Abstract
Our understanding of the phylogenetic relationships among eukaryotic lineages has improved dramatically over the few past decades thanks to the development of sophisticated phylogenetic methods and models of evolution, in combination with the increasing availability of sequence data for a variety of eukaryotic lineages. Concurrently, efforts have been made to infer the age of major evolutionary events along the tree of eukaryotes using fossil-calibrated molecular clock-based methods. Here, we review the progress and pitfalls in estimating the age of the last eukaryotic common ancestor (LECA) and major lineages. After reviewing previous attempts to date deep eukaryote divergences, we present the results of a Bayesian relaxed-molecular clock analysis of a large dataset (159 proteins, 85 taxa) using 19 fossil calibrations. We show that for major eukaryote groups estimated dates of divergence, as well as their credible intervals, are heavily influenced by the relaxed molecular clock models and methods used, and by the nature and treatment of fossil calibrations. Whereas the estimated age of LECA varied widely, ranging from 1007 (943–1102) Ma to 1898 (1655–2094) Ma, all analyses suggested that the eukaryotic supergroups subsequently diverged rapidly (i.e., within 300 Ma of LECA). The extreme variability of these and previously published analyses preclude definitive conclusions regarding the age of major eukaryote clades at this time. As more reliable fossil data on eukaryotes from the Proterozoic become available and improvements are made in relaxed molecular clock modeling, we may be able to date the age of extant eukaryotes more precisely.
Fossil-calibrated molecular clock-based approaches are being used to estimate the age of major evolutionary events in eukaryotes. The estimates are heavily influenced by the methods and models used, but are steadily improving.
Our conception of the tree of eukaryotes has changed dramatically over the last few decades. In the 1980s and early 1990s, prevailing views were based on small subunit ribosomal RNA (SSU rRNA) gene phylogenies (e.g., Sogin 1991). However, as multiple protein-coding gene datasets were developed and more sophisticated phylogenetic methods were used, it became clear that the deep structure of the rRNA tree was the result of a methodological artifact known as long branch attraction (LBA) (Budin and Philippe 1998; Roger et al. 1999; Philippe et al. 2000a,b). Analyses based on multiple protein genes instead hinted at the existence of higher-level eukaryotic “supergroups” that encompassed both protistan and multicellular eukaryotic lineages (Baldauf et al. 2000). More recently, a better understanding of protistan ultrastructural diversity and the development of phylogenomic approaches have refined this picture and further delineated these groups (see also Fig. 1) (Bapteste et al. 2002; Burki et al. 2007; Hampl et al. 2009; Brown et al. 2012; Zhao et al. 2012).
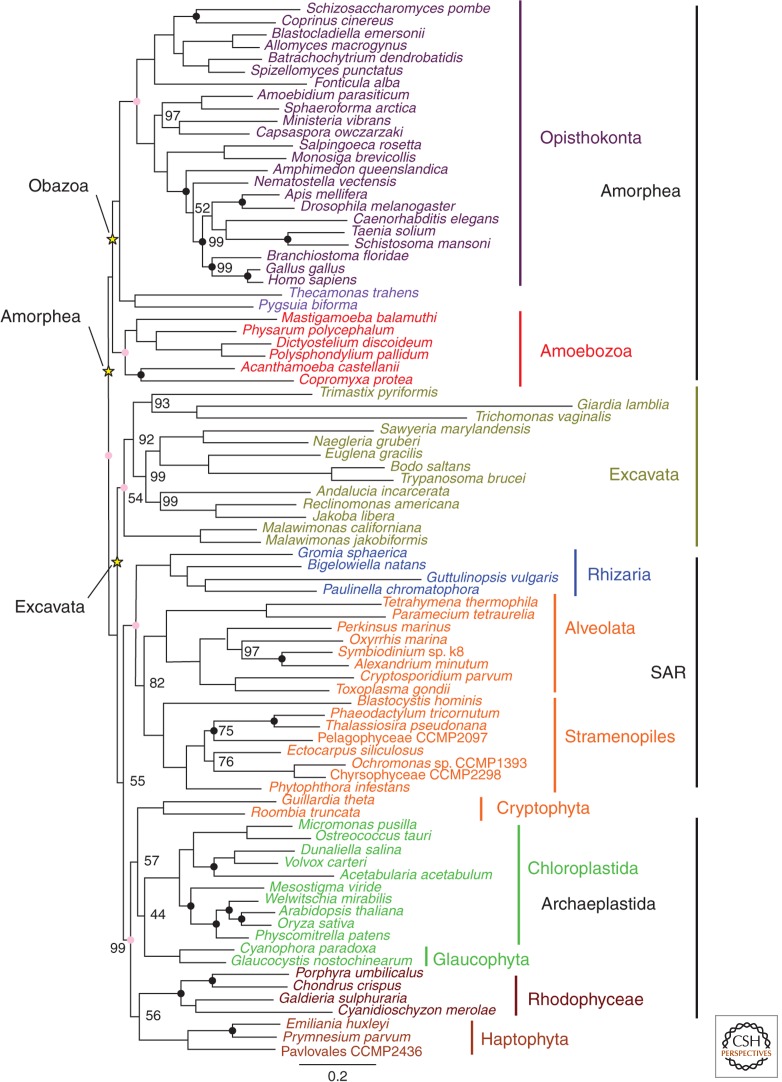
Figure 1.
Maximum likelihood phylogenetic tree of eukaryotes based on a phylogenomic dataset. Additional taxa were added to the original 159-gene Brown et al. (2013) dataset to maximize available fossil calibrations (total of 85 taxa, 43,099 sites). Black dots represent nodes on which fossil calibration constraints were imposed; yellow stars indicate the various positions of the root of the eukaryote tree considered; pink dots indicate the origin of major eukaryotic groups discussed here. A maximum likelihood (ML) phylogenetic tree was obtained from 60 heuristic searches using RAxML version 7.2.6 (Stamatakis 2006) under the Le and Gascuel (LG) + Γ +F amino acid substitution model (Le and Gascuel 2008). Numbers at nodes indicate bootstrap support (BS) for splits estimated from 500 bootstrap replicates. Most splits received maximum support and only BS < 100% is reported. Tree is shown rooted at the base of Amorphea, although roots at the base of either Obazoa or Excavates were also explored. Bayesian inference was also conducted using PhyloBayes 3.2 (Lartillot et al. 2009) by running four chains under either the CAT-GTR, CAT-Poisson, or the catfix C60-Poisson models of evolution, all combined with a gamma rates model. Bayesian calculations were not completed because of lack of convergence between chains, although the postburn-in consensus phylogeny from all runs was identical to the ML tree except for an unresolved multifurcation at the base of Excavata. Relaxed molecular clock (RMC) analyses were conducted with Phylobayes using the ML tree as a fixed topology. For all analyses, a birth–death tree prior was applied. Two chains were run until diagnostic statistics indicated convergence or estimated dates on nodes of interest for the two chains were <5% different. Fossil calibrations were taken from Parfrey et al. (2011) with the following modifications: four calibrations (Gonyaulacales, Spirotrichs, Foraminifera, Euglenids) were removed because of insufficient gene coverage within the clade of interest; the “Ciliate” calibration based on the tetrahymenol biomarker was removed (see text); as insufficient gene data was available from the haptophyte Isochrysis galbana, the upper bound on the coccolithophorid calibration was adjusted to an uninformative maximum (3000 Ma); the oldest cestode fossil (tapeworm) (Dentzien-Dias et al. 2013) was added as a calibration for Platyhelminths. The minimum age (250 Ma) was taken from the youngest possible age of the fossil and the upper boundary was set equal to the next-oldest calibrated node (Bilateria).
