Abstract
RecA/Rad51 catalyzed pairing of homologous DNA strands, initiated by polymerization of the recombinase on single-stranded DNA (ssDNA), is a universal feature of homologous recombination (HR). Generation of ssDNA from a double-strand break (DSB) requires nucleolytic degradation of the 5′-terminated strands to generate 3′-ssDNA tails, a process referred to as 5′–3′ end resection. The RecBCD helicase–nuclease complex is the main end-processing machine in Gram-negative bacteria. Mre11-Rad50 and Mre11-Rad50-Xrs2/Nbs1 can play a direct role in end resection in archaea and eukaryota, respectively, by removing end-blocking lesions and act indirectly by recruiting the helicases and nucleases responsible for extensive resection. In eukaryotic cells, the initiation of end resection has emerged as a critical regulatory step to differentiate between homology-dependent and end-joining repair of DSBs.
For double-strand breaks to be repaired by homologous recombination, the ends must be degraded to form long 3′ single-stranded DNA tails. Although this step is conserved across all domains of life, the mechanisms are distinct.
DSBs can arise accidentally during normal cell metabolism or after exposure of cells to DNA-damaging agents, and also serve as intermediates in a number of programmed recombination events in eukaryotic cells (Mehta and Haber 2014). The repair of DSBs is critical for maintenance of genome integrity, and misrepair, or failure to repair, is associated with chromosome rearrangements, chromosome loss, or even cell death. Both prokaryotic and eukaryotic cells have evolved elaborate mechanisms for the recognition and repair of DSBs. The two predominant repair mechanisms are HR and non-homologous end joining (NHEJ). HR relies on the presence of an intact homologous duplex to template repair of the broken strands, whereas NHEJ repairs DSBs by direct ligation of the DNA ends. For DSBs to be repaired by HR, the ends must first be degraded to generate long 3′-ssDNA tails, a process referred to as 5′–3′ end resection. The 3′-ssDNA tails are then bound by a member of the RecA/Rad51 family of proteins to initiate homologous pairing and serve as primers for DNA synthesis following strand invasion. Strand invasion intermediates are further processed by helicases and/or nucleases (Bizard and Hickson 2014; Wyatt and West 2014), and ultimately by gap-filling DNA synthesis and ligation, to generate mature recombinant products. The DNA end-resection step of HR is conserved in all domains of life, but the mechanisms used for generating ssDNA are distinct. Here, we review the basic machinery for DNA end resection in bacteria, archaea, and eukaryota and the regulation of end resection in eukaryotic cells.
END RESECTION IN BACTERIA
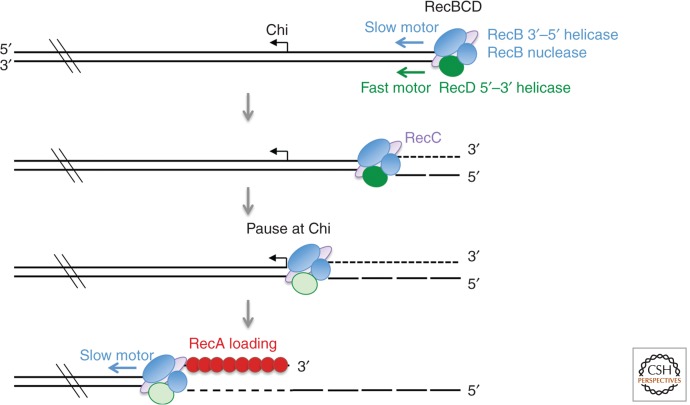
The heterotrimeric RecBCD nuclease is the major end-processing machine in Escherichia coli and is conserved across the majority of Gram-negative bacteria (Dillingham and Kowalczykowski 2008). RecBCD is a complex enzyme that couples ATP-dependent unwinding to DNA degradation (Smith 2001; Dillingham and Kowalczykowski 2008). The potent exonuclease activity of RecBCD can degrade thousands of bases per second. This destructive activity of RecBCD plays an important role in protecting bacteria from invading bacteriophages with linear genomes. Nuclease activity resides in the carboxy-terminal region of the RecB subunit and is regulated by RecC and by interaction with a specific sequence called Chi (5′-GCTGGTGG-3′) (Wang et al. 2000). Chi sites suppress the nuclease activity of RecBCD and stimulate recombination locally (Lam et al. 1974; Dixon and Kowalczykowski 1993). The 8-bp nonpalindromic Chi sites are overrepresented in the E. coli genome and are oriented toward the replication origin such that loading of RecBCD at a collapsed replication fork would lead to suppression of DNA degradation upon Chi recognition by RecBCD and activation of HR (Blattner et al. 1997).
Our current view of how RecBCD promotes recombination derives from a combination of bulk-phase biochemistry, single-DNA molecule imaging, electron microscopy (EM), and structural studies. RecBCD binds with high affinity to blunt or nearly blunt-ended linear duplex DNA (Taylor and Smith 1985). Unwinding is driven by the RecB and RecD subunits, which are helicases with opposite polarities and thus translocate the complex on both strands of duplex DNA in the same direction (Dillingham et al. 2003; Taylor and Smith 2003). The robust translocase activity of the RecBCD complex is able to displace tightly bound proteins from duplex DNA (Finkelstein et al. 2010). Under conditions in which the nuclease activity of the complex is minimized, the enzyme unwinds duplex DNA to produce one long 5′-ssDNA tail and an ssDNA loop associated with a short 3′-ssDNA tail owing to the two helicases operating at different speeds (Taylor and Smith 2003). RecD is the fast, or lead, motor on the 5′-terminated strand, whereas RecB translocates more slowly on the 3′-terminated strand until the complex encounters a Chi site (Fig. 1). Upon Chi recognition, the enzyme pauses, the RecD subunit is inactivated, and continued unwinding is driven by the RecB helicase, resulting in a slower translocation rate (Spies et al. 2003). Before encountering Chi, the 3′ end is more extensively cleaved by the RecB endonuclease than the 5-terminated strand, but after Chi recognition, degradation of the 3′ end is suppressed, and cleavage of the 5′-terminated strand is stimulated, generating a 3′-ssDNA tail (Anderson and Kowalczykowski 1997a). In addition, RecB facilitates loading of RecA onto the 3′-terminated strand after Chi recognition (Anderson and Kowalczykowski 1997b). How does Chi regulate the nuclease activities of the RecBCD complex? Structural studies indicate that a “pin” in RecC separates the strands of duplex DNA entering the complex driven by the RecB and RecD translocases (Singleton et al. 2004). As the separated strands pass through the RecBCD complex, the RecC subunit recognizes Chi, resulting in a conformational change that opens a molecular latch allowing the 3′-terminated strand to bypass the RecB nuclease domain and exit the complex (Handa et al. 2012; Yang et al. 2012).
Figure 1.
End processing by the RecBCD complex. RecBCD loads at ends and translocates on both strands using the RecD and RecB helicase subunits. RecB degrades both DNA strands exiting the complex, but with more incisions on the 3′ strand than the 5′-terminated strand. RecBCD pauses at a Chi site, and the RecD subunit is modified; continued translocation is driven by the RecB helicase. After Chi recognition, RecB directs loading of RecA onto the 3′ end and degrades only the 5′ strand.
