Abstract
A particularly relevant phenomenon in cell physiology and proliferation is the fact that spontaneous mitotic recombination is strongly enhanced by transcription. The most accepted view is that transcription increases the occurrence of double-strand breaks and/or single-stranded DNA gaps that are repaired by recombination. Most breaks would arise as a consequence of the impact that transcription has on replication fork progression, provoking its stalling and/or breakage. Here, we discuss the mechanisms responsible for the cross talk between transcription and recombination, with emphasis on (1) the transcription–replication conflicts as the main source of recombinogenic DNA breaks, and (2) the formation of cotranscriptional R-loops as a major cause of such breaks. The new emerging questions and perspectives are discussed on the basis of the interference between transcription and replication, as well as the way RNA influences genome dynamics.
Transcription–replication conflicts may generate DNA breaks or gaps that are repaired by recombination. R-loops, DNA–RNA hybrids formed during transcription, may play a central role in their occurrence.
Homologous recombination (HR) is a conserved pathway responsible for the repair of double-strand breaks (DSBs). In mitotic cells, DSBs may be induced by genotoxic agents, such as γ irradiation or may occur spontaneously, in most of the cases in association with replication. Although HR represents one of the two main mechanisms of DSB repair, the other being nonhomologous end joining (NHEJ), eukaryotic cells favor HR as the preferential DSB repair pathway during the S/G2 phases of the cell cycle, when the sister chromatid is available as template for error-free repair. Thus, HR events are regulated at different steps along the cell cycle, among others by the cyclin-dependent kinase 1 during 5′-end resection to guarantee its occurrence at S/G2 (Heyer et al. 2010; Huertas 2010). Consequently, spontaneous mitotic recombination events are generally interpreted as the result of DSB repair during S/G2, although it cannot be disregarded that recombination could also be initiated by single-stranded DNA (ssDNA) gaps generated during replication.
Spontaneous mitotic recombination might, in principle, take place anywhere in the genome with similar probability. However, as for mutations, both recombination and chromosome breakages occur more frequently in particular regions, referred to as hot spots. Those hot spots might arise from different local features, including the formation of non-B secondary structures, chromatin compaction, DNA–protein barriers to replication, or low-replication initiation density (Aguilera and Garcia-Muse 2013). Nevertheless, the most extended and physiological relevant feature enhancing the probability of recombination is likely to be transcription (Aguilera 2002; Kim and Jinks-Robertson 2012; Gaillard et al. 2013). From bacteria to humans, a large body of evidence has accumulated showing that transcription stimulates spontaneous recombination, a phenomenon referred to as transcription-associated recombination (TAR). As replication failures seem to be the main source of recombinogenic DSBs, our actual view is that the major mechanism by which transcription stimulates recombination is via DSBs or ssDNA gaps potentially generated by the difficulties of the DNA replication fork to progress through transcribed DNA sequences (Aguilera and Gomez-Gonzalez 2008; Bermejo et al. 2012). Here, we review our actual knowledge of the cross talk between transcription and recombination with the aim of providing mechanistic insights into the relevance for TAR as a natural source of genome instability along with discussing emerging questions and perspectives.
TRANSCRIPTION STIMULATES RECOMBINATION FROM BACTERIA TO HUMAN CELLS
The first evidence of TAR comes from λ-phage studies showing that recombination occurs in transcribed DNA regions and depends on Escherichia coli RNA polymerase (RNAP) activity but not on RecA (Ikeda and Kobayashi 1977; Ikeda and Matsumoto 1979). Probably, the most influential reports on TAR were those showing the identification of the recombinant DNA (rDNA) sequence HOT1 as a hot spot of recombination in Saccharomyces cerevisiae (Keil and Roeder 1984), and the following demonstration that the ability of HOT1 to stimulate Rad52-dependent ectopic recombination of non-rDNA sequences depends on the RNAPI activity (Voelkel-Meiman et al. 1987; Stewart and Roeder 1989). Subsequent work on phage transduction (Dul and Drexler 1988), illegitimate recombination in bacterial plasmids (Vilette et al. 1992), RNAPII-mediated recombination between DNA repeats in S. cerevisiae (Thomas and Rothstein 1989), at the ADE6 locus of Schizosaccharomyces pombe (Grimm et al. 1991) and in rodent cells (Nickoloff and Reynolds 1990) showed that TAR is a conserved feature. A survey of these and later examples of TAR from bacteria to mammals can be found in a recent review (Gaillard et al. 2013).