As our understanding of eukaryote phylogeny improved, fossil-calibrated molecular clock-based methods were beginning to be applied to date the major diversification events in this domain (Hedges et al. 2001; Douzery et al. 2004; Hedges and Kumar 2004; Berney and Pawlowski 2006; Parfrey et al. 2011). Molecular clock analyses were first introduced by Zuckerkandl and Pauling (1965). They showed that the differences between homologous proteins of different species are approximately proportional to their divergence time. Since then, sophisticated RMC methods have been developed that combine fossil data with molecular phylogenies for the inference of divergence times. However, attempts to estimate the age of deep divisions within eukaryotes using these methods have yielded vastly different estimates (e.g., see Douzery et al. 2004 vs. Hedges et al. 2004). These discrepancies can be explained by a myriad of sources of variability and error including (1) the assumed phylogeny of eukaryotes, (2) the sparse fossil record of protists and other organisms lacking hard structures for fossilization, (3) how fossil constraints are applied to phylogenetic trees, (4) methods and models used in RMC analysis, and (5) the selection of taxa and genes included.
Here, we review the progress and pitfalls in estimating the age of the last eukaryotic common ancestor (LECA) and supergroups using molecular clock-based analyses. We first discuss recent progress in our understanding of eukaryotic phylogeny and the ancient eukaryotic fossil record, and then we review the development of molecular clock-based methods and how fossil constraints are treated. Next, we describe attempts to date ancient eukaryotic divergences using RMC methods. Finally, we present an RMC analysis of a very large dataset comprised of 159 proteins and 85 taxa, using 19 fossil calibrations.
THE EUKARYOTIC TREE OF LIFE
Estimates of divergence dates from molecular clock analyses are only meaningful if the phylogeny on which they are based is correct. However, recovering deep phylogenetic relationships among eukaryotes has proven to be an extremely challenging task.
The most recent conceptions of the eukaryotic tree of life feature five or six “supergroups” (Keeling et al. 2005; Roger and Simpson 2009; Burki 2014) including Opisthokonta, Amoebozoa, Excavata, the SAR group, Archaeplastida, and Hacrobia (Haptophyta and Cryptophyta). Whereas phylogenomic analyses robustly recover the monophyly of Opisthokonta, Amoebozoa and SAR, the phylogenetic coherence of Excavata, Archaeplastida, and Hacrobia is less certain (see also Fig. 1) (Burki et al. 2008, 2012; Hampl et al. 2009; Parfrey et al. 2010; Brown et al. 2012; Zhao et al. 2012; Burki 2014).
Another challenge inherent to dating ancient events in eukaryotic evolution is the current uncertainty regarding the location of the root (Stechmann and Cavalier-Smith 2002, 2003a,b; Cavalier-Smith 2010; Derelle and Lang 2012; Katz et al. 2012). Tackling this question is made especially difficult by the absence of closely related outgroups to eukaryotes. The large phylogenetic distance between sequences from eukaryotes and their archaeal or bacterial orthologs makes their use as outgroups highly prone to phylogenetic artefacts like LBA (Felsenstein 1978; Roger and Hug 2006). Consequently, various alternative methods have been used in the last decade, yielding different results regarding the placement of the root; between Amorphea (or “unikonts”) and all other eukaryotes (i.e., “bikonts”) (Richards and Cavalier-Smith 2005; Roger and Simpson 2009; Cavalier-Smith 2010), which, despite being the current leading working hypothesis, is challenged by several lines of evidence (Arisue et al. 2005; Roger and Simpson 2009): at the base of an Excavata lineage, Euglenozoa (Cavalier-Smith 2010); between Archaeplastida and other eukaryotes (Rogozin et al. 2009; Koonin 2010); and between Opisthokonta and other eukaryotes (Katz et al. 2012). Therefore, although the unikont/bikont root position remains the most popular, the evidence supporting it is not definitive and the debate remains open.
CALIBRATING ESTIMATES OF EVOLUTIONARY RATES: BIOMARKERS AND FOSSILS
The evolutionary distance between sequences is the product of their evolutionary rate and the time that passed since they diverged. Therefore, to estimate the molecular rate, and thus to infer the precise timing of an evolutionary event, it is necessary to calibrate the tree of life with “known” dates associated with the available paleobiological data. For ancient evolutionary events, calibrations are commonly based on the fossil record and, to a lesser extent, on biomarkers (organic molecules in the rock record that are characteristic of particular organismal groups) (for a more comprehensive discussion, see Knoll 2013). When considering fossil evidence for ancient eukaryotes, it is important to distinguish between “stem” and “crown” lineages. Briefly, all lineages that descend from LECA are known as crown eukaryotes, whereas stem groups are extinct lineages that emerged before LECA, but diverged from the eukaryote nucleocytoplasmic lineage after the eukaryote/archaeal split. Any characteristic feature of eukaryotes evident in ancient microfossils or biomarkers can, in principle, be a property of either stem or crown organisms. Therefore, unless there are specific features that definitively associate the fossils/biomarkers with particular crown eukaryote groups, they cannot be used as divergence time constraints in molecular clock analyses.
The oldest proposed biomarker evidence for the existence of eukaryotes is ∼2.7-Ga-old steranes (i.e., breakdown products of sterols) (Brocks 1999). It was later shown that these sterol biomarkers were younger than the rocks in which they were found (Fischer 2008; Rasmussen et al. 2008), although there have been new strictly controlled studies reporting steranes in similar aged sediments (Waldbauer et al. 2009). A bacterial origin of these biomarkers cannot be excluded as some bacteria make sterols. However, these differ from many that are characteristic of specific eukaryotes (Summons et al. 2006; Desmond and Gribaldo 2009). In general, biomarkers need to be considered carefully as contamination is difficult to rule out, and their specificity to one lineage cannot be guaranteed, especially for microbes in which lateral gene transfer is prevalent. For example, gammacerane (a breakdown product of the triterpenoid tetrahymenol) has been suggested to indicate the presence of ciliates (e.g., Tetrahymena) that produce tetrahymenol (Summons and Walter 1990). However, the gene responsible for tetrahymenol synthesis was shown to have been transferred between a wide range of microaerophilic eukaryotes (Takishita et al. 2012).