The RecBC enzyme behaves similarly to Chi-modified RecBCD. RecBC unwinds double-stranded DNA (dsDNA) more slowly than RecBCD and constitutively loads RecA onto the 3′ end of the unwound strands. Consistent with the in vitro studies, recD mutants are recombination proficient and recombination is stimulated at ends instead of in the vicinity of Chi sites (Thaler et al. 1989; Churchill et al. 1999). By contrast, recB and recC mutants show high sensitivity to X rays and low frequency of recombination as measured by conjugation or transduction (Persky and Lovett 2008). However, these defects can be suppressed by inactivation of the 3′ exonucleases ExoI and SbcCD, suggesting that an alternative mechanism is able to generate 3′-ssDNA tailed intermediates in the absence of RecBCD, but the ends are unstable because of 3′ nuclease activity. Recombination in the recBC-suppressed strains is caused by the RecF pathway of recombination, which normally functions during ssDNA gap repair (Persky and Lovett 2008). Resection by the RecF pathway requires the 5′–3′ exonuclease, RecJ, and is stimulated by the RecQ 3′–5′ helicase and the ssDNA-binding protein, SSB (Han et al. 2006; Handa et al. 2009). RecJ requires an ssDNA tail of >6 nucleotides for binding and degrades to the ssDNA–dsDNA junction, releasing mononucleotide products (Han et al. 2006). Although originally characterized biochemically as an ssDNA-specific exonuclease, RecJ shows limited degradation of dsDNA (Lovett and Kolodner 1989; Handa et al. 2009). In a reconstituted reaction with other RecF pathway proteins, RecJ was shown to generate sufficient ssDNA to promote RecA-catalyzed strand invasion, although the extent of degradation and joint molecule formation was less than observed in the presence of RecQ (Handa et al. 2009).
The normal function of the RecF pathway is to promote recombination at ssDNA gaps formed during replication—for example, when a UV-induced pyrimidine dimer prevents primer extension by DNA polymerase III (see Syeda et al. 2014). recJ and recQ mutants show UV sensitivity and may be required to expand ssDNA gaps to facilitate RecA binding (Persky and Lovett 2008). RecJ can also cooperate with RecB and RecC in the absence of RecD (Lovett et al. 1988; Dermic 2006). The high frequency of conjugal recombination observed in recD mutants is reduced by mutation of recJ, but not by recQ. The residual recombination observed in the recD recJ mutant requires ExoVII, which degrades ssDNA from 5′ or 3′ ends, but the recD xseA mutant is recombination proficient, indicating that RecJ is the main activity with ExoVII serving as a backup function (Dermic 2006).
Ironically, there appears to be no role for SbcCD in end resection in bacteria, in contrast to archaea and eukaryota, where the SbcC and SbcD orthologs, Rad50 and Mre11, respectively, play an important role in coordinating DNA end processing (see below). Instead, the main function of SbcCD is to resolve hairpin structures formed by annealing between closely spaced inverted repeats, a role that is conserved in yeast (Lobachev et al. 2002; Rattray et al. 2005; Eykelenboom et al. 2008).
END RESECTION IN ARCHAEA
Homologs of the helicases and nucleases responsible for end resection in bacteria have not been identified in any of the archaeal genomes examined to date; RecQ-like helicases have been found but have no characterized role in end resection (Guy and Bolt 2005; Fujikane et al. 2006; Oyama et al. 2009). Mre11 and Rad50 are present in archaea, and biochemical studies suggest a role in end processing. Most of the structural analyses have been performed with the archaeal proteins; however, the functional analysis of catalytic and architectural motifs has mainly been performed in yeast (see below). Mre11 shows Mn2+-dependent 3′–5′ exonuclease activity in vitro and an endonuclease activity that acts at the dsDNA–ssDNA transition of secondary structures within ssDNA (Hopfner et al. 2000a; Trujillo and Sung 2001). The genes encoding Mre11 and Rad50 are within the same operon as the HerA helicase and the NurA nuclease in thermophilic archaea, suggesting they might functionally cooperate to promote end resection (Hopkins and Paull 2008). HerA is a member of the FtsK superfamily of hexameric translocases and helicases, and NurA forms a dimer with RNaseH-like domains (Blackwood et al. 2012). HerA and NurA physically interact, and the catalytic activities are mutually interdependent (Hopkins and Paull 2008; Blackwood et al. 2012). In assays with limiting amounts of HerA and NurA, addition of Mre11 and Rad50 strongly stimulates ATP-dependent DNA degradation, requiring the helicase and nuclease activities of HerA and NurA, respectively (Hopkins and Paull 2008). Interestingly, the Mre11–Rad50 (MR) complex alone is able to remove 15–55 nt from the 5′ ends of long linear substrates in a reaction dependent on the Mre11 nuclease and Mg2+. The initial processing by MR stimulates degradation by NurA but is not essential for end resection in the reconstituted reaction (Hopkins and Paull 2008).
END RESECTION IN EUKARYOTES
Much of our knowledge of the factors involved in eukaryotic end resection has come from genetic analysis in Saccharomyces cerevisiae, where DNA end processing can be followed physically at sites of endonuclease-generated DSBs in vegetatively dividing (mitotic) cells or Spo11-induced DSBs in meiosis. These studies identified the Mre11-Rad50-Xrs2 (MRX) complex, Sae2, Exo1, Replication Protein A (RPA), Sgs1, and Dna2 as key factors for 5′–3′ end resection, and their activities are conserved in other eukaryotes investigated (human NBS1, CtIP, and BLM are the functional orthologs of Xrs2, Sae2, and Sgs1, respectively) (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008; Nimonkar et al. 2011; Peterson et al. 2011; Karanja et al. 2012; Chen et al. 2013). A widely accepted view is for MRX/N and Sae2/CtIP to initiate end resection by endonucleolytic cleavage of the 5′ ends internal to break ends releasing oligonucleotides. The short 3′-ssDNA tails formed are then subject to extensive resection executed via two parallel pathways. One is dependent on the 5′–3′ exonuclease, Exo1, whereas the other depends on the concerted action of the Sgs1/BLM-Top3-Rmi1 complex with the Dna2 endonuclease, hereafter referred to as STR-Dna2. The extensively resected ssDNA tracts formed vary in length from a few hundred nucleotides to tens of kilobases, depending on the availability and location of the homologous template, and correlate with the kinetics of repair (Chung et al. 2010).