Although the possibility that the impact of transcription on features such as chromatin structure or the channeling of DNA breaks into different types of repair has been considered an explanation of TAR, our current understanding favors TAR being the consequence of an increase in recombinogenic DNA damage caused by transcription. In yeast, DSBs generated by the homothallic switching (HO) endonuclease induce recombination to similar levels regardless of transcription being active or not (Weng et al. 2000; Gonzalez-Barrera et al. 2002). In mammalian cells, transcription did not stimulate DSB-induced recombination (Taghian and Nickoloff 1997). DSB-induced recombination and TAR events share similar features of reciprocal versus nonreciprocal events and are dependent on recombinational DSB repair functions such as Rad52 and Rad51 in yeast (Gonzalez-Barrera et al. 2002) or BRCA2 in mammals (Savolainen and Helleday 2009). Recently, human cell lines defective in XPD, a DNA helicase subunit of transcription factor II H (TFIIH) required for transcription initiation and nucleotide excision repair, revealed that XPD is required for TAR, but not for DSB-induced recombination (Savolainen et al. 2010). Although it is possible that a fraction of TAR events does not initiate via DSBs, we cannot exclude the possibility that some TAR events may also require XPD to be initiated. All data, therefore, strongly support the idea that transcription stimulates recombination by inducing recombinogenic damage at the transcribed DNA sequence.
In support of this idea, it has been shown that genotoxic agents such as 4-nitroquinoline 1-oxide or methyl methanesulfonate induce gene conversion synergistically with transcription (Garcia-Rubio et al. 2003). Although the looser chromatin state of transcribed regions has been proposed to provide a better substrate for the action of genotoxic agents, there is no definitive evidence for this possibility. Also, it does not seem that transcription by itself increases DNA accessibility to endonucleases or other recombination enzymes, as high transcription reduced both HO cleavage efficiency and strand invasion (Saxe et al. 2000; Gonzalez-Barrera et al. 2002). Instead, and according to the “twin-supercoiled domain” model (Liu and Wang 1987) in which transcription creates negative and positive supercoils behind and ahead of the elongating RNAP, respectively, the proneness of the strands of negatively supercoiled double-stranded DNA (dsDNA) to separate would make DNA more accessible to genotoxic agents (Fig. 1A).
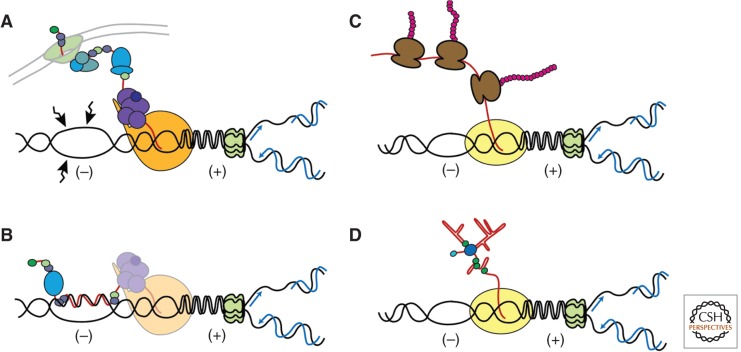
Figure 1.
Transcription intermediates can compromise genome integrity in prokaryotes and eukaryotes. (A) In eukaryotes, mRNP biogenesis and export are coupled to transcription. Transcribed genes may also be anchored to the nuclear pore. Negative supercoiling (−) accumulates behind the elongating RNAPII and facilitates the separation of both strands, making the DNA more susceptible to genotoxic agents. Positive supercoiling (+) accumulates ahead of the transcription machinery, in front of a head-on oncoming replication fork. (B) The nascent messenger RNA (mRNA) might hybridize back to its DNA template behind the RNAPII, forming an R-loop. (C) In prokaryotes, cotranscriptional translation of the nascent mRNA may impede the formation of R-loop. (D) Cotranscriptional folding of nonprotein coding RNAs may reduce the probability of R-loop formation in both eukaryotes and prokaryotes.
This model is supported by the reports on transcription-associated mutagenesis. Mutagenicity of a variety of external genotoxic agents is strongly stimulated in actively transcribed DNA sequences (Aguilera 2002; Kim and Jinks-Robertson 2012; Gaillard et al. 2013). Moreover, S. cerevisiae lacking topoisomerase 1 (Top1) activity undergoes a strong increase in recombination leading to the loss of a marker inserted in the rDNA (Christman et al. 1988), which has been recently extended to inverted- and direct-repeat systems under the control of RNAPII (Garcia-Rubio and Aguilera 2012). These observations, as well as the hypermutation engendered by the top1 mutation in yeast (Kim et al. 2011; Lippert et al. 2011; Takahashi et al. 2011), would be consistent with better accessibility of genotoxic agents facilitated by negative DNA supercoiling. Besides, nonrandom mutation clusters probably associated with long ssDNA were found both in yeast grown under chronic aklylation damage and in human malignant tumors (Roberts et al. 2012, 2013; Alexandrov et al. 2013; Burns et al. 2013). These results suggest that ssDNA patches are formed in proliferative cells, possibly during DSB repair, replication, or R-loop formation (see below). Yet, the fact that recombination is tightly regulated during the cell cycle, having its peak during the S/G2 phases, suggests that TAR, like any other form of mitotic recombination, is associated with replication.
