiated and, hence, provided already complex protein targeting and transport machineries as well as a sophisticated vesicle system. It is to be expected that the design of new mechanisms for the organelle in-the-making look completely different from what we know so far.
ACKNOWLEDGMENTS
We thank Uwe G. Maier (Marburg) and Kathrin Bolte (Marburg) for critical comments and useful suggestions for this manuscript. The work is supported by grants from the Deutsche Forschungsgemeinschaft SFB807.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org.
REFERENCES
- Agrawal S, Striepen B 2010. More membranes, more proteins: Complex protein import mechanisms into secondary plastids. Protist 161: 672–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan SO 2013. Signal recognition particle: An essential protein-targeting machine. Ann Rev Biochem 82: 693–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiniak AM, Baglieri J, Robinson C 2012. Targeting of lumenal proteins across the thylakoid membrane. J Exp Bot 63: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Andersson SGE, Karlberg O, Canback B, Kurland CG 2003. On the origin of mitochondria: A genomics perspective. Philos Trans R Soc Lond B Biol Sci 358: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Schatz G 1987. Evolution of mitochondrial targeting sequences. J Cell Biochem: 286–286 [Google Scholar]
- Bapteste E, Charlebois RL, MacLeod D, Brochier C 2005. The two tempos of nuclear pore complex evolution: Highly adapting proteins in an ancient frozen structure. Genome Biol 6: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Archibald JM, Weber AP, Reyes-Prieto A 2007. How do endosymbionts become organelles? Understanding early events in plastid evolution. BioEssays 29: 1239–1246 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Schleiff E 2010. The evolution of protein targeting and translocation systems. Biochim Biophys Acta 1803: 1115–1130 [DOI] [PubMed] [Google Scholar]
- Bolte K, Bullmann L, Hempel F, Bozarth A, Zauner S, Maier UG 2009. Protein targeting into secondary plastids. J Eukaryot Microbiol 56: 9–15 [DOI] [PubMed] [Google Scholar]
- Bolte K, Gruenheit N, Felsner G, Sommer MS, Maier UG, Hempel F 2011. Making new out of old: Recycling and modification of an ancient protein translocation system during eukaryotic evolution. Mechanistic comparison and phylogenetic analysis of ERAD, SELMA and the peroxisomal importomer. BioEssays 33: 368–376 [DOI] [PubMed] [Google Scholar]
- Braulke T, Bonifacino JS 2009. Sorting of lysosomal proteins. Biochim Biophys Acta 1793: 605–614 [DOI] [PubMed] [Google Scholar]
- Bredemeier R, Schlegel T, Ertel F, Vojta A, Borissenko L, Bohnsack MT, Groll M, von Haeseler A, Schleiff E 2007. Functional and phylogenetic properties of the pore-forming β-barrel transporters of the Omp85 family. J Biol Chem 282: 1882–1890 [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52: 297–354 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2006. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc Biol Sci 273: 1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 2009. Predation and eukaryote cell origins: A coevolutionary perspective. Int J Biochem Cell Biol 41: 307–322 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2010. Origin of the cell nucleus, mitosis and sex: Roles of intracellular coevolution. Biol Direct 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli EA, Viale AM, Krapp AR, Carrillo N 1991. Expression, assembly, and processing of an active plant ferredoxin-NADP+ oxidoreductase and its precursor protein in Escherichia coli. J Biol Chem 266: 14283–14287 [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V 2007. Structure of the membrane protein FhaC: A member of the Omp85–TpsB transporter superfamily. Science 317: 957–961 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Field MC 2007. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J Cell Sci 120: 2977–2985 [DOI] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A 2008. A fence-like coat for the nuclear pore membrane. Mol Cell 32: 815–826 [DOI] [PubMed] [Google Scholar]
- de Duve C 2007. The origin of eukaryotes: A reappraisal. Nat Rev Genet 8: 395–403 [DOI] [PubMed] [Google Scholar]
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JH, Wen JF, Tian HF 2007. Homologs of eukaryotic Ras superfamily proteins in prokaryotes and their novel phylogenetic correlation with their eukaryotic analogs. Gene 396: 116–124 [DOI] [PubMed] [Google Scholar]
- du Plessis DJ, Nouwen N, Driessen AJ 2011. The Sec translocase. Biochim Biophys Acta 1808: 851–865 [DOI] [PubMed] [Google Scholar]
- Ellis RJ 2001. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr Opin Struct Biol 11: 114–119 [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W 2006. Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Kunau WH 1997. Peroxisomes: Organelles at the crossroads. Trend Cell Biol 7: 400–407 [DOI] [PubMed] [Google Scholar]