Fossilized features that are consistent with eukaryotic affinity include a combination of a large size, the presence of complex morphological features, wall ultrastructure or ornamentation, or typical excystment structures unknown in prokaryotic organisms (Javaux et al. 2001, 2003, 2004; Knoll et al. 2006; Strother et al. 2011). Many microfossils of suggested eukaryotic origin are large (>50 μm) organic-walled structures known as acritarchs (Buick and Young 2010; Javaux et al. 2010). The oldest of these are microfossils described from the ∼3.2-billion-year-old (Ga) Moodies group (Buick and Young 2010; Javaux et al. 2010), far older than most acritarchs described to date (Javaux 2007). Javaux and colleagues were very cautious in their interpretation and argue that a prokaryote origin could not be definitively excluded. The next oldest remains of possible eukaryotes are large spiral ribbon-shaped fossils identified as the most ancient putative representatives of Grypania spiralis and found in 1800–2100-million-year-old iron formations (Han and Runnegar 1992), although their eukaryotic origin has been challenged (Samuelsson and Butterfield 2001; for review, see Javaux et al. 2003; Porter et al. 2003; Knoll et al. 2006). Additionally, large acritarchs with possible eukaryotic attributes were discovered in 1.8-Ga formations from China (Zhongying 1986), although their eukaryotic affiliation is still debated (Javaux et al. 2003). In fact, most of the ancient Proterozoic assemblages (i.e., late Paleoproterozoic and Mesoproterozoic rocks, 1800–1000 million years [Ma]) include fossils that are difficult, if not impossible, to associate with crown group eukaryotes. One of the most convincingly “eukaryotic” of these is Shuiyousphaeridium macroreticulatum from the ∼1.7-Ga Ruyang group, an ornamented acritarch with ridged walls made up of regularly packed hexagonal plates (Pang et al. 2013). Although an affinity to dinoflagellates (Alveolata) has been suggested (Leiming et al. 2005), there is little evidence for a crown group affiliation and they could represent a stem eukaryote lineage (Javaux 2006). Another example are microfossils of Tappania plana from the early Mesoproterozoic (Yin 1997) that were proposed to be of fungal affinity (Butterfield 2005), but which are still viewed as uncertain because of the limited number of systematically informative characters (Knoll et al. 2006). A notable exception is Bangiomorpha pubescens from the 1.2-Ga hunting formation (Butterfield 2000) that represent the oldest fossils confidently assigned to a crown eukaryotic lineage, the bangiophyte red algae. Many unambiguously eukaryotic fossils from the Neoproterozoic (1000–543 Ma) are also difficult to assign to specific extant eukaryotic clades (Javaux et al. 2003). For example, although some vase-shaped microfossils found in >742-Ma rocks have been relatively confidently identified as arcellinids (Amoebozoa), others from this assemblage could correspond to either amoebozoan or euglyphid (Rhizaria) amoebae (Porter and Knoll 2000; Porter et al. 2003). Based on an apparent increase in the diversity of complex organic-walled microfossils, a number of paleontologists suggest that the diversification of most eukaryotic kingdoms occurred at ∼800 Ma (Porter et al. 2003; Knoll et al. 2006; Porter 2006; Javaux 2007; Knoll 2013).
APPLYING FOSSIL CONSTRAINTS
There has been much debate in the last decade over how fossil dates should be treated in the context of molecular clock analyses (Graur and Martin 2004; Hedges and Kumar 2004; Reisz and Müller 2004; Blair and Hedges 2005; Glazko et al. 2005; reviewed in detail by Parham et al. 2012; Ronquist et al. 2012; Warnock et al. 2012).
First, there is inherent uncertainty associated with the dating of the rocks in which the fossils are found. Second, a systematic bias is introduced by the fact that the true divergence date must be older than the age of the fossil itself, and the time gap between the two is often unclear (Hedges and Kumar 2004). Because genetic divergence precedes detectable morphological variation, genetic divergence times are commonly underestimated by paleontological evidence, leading to overestimates of molecular rates (van Tuinen and Hadly 2004; Near et al. 2005; Roger and Hug 2006; Ho et al. 2011). Consequently, how fossil calibrations are applied on a phylogenetic tree has a significant impact on age estimates, affecting the age of the deepest nodes of the eukaryotic tree by hundreds of millions of years (Hug and Roger 2007). In some cases, fossil calibration methods and the presence of particular fossil calibrations has a greater impact on age estimates than taxonomic sampling (Hug and Roger 2007).
There are several approaches to applying fossil constraints. “Hard constraints” (or “hard bounds”) treat calibration points as fixed and accurate time intervals (specified by the paleontological evidence) assigned to particular nodes on the tree (Kishino et al. 2001). Hard bounds disallow the estimated age of a constrained node to be outside the specified interval—an assumption that is hardly justified given the uncertainties discussed above. If the fossil of interest is accurately dated, it can provide a realistic lower bound on the age of a divergence, but rarely a good upper bound. To overcome this problem, extremely large hard upper bounds can be used, although this is likely to bias time estimates to be too old (Yang 2006). In contrast, the “soft bound” approach allows for a smoothly decreasing probability of the node age falling outside the interval (Drummond et al. 2006; Yang and Rannala 2006; Rannala and Yang 2007; Inoue et al. 2010). However, the nature of these probability distributions and how they are applied (i.e., with or without maximum constraints) can significantly alter the resulting estimates (Warnock et al. 2012). In the best-case scenario, several fossils are available that display apomorphies corresponding to different clades within a group, allowing for sequential constraints on minimum and maximum ages of the nodes in that part of the phylogeny. Unfortunately, the patchy and difficult-to-interpret ancient fossil record corresponding to the deeper eukaryote divergences is extremely limiting for the accuracy of this type of upper bound implementation. Some progress has been made in the recent years through the greater use of the taxonomically diverse older microfossil record that documents the appearance of a variety of protistan groups (Berney and Pawlowski 2006; Parfrey et al. 2011).
RMC METHODS
Molecular dating approaches theoretically allow divergence times to be estimated from genetic distances. To do this, the phylogenetic tree is calibrated with one or more known dates, usually based on fossil or biomarker records, and divergence times are extrapolated throughout the tree. Originally, molecular dating relied on the assumption of a strict molecular clock postulating a constant rate of evolution over the whole tree (Zuckerkandl and Pauling 1965). However, variation in substitution rates has been widely documented (Smith and Peterson 2002; Bromham and Penny 2003; Davies et al. 2004). Consequently, RMC methods were developed that allow the rate of sequence evolution to vary across different branches (for reviews, see Welch and Bromham 2005; Lepage et al. 2007; Ho and Phillips 2009). A number of different methods have been developed and debate continues as to which best captures biological reality (Drummond et al. 2006; Lepage et al. 2007; Ho and Phillips 2009; Linder et al. 2011).
Local clock implementations estimate a separate molecular rate for each user-defined part of the tree (Yoder and Yang 2000). This requires arbitrary choices to be made regarding the number of different rate classes and how they should be assigned to branches; it is unclear how this can be achieved in a rigorous way in the absence of substantial prior information.
Rate-smoothing algorithms (Sanderson 1997, 2002) assume that the rate of evolution itself is evolving and is correlated across adjacent branches on the phylogeny such that related lineages have similar rates. For nonparametric rate smoothing (NPRS), rates are optimized to minimize a smoothing function that summarizes differences between rates on adjacent branches, taking into account fossil constraints (Sanderson 1997). The penalized-likelihood approach (Sanderson 2002), in contrast, combines a probabilistic model of sequence evolution with a penalty function (similar to the “smoothing function” of NPRS methods). These rate-smoothing methods require arbitrary decisions to be made regarding the parameters of the smoothing/penalty functions, and doubts persist regarding their statistical properties (Yang 2006).
The most sophisticated RMC methods are Bayesian approaches that probabilistically model every feature of molecular evolution over the tree, including the substitution process, tree generation processes, and substitution rate changes across branches (i.e., the RMC process). The various RMC process models can be divided in two main classes differing in whether temporal autocorrelation among adjacent lineages is assumed (Kishino et al. 2001; Aris-Brosou and Yang 2003; Rannala and Yang 2007) or not (Drummond et al. 2006; Akerborg et al. 2008). When rates are not autocorrelated, each rate is independent from the neighboring ones and randomly sampled from a probability distribution (e.g., the uncorrelated models introduced by Drummond et al. 2006). In autocorrelated models, rates follow a diffusion process along lineages, with the rate in each branch being drawn a priori from a parametric distribution whose mean is a function of the rate on the parent branch (e.g., the log-normal [LogN] distribution) (Thorne et al. 1998; Kishino et al. 2001), in which the evolutionary rate varies according to a lognormal distribution; or the Cox–Ingersoll–Rand distribution (CIR) (Lepage et al. 2007), which possesses a stationary distribution, contrasting with the linearly increasing variance of the LogN model.