R-LOOPS AS MEDIATORS OF TAR
R-loops are structures in which an RNA segment hybridizes with its DNA template, thereby displacing the complementary strand, which remains as a single-stranded loop (Fig. 1B). Its existence as a physiologically relevant intermediate has been shown in many systems, including origin-independent replication in bacteria (Kogoma 1997), class-switching recombination (CSR) (Yu et al. 2003), or E. coli topA mutants in which transcription-dependent formation of R-loops behind the RNAP have been genetically inferred (Drolet et al. 1995). More recently, R-loops were found at CpG island promoters and G-rich termination elements in human cells (Skourti-Stathaki et al. 2011; Ginno et al. 2012, 2013) and at an Arabidopsis antisense promoter, where it regulates the expression of a long noncoding RNA (Sun et al. 2013).
The first demonstration that R-loops generated during transcription induce genome instability in the form of recombination was provided in S. cerevisiae with mutants of the messenger RNA particle (mRNP) biogenesis and export factor THO (Huertas and Aguilera 2003). The hyper-recombination phenotype of these mutants not only is transcription-dependent and can be suppressed by mutations in the RNAPII machinery that diminish transcription (Santos-Rosa and Aguilera 1995; Fan et al. 1996; Piruat and Aguilera 1996, 1998), but also relies on the nascent RNA molecule and its capacity to form an R-loop behind the elongating DNA polymerase (Huertas and Aguilera 2003). The relevance of R-loops as an intermediate responsible for different forms of instability is, nowadays, supported by an increasing number of reports. These include the demonstration of the formation of R-loops in DT40 chicken and HeLa cells depleted of the splicing factor ASF/SF2 (Li and Manley 2005) or the transcription-dependent instability manifested as gross-chromosomal rearrangements, γ-H2AX foci, or hyper-recombination in yeast, Caenorhabditis elegans, and human cell lines mutated in or depleted of a number of factors involved in RNA metabolism (Gomez-Gonzalez and Aguilera 2007; Gonzalez-Aguilera et al. 2008; Paulsen et al. 2009; El Hage et al. 2010; Dominguez-Sanchez et al. 2011; Wahba et al. 2011; Castellano-Pozo et al. 2012; Stirling et al. 2012; Gavalda et al. 2013; Santos-Pereira et al. 2013). Interestingly, different reports have provided evidence that genome instability associated with DNA repeats with the potential to form non-B DNA structures, such as trinucleotide repeats (Grabczyk et al. 2007; Lin et al. 2010), or common fragile sites (CFSs) in mammalian cells (Helmrich et al. 2011) are also dependent on transcription and associates with the formation of R-loops. Regardless of whether recombination may be triggered in such regions or not, the results confirm that known hot spots of DNA fragility owe their instability to a process dependent on transcription that can be, either partially or completely, related to R-loop accumulation. The RNA, and specifically the R-loop, has thus become a key element in all studies aimed at understanding the cross talk between transcription and recombination (Aguilera and Garcia-Muse 2012).
Top1 restrains R-loop formation by counteracting the transient local accumulation of negative supercoils at transcribed regions that facilitate DNA strand opening (Drolet et al. 1995; Tuduri et al. 2009; El Hage et al. 2010; Marinello et al. 2013; Teves and Henikoff 2014). Cotranscriptional mRNP biogenesis and processing in eukaryotes may also diminish the chances of the nascent RNA to hybridize back with the template DNA behind the RNAP (Fig. 1A,B). The suppression of the transcription-dependent hyper-recombination phenotypes of THO and THSC/TREX-2 mutants by overexpression of heterogeneous nuclear ribonucleoproteins able to bind RNA, such as Tho1, Nab2, or the RNA dependent on ATPase Sub2/UAP56, whose deletion also causes a strong transcription-dependent hyper-recombination (Fan et al. 2001; Jimeno et al. 2002), would support this view. We still know little about the structure of the eukaryotic mRNP, the posttranslational modifications that RNA-binding factors undergo during the RNA cycle, and how they can affect the flexibility or hybridizing properties of the nascent RNA. A speculative view based on the known coupling of transcription with RNA processing could be that improper mRNP biogenesis triggers a transcription checkpoint signal that might affect transcription elongation therefore causing a more stably bound RNAP to the DNA, interfering with other DNA processes such as transcription-coupled repair or replication. In bacteria, it has been postulated that the nascent RNA is prevented from hybridizing with the DNA template by the cotranscriptional nature of translation that disposes ribosomes on the nascent mRNA in protein-encoding genes (Fig. 1C) (Gowrishankar and Harinarayanan 2004). The folding of the nascent RNA into secondary structures in nonprotein-encoding genes may fulfill the same function (Fig. 1D).