Biochemical and Structural Characterization of Resection Nucleases
MRX/N
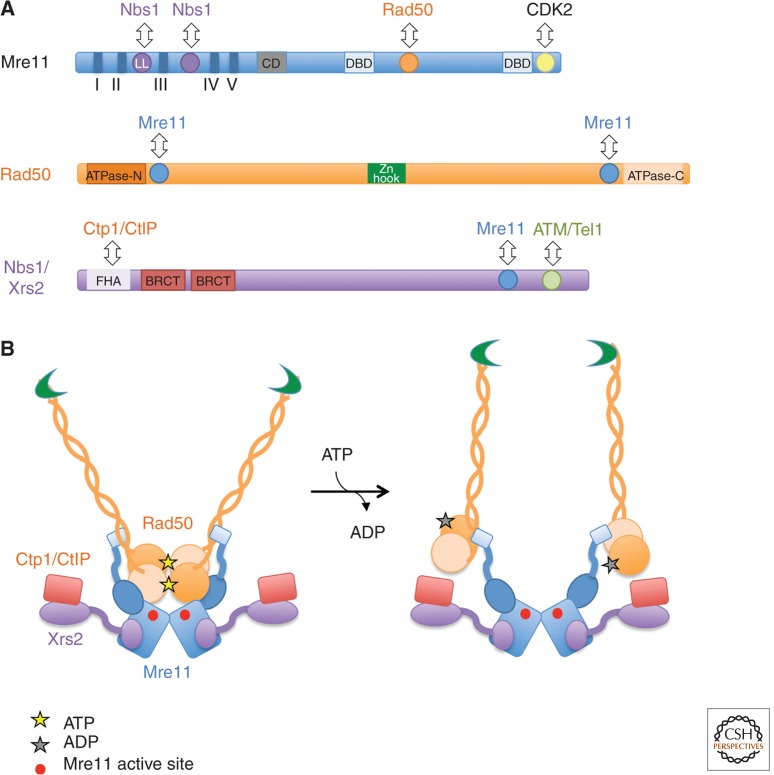
The MRE11, RAD50, and XRS2 genes were originally identified by their essential roles for ionizing radiation (IR) resistance and meiotic recombination, and null mutations confer similar phenotypes (Mimitou and Symington 2009). Mre11, Rad50, and Xrs2/Nbs1 interact and copurify as a complex (Trujillo et al. 1998; Usui et al. 1998; Paull and Gellert 1999). Mre11 has five conserved phosphoesterase motifs in the amino-terminal half of the protein that are required for Mn2+-dependent 3′–5′ dsDNA exonuclease and ssDNA endonuclease activities in vitro (Fig. 2A) (Bressan et al. 1998; Furuse et al. 1998; Usui et al. 1998; Moreau et al. 1999; Trujillo and Sung 2001). Substitution of conserved Asp or His residues within the nuclease motifs (e.g., D16, D56, H125, or H213 of ScMre11) with Asn or Ala abolishes exo- and endonuclease activities in vitro (Furuse et al. 1998; Usui et al. 1998; Moreau et al. 1999); hereafter, nuclease-deficient mre11 alleles are referred to as mre11-nd. Mutation of His59 reduces the exonuclease activity to a greater extent than the endonuclease activity and has been used to evaluate the role of the exonuclease activity in vivo (Williams et al. 2008; Garcia et al. 2011). Two α helices of Mre11 located carboxy terminal to the nuclease core domain are responsible for interaction with the Rad50 coiled-coil base (Lammens et al. 2011; Lim et al. 2011; Williams et al. 2011). Schizosaccharomyces pombe Mre11 interacts with Nbs1 via a eukaryotic-specific insertion between phosphoesterases motifs II and III, referred to as the latching loop, and through additional residues in the amino-terminal region (Schiller et al. 2012). Mutations within the latching loop that are found in individuals with ataxia-telangiectasia-like disorder (ATLD) or Nijmegen breakage syndrome (NBS)-like disorder disrupt the interaction with Nbs1. In the absence of Xrs2/Nbs1, or point mutations that abolish the interaction with Nbs1, Mre11 fails to localize to the nucleus. Interestingly, addition of an nuclear localization signal (NLS) to Mre11 can partially suppress the DNA damage sensitivity of the xrs2Δ mutant, indicating that one of the main functions for Xrs2 is Mre11 localization to the nucleus (Tsukamoto et al. 2005). The carboxy-terminal 54 residues of murine Mre11 interact with cyclin-dependent kinase 2 (CDK2) to facilitate CtIP phosphorylation and stability (Buis et al. 2012).
Figure 2.
Structural organization of the MRX/N complex. (A) Schematic showing domains of the Mre11, Rad50, and Nbs1/Xrs2 proteins. Interaction domains are shown as color-coded circles, and other functional domains are indicated by rectangles. LL, latching loop; CD, capping domain; DBD, DNA-binding domain; FHA, forkhead associated; BRCT, BRCA1 carboxy-terminal domain. (B) Panel created from data in Lim et al. (2011) to show how ATP hydrolysis by Rad50 causes a conformational change exposing the Mre11 active site. Note that there are no structures of the entire MRX/N complex, or the complex with Sae2/Ctp1/CtIP, and the cartoon depicts the known interactions based on partial complexes.
Rad50 has a similar domain organization to the structural maintenance of chromosomes family of proteins, which are characterized by Walker A and B ATP-binding cassettes located at the amino- and carboxy-terminal regions of the primary sequence that come together by collapse of the intervening sequence to form a long antiparallel coiled-coil (Fig. 2) (Hopfner et al. 2000b). Two Rad50 ATP-binding cassettes interact with an Mre11 dimer to form a “head” domain with DNA-binding and ATP-regulated nuclease activity (Fig. 2B). The Rad50 coiled-coil domains emanate from the head and can interact with other MR complexes by Zn2+-mediated dimerization of the hook domains at the apexes of the coiled-coils intramolecularly, or intermolecularly to tether linear DNA molecules (Anderson et al. 2001; Chen et al. 2001; de Jager et al. 2001; Hopfner et al. 2002; Wiltzius et al. 2005). Mre11 stabilizes dimerization of Rad50 and stimulates Rad50 ATP hydrolysis. The ATP-bound form of Rad50 negatively regulates the Mre11 nuclease activity by masking the active site of Mre11 (Lim et al. 2011). ATP hydrolysis triggers substantial conformational changes of both Rad50 and Mre11 within the MR complex, resulting in exposure of the Mre11 nuclease site and activation of DNA degradation (Lim et al. 2011; Mockel et al. 2012). Mutation of conserved residues in the Walker A-type ATPase domain confer a rad50 null phenotype, whereas a class of mutations located close to the ATPase domain called rad50S behave similarly to mre11-nd alleles (Alani et al. 1990). Exactly how the rad50S mutations affect the in vitro functions of the Mre11 complex has not been determined.
Although Mre11 and Rad50 are conserved in bacteria, bacteriophage T4, and archaea, Xrs2 and Nbs1 are unique to eukaryotes and are more diverged. The amino-terminal region of Xrs2/Nbs1 has phosphoprotein-binding motifs that are separated from the Mre11 and Tel1/ATM interaction regions in the carboxyl terminus by a flexible linker (Lloyd et al. 2009; Williams et al. 2009). Xrs2 has only the conserved FHA domain, whereas Nbs1 has two BRCT domains adjacent to the FHA domain. Diphosphorylated pSDpTD motifs are Nbs1 FHA domain-binding targets and direct the interaction between Ctp1 (S. pombe Sae2 ortholog) and Nbs1; this interaction is important for resistance to IR and the topoisomerase I poison, camptothecin (CPT) in fission yeast (Lloyd et al. 2009; Williams et al. 2009). A conserved region within the carboxy-terminal region of Xrs2/Nbs1 is responsible for Mre11 interaction (Tsukamoto et al. 2005).