RECENT ATTEMPTS TO DATE LECA AND MAJOR EUKARYOTIC GROUPS
In 2004, Hedges and colleagues analyzed multiprotein datasets with both constant rate and RMC methods to estimate the age of a number of deep eukaryote divergences (Hedges et al. 2004). After screening out proteins with detectable departures from the strict molecular clock, they obtained similar results with all methods, estimating LECA to be 2309 (2115–2503) Ma old, although several methodological aspects of this work have been debated (Graur and Martin 2004; Hedges and Kumar 2004; Roger and Hug 2006). Another potential problem with these analyses is that inferences regarding LECA were based on rooting the eukaryote tree on the lineage leading to the diplomonad Giardia. As discussed earlier, this rooting position is likely to be an artefactual result of LBA.
Douzery et al. (2004) estimated the age of LECA and other major groups using a Bayesian RMC approach calibrated with, for the first time, six paleontological constraints from the Phanerozoic. To reduce the impact of stochastic error resulting from the variation in evolutionary rates among individual genes, they analyzed a large supermatrix of 129 proteins and a tree rooted on the branch leading to Dictyostelium (close to the unikont/bikont root). They estimated a substantially younger age for LECA, dated at 1085 Ma (950–1259 Ma), and inferred that the subsequent diversification of all major groups occurred within ∼200 Myr.
To avoid some sources of error associated with the fossil record, Berney and Pawlowski (2006) used many more fossil constraints including Phanerozoic protist microfossils as a source of 26 calibration points (four maximum and 22 minimum time constraints). Their phylogeny, inferred from a single gene (SSU rRNA), was analyzed with fossil constraints in a Bayesian RMC framework. Based on a unikont/bikonts rooting, they estimated the age of LECA to be 1126 (948–1357) Ma. They concluded that most Proterozoic fossils suggested to be eukaryotes should not be assumed to belong to extant groups or used as calibration points for molecular dating analyses.
Most recently, Parfrey et al. (2011) performed Bayesian RMC analyses of a taxon-rich multigene dataset (91–109 taxa and 15 proteins) combined with 23 calibration points derived from Proterozoic and Phanerozoic eukaryotic fossils and biomarkers. They concluded that LECA was 1679–1866 Ma old. In contrast to Berney and Pawlowski (2006), Parfrey et al. (2011) argue that estimated ages of major groups are consistent with the tentative taxonomic assignments of many of the putative eukaryotic fossils from the Proterozoic.
ON THE DIFFICULTY OF DATING DEEP EVENTS IN EUKARYOTIC EVOLUTION
As discussed in the foregoing sections, the discrepancies between date estimates from these previous RMC analyses can be attributed to a myriad of different causes. Roger and Hug (Roger and Hug 2006; Hug and Roger 2007) examined a number of these sources of variation for dates estimated from two published datasets (Douzery et al. 2004; Peterson and Butterfield 2005). They found that both the estimated ages of nodes and size of the confidence (or credible) intervals associated with them were extremely sensitive to the RMC method used, the number and nature of fossils chosen for constraints, and how these constraints were applied.
Here, we further explore sources of variation in molecular clock analyses focusing on the chosen root position for eukaryotes, the amino acid substitution model, the RMC model, and the manner in which calibration constraints are applied. In these analyses, we rely on a sophisticated Bayesian implementation of RMC models (Lartillot et al. 2009) and analyze a large phylogenomic dataset (159 proteins, 85 taxa). Nineteen calibration points were used, the majority of them taken from Parfrey et al. (2011) (for details, see Fig. 1).
Impact of Calibration Constraints
First, we investigated the impact of the way of applying fossil calibrations by using both “hard-” and “soft-” bounds constraints. Strikingly, hard bounds gave notably older estimates for all nodes over a range of RMC models and root positions (9%–41% older) (compare filled and open shapes in Figs. 2 and 3). This suggests that at least one of the fossil calibrations is not consistent with the others (i.e., the estimated date of the fossil itself or its taxonomic affiliation/node assignment is incorrect). Closer inspection revealed that, under soft bounds, the fossil Bangiomorpha pubescens (assigned to the basal red algal split) was estimated to be 690 (639–771)–1026 (835–1206) Ma old, departing hugely from the age constraints applied (1174–3000 Myr). This was the only Proterozoic fossil in our analyses that was estimated to be younger than its lower age bound when soft-bounds constraints were applied. That the age or taxonomic assignment of Bangiomorpha is not consistent with other fossil dates in our analyses is similar to the observations of Berney and Pawlowski (2006). Similarly, Parfrey et al. (2011) found that excluding Proterozoic fossil constraints shifted the age of major groups to 200–300 Ma younger and also noted a marked change in the estimated age of red algae. Therefore, all recent molecular clock analyses seem to concur that the Bangiomorpha fossil date or taxonomic assignment is problematic. An alternative explanation is that the rates of evolution have dramatically shifted along the red algal lineage and this phenomenon is not well captured by relaxed clock models, in general.
Figure 2.
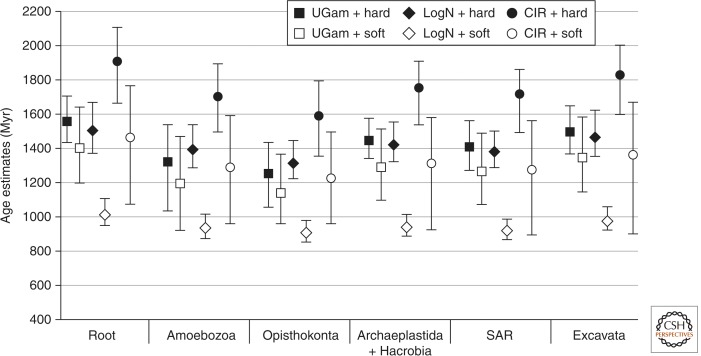
Impact of the root position and calibration constraints on estimated divergence times. Estimated age (in Myr) of LECA and major eukaryotic groups when the root was placed at the base of Amorphea (squares), Obazoa (diamonds), and Excavata (circles), using hard- (filled shapes) and soft- (open shapes) bounds constraints. Error bars represent 95% credible intervals. All estimations were performed using the C60 substitution model and the uncorrelated γ distribution of rates (UGam) clock model.
Figure 3.
Impact of the molecular clock models and calibration constraints on estimated divergence times. Estimated age (in Myr) of LECA and major eukaryotic groups using the UGam (squares), LogN (diamonds), and CIR (circles) clock models, applying hard- (filled shapes) and soft- (open shapes) bounds constraints. Error bars represent 95% credible intervals. All analyses are based on the Amorphea root, using the C60 substitution model.
Impact of the Root Position
During our analyses, we considered three possible positions for the eukaryote root. We first placed the root between Amorphea and the remainder of eukaryotes (i.e., unikont-bikont rooting). We tested a second rooting position between the Obazoa (Opisthokonta + apusomonads + breviates) and all other taxa—a variation of the hypothesis by Katz et al. (2012) in which they located the root at the base of Opisthokonta. This modification is based on recent work presenting evidence for the monophyly of Obazoa (Brown et al. 2013a). Finally, we tested a root at the base of Excavata as many excavate protists (e.g., diplomonads [Hedges et al. 2004], Euglenozoa [Cavalier-Smith 2010], or jakobids [Brinkmann et al. 2007]) have variously been suggested to be “basal” eukaryote lineages.