It is possible that transcription elongation impairment linked to the formation of R-loops might cause TAR. However, the fact that no transcription defect was observed in ASF/SF2-depleted chicken DT40 cells (Li and Manley 2005), the existence of THO mutants with a transcription defect that does not lead to increased TAR (Huertas et al. 2006), or the lack of correlation between transcription elongation defects and TAR in a number of mRNA processing and export factors (Luna et al. 2005) would argue against such a possibility as a general mechanism. Recently, it has also been concluded that Rad51 is required for DNA–RNA hybrids formed in trans (Wahba et al. 2013), but it is unclear at this point whether this is a mechanism related to TAR.
The increasing evidence accumulated on the relevance of R-loops in different forms of genome instability and existence of proteins with the potential to remove the RNA moiety of R-loops suggest that cotranscriptional R-loops may be formed in wild-type cells proficient in mRNP biogenesis and surveillance. The observation that the sen1-1 mutation of the Sen1/SETX putative RNA–DNA helicase, as reported for the S. pombe Sen1 (Kim et al. 1999), shows a strong TAR phenotype that correlates with transcription and R-loop accumulation argues in favor of this conclusion (Mischo et al. 2011). The questions remaining are whether all TAR events, occurring in both wild-type and mutant cells, are mediated by R-loops, and whether an R-loop by itself can trigger the formation of a DSB and/or a ssDNA gap responsible for the recombination event.
Recently, a link between R-loops and the H3 S10 phosphorylation chromatin condensation mark was uncovered in yeast, nematodes, and human cells (Castellano-Pozo et al. 2013), suggesting an additional possible mechanism by which R-loop can modulate genome dynamics. In the same study, the H3 K9me2 heterochromatin mark has also been associated with R-loops in C. elegans. In addition, DNA–RNA hybrids have been shown to mediate RNA interference–directed heterochromatin formation at pericentromeric regions of S. pombe (Nakama et al. 2012). Hence, chromatin condensation or heterochromatin taking place at or around R-loops could interfere with replication and/or transcription, thus triggering genomic instability (Castellano-Pozo et al. 2013). In agreement with this model, CFSs correlate with slow or incomplete replication at regions of chromatin condensation and cotranscriptional R-loops (Helmrich et al. 2011; Debatisse et al. 2012). It is worth noting, however, that R-loops coincide with increased chromatin decondensation in neurons (Powell et al. 2013). Thus, it seems likely that although the R-loop itself might destabilize nucleosomes, chromatin condensation would occur around and extend from the R-loop. Further work will be required to unravel the mechanism by which R-loops alter chromatin structure and how this chromatin connection modulates TAR.
THE SPECIFIC CASE OF CSR
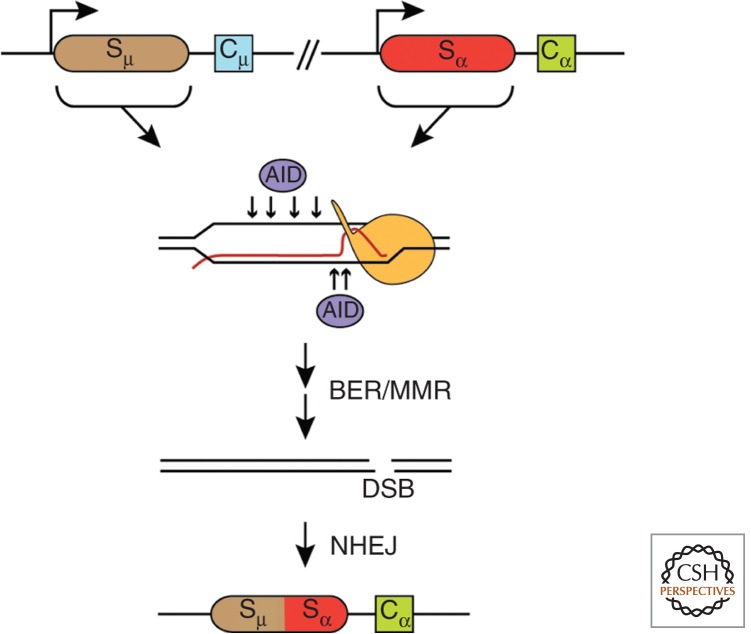
Whereas TAR represents a global phenomenon of HR, CSR defines a specific type of developmentally controlled recombination occurring by NHEJ, not by HR, in vertebrate B cells, which is worth mentioning here. CSR occurs between the S regions located beside each C segment of the immunoglobulin (Ig) genes. Only when the S region is transcribed does it undergo a DSB event that triggers the NHEJ process responsible for CSR (Fig. 2). The first indications that S regions had the potential to form R-loops were gained using in vitro transcription assays (Reaban and Griffin 1990; Reaban et al. 1994; Daniels and Lieber 1995); definitive evidence of its existence in B cells was provided later by direct mapping (Yu et al. 2003). Significantly, however, CSR differs from TAR in important aspects. CSR is a process catalyzed by the activation-induced cytidine deaminase (AID) enzyme, which deaminates deoxycytidines (dCs) generating high levels of deoxyuracils (dUs) that are processed by multiple base excision and mismatch repair