Sae2/Ctp1/CtIP
Sae2 (also known as Com1) was originally identified by its requirement to process meiotic DSBs, and the phenotype conferred by sae2Δ is very similar to mre11-nd and rad50S mutations (McKee and Kleckner 1997; Prinz et al. 1997; Mimitou and Symington 2009). Mammalian CtIP is thought to be the ortholog of Sae2, but sequence homology is limited to a small region of the carboxyl terminus that includes sites for cyclin-dependent kinase (CDK) and Mec1/ATR and Tel1/ATM phosphorylation, and an oligomerization motif (LKEX4EV/L) close to the amino terminus (Sartori et al. 2007; Kim et al. 2008; Wang et al. 2012). Although Sae2 does not form a stable complex with MRX in solution, together they form a higher-order complex in association with DNA (Lengsfeld et al. 2007). The similarity of the sae2Δ and mre11-nd phenotypes initially led to speculation that Sae2 activates the Mre11 nuclease, and recombinant Sae2 does indeed stimulate the 3′–5′ Mre11 exonuclease activity. In addition, Sae2 alone functions as an endonuclease and cuts a variety of branched DNA structures with a preference for cleavage within an ssDNA region near a branch or hairpin-capped end, and the activity toward hairpin structures is stimulated by MRX (Lengsfeld et al. 2007).
Exo1
Exo1 is a member of the XPG family of nucleases, which includes Rad2/XPG, Rad27/FEN-1, and Yen1/GEN1 in eukaryotes (Szankasi and Smith 1995). These proteins have conserved nuclease motifs in the amino-terminal region but have distinct substrate preferences. Exo1 shows 5′–3′ dsDNA exonuclease and 5′ flap endonuclease activities in vitro and is able to degrade from a dsDNA end or an internal nick releasing mononucleotide products (Szankasi and Smith 1992; Tran et al. 2004). Exo1 acts preferentially on dsDNA substrates with recessed 5′ ends, analogous to the ends produced by MRX and Sae2 in vivo (Cannavo et al. 2013). RPA stimulates Exo1 nucleolytic processing by binding to the ssDNA produced by end resection and preventing formation of nonproductive Exo1-ssDNA complexes (Cannavo et al. 2013). MRX and Sae2 also stimulate Exo1-catalyzed degradation, which could occur by MRX-mediated unwinding of duplex ends to create the preferred substrate for Exo1 binding and flap endonuclease activity, or by clipping the 5′ strand to generate a recessed 5′ end for the exonuclease activity (Nicolette et al. 2010; Cannavo et al. 2013). Although no direct interaction between MRX or Sae2 and Exo1 has been reported, human CtIP and EXO1 do interact, and CtIP is required for recruitment of EXO1 to damaged sites in vivo (Eid et al. 2010). In yeast, MRX is required for Exo1 localization to DSBs, but Sae2 and Mre11 nuclease activity are not, suggesting the strand-separation function of MRX might be more important for recruitment than end clipping (Paull and Gellert 1999; Shim et al. 2010; Cannon et al. 2013). BLM is also able to stimulate EXO1 degradation by increasing the affinity of EXO1 for DNA ends, but this function is independent of the ATPase activity and is not conserved in yeast (Nimonkar et al. 2011; Cannavo et al. 2013).
Sgs1/BLM-Dna2-RPA
Sgs1, BLM, and WRN are members of the RecQ family of helicases that unwind DNA by ATP-dependent 3′–5′ translocation on the 3′-terminated strand (Bernstein et al. 2010). Dna2, which is related to bacterial RecB proteins, shows helicase and ssDNA endonuclease activities in vitro (Bae et al. 1998; Budd et al. 2000). The nuclease activity of Dna2 is essential for end resection, but the helicase activity is dispensable, and Dna2 is dependent on Sgs1/BLM to generate the ssDNA substrate for degradation (Zhu et al. 2008; Cejka et al. 2010; Niu et al. 2010; Nimonkar et al. 2011). Sgs1-Dna2-catalyzed end resection is completely dependent on RPA (Cejka et al. 2010; Niu et al. 2010; Nimonkar et al. 2011; Chen et al. 2013). RPA directly interacts with Sgs1 and stimulates Sgs1 unwinding. The function of yRPA is only partially substituted by hRPA or E. coli SSB, suggesting a species-specific interaction is important and the role of RPA is not restricted to stabilizing the unwound strands. This is in contrast to the role of RPA in stimulation of Exo1 resection, which can be substituted by E. coli SSB (Cannavo et al. 2013). The Dna2 endonuclease can degrade either 3′- or 5′-terminated ssDNA; however, in the presence of RPA, the 3′ nuclease activity is attenuated and the 5′ endonuclease activity is stimulated, explaining the strand bias of end resection (Cejka et al. 2010; Niu et al. 2010). Dna2 fails to localize to DSBs in the absence of RPA, which could account for the strict RPA requirement for Dna2-catalyzed resection in vivo (Chen et al. 2013).
Top3 and Rmi1, which function with Sgs1 to dissolve double Holliday intermediates (Bizard and Hickson 2014), stimulate end resection by increasing the affinity of Sgs1 for DNA ends (Cejka et al. 2010). Unlike dissolution, the role of Top3 in end resection is independent of its catalytic activity (Niu et al. 2010). Although Top3 and Rmi1 are not essential for Sgs1-Dna2-RPA end resection in vitro, they are necessary in vivo (Zhu et al. 2008). Similarly, MRX stimulates end resection by Sgs1-Dna1-RPA by increasing Sgs1 helicase activity. The MRX stimulation can be bypassed by providing a dsDNA substrate with 5′ overhangs, suggesting MRX recruits Sgs1 to DNA ends or creates an unwound end that is the preferred substrate for Sgs1 binding (Niu et al. 2010). Sgs1 and Mre11 cofractionate after DNA damage, and MRX is required for Sgs1 and Dna2 recruitment to DSBs in vivo (Chiolo et al. 2005; Niu et al. 2010; Shim et al. 2010).