Interestingly, these three alternatives for the position of the root did not have much impact on the estimated age of the root itself and the supergroups under the uncorrelated gamma (UGam) RMC model (this model setting is discussed further below) (Fig. 2). For fixed model settings, the three tested positions gave similar date estimates for a given specific timepoint and varied by less than 8%, with the 95% credible intervals largely overlapping (Fig. 2) (compare the variation among filled shapes and open shapes). Berney and Pawlowski also found that four different placements of the root (at the base of unikonts, Opisthokonta, Amoebozoa, and Excavata, respectively) yielded similar time estimates (Berney and Pawlowski 2006). Parfrey et al. (2011) also reached similar conclusions for the unikont, Opisthokonta, Excavata, and Excavata + unikont roots.
For all sets of parameters, our results show that Opisthokonta is estimated as being the youngest of all major groups. Nevertheless, the estimated age difference between LECA and the last common ancestor of Opisthokonta is relatively small, and ranges between 23 and 334 Ma with LECA estimated to be between 1007 and 1898 Ma old, and Opisthokonta between 904 and 1579 Ma old (Fig. 3, filled circles and open diamonds). This suggests that the six groups considered here diversified over a relatively short period of time after LECA and explains why the various placements of the root in this region of the tree do not have a major impact on its estimated age. This observation is in agreement with the “big-bang hypothesis” for eukaryotic evolution, according to which major eukaryotic groups emerged rapidly, virtually leading to a massive multifurcation (Philippe et al. 2000a,b). However, caution is warranted as problems with saturation of sequence changes, model misspecification, and other sources of conflict within the data could also lead to the estimation of extremely short internal branches deep in the eukaryote tree, artefactually generating a multifurcating topology (see Roger and Hug 2006). It should be noted that the roots considered here and in the other aforementioned studies (Berney and Pawlowski 2006; Parfrey et al. 2011) are all topologically “close”; we expect that more distantly placed putative roots (i.e., well within one of the supergroups) would give significantly different estimates for the age of LECA, although we did not test this in our analyses.
Impact of the Substitution Model and Relaxed Clock Model
To test the impact of the substitution and the relaxed clock models, we estimated the age of each major group by applying six model combinations (Fig. 4).
Figure 4.
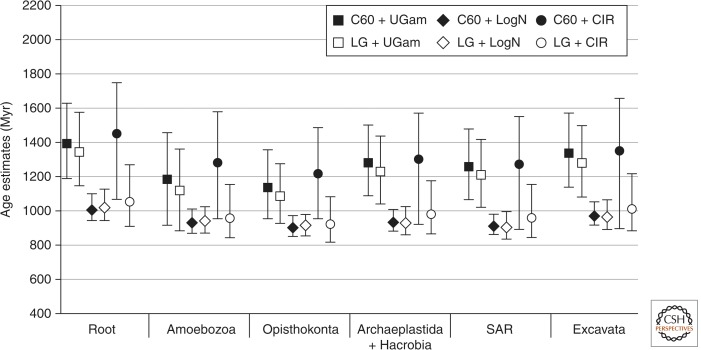
Impact of the substitution and molecular clock models on estimated divergence times. Estimated age (in Myr) of LECA and major eukaryotic groups using the UGam (squares), LogN (diamonds), and CIR (circles) clock models. Substitution model was either C60 (filled shapes) or LG (open shapes). Error bars represent 95% credible intervals. All estimations were calculated with the Amorphea root, using soft bounds calibration constraints.
The two substitution models used were either the site-heterogeneous empirical profile mixture model C60-Poisson (C60) (Le et al. 2008) or the more classical site-homogeneous LG substitution matrix (Le and Gascuel 2008). The three RMC models included the UGam and the autocorrelated LogN and CIR models.
Isolating the impact of the substitution model and the RMC model turns out to be a difficult task. C60 + UGam and LG + UGam both gave similar date estimates (difference <7%) (Fig. 4, filled and empty squares) as did C60 + LogN and LG + LogN (<2%) (Fig. 4, filled and empty diamonds). In contrast, C60 + CIR yielded very different results from LG + CIR (27%–32% difference) (Fig. 4, filled and empty squares).
However, comparing the estimated dates under the LG substitution model showed that they were similar when the LogN or CIR relaxed clock models were used (1%–6% difference between LG + LogN and LG + CIR) (Fig. 4, open diamonds and circles), whereas LG + UGam gave much older estimates (difference up to 30% with LG + LogN and LG + CIR) (Fig. 4, compare open squares with open diamonds and circles, respectively). Finally, C60 + UGam and C60 + CIR models led to similar date estimates (1%–8% difference) (Fig. 4, filled squares and circles), whereas C60 + LogN gave much younger ages (22%–36% younger) (Fig. 4, compare filled diamonds with filled squares and circles).
The complexity of the relationship between these parameters is further amplified by the choice of hard- versus “soft-bounds calibration constraints. All previous comparisons used soft bounds and application of hard bounds led to very different conclusions. For example, with hard bounds, the C60 + UGam and C60 + LogN models gave similar estimates (<5% difference) (Fig. 3, filled squares and diamonds), that were drastically younger than estimates of the C60 + CIR model (19%–23%) (Fig. 3, compare filled squares and diamonds with filled circles). In contrast, with soft bounds, the C60 + CIR model gave estimates similar to the C60 + UGam model (<8% variation) (Fig. 3, open circles and squares), whereas C60 + LogN yielded much younger dates (24%–36%) (Fig. 3, compare open circles and squares to open diamonds).
Overall, the largest differences among estimates is observed between the CIR clock model with hard bounds calibrations combination (which gave the oldest dates) (Fig. 3, filled circles), and the “LogN + soft bounds” combination (estimating the youngest ones) (Fig. 3, open diamonds). Using these two settings, the eukaryotic root was estimated to be 1898 versus 1007 Ma old, respectively. Because the soft-bounds approach is a more realistic way to treat fossil constraints, the younger date estimates obtained under these constraints are likely more reasonable. Restricting attention to soft-bounds analyses, combinations of RMC models and calibration constraints gave estimates for LECA between 1007–1456 Ma (Fig. 3, open shapes). It is important to note, however, that credible intervals on the age of LECA and most other nodes can often be quite large, sometimes spanning up to ∼650 Ma (e.g., CIR model) (Fig. 3).
MODEL COMPARISON AND ASSESSMENT
As suggested by previous studies and the foregoing analysis, the various features of RMC analyses interact in a complex manner in influencing the age estimates obtained. This inevitably leads to the question of which models or settings should be trusted over others. Generally, this involves model selection via the assessment of the relative fit of various models used in the analysis (e.g., the substitution or molecular clock models); better fitting models should, ceteris paribus, provide better estimates. Therefore, we suggest that model selection techniques be used to choose among alternative RMC and substitution models. Below, we briefly review the various model selection techniques in the Bayesian context, the statistical inference paradigm in which most RMC models have been implemented to date.