events, resulting in the occurrence of DSBs (Xu et al. 2012b). During transcription-dependent R-loop formation in the S region, the high G content of the displaced DNA strand would favor G-quadruplex (G4) formation, thus stabilizing the R-loop (Duquette et al. 2004). It has also been argued that the high G content of the nascent RNA molecule may confer a structural difficulty on its assembly into a proper mRNP, enhancing their options to back hybridize with the DNA template and form an R-loop (Gomez-Gonzalez and Aguilera 2007). Finally, in contrast to TAR (see below), there is no evidence so far that CSR requires replication. It is also worth noting that AID can induce DSB-mediated translocations like those responsible for Burkitt’s lymphoma, with breakpoints in the Ig S region and a G-rich region of c-MYC (Ramiro et al. 2004; Robbiani et al. 2008; Klein et al. 2011), a phenomenon that has also been reproduced in yeast THO mutants (Ruiz et al. 2011). Therefore, although CSR is a paradigmatic case in which cotranscriptional R-loops become a key substrate for the origin of instability, it is not a model system to understand TAR as a major manifestation of the cross talk between transcription and HR.
Figure 2.
CSR. Transcription of switch (S) regions of the Ig genes generates R-loops, providing ssDNA substrates for the activation-induced cytidine deaminase (AID). Subsequent sequential enzymatic activities of different DNA repair pathways, including base excision and mismatch repair, would lead to DSBs that, by NHEJ, would complete the CSR event.
REPLICATION–TRANSCRIPTION CONFLICTS AS A SOURCE OF TAR
The way replication–transcription conflicts are resolved has been a main question in biology since it became clear that most genes are disposed codirectionally with replication along the E. coli chromosome (Ellwood and Nomura 1982). French (1992) reported that the movement of the replication fork was reduced in the ribosomal operon of E. coli as a result of its high transcriptional activity. Using an in vitro reconstituted T4 phage replication system, the laboratory of B. Alberts showed that, whereas the bacteriophage replication machinery could overtake a codirectionally moving E. coli RNAP without major consequences, a stalled RNAP caused the pausing of a head-on colliding DNA polymerase (Liu et al. 1993; Liu and Alberts 1995). In the Φ29 bacteriophage, the replication fork also stalled when the RNAP was arrested in a head-on orientation (Elias-Arnanz and Salas 1999). In E. coli, the replication machinery was able to displace a codirectionally elongating RNAP, but not an oncoming RNAP (Pomerantz and O’Donnell 2008). In the first case, the transient RNA–DNA hybrid formed during transcription could serve to reinitiate replication (Pomerantz and O’Donnell 2010). The ability of transcription to pause or stall replication in eukaryotes was first shown by 2D-gel electrophoresis at RNAPIII-driven transcription in transfer RNA (tRNA) genes (Deshpande and Newlon 1996). RNAPII transcription of a GAL fusion gene has a stronger effect on head-on replication than codirectional replication encounters (Prado and Aguilera 2005). Genome-wide evidence that transcription supposes an obstacle for the progression of forks, regardless of orientation, has been provided in yeast by determining the distribution of the Rrm3 helicase, which works in association with the replication fork (Azvolinsky et al. 2009). This study does not distinguish whether collisions might be stronger or more persistent under codirectional versus head-on orientation, but it certainly indicates that in vivo transcription in either direction may suppose an obstacle to replication fork progression. These and other observations, such as that E. coli replication fork stop at a dG (deoxiguanosine)-dC tandem repeat in which, normally, the RNAP stalls (Krasilnikova et al. 1998), support the general conclusion that transcription has a negative impact on replication, especially when both processes occur in head-on orientation.