Resection of Meiotic DSBs
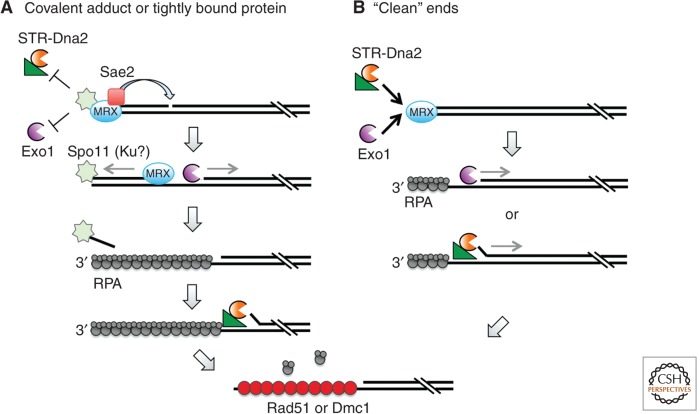
The Spo11 transesterase generates meiotic DSBs by forming a covalent linkage between a conserved tyrosine residue and the 5′ end of the cleaved strand (Keeney et al. 1997; Lam and Keeney 2014). A dimer of Spo11 acts to cut both DNA strands in concert. Spo11 is then removed endonucleolytically, releasing it with a short (12- to 40-nt) oligonucleotide attached (Neale et al. 2005). The sae2Δ/ctp1Δ, rad50S, and mre11-nd mutants of budding and fission yeast generate meiotic DSBs with Spo11 stably bound to the 5′ ends, suggesting the endonuclease activity of the MRX/N complex and/or Sae2/Ctp1 incises DNA internal to the DSB ends to liberate Spo11-oligonucleotides (Neale et al. 2005; Hartsuiker et al. 2009; Milman et al. 2009). Mutation of the Mre11 exonuclease activity (mre11-H59S) results in release of longer oligonucleotides attached to Spo11 (Garcia et al. 2011). In wild-type cells, the average length of 3′-ssDNA tails formed by end resection is ∼800 nt but is reduced to ∼270 nt in the exo1Δ mutant (Zakharyevich et al. 2010; Hodgson et al. 2011; Keelagher et al. 2011). These findings are consistent with a model whereby MRX and Sae2 incise the 5′ strand 250–300 nt from the Spo11-bound end and the Mre11 3′–5′ exonuclease degrades from the nick toward Spo11, whereas Exo1 degrades in the opposite direction, removing an additional ∼500 nt (Fig. 3) (Zakharyevich et al. 2010; Garcia et al. 2011). DSB formation and processing are highly coordinated events during meiosis, and the intermediates with Spo11 attached to ends, or products of MRX-Sae2 processing are not observed in wild-type cells (Zakharyevich et al. 2010). STR-Dna2 does not contribute to resection in meiosis, except in the absence of the Dmc1 recombinase (Manfrini et al. 2010; Zakharyevich et al. 2010). Loss of Exo1 nuclease activity does not significantly impair meiotic recombination, indicating that the short ssDNA tails generated by MRX and Sae2 are sufficient for homologous pairing (Zakharyevich et al. 2010).
Figure 3.
End processing in eukaryotes. (A) Resection of meiotic DSBs involves MRX- and Sae2-dependent incision of the 5′ strand ∼270 nt internal to the Spo11-bound end. MRX and Exo1 degrade bidirectionally from the nick to generate an ssDNA tail of ∼800 nt, and Spo11 is released from ends with a short (15- to 40-nt) oligonucleotide attached. The resulting ssDNA is bound by RPA, which is rapidly replaced by Rad51 and Dmc1 to promote strand invasion. In the absence of Dmc1, STR and Dna2 carry out more extensive end processing. This model could also apply to resection of DSBs blocked by a covalent adduct, or tightly bound protein such as Ku, in vegetatively dividing cells. (B) Resection of ends with no covalent modification (e.g., ends produced by the HO and I-SceI endonucleases) can initiate directly by STR-Dna2 or Exo1 via MRX recruitment. The two extensive resection mechanisms appear to function independently and redundantly at endonuclease-induced DSBs.
Resection of Endonuclease-Generated DSBs
MRX/N rapidly localizes to DSBs and precedes recruitment of RPA and Rad51 (Nelms et al. 1998; Lisby et al. 2004). MRX localizes very close to a DSB and does not spread from the break site, consistent with a role in resection initiation but not in extensive resection (Shroff et al. 2004). In the absence of MRX, DSBs generated by the HO or I-SceI endonucleases remain stable for several hours (Ivanov et al. 1994; Tsubouchi and Ogawa 1998). Resection can be initiated by Exo1, but is inhibited by Ku binding to DNA ends; the absence of Ku, or Exo1 overexpression, results in suppression of the mre11Δ resection initiation defect (Bressan et al. 1999; Lee et al. 2002; Tomita et al. 2003; Williams et al. 2008; Mimitou and Symington 2010; Shim et al. 2010). STR-Dna2 is unable to initiate end resection without MRX, even in the absence of Ku (Mimitou and Symington 2010). Loss of the Mre11 nuclease activity or Sae2 results in a much shorter delay in resection initiation than observed in the absence of the MRX complex, attributed to the role of MRX in recruiting Exo1, Sgs1, and Dna2 to DSBs (Fig. 3) (Llorente and Symington 2004; Mimitou and Symington 2008; Shim et al. 2010). However, the Mre11 nuclease and Sae2 are essential for processing DSBs that have covalent adducts at the 5′ ends, such as Spo11-induced DSBs (see above) or hairpin-capped ends; these phenotypes are shared by rad50S mutants (Mimitou and Symington 2009). It is possible that the mechanism of end resection envisioned during meiosis occurs during DSB processing in mitotic cells, with Exo1 and Mre11 degrading bidirectionally from a nick created internal to the ends. Having Exo1 initiate resection from a nick would overcome the problem of Ku inhibiting Exo1 at DNA ends.
In fission yeast and mammalian cells, the initial processing step by Mre11 nuclease and Sae2/Ctp1/CtIP appears to be more important for homology-dependent repair than in budding yeast (Limbo et al. 2007; Sartori et al. 2007; Buis et al. 2008; Langerak et al. 2011; Truong et al. 2013). The fission yeast ctp1Δ and mre11-H134S mutants show similar DNA damage sensitivity to the mre11Δ mutant, and recruitment of RPA adjacent to an HO-induced DSB is greatly reduced (Limbo et al. 2007; Williams et al. 2008). Interestingly, null mutation of CtIP or the Mre11H129N/H129N mutation (nuclease defective) causes mouse embryonic lethality, highlighting the importance of MRN-CtIP regulated processing in mammalian cells (Chen et al. 2005; Buis et al. 2008). Knockdown of CtIP in human cells, or use of a conditional Mre11H129N/H129N cell line, results in a dramatic reduction in resection as determined by formation of IR-induced RPA or Rad51 foci (Sartori et al. 2007; Buis et al. 2008).
In the absence of Exo1 resection initiation occurs with normal kinetics, but resection 1–5 kb from the DSB is reduced (Llorente and Symington 2004; Mimitou and Symington 2008). DNA ends are stable for ∼6 h in the mre11Δ exo1Δ double mutant, but some end processing eventually occurs that must be caused by low residual STR-Dna2 activity (Tsubouchi and Ogawa 2000; Moreau et al. 2001). STR-Dna2 is mainly responsible for resection >5 kb from DSB ends and acts redundantly with Exo1 in early resection (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008). The mre11-nd sgs1Δ double mutant shows synergistic sensitivity to IR and CPT, and delayed resection initiation as compared to the single mutants, suggesting STR-Dna2 is able to initiate end resection at clean ends in the absence of the Mre11 nuclease (Budd and Campbell 2009; Mimitou and Symington 2010; Shim et al. 2010). In the absence of Sgs1-Dna2 and Exo1, resection initiates by an endonucleolytic mechanism removing nucleotides from the 5′ end in increments of ∼100 nt; however, the length of ssDNA tails rarely exceeds 700 nt (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008). Depletion of RPA from cells prevents extensive resection, similar to the phenotype of the exo1Δ sgs1Δ double mutant; furthermore, the 3′ tails formed by MRX-Sae2 are unstable because of formation of secondary structures and degradation (Chen et al. 2013).