The Bayes factor is the most widely used Bayesian model selection criterion and is defined as the ratio of the marginal likelihoods under the two models of interest (Linder et al. 2011). Bayes factors measure how strongly the data support a given model and have the advantages of implicitly penalizing more complex models as well as allowing for a general comparison among models that are not necessarily nested (Lepage et al. 2007). Unfortunately, in the phylogenetic context, exact calculation of Bayes factors is computationally infeasible and approximation techniques must be used. Although the harmonic mean estimator has been used for this purpose, it turns out to be extremely unstable (Lartillot and Philippe 2006). More reliable estimators are complex approximation methods such as thermodynamic integration (path-sampling) (Ogata 1989; Lartillot and Philippe 2004, 2006; Blanquart and Lartillot 2006; Rodrigue et al. 2006) and the stepping-stone method (Xie et al. 2011) that have been implemented in programs like Phylobayes (Lartillot et al. 2009) and BEAST (Baele et al. 2012), respectively. Note that these methods are generally used for RMC model selection but can be difficult to implement for selecting among substitution models.
Several other methods have been developed for substitution model selection in a Bayesian context. Lartillot and colleagues have developed cross-validation (CV) (Lartillot et al. 2007) methods that calculate the ability of one part of the data (the learning set) to predict the remaining data (the test set) after the dataset is randomly split into two uneven parts. In Phylobayes, Markov chain Monte Carlo (MCMC) is run on the learning set and, after burn-in is discarded, the average likelihood of the topology/parameters/model visited in the chain is evaluated in the test set. The procedure is repeated 10 times for each model, and the model with the greatest average likelihood over the test sets is the one selected as better fitting. CV analyses virtually always select site-heterogeneous substitution models (e.g., CAT or C60) over the simpler single-matrix models (e.g., LG) (Lartillot et al. 2009).
Posterior predictive simulation is another way of evaluating relative fit of substitution models (Bollback 2002; Nielsen and Huelsenbeck 2002; Lartillot and Philippe 2004; Blanquart and Lartillot 2008). From each postburn-in MCMC sample, a dataset is simulated from the model parameters. A statistic measuring some property (e.g., mean number of distinct residues observed at each column of the alignment) can be calculated for the real dataset and compared with the distribution of this statistic from the simulated datasets (e.g., Lartillot et al. 2009). A significant deviation between the statistics evaluated on real and simulated data implies that some of the model assumptions are unrealistic, indicating misspecification. The model in which the statistic evaluated on the simulated distribution falls closest to that of the real dataset is then considered the better fitting model.
FOSSIL ACCURACY ASSESSMENT
CV procedures were also developed to assess consistency between fossil and molecular age estimates (Near and Sanderson 2004; Near et al. 2005; Rutschmann et al. 2007). They aim to detect point calibrations that poorly predict other calibration dates in the dataset, and are therefore suspected to be erroneous. The main pitfall of these approaches stems from their tendency to discard “outliers” to obtain an internally consistent set without taking into account the relative credibility of the corresponding fossils. Another weakness comes from the fact that they consider point calibrations and not minimum age constraints (Parham and Irmis 2008).
As point calibrations were progressively being replaced by distributions that better represent palaeontological uncertainty (Drummond et al. 2006; Yang and Rannala 2006), Sanders and Lee (2007) proposed to test for calibration accuracy in a Bayesian context through the comparison of the posterior age distribution of a calibration node with the prior specified for it.
CONCLUSION
Molecular clock estimates of ancient divergence times in the tree of life are affected by numerous sources of errors and uncertainties. Although resolution in the tree of eukaryotes appears to be steadily improving, the location of the root (i.e., LECA) remains uncertain. Moreover, controversy in assigning Proterozoic fossils to extant eukaryote groups means that molecular clock analyses must rely heavily on extrapolation from the younger, but richer, Phanerozoic fossil record. There are also inherent biases and uncertainties associated with assigning fossil calibrations to nodes in molecular phylogenies. These factors, combined with variability in estimates and credible intervals yielded by different molecular clock model assumptions, have led to the wide ranges of estimated ages of LECA and major eukaryote supergroups published in the last decade. The analyses of a taxon-rich supermatrix with 19 fossil calibrations presented here provide estimates for the age of LECA in the range of ∼1000–1900 Ma depending on the methods used, with the credibility intervals on estimates in some cases spanning up to ∼650 Myr. Despite this uncertainty about precise ages, both our and other recent molecular clock analyses recover a relatively short time interval (<300 Myr) between the age of LECA and emergence of all of the various supergroups, consistent with a rapid big-bang diversification of eukaryotes (Philippe et al. 2000a).
It is unclear how much more precision we will be able to achieve in dating the extant eukaryote clade. For molecular phylogenetics, difficult to resolve parts of trees can be clarified by the addition of more taxa and more genes (sites) because, in theory, likelihood-based methods are statistically consistent (see discussion in Yang 2006). However, this is not the case for molecular clock analyses because inherent uncertainties in fossil ages and biases in their node assignments imply that molecular clock age estimates will always be associated with error. Nevertheless, the addition of more fossils and genes may help narrow confidence or credible intervals, and steady progress is being made in the development of better RMC methods, model selection, and fossil assignment validation. These methods, combined with novel sources of time constraints such as those provided by horizontal or endosymbiotic gene transfer (see Shih and Matzke 2013), may ultimately help in the quest to determine the age of extant eukaryotes.
ACKNOWLEDGMENTS
This work and M.W.B.'s postdoctoral fellowship is supported by Discovery grant 227085-11 and an Accelerator grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) awarded to A.J.R. L.E. is supported by a Centre for Comparative Genomics and Evolutionary Bioinformatics postdoctoral fellowship from the Tula Foundation; S.C.S. is supported by graduate scholarships from NSERC and Killam trusts. We thank Nicolas Lartillot, Laura Wegener Parfrey, and Andrew H. Knoll for very stimulating discussions and insights. Computations were partially performed on the supercomputers at the SciNet HPC Consortium. SciNet is funded by the Canada Foundation for Innovation under the auspices of Compute Canada, the Government of Ontario, Ontario Research Fund—Research Excellence, and the University of Toronto (Loken et al. 2010).