The identification of DNA helicases, such as T4 dda and gp41 (Bedinger et al. 1983; Liu and Alberts 1995), E. coli DinG, Rep, and UvrD (Boubakri et al. 2010), yeast Rrm3 (Azvolinsky et al. 2006, 2009), and human RecQL5 (Li et al. 2011), as well as the E. coli transcription-coupled repair factor Mfd (Pomerantz and O’Donnell 2010), as factors that help bypass head-on stalled RNA polymerases confirms that cells use a battery of proteins to either prevent or resolve putatively harmful collisions. Removal of these proteins has been shown to enhance recombination in a transcription-dependent manner, supporting the idea that at least one major mechanism of TAR is mediated by transcription–replication collisions.
Clearly, in vivo evidence that replication is linked to TAR was provided by the observation that transcription from an S-phase active promoter causes hyper-recombination in yeast, whereas this is not the case of transcription driven from G1-specific promoters (Prado and Aguilera 2005). In such assays, the effect was stronger in head-on collisions systems as compared with codirectional ones and stimulated in the absence of the Rrm3 helicase. Accordingly, inversion of the E. coli rrn operon from a codirectional to a head-on disposition with respect to replication makes cell viability strictly dependent on the RecBC DSB repair protein (Boubakri et al. 2010). Evidence for the replication dependency of TAR in mammals has been provided in Chinese hamster cells, in which replication stress generated by thymidine, which slows down replication fork progression, enhances TAR at the HPRT gene (Gottipati et al. 2008). These and other data (Gaillard et al. 2013) suggest that recombination stimulated by transcription is a consequence of the difficulties of the replication fork to traverse through transcribed DNA regions. This phenomenon has also been observed in tRNA genes flanked by direct repeats (de la Loza et al. 2009), as well as in TAR mediated by R-loops. Thus, TAR in yeast THO complex mutants is only observed under S-phase transcription, and replication forks pause or stall in the DNA region in which a hot spot of RNA–DNA hybrid formation at the end of a lacZ gene has been identified (Huertas et al. 2006; Wellinger et al. 2006). Indeed, the ability of R-loops to negatively affect progression of replication forks has been confirmed in yeast, C. elegans, and bacteria (Gan et al. 2011; Castellano-Pozo et al. 2012; Santos-Pereira et al. 2013). Therefore, it seems that regardless of the type of TAR detected, whether or not stimulated in mRNP biogenesis and export mutants or mediated by R-loops, its origin is linked to a defective replication progression.
THE rDNA REGION AS A PARADIGM OF TAR EVENTS
TAR takes place in the rDNA region, which is organized in tandem repeats in most organisms to ensure the maintenance of its integrity. Interestingly, many features underlying TAR are recapitulated in the rDNA, such as the dependency on topoisomerase activities to restrain recombination and R-loop formation (Kim and Wang 1989; El Hage et al. 2010). In eukaryotes, DNA elements with replication fork barrier properties (RFBs) are found in rDNA regions, as first shown in yeast (Brewer and Fangman 1988). They associate with specialized proteins to exert a programmed polar pausing of replication, thus avoiding frontal collisions with the transcriptional machinery (Tsang and Carr 2008). Structures with RFB capacity are also found at other genomic locations, including replication termination sites in E. coli (Khatri et al. 1989; Hill and Marians 1990), the mat locus in S. pombe (Dalgaard and Klar 2001), or transposons (Zaratiegui et al. 2011), indicating that programmed fork block to avoid frontal encounters with transcriptional machineries is a broadly used cellular strategy to avoid genomic instability.
In addition to the RFB, another important mechanism contributing to rDNA stability involves the regulated recruitment of cohesin, a multifunctional protein complex that ensures sister chromatid cohesion during replication and at sites of DSB, thus contributing to prevent genomic instability by favoring equal recombination events (Nasmyth 2011; Dorsett and Strom 2012). In the yeast S. cerevisiae, only about half of the rDNA repeats are actively transcribed, and repressed rDNA copies have been shown to be important for sister chromatid cohesion (Ide et al. 2010). rDNA silencing is mediated by the Sir2 protein and appears to control transcription from E-pro, a bidirectional RNAPII promoter located adjacent to the RFB (Santangelo et al. 1988; Ganley et al. 2005). Interestingly, constitutive transcription from E-pro results in dissociation of cohesin from the rDNA spacer, and the proportion of unequal recombination events was shown to increase in a conditional cohesin mutant, leading to an increase in rDNA repeats expansion, marker loss, and extrachromosomal rDNA circle formation (Kobayashi et al. 2004; Kobayashi and Ganley 2005). Hence, the activation of E-pro transcription has been shown to lead to cohesin dissociation at the rDNA. Similarly, transcription activation leads to cohesin delocalization at other loci in yeast (Lengronne et al. 2004; Bausch et al. 2007). It is thus tempting to speculate that transcription-dependent changes of sister chromatid cohesion may also contribute to TAR. Indeed, the hyper-recombination associated with cohesion mutations has been observed at different loci (De Piccoli et al. 2006; Alvaro et al. 2007).