Cell Cycle, Ku, and DNA Damage Checkpoint Regulation of End Resection
HR is generally restricted to the S and G2 phases of the cell cycle, when a sister chromatid is available as a repair template. This restriction is mainly caused by reduced end resection in G1 compared with cycling or G2-arrested cells (Aylon et al. 2004; Ira et al. 2004; Jazayeri et al. 2006; Barlow et al. 2008; Zierhut and Diffley 2008). Indeed, restoration of end resection in G1 permits HR repair if a donor sequence is available (Zhang et al. 2009; Trovesi et al. 2011). Reduced resection in G1 results from Ku binding to DNA ends, NHEJ, and low CDK (Cdc28) activity. Elimination of Ku or Dnl4 restores resection initiation to G1-phase cells, but extensive resection is still partially defective (Clerici et al. 2008; Zierhut and Diffley 2008). Activation of Cdc28 in G1 restores resection, whereas inhibition of Cdc28 in G2 cells blocks resection (Aylon et al. 2004; Ira et al. 2004; Clerici et al. 2008).
Sae2 and Dna2 show S-phase-specific phosphorylation and are targets for Cdc28-mediated regulation of end resection (Baroni et al. 2004; Huertas et al. 2008; Chen et al. 2011). Mutation of Ser267 of Sae2 to a nonphosphorylatable residue, S267A, phenocopies sae2Δ, including hypersensitivity to camptothecin, defective sporulation, reduced hairpin-induced recombination, impaired DSB processing, and delayed Rad52 recruitment (Huertas et al. 2008). Similarly, substitution of the equivalent CDK site in human CtIP (Thr847) to alanine impairs resection in human cells (Huertas and Jackson 2009). Cell-cycle regulation of S. pombe Ctp1 is mainly transcriptional (Limbo et al. 2007). Mutation of CDK consensus site residues in the amino-terminal region of Dna2 abolishes Cdc28-dependent phosphorylation (Chen et al. 2011). Substitution of Thr4, Ser17, and Ser327 with alanine (dna2-3A mutant) reduces extensive resection but not to the same extent as dna2Δ or by inhibition of Cdc28. Dna2 has a bipartite nuclear localization sequence overlapping Ser17, and the dna2-S17A mutation reduces nuclear entry during S phase and Dna2 localization to an HO-induced DSB (Kosugi et al. 2009; Chen et al. 2011); however, extensive resection is unaffected by the dna2-S17A mutant, suggesting the pool of nuclear Dna2 is sufficient for end processing. Expression of Dna2 with phosphomimetic substitutions of Thr4, Ser17, and Ser327 does not override Cdc28 regulation of extensive resection, indicating that there must be other Cdc28 targets.
The DNA damage sensitivity of the sae2Δ/ctp1Δ mutant is suppressed by elimination of Ku, and the suppression requires both Exo1 and Sgs1, suggesting CDK activation of Sae2 removes Ku from DNA ends to allow access to Exo1 or STR-Dna2 to DSBs (Fig. 3) (Limbo et al. 2007; Mimitou and Symington 2010; Langerak et al. 2011). It is possible that Ku is removed from ends by MRX-Sae2 clipping, similar to Spo11, or that a dynamic equilibrium exists between MRX-Sae2 and Ku binding, and that once MRX-Sae2 initiate resection, the preferred substrate for Ku binding is no longer available. Sae2 is still required for meiosis and hairpin cleavage in the absence of Ku, indicating an essential role for Sae2 nuclease, or activation of the Mre11 endonuclease by Sae2, to process these ends (Rattray et al. 2005; Mimitou and Symington 2010). Interestingly, the meiotic defect of the Caenorhabditis elegans com-1 (Sae2/CtIP ortholog) mutant is suppressed in the absence of Ku, suggesting that Com1/Sae2 may not play a direct role in the endonucleolytic removal of Spo11 in all organisms (Lemmens et al. 2013).
Mec1/ATR and/or Tel1/ATM phosphorylate many of the proteins involved in end resection after DNA damage. Cell-cycle and DNA damage–dependent phosphorylation of Sae2 require Mec1 and Tel1, and mutations altering the main phosphorylation sites cause DNA damage sensitivity (Baroni et al. 2004). A highly conserved ATM/ATR site in the carboxy-terminal region of CtIP (Thr859 of hCtIP or Thr818 of XCtIP) is phosphorylated in response to DSBs and is required for CtIP association with chromatin, DNA end resection, and HR repair (Peterson et al. 2013; Wang et al. 2013).
Regulation of End Resection by Chromatin Binding and Remodeling Proteins
Rad9
Rad9 is considered as an adaptor protein in the DNA damage checkpoint linking the upstream Mec1 kinase to the effector kinases, Rad53 and Chk1 (Harrison and Haber 2006). The Tudor domain of Rad9 interacts with methylated K79 of histone H3, and the BRCT domain binds to H2A sites phosphorylated by Mec1/ATR or Tel1/ATM following DNA damage (γH2A). In addition to its role in DNA damage checkpoint signaling, several studies have shown that Rad9 prevents the accumulation of ssDNA at uncapped telomeres and slows resection of endonuclease-induced DSBs (Zubko et al. 2004; Lazzaro et al. 2008; Doksani and de Lange 2014). End resection is increased in the absence of Dot1 (encodes the methyltransferase for histone H3 K79) and in the nonphosphorylatable h2a-S129A mutant, indicating that the end-protection function of Rad9 requires chromatin association (Lazzaro et al. 2008; Chen et al. 2012; Eapen et al. 2012). Resection of uncapped subtelomeric sequences is mostly Exo1-dependent in the presence or absence of Rad9, with only a small contribution from the Sgs1 pathway (Ngo and Lydall 2010). However, the increased resection of sequences further from uncapped ends that is seen in the rad9Δ mutant is mainly caused by Sgs1 activity (Ngo and Lydall 2010).
53BP1 and RIF1
53BP1 (p53 binding protein 1) shares a similar domain organization to Rad9 and plays a comparable end-protection role at telomeres and DSBs. Like Rad9, 53BP1 binds chromatin constitutively through the Tudor domain and forms γ-H2AX-dependent foci in response to IR. Simultaneous loss of the mammalian telomere-binding complex, Shelterin, and 53BP1 causes extensive resection of telomeres, which is partially dependent on CtIP, BLM, and EXO1 (Sfeir and de Lange 2012). The association of 53BP1 with DSBs in G1 prevents ATM-dependent resection of AID- (activation-induced cytidine deaminase) or IR-induced DSBs. BRCA1 competes with 53BP1, binding to ends when cells are in the S and G2 phases of the cell cycle to promote end resection and homology-directed repair (Escribano-Diaz et al. 2013). Interestingly, the HR defect and chromosomal instability associated with loss of BRCA1 are abrogated in the absence of 53BP1 by restoration of CtIP and ATM-dependent end resection (Bouwman et al. 2010; Bunting et al. 2010). The role of 53BP1 in preventing end resection requires RIF1 (Chapman et al. 2013; Di Virgilio et al. 2013; Feng et al. 2013; Zimmermann et al. 2013). Rif1 was originally identified as a Rap1-interacting protein and modulates telomere length in budding yeast (Hardy et al. 1992). RIF1 has no obvious telomere function in mammals, but was shown to interact with the amino-terminal domain of 53BP1 (Silverman et al. 2004). Accumulation of RIF1 at DSBs is ATM- and 53BP1-dependent and requires ATM/ATR target sites (S/TQ) within the 53BP1 amino terminus.