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Akerborg O, Sennblad B, Lagergren J 2008. Birth-death prior on phylogeny and speed dating. BMC Evol Biol 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris-Brosou S, Yang Z 2003. Bayesian models of episodic evolution support a late Precambrian explosive diversification of the Metazoa. Mol Biol Evol 20: 1947–1954 [DOI] [PubMed] [Google Scholar]
- Arisue N, Hasegawa M, Hashimoto T 2005. Root of the Eukaryota tree as inferred from combined maximum likelihood analyses of multiple molecular sequence data. Mol Biol Evol 22: 409–420 [DOI] [PubMed] [Google Scholar]
- Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977. [DOI] [PubMed] [Google Scholar]
- Bapteste E, Brinkmann H, Lee Ja, Moore DV, Sensen CW, Gordon P, Duruflé L, Gaasterland T, Lopez P, Müller M, et al. 2002. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci 99: 1414–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney C, Pawlowski J 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc Biol Sci 273: 1867–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Hedges SB 2005. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol 22: 2275–2284 [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N 2006. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol Biol Evol 23: 2058–2071. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N 2008. A site- and time-heterogeneous model of amino acid replacement. Mol Biol Evol 25: 842–858 [DOI] [PubMed] [Google Scholar]
- Bollback JP 2002. Bayesian model adequacy and choice in phylogenetics. Mol Biol Evol 19: 1171–1180 [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Burger G, Roger AJ, Gray MW, Lang BF, Rodríguez-Ezpeleta N, Philippe H 2007. Toward resolving the eukaryotic tree: The phylogenetic positions of jakobids and cercozoans. Curr Biol 17: 1420–1425 [DOI] [PubMed] [Google Scholar]
- Brocks JJ 1999. Archean molecular fossils and the early rise of eukaryotes. Science 285: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Bromham L, Penny D 2003. The modern molecular clock. Nat Rev Genet 4: 216–224 [DOI] [PubMed] [Google Scholar]
- Brown MW, Kolisko M, Silberman JD, Roger AJ 2012. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr Biol 22: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Brown MW, Sharpe SC, Silberman JD, Heiss AA, Simpson AG, Roger AJ 2013. Phylogenomics demonstrate that breviate flagellates are related to opisthokonts and apusomonads. Proc Biol Sci 280: 20131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budin K, Philippe H 1998. New insights into the phylogeny of eukaryotes based on ciliate Hsp70 sequences. Mol Biol Evol 15: 943–956 [DOI] [PubMed] [Google Scholar]
- Buick R, Young A 2010. Ancient acritarchs. 463: 885–886 [DOI] [PubMed] [Google Scholar]
- *.Burki F 2014. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J 2007. Phylogenomics reshuffles the eukaryotic supergroups. PloS ONE 2: e790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J 2008. Phylogenomics reveals a new “megagroup” including most photosynthetic eukaryotes. Biol Lett 4: 366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert J-F, Keeling PJ 2012. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc Biol Sci 279: 2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield NJ 2000. Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26: 386–404 [Google Scholar]
- Butterfield NJ 2005. Probable Proterozoic fungi. Paleobiology 31: 165–182 [Google Scholar]
- Cavalier-Smith T 2010. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett 6: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TJ, Savolainen V, Chase MW, Moat J, Barraclough TG 2004. Environmental energy and evolutionary rates in flowering plants. Proc Biol Sci 271: 2195–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentzien-Dias PC, Poinar G Jr, de Figueiredo AEQ, Pacheco ACL, Horn BLD, Schultz CL 2013. Tapeworm eggs in a 270 million-year-old shark coprolite. PLoS ONE 8: e55007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lang BF 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol 29: 1277–1289 [DOI] [PubMed] [Google Scholar]
- Desmond E, Gribaldo S 2009. Phylogenomics of sterol synthesis: Insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol Evol 1: 364–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H 2004. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci 101: 15386–15391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool 27: 401–410. [Google Scholar]
- Fischer WW 2008. Biogeochemistry: Life before the rise of oxygen. Nature 455: 1051–1052 [DOI] [PubMed] [Google Scholar]
- Glazko GV, Koonin EV, Rogozin IB 2005. Molecular dating: Ape bones agree with chicken entrails. Trends Genet 21: 89–92 [DOI] [PubMed] [Google Scholar]
- Graur D, Martin W 2004. Reading the entrails of chickens: Molecular timescales of evolution and the illusion of precision. Trends Genet 20: 80–86 [DOI] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.” Proc Natl Acad Sci 106: 3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Runnegar B 1992. Megascopic eukaryotic algae from the 2.1-billion-year-old negaunee iron-formation, Michigan. Science 257: 232–235 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S 2004. Precision of molecular time estimates. Trends Genet 20: 242–247 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Chen H, Kumar S, Wang DY, Thompson AS, Watanabe H 2001. A genomic timescale for the origin of eukaryotes. BMC Evol Biol 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol 58: 367–380 [DOI] [PubMed] [Google Scholar]
- Ho SYW, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A 2011. Time-dependent rates of molecular evolution. Mol Ecol 20: 3087–3101 [DOI] [PubMed] [Google Scholar]
- Hug LA, Roger AJ 2007. The impact of fossils and taxon sampling on ancient molecular dating analyses. Mol Biol Evol 24: 1889–1897 [DOI] [PubMed] [Google Scholar]
- Inoue J, Donoghue PCJ, Yang Z 2010. The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst Biol 59: 74–89 [DOI] [PubMed] [Google Scholar]
- Javaux EJ 2006. Extreme life on Earth—Past, present and possibly beyond. Res Microbiol 157: 37–48 [DOI] [PubMed] [Google Scholar]
- Javaux EJ 2007. The early eukaryotic fossil record. Adv Exp Med Biol 607: 1–19 [DOI] [PubMed] [Google Scholar]
- Javaux E, Knoll A, Walter M 2001. Morphological and ecological complexity in early eukaryotic ecosystems. Nature 412: 66–69. [DOI] [PubMed] [Google Scholar]
- Javaux EJ, Knoll AH, Walter M 2003. Recognizing and interpreting the fossils of early eukaryotes. Orig Life Evol Biosph 33: 75–94 [DOI] [PubMed] [Google Scholar]
- Javaux E, Knoll A, Walter M 2004. TEM evidence for eukaryotic diversity in mid-Proterozoic oceans. Geobiology 2: 121–132. [Google Scholar]
- Javaux EJ, Marshall CP, Bekker A 2010. Organic-walled microfossils in 3.2-billion-year-old shallow-marine siliciclastic deposits. Nature 463: 934–938 [DOI] [PubMed] [Google Scholar]
- Katz LA, Grant JR, Parfrey LW, Burleigh JG 2012. Turning the crown upside down: Gene tree parsimony roots the eukaryotic tree of life. Syst Biol 61: 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW 2005. The tree of eukaryotes. Trends Ecol Evol 20: 670–676 [DOI] [PubMed] [Google Scholar]
- Kishino H, Thorne JL, Bruno WJ 2001. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol Biol Evol 18: 352–361 [DOI] [PubMed] [Google Scholar]
- *.Knoll AH 2013. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb Perspect Biol 6: a016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH, Javaux EJ, Hewitt D, Cohen P 2006. Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci 361: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21: 1095–1109 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H 2006. Computing Bayes factors using thermodynamic integration. Syst Biol 55: 195–207. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S 2009. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O, Lartillot N 2008. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24: 2317–2323 [DOI] [PubMed] [Google Scholar]
- Leiming Y, Xunlai Y, Fanwei M, Jie H 2005. Protists of the upper Mesoproterozoic Ruyang group in Shanxi Province, China. Precambrian Res 141: 49–66 [Google Scholar]
- Lepage T, Bryant D, Philippe H, Lartillot N 2007. A general comparison of relaxed molecular clock models. Mol Biol Evol 24: 2669–2680 [DOI] [PubMed] [Google Scholar]
- Linder M, Britton T, Sennblad B 2011. Evaluation of Bayesian models of substitution rate evolution—Parental guidance versus mutual independence. Syst Biol 60: 329–342 [DOI] [PubMed] [Google Scholar]
- Loken C, Gruner D, Groer L, Peltier R, Bunn N, Craig M, Henriques T, Dempsey J, Yu C-H, Chen J, et al. 2010. SciNet: Lessons learned from building a power-efficient top-20 system and data centre. J Phys Conf Ser 256: 012026 [Google Scholar]