- Ertel F, Bredemeier R, Schleiff E 2005. Die Grenze zwischen Prokaryont und Eukaryont. BioForum 10: 35–37 [Google Scholar]
- Flinner N, Schleiff E, Mirus O 2012. Identification of two voltage-dependent anion channel-like protein sequences conserved in Kinetoplastida. Biol Lett 8: 446–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröbel J, Rose P, Müller M 2012. Twin-arginine-dependent translocation of folded proteins. Philos Trans R Soc Lond B Biol Sci 367: 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA 2006. Origin and evolution of the peroxisomal proteome. Biol Direct 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagat P, Bodyl A, Mackiewicz P 2013. How protein targeting to primary plastids via the endomembrane system could have evolved? A new hypothesis based on phylogenetic studies. Biol Direct 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessmann D, Flinner N, Pfannstiel J, Schlosinger A, Schleiff E, Nussberger S, Mirus O 2011. Structural elements of the mitochondrial preprotein-conducting channel Tom40 dissolved by bioinformatics and mass spectrometry. Biochim Biophys Acta 1807: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Gould SB, Fan E, Hempel F, Maier UG, Klosgen RB 2007. Translocation of a phycoerythrin α subunit across five biological membranes. J Biol Chem 282: 30295–30302 [DOI] [PubMed] [Google Scholar]
- Gray MW 1992. The endosymbiont hypothesis revisited. Int Rev Cytol 141: 233–357 [DOI] [PubMed] [Google Scholar]
- Gunkel K, Veenhuis M, van der Klei IJ 2005. Protein translocation machineries: How organelles bring in matrix proteins. Fems Yeast Res 5: 1037–1045 [DOI] [PubMed] [Google Scholar]
- Hackstein JH, Tjaden J, Huynen M 2006. Mitochondria, hydrogenosomes and mitosomes: Products of evolutionary tinkering! Curr Genet 50: 225–245 [DOI] [PubMed] [Google Scholar]
- Harsman A, Niemann M, Pusnik M, Schmidt O, Burmann BM, Hiller S, Meisinger C, Schneider A, Wagner R 2012. Bacterial origin of a mitochondrial outer membrane protein translocase: New perspectives from comparative single channel electrophysiology. J Biol Chem 287: 31437–31445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harter C, Reinhard C 2000. The secretory pathway from history to the state of the art. Subcell Biochem 34: 1–38 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2002. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hempel F, Bozarth A, Sommer MS, Zauner S, Przyborski JM, Maier UG 2007. Transport of nuclear-encoded proteins into secondarily evolved plastids. Biol Chem 388: 899–906 [DOI] [PubMed] [Google Scholar]
- Hewitt V, Alcock F, Lithgow T 2011. Minor modifications and major adaptations: The evolution of molecular machines driving mitochondrial protein import. Biochim Biophysica Acta 1808: 947–954 [DOI] [PubMed] [Google Scholar]
- Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G 2008. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321: 1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland HD 2006. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci 361: 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M, Blobel G 2009. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol 16: 782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ 2010. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Miura R 2004. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences. Eur J Biochem 271: 483–493 [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, et al. 2009. The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D 2009. DNA transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Ann Rev Plant Biol 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Koenig P, Mirus O, Haarmann R, Sommer MS, Sinning I, Schleiff E, Tews I 2010. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J Biol Chem 285: 18016–18024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N 2009. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell 34: 344–353 [DOI] [PubMed] [Google Scholar]
- Koonin EV 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L 2002. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317: 41–72 [DOI] [PubMed] [Google Scholar]
- Lithgow T, Schneider A 2010. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B Biol Sci 365: 799–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffelhardt W, von Haeseler A, Schleiff E 2007. The β-barrel shaped polypeptide transporter, an old concept for precursor protein transfer across membranes. Symbiosis 44: 33–42 [Google Scholar]
- Lopez-Garcia P, Moreira D 2006. Selective forces for the origin of the eukaryotic nucleus. BioEssays 28: 525–533 [DOI] [PubMed] [Google Scholar]
- Luirink J, Samuelsson T, de Gier JW 2001. YidC/Oxa1p/Alb3: Evolutionarily conserved mediators of membrane protein assembly. FEBS Lett 501: 1–5 [DOI] [PubMed] [Google Scholar]