Chromatin-Remodeling Complexes
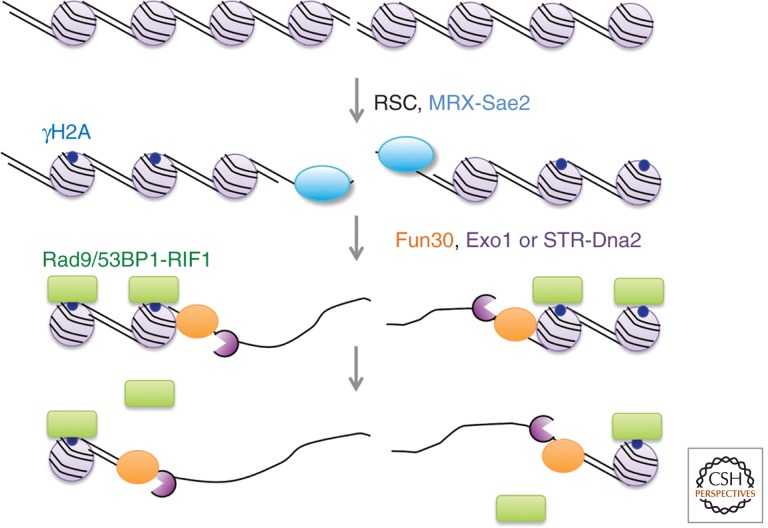
How the resection machinery navigates nucleosomal DNA and nonhistone protein–DNA complexes is not well understood. Nucleosomes assembled on a linear dsDNA template impede resection by Exo1 in vitro, but the inhibitory effect is less for Sgs1-Dna2, particularly if a nucleosome-free gap is adjacent to the DNA ends (Adkins et al. 2013). ATP-dependent chromatin remodeling complexes translocate on dsDNA disrupting histone-DNA contacts by nucleosome sliding, eviction, or histone exchange. In budding yeast, the RSC, SWI/SNF, INO80, SWR-C, and Fun30 remodeling enzymes are all recruited to HO-induced DSBs (Bennett et al. 2013). The RSC complex is required for early resection and promotes recruitment of the MRX complex to DSBs, whereas Fun30 is important for extensive resection (Fig. 4) (Shim et al. 2007; Chen et al. 2012; Costelloe et al. 2012; Eapen et al. 2012). A role for the INO80 complex in early resection is only apparent in the absence of Fun30 and the RSC complex (Chen et al. 2012). Although Fun30 facilitates both extensive resection mechanisms, the phenotype of fun30Δ is similar to exo1Δ and overexpression of Exo1 suppresses the DNA damage sensitivity of the fun30Δ mutant (Chen et al. 2012; Costelloe et al. 2012). Additionally, SMARCAD1, the human ortholog of Fun30, is required for RPA localization to laser-induced DNA damage, similar to the role of EXO1 (Costelloe et al. 2012; Tomimatsu et al. 2012). Although the recruitment of RSC, INO80, and Fun30 would be expected to precede resection, localization of these factors to DSBs is reduced in exo1Δ sgs1Δ cells; furthermore, recruitment of Sgs1, Dna2, and Exo1 is reduced in the fun30Δ mutant, indicating a complex interdependency. Interestingly, the extensive resection defect of the fun30Δ mutant is completely suppressed by elimination of Rad9, suggesting Fun30 helps to overcome the resection barrier formed by Rad9 (Chen et al. 2012; Eapen et al. 2012).
Figure 4.
Chromatin and chromatin-bound proteins are barriers to end resection. The RSC complex is required for early end resection in collaboration with MRX, whereas more extensive end resection requires Fun30 acting with Exo1 and STR-Dna2. Chromatin-bound Rad9 or 53BP1-RIF1 complexes impose an additional barrier that requires Fun30.
CONCLUDING REMARKS
Considerable progress has been made in identifying components of the end-resection machinery in eukaryotes, and the extensive resection pathways have been reconstituted in vitro. However, a detailed mechanistic understanding of resection initiation is lacking, in particular, how the Mre11 nuclease and Sae2/CtIP collaborate to initiate resection is poorly understood. NHEJ is a prominent repair pathway during the G1 phase of the cell cycle in eukaryotes, and the initiation of end resection has emerged as a key regulatory step to differentiate between repair mechanisms. How the DNA damage checkpoint and CDKs coordinate to regulate resection, in particular in the chromatin context and during DNA replication, is an important issue for future studies.
ACKNOWLEDGMENTS
Research in the Symington laboratory is supported by grants from the National Institutes of Health (R01 GM041784 and R01 GM094386).
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adkins NL, Niu H, Sung P, Peterson CL 2013. Nucleosome dynamics regulates DNA processing. Nat Struct Mol Biol 20: 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436 [DOI] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC 1997a. The recombination hot spot Chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev 11: 571–581 [DOI] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC 1997b. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a Chi-regulated manner. Cell 90: 77–86 [DOI] [PubMed] [Google Scholar]
- Anderson DE, Trujillo KM, Sung P, Erickson HP 2001. Structure of the Rad50 x Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J Biol Chem 276: 37027–37033 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Choi E, Lee KH, Park JS, Lee SH, Seo YS 1998. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem 273: 26880–26890 [DOI] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP 2004. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol 24: 4151–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett G, Papamichos-Chronakis M, Peterson CL 2013. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat Commun 4: 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R 2010. The RecQ DNA helicases in DNA repair. Annu Rev Genet 44: 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood JK, Rzechorzek NJ, Abrams AS, Maman JD, Pellegrini L, Robinson NP 2012. Structural and functional insights into DNA-end processing by the archaeal HerA helicase-NurA nuclease complex. Nucl Acids Res 40: 3183–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Baxter BK, Petrini JH 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol 19: 7681–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL 2009. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE 4: e4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Choe W, Campbell JL 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem 275: 16518–16529 [DOI] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Stoneham T, Spehalski E, Ferguson DO 2012. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat Struct Mol Biol 19: 246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P, Kowalczykowski SC 2013. Relationship of DNA degradation by Saccharomyces cerevisiae Exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci 110: E1661–E1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Kuhnlein J, Yang SH, Cheng A, Schindler D, Stark JM, Russell R, Paull TT 2013. Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET. Proc Natl Acad Sci 110: 18868–18873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49: 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH 2005. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol 25: 3535–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol 18: 1015–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G 2012. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489: 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lisby M, Symington LS 2013. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell 50: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G 2005. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol 25: 5738–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G 2010. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet 6: e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill JJ, Anderson DG, Kowalczykowski SC 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev 13: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP 2008. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. 2012. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489: 581–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Dermic D 2006. Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli. Genetics 172: 2057–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72: 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Spies M, Kowalczykowski SC 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423: 893–897 [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Kowalczykowski SC 1993. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell 73: 87–96 [DOI] [PubMed] [Google Scholar]
- *.Doksani Y, de Lange T 2014. The role of DSB repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE 2012. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol 32: 4727–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S 2010. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep 11: 962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 49: 872–883 [DOI] [PubMed] [Google Scholar]
- Eykelenboom JK, Blackwood JK, Okely E, Leach DR 2008. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell 29: 644–651 [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem 288: 11135–11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein IJ, Visnapuu ML, Greene EC 2010. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature 468: 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikane R, Shinagawa H, Ishino Y 2006. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells 11: 99–110 [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J 17: 6412–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CP, Bolt EL 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucl Acids Res 33: 3678–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ES, Cooper DL, Persky NS, Sutera VA Jr, Whitaker RD, Montello ML, Lovett ST 2006. RecJ exonuclease: Substrates, products and interaction with SSB. Nucl Acids Res 34: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC 2009. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev 23: 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Yang L, Dillingham MS, Kobayashi I, Wigley DB, Kowalczykowski SC 2012. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, chi, by RecBCD enzyme. Proc Natl Acad Sci 109: 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE 2006. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet 40: 209–235 [DOI] [PubMed] [Google Scholar]
- Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM 2009. Ctp1/CtIP and Rad32/Mre11 nuclease activity are required for Rec12/Spo11 removal, but Rec12/Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol 29: 1671–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson A, Terentyev Y, Johnson RA, Bishop-Bailey A, Angevin T, Croucher A, Goldman AS 2011. Mre11 and Exo1 contribute to the initiation and processivity of resection at meiotic double-strand breaks made independently of Spo11. DNA Repair (Amst) 10: 138–148 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP 2000a. Mre11 and Rad50 from Pyrococcus furiosus: Cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol 182: 6036–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000b. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562–566 [DOI] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT 2008. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol 14: 3414–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL 2012. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 11: 3983–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keelagher RE, Cotton VE, Goldman AS, Borts RH 2011. Separable roles for Exonuclease I in meiotic DNA double-strand break repair. DNA Repair (Amst) 10: 126–137 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, Petrini JH 2008. Functional interactions between Sae2 and the Mre11 complex. Genetics 178: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H 2009. Systematic identification of cell cycle–dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci 106: 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lam I, Keeney S 2015. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam ST, Stahl MM, McMilin KD, Stahl FW 1974. Rec-mediated recombinational hot spot activity in bacteriophage λ II. A mutation which causes hot spot activity. Genetics 77: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, et al. 2011. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet 7: e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27: 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Bressan DA, Petrini JH, Haber JE 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 1: 27–40 [DOI] [PubMed] [Google Scholar]
- Lemmens BB, Johnson NM, Tijsterman M 2013. COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet 9: e1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y 2011. Crystal structure of the Mre11-Rad50-ATPγS complex: Understanding the interplay between Mre11 and Rad50. Genes Dev 25: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Sspatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139: 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Lovett ST, Kolodner RD 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci 86: 2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST, Luisi-DeLuca C, Kolodner RD 1988. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics 120: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrini N, Guerini I, Citterio A, Lucchini G, Longhese MP 2010. Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J Biol Chem 285: 11628–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AH, Kleckner N 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR 2009. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol 29: 5998–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29: 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockel C, Lammens K, Schele A, Hopfner KP 2012. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucl Acids Res 40: 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280: 590–592 [DOI] [PubMed] [Google Scholar]
- Ngo HP, Lydall D 2010. Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9. PLoS Genet 6: e1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT 2010. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol 17: 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Oka H, Mayanagi K, Shirai T, Matoba K, Fujikane R, Ishino Y, Morikawa K 2009. Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm. BMC Struct Biol 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky NS, Lovett ST 2008. Mechanisms of recombination: Lessons from E. coli. Crit Rev Biochem Mol Biol 43: 347–370 [DOI] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Chait BT, Gottesman ME, Baer R, Gautier J 2011. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J Cell Biol 194: 705–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, Gottesman ME, Gautier J 2013. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol Cell 49: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Amon A, Klein F 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray AJ, Shafer BK, Neelam B, Strathern JN 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev 19: 1390–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. 2012. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol 19: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T 2012. Removal of shelterin reveals the telomere end-protection problem. Science 336: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol 27: 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29: 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T 2004. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432: 187–193 [DOI] [PubMed] [Google Scholar]
- Smith GR 2001. Homologous recombination near and far from DNA breaks: Alternative roles and contrasting views. Annu Rev Genet 35: 243–274 [DOI] [PubMed] [Google Scholar]
- Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC 2003. A molecular throttle: The recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell 114: 647–654 [DOI] [PubMed] [Google Scholar]
- *.Syeda AH, Hawkins M, McGlynn P 2014. Recombination and replication. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P, Smith GR 1992. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem 267: 3014–3023 [PubMed] [Google Scholar]
- Szankasi P, Smith GR 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267: 1166–1169 [DOI] [PubMed] [Google Scholar]
- Taylor AF, Smith GR 1985. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol 185: 431–443 [DOI] [PubMed] [Google Scholar]
- Taylor AF, Smith GR 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423: 889–893 [DOI] [PubMed] [Google Scholar]
- Thaler DS, Sampson E, Siddiqi I, Rosenberg SM, Thomason LC, Stahl FW, Stahl MM 1989. Recombination of bacteriophage λ in recD mutants of Escherichia coli. Genome 31: 53–67 [DOI] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, Burma S 2012. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) 11: 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, et al. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Erdeniz N, Symington LS, Liskay RM 2004. EXO1—A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 3: 1549–1559 [DOI] [PubMed] [Google Scholar]
- Trovesi C, Falcettoni M, Lucchini G, Clerici M, Longhese MP 2011. Distinct Cdk1 requirements during single-strand annealing, noncrossover, and crossover recombination. PLoS Genet 7: e1002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem 273: 21447–21450 [DOI] [PubMed] [Google Scholar]
- Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, Wu X 2013. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci 110: 7720–7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H 1998. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol 18: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T 2005. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol Biol Cell 16: 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716 [DOI] [PubMed] [Google Scholar]
- Wang J, Chen R, Julin DA 2000. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J Biol Chem 275: 507–513 [DOI] [PubMed] [Google Scholar]
- Wang H, Shao Z, Shi LZ, Hwang PY, Truong LN, Berns MW, Chen DJ, Wu X 2012. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J Biol Chem 287: 21471–21480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, et al. 2013. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet 9: e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, Sildas S, Hammel M, Russell P, et al. 2011. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol 18: 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius JJ, Hohl M, Fleming JC, Petrini JH 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol 12: 403–407 [DOI] [PubMed] [Google Scholar]
- *.Wyatt HDM, West SC 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Handa N, Liu B, Dillingham MS, Wigley DB, Kowalczykowski SC 2012. Alteration of chi recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. Proc Natl Acad Sci 109: 8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol Cell 40: 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shim EY, Davis M, Lee SE 2009. Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair (Amst) 8: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339: 700–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Guillard S, Lydall D 2004. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 168: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]