- Near TJ, Sanderson MJ 2004. Assessing the quality of molecular divergence time estimates by fossil calibrations and fossil-based model selection. Philos Trans R Soc Lond B Biol Sci 359: 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Meylan Pa, Shaffer HB 2005. Assessing concordance of fossil calibration points in molecular clock studies: An example using turtles. Am Nat 165: 137–146 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Huelsenbeck JP 2002. Detecting positively selected amino acid sites using posterior predictive P-values. Pac Symp Biocomput 588: 576–588 [DOI] [PubMed] [Google Scholar]
- Ogata Y 1989. A Monte Carlo method for high dimensional integration. Numerische Mathematik 157: 137–157 [Google Scholar]
- Pang K, Tang Q, Schiffbauer JD, Yao J, Yuan X, Wan B, Chen L, Ou Z, Xiao S 2013. The nature and origin of nucleus-like intracellular inclusions in Paleoproterozoic eukaryote microfossils. Geobiology 11: 499–510. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA 2010. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol 59: 518–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Knoll AH, Katz LA 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci 108: 13624–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham JF, Irmis RB 2008. Caveats on the use of fossil calibrations for molecular dating: A comment on Near et al. Am Nat 171: 132–136; author reply 137–140 [DOI] [PubMed] [Google Scholar]
- Parham JF, Donoghue PCJ, Bell CJ, Calway TD, Head JJ, Holroyd Pa, Inoue JG, Irmis RB, Joyce WG, Ksepka DT, et al. 2012. Best practices for justifying fossil calibrations. Syst Biol 61: 346–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Butterfield NJ 2005. Origin of the Eumetazoa: Testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc Natl Acad Sci 102: 9547–9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Germot A, Moreira D 2000a. The new phylogeny of eukaryotes. Curr Opin Genet Dev 10: 596–601 [DOI] [PubMed] [Google Scholar]
- Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Müller M, Le Guyader H 2000b. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc Biol Sci 267: 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SM 2006. The Proterozoic fossil record of heterotrophic eukaryotes. In Neoproterozoic geobiology and paleobiology, pp. 1–21. Springer, New York [Google Scholar]
- Porter SM, Knoll AH 2000. Testate amoebae in the Neoproterozoic era: Evidence from vase-shaped microfossils in the Chuar group, Grand Canyon. Paleobiology 26: 360–385 [Google Scholar]
- Porter S, Meisterfeld R, Knoll A 2003. Vase-shaped microfossils from the Neoproterozoic Chuar group, Grand Canyon: A classification guided by modern testate amoebae. J Paleont 77: 409–429 [Google Scholar]
- Rannala B, Yang Z 2007. Inferring speciation times under an episodic molecular clock. Syst Biol 56: 453–466 [DOI] [PubMed] [Google Scholar]
- Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR 2008. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455: 1101–1104 [DOI] [PubMed] [Google Scholar]
- Reisz RR, Müller J 2004. Molecular timescales and the fossil record: A paleontological perspective. Trends Genet 20: 237–241 [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436: 1113–1118 [DOI] [PubMed] [Google Scholar]
- Rodrigue N, Philippe H, Lartillot N 2006. Assessing site-interdependent phylogenetic models of sequence evolution. Mol Biol Evol 23: 1762–1775. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Hug LA 2006. The origin and diversification of eukaryotes: Problems with molecular phylogenetics and molecular clock estimation. Philos Trans R Soc Lond B Biol Sci 361: 1039–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Simpson AGB 2009. Evolution: Revisiting the root of the eukaryote tree. Curr Biol 19: R165–R167 [DOI] [PubMed] [Google Scholar]
- Roger AJ, Sandblom O, Doolittle WF, Philippe H 1999. An evaluation of elongation factor 1α as a phylogenetic marker for eukaryotes. Mol Biol Evol 16: 218–233 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Basu MK, Csürös M, Koonin EV 2009. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol 1: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the hymenoptera. Syst Biol 61: 973–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann F, Eriksson T, Salim KA, Conti E 2007. Assessing calibration uncertainty in molecular dating: The assignment of fossils to alternative calibration points. Syst Biol 56: 591–608 [DOI] [PubMed] [Google Scholar]
- Samuelsson J, Butterfield NJ 2001. Neoproterozoic fossils from the Franklin Mountains, northwestern Canada: Stratigraphic and palaeobiological implications. Precambrian Research 107: 235–251 [Google Scholar]
- Sanders KL, Lee MSY 2007. Evaluating molecular clock calibrations using Bayesian analyses with soft and hard bounds. Biol Lett 3: 275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ 1997. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol 14: 1218–1231 [Google Scholar]
- Sanderson MJ 2002. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol 19: 101–109 [DOI] [PubMed] [Google Scholar]
- Shih PM, Matzke NJ 2013. Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc Natl Acad Sci 110: 12355–12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB, Peterson KJ 2002. Dating the time of origin of major clades: Molecular clocks and the fossil record. Annu Rev Earth Planet Sci 30: 65–88 [Google Scholar]
- Sogin ML 1991. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev 1: 457–463 [DOI] [PubMed] [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T 2002. Rooting the eukaryote tree by using a derived gene fusion. Science 297: 89–91 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T 2003a. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol 57: 408–419 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T 2003b. The root of the eukaryote tree pinpointed. Curr Biol 13: R665–R666 [DOI] [PubMed] [Google Scholar]
- Strother PK, Battison L, Brasier MD, Wellman CH 2011. Earth’s earliest non-marine eukaryotes. Nature 473: 505–509 [DOI] [PubMed] [Google Scholar]
- Summons RE, Walter MR 1990. Molecular fossils and microfossils of prokaryotes and protists from Proterozoic sediments. Am J Sci 290: 212–244. [Google Scholar]
- Summons RE, Bradley AS, Jahnke LL, Waldbauer JR 2006. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc Lond B Biol Sci 361: 951–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takishita K, Chikaraishi Y, Leger MM, Kim E, Yabuki A, Ohkouchi N, Roger AJ 2012. Lateral transfer of tetrahymanol-synthesizing genes has allowed multiple diverse eukaryote lineages to independently adapt to environments without oxygen. Biol Direct 7: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne JL, Kishino H, Painter IS 1998. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol 15: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Van Tuinen M, Hadly EA 2004. Error in estimation of rate and time inferred from the early amniote fossil record and avian molecular clocks. J Mol Evol 59: 267–276 [DOI] [PubMed] [Google Scholar]
- Waldbauer JR, Sherman LS, Sumner DY, Summons RE 2009. Late Archean molecular fossils from the Transvaal Supergroup record the antiquity of microbial diversity and aerobiosis. Precambrian Research 169: 28–47 [Google Scholar]
- Warnock RCM, Yang Z, Donoghue PCJ 2012. Exploring uncertainty in the calibration of the molecular clock. Biol Lett 8: 156–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Bromham L 2005. Molecular dating when rates vary. Trends Ecol Evol 20: 320–327 [DOI] [PubMed] [Google Scholar]
- Xie W, Lewis PO, Fan Y, Kuo L, Chen M-H 2011. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol 60: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 2006. Computational molecular evolution. Oxford University Press, Oxford [Google Scholar]
- Yang Z, Rannala B 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol 23: 212–226 [DOI] [PubMed] [Google Scholar]
- Yin L 1997. Acanthomorphic acritarchs from Meso-Neoproterozoic shales of the Ruyang group, Shanxi, China. Rev Palaeobot Palynol 98: 15–25 [Google Scholar]
- Yoder AD, Yang Z 2000. Estimation of primate speciation dates using local molecular clocks. Mol Biol Evol 17: 1081–1090 [DOI] [PubMed] [Google Scholar]
- Zhao S, Burki F, Bråte J, Keeling PJ, Klaveness D, Shalchian-Tabrizi K 2012. Collodictyon—An ancient lineage in the tree of eukaryotes. Mol Biol Evol 29: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhongying Z 1986. Clastic facies microfossils from the Chuanlinggou Formation (1800 Ma) near Jixian, North China. J Micropalaeontology 5: 9–16 [Google Scholar]
- Zuckerkandl E, Pauling L 1965. Evolutionary divergence and convergence in proteins. Evol Genes Proteins 97: 97–166. [Google Scholar]