TELOMERES AS A PUTATIVE REGION PRONE TO TERRA-MEDIATED TAR
The ends of eukaryotic chromosomes not only pose the “end-replication” problem, solved in yeast and proliferative tissues by the action of the telomerase, but they also need to be protected by the association with specialized proteins and formation of a lariat structure, the T-loop, created by the invasion of the 3′ ssDNA end into the duplex region to prevent the activation of the DNA damage response (de Lange 2009; O’Sullivan and Karlseder 2010). Replication is challenged at telomeres, which, in this sense, resemble fragile sites (Sfeir et al. 2009). Although telomeres are heterochromatin structures, they are transcribed by RNAPII into telomeric repeat-containing RNA (TERRA) molecules in several eukaryotes from yeast to mammals (Feuerhahn et al. 2010; Bah and Azzalin 2012). TERRA might interfere with replication, either through collision between the transcription and replication machineries or because TERRA might form R-loops with telomeric DNA, which could be further stabilized by G4-DNA structures in the displaced strand or G4-hybrid structures. Evidence in support of this idea includes the observation that a fraction of TERRA remains associated with telomeric chromatin (Azzalin et al. 2007; Schoeftner and Blasco 2008; Deng et al. 2009) and dysfunction of the Rat1 ribonuclease leads to TERRA accumulation and telomere elongation defects in S. cerevisiae, both phenotypes being suppressed by RNase H overexpression (Luke et al. 2008). Furthermore, both telomeric DNA and TERRA adopt G4-DNA structures (Phan 2010; Xu et al. 2010; Lu et al. 2013), and RNA–DNA hybrid G4 structures may represent a telomere component according to recent modeling studies (Xu et al. 2012a). The existence of telomeric R-loops has been recently shown in budding yeast in two independent studies. Interestingly, those telomeric R-loops were significantly increased in THO mutants, which suffer from short telomeres (Pfeiffer et al. 2013), indicating that proper TERRA biogenesis is important for telomere maintenance. Noticeably, THO subunits were identified at human telomeres as well (Grolimund et al. 2013). On the other hand, telomeric R-loop accumulation was shown to promote recombination-mediated elongation events in the absence of telomerase (Balk et al. 2013), thus being an important player in telomere dynamics. Further work will be required to dissect the role of telomeric R-loops in the regulation of telomere function.
FROM TRANSCRIPTION–REPLICATION CONFLICTS TO RECOMBINATION
The concept that the stalling of the replication fork by transcription, whether or not mediated by the formation of R-loops, is a major cause of TAR seems to be generally accepted. However, the mechanism by which a stalled replication fork can lead to the DSBs or ssDNA gaps responsible for recombination remains unclear. In principle, different mutually nonexclusive options could explain this phenomenon, of which we review three.
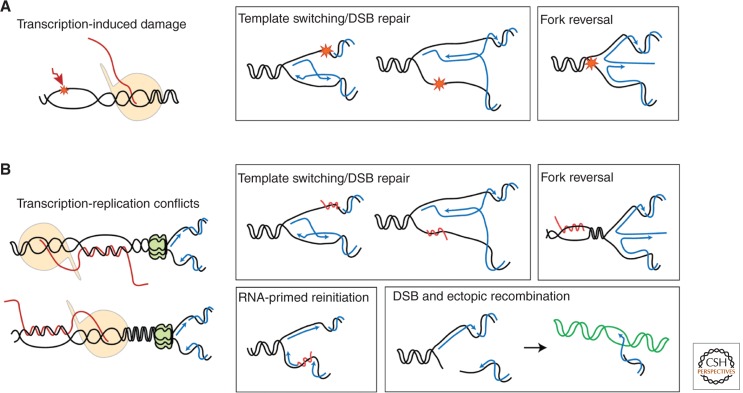
First, as discussed above, the accumulation of negative supercoiled DNA during transcription may increase its susceptibility to endogenous genotoxic agents. The resulting DNA lesions, unless properly repaired, will constitute physical obstacles for the progression of the replication fork. As a consequence, an ssDNA stretch would be created in front of the lagging or leading DNA polymerases, and replication of the gapped region completed by recombinational template switching (Fig. 3A). In this case, although the recombination event would be replication-dependent, the transcription machinery does not need to be present in the DNA during replication, and a DSB may not necessarily be the initiation intermediate.