- Lycklama ANJA, Driessen AJ 2012. The bacterial Sec-translocase: Structure and mechanism. Philos Trans R Soc Lond B Biol Sci 367: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Haslbeck M, Babujee L, Jahn O, Reumann S 2006. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol 141: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV 2005. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acid Res 33: 4626–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3: 1612–1637 [DOI] [PubMed] [Google Scholar]
- Martijn J, Ettema TJ 2013. From archaeon to eukaryote: The evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans 41: 451–457 [DOI] [PubMed] [Google Scholar]
- Martin W 2010. Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond B Biol Sci 365: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus–cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Martin W, Kowallik KV 1999. Annotated English translation of Mereschkowsky’s 1905 paper “Uber Natur und Ursprung der Chromatophoren im Pflanzenreiche.” Eur J Phycol 34: 287–295 [Google Scholar]
- Martoglio B, Dobberstein B 1998. Signal sequences: More than just greasy peptides. Trend Cell Biol 8: 410–415 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B 2005. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Warren G 2000. The road taken: Past and future foundations of membrane traffic. Cell 100: 99–112 [DOI] [PubMed] [Google Scholar]
- Mirus O, Schleiff E 2012. Recycling and tinkering—The evolution of protein transport to and into endosymbiotically derived organelles. In Organelle genetics: Evolution of organelle genomes and gene expression (ed. Bullerwell ECE), pp. 175–216 Springer, Berlin [Google Scholar]
- Mirus O, Bionda T, von Haeseler A, Schleiff E 2009. Evolutionarily evolved discriminators in the 3-TPR domain of the Toc64 family involved in protein translocation at the outer membrane of chloroplasts and mitochondria. J Mol Model 15: 971–982 [DOI] [PubMed] [Google Scholar]
- Mirus O, Hahn A, Schleiff E 2010. Outer membrane proteins. In Prokaryotic cell wall compounds: Structure and biochemistry (ed. König H, et al. ), pp. 175–230 Springer, New York [Google Scholar]
- Mori H, Sugiyama H, Yamanaka M, Sato K, Tagaya M, Mizushima S 1998. Amino-terminal region of SecA is involved in the function of SecG for protein translocation into Escherichia coli membrane vesicles. J Biochem 124: 122–129 [DOI] [PubMed] [Google Scholar]
- Morimoto K, Nishio K, Nakai M 2002. Identification of a novel prokaryotic HEAT-repeats-containing protein which interacts with a cyanobacterial IscA homolog. FEBS Lett 519: 123–127 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Ishida K 2009. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr Biol 19: R284–R285 [DOI] [PubMed] [Google Scholar]
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Munoz FJ, Rodriguez-Lopez M, Baroja-Fernandez E, Pozueta-Romero J 2006. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER–Golgi to the chloroplast through the secretory pathway. Plant Cell 18: 2582–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr A 2002. Vesicle traffic in the endomembrane system: A tale of COPs, Rabs and SNAREs. Curr Opin Plant Biol 5: 507–512 [DOI] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O 2011. Translation-independent localization of mRNA in E. coli. Science 331: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Lowe J 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5: 451–458 [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Platta HW, Hagen S, Erdmann R 2013. The exportomer: The peroxisomal receptor export machinery. Cell Mol Life Sci 70: 1393–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AM, Penny D 2007. Evaluating hypotheses for the origin of eukaryotes. BioEssays 29: 74–84 [DOI] [PubMed] [Google Scholar]
- Pusnik M, Charriere F, Maser P, Waller RF, Dagley MJ, Lithgow T, Schneider A 2009. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol 26: 671–680 [DOI] [PubMed] [Google Scholar]
- Pusnik M, Schmidt O, Perry AJ, Oeljeklaus S, Niemann M, Warscheid B, Lithgow T, Meisinger C, Schneider A 2011. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol 21: 1738–1743 [DOI] [PubMed] [Google Scholar]
- Rabu C, High S 2007. Membrane protein chaperones: A new twist in the tail? Curr Biol 17: R472–R474 [DOI] [PubMed] [Google Scholar]
- Rapoport TA 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J 1999. The preprotein translocase of the mitochondrial inner membrane: Function and evolution. J Mol Biol 286: 105–120 [DOI] [PubMed] [Google Scholar]
- Robinson C, Thompson SJ, Woolhead C 2001. Multiple pathways used for the targeting of thylakoid proteins in chloroplasts. Traffic 2: 245–251 [DOI] [PubMed] [Google Scholar]
- Rucktäschel R, Girzalsky W, Erdmann R 2011. Protein import machineries of peroxisomes. Biochim Biophys Acta 1808: 892–900 [DOI] [PubMed] [Google Scholar]
- Sagan L 1967. On the origin of mitosing cells. J Theoret Biol 14: 255–274 [DOI] [PubMed] [Google Scholar]
- Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, et al. 2013. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 21: 560–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B 1996. Common principles of protein translocation across membranes. Science 271: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Schledzewski K, Brinkmann H, Mendel RR 1999. Phylogenetic analysis of components of the eukaryotic vesicle transport system reveals a common origin of adaptor protein complexes 1, 2, and 3 and the F subcomplex of the coatomer COPI. J Mol Evol 48: 770–778 [DOI] [PubMed] [Google Scholar]
- Schlegel T, Mirus O, von Haeseler A, Schleiff E 2007. The tetratricopeptide repeats of receptors involved in protein translocation across membranes. Mol Biol Evol 24: 2763–2774 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Becker T 2011. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol 12: 48–59 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J 2005. Membrane protein insertion: Mixing eukaryotic and prokaryotic concepts. EMBO Rep 6: 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Maier UG, Becker T 2011. Omp85 in eukaryotic systems: One protein family with distinct functions. Biol Chem 392: 21–27 [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, Silhavy TJ 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer MS, Schleiff E 2014. Evolution of the protein translocon at the envelopes of plastids. In Endosymbiosis (ed. Löffelhardt W). Springer, New York [Google Scholar]
- Sommer MS, Daum B, Gross LE, Weis BL, Mirus O, Abram L, Maier UG, Kuhlbrandt W, Schleiff E 2011. Chloroplast Omp85 proteins change orientation during evolution. Proc Natl Acad Sci 108: 13841–13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS 2007. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Stoebe B, Maier UG 2002. One, two, three: Nature’s tool box for building plastids. Protoplasma 219: 123–130 [DOI] [PubMed] [Google Scholar]
- Struyve M, Moons M, Tommassen J 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer-membrane protein. J Mol Biol 218: 141–148 [DOI] [PubMed] [Google Scholar]
- Tripp J, Hahn A, Koenig P, Flinner N, Bublak D, Brouwer EM, Ertel F, Mirus O, Sinning I, Tews I, et al. 2012. Structure and conservation of the periplasmic targeting factor Tic22 protein from plants and cyanobacteria. J Biol Chem 287: 24164–24173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping PP, Abramson J 2008. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci 105: 17742–17747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- van der Zand A, Braakman I, Tabak HF 2010. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell 21: 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A, Gent J, Braakman I, Tabak HF 2012. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW 2008. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Bio 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A, Buren S, Larsson S, Dejardin A, Monne M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, et al. 2005. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol 7: 1224–1231 [DOI] [PubMed] [Google Scholar]
- Walter P, Keenan R, Schmitz U 2000. Perspectives: Structural biology. SRP—Where the RNA and membrane worlds meet. Science 287: 1212–1213 [DOI] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J 2004. Hsp70 and Hsp90—A relay team for protein folding. Rev Physiol Biochem Pharmacol 151: 1–44 [DOI] [PubMed] [Google Scholar]
- Weis BL, Schleiff E, Zerges W 2013. Protein targeting to subcellular organelles via MRNA localization. Biochim Biophys Acta 1833: 260–273 [DOI] [PubMed] [Google Scholar]
- Woese CR 2002. On the evolution of cells. Proc Natl Acad Sci 99: 8742–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T, Bredemeier R, Ruprecht M, Schleiff E 2009. The Omp85 proteins of Anabaenasp. PCC 7120 and E. coli are functionally distinct. J Endocytobiosis Cell Res 19: 20–30 [Google Scholar]
- Xie K, Kiefer D, Nagler G, Dalbey RE, Kuhn A 2006. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45: 13401–13408 [DOI] [PubMed] [Google Scholar]
- Yano M, Nakamuta S, Wu XJ, Okumura Y, Kido H 2006. A novel function of 14–3–3 protein: 14–3–3ζ is a heat-shock-related molecular chaperone that dissolves thermal-aggregated proteins. Mol Biol Cell 17: 4769–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Nakayama T, Reyes-Prieto A, Andersen RA, Boo SM, Ishida K, Bhattacharya D 2009. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol Biol 9: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf MY, Wolf YI, Koonin EV 2009. The origins of phagocytosis and eukaryogenesis. Biol Direct 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamocky M, Furtmuller PG, Obinger C 2008. Evolution of catalases from bacteria to humans. Antioxid Redox Signal 10: 1527–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K, Thein M 2010. Porins in prokaryotes and eukaryotes: Common themes and variations. Biochem J 431: 13–22 [DOI] [PubMed] [Google Scholar]