Figure 3.
Putative molecular intermediates and mechanisms responsible for TAR. (A) The negative supercoiled DNA accumulating behind the elongating transcription machinery would expose ssDNA segments that would be more susceptible to DNA damage. Unless repaired, such transcription-dependent DNA lesions could block the replicative DNA polymerase. Template switching, DSB repair (in this case, the original damage was a ssDNA nick that, after replication, is converted to a DSB), or fork reversal would be required to complete replication. Postreplicative repair by translesion synthesis polymerases is not drawn for simplification. (B) The transcription machinery can be a direct obstacle for the progression of the replication fork, whether or not mediated by a cotranscriptional R-loop. An R-loop, or any other kind of transcription-dependent obstacle, could block DNA synthesis so that template switching would be needed for replication completion. Similarly, a head-on transcription-replication collision might cause replication fork arrest and reversal. Alternatively, R-loop-primed reinitiation of DNA synthesis might occur. Replication fork arrest could also result in DSBs, whether or not catalyzed by endonucleases, that would use recombination either with the sister chromatid or homologous chromosome or an ectopically located homologous DNA sequence, which can result in a transcription-dependent hyper-recombination phenotype.
Second, during codirectional collisions, the replication fork hitting the back of the RNAP would probably pause or strongly reduce its speed, presumably without arrest in most cases. Once the elongating RNAP finishes its transcription cycle, replication could, in principle, resume normally. Alternatively, it is also possible that the replication fork coming from the other side finishes replication. However, the presence of a DNA lesion with the ability to block RNAP elongation or requiring RNAP backtracking to resume elongation (Dutta et al. 2011), or mutants defective in 3′-end RNA processing or transcription termination (Mischo et al. 2011), may make this codirectional fork pausing permanent, leading to a replication arrest. A similar situation could occur if an R-loop would have formed behind the RNAP, in which case the replication fork would hit the RNA:DNA hybrid directly, presumably causing fork stalling (Fig. 3B). Resumption of replication may then require fork breakage, whether or not mediated by a nuclease, replication restart thus occurring by a recombination-mediated mechanism. Alternatively, the hybrid might be used as primer to reinitiate DNA synthesis.
Third, during head-on collisions, replication arrest likely becomes inevitable. It remains unclear, however, whether or not a physical collision between the two polymerases takes place. The positive supercoiling generated between the two machineries, and its possible impact on nucleosome organization, may produce some signals in response to which cells could block replication fork progression avoiding physical collision (Fig. 3B). The observations that the human RECQL5 helicase interacts with RNAPII and functions in transcription repression (Aygun et al. 2008, 2009; Kassube et al. 2013) suggest that direct inhibition of transcription elongation might provide a means to prevent collisions. RecQL5-depleted mammalian cells suffer from strong increase in sister chromatid exchanges (Hu et al. 2005). Whether this phenotype is caused by the RNAPII-regulating activity of RECQL5, a consequence of a more direct role of RecQL5 in DNA metabolism (Hu et al. 2007), or a combination of both (Islam et al. 2010) remains yet an open question. It is thus possible that transcription may not behave just as a protein–DNA barrier to replication like natural or even artificial recombinogenic RFBs (Horiuchi and Fujimura 1995; Lambert et al. 2005; Jacome and Fernandez-Capetillo 2011). In this sense, genome-wide chromatin immuno-precipitation (ChIP)-chip analyses have revealed that Top2 accumulates in regions in which replication forks encounter highly transcribed genes (Bermejo et al. 2009). Also, a transcription-dependent effect of DNA topoisomerases on recombination between DNA repeats has been described (Garcia-Rubio and Aguilera 2012). Essentially, a DNA region attached to the nuclear periphery would hinder torsional stress spreading and, thus, further enhance the transcriptional barrier to replication. The DNA-damage checkpoint would release the DNA from the nuclear pore, providing the flexibility required to solve the topological constraints generated by the replication-transcription encounter (Bermejo et al. 2011). In this scenario, an R-loop would make the topological constraint more accused, as it would prevent the release of the topological stress generated by the progression of the fork, retaining a structural feature that would contribute further to replication arrest (Alzu et al. 2012).
An additional consequence of a head-on encounter could be the extrusion of the replication fork backward, generating a chicken-foot structure (Fig. 3B) (Postow et al. 2001; Olavarrieta et al. 2002). This structure, by itself, contains a dsDNA end that can initiate recombination or constitute the substrate for structure-selective endonucleases, such as Mus81-Mms4/MUS81-EME1, Slx1-Slx4/SLX1-SLX4, Yen1/GEN1, Rad2/XPG, etc., with the potential to initiate recombination events as potential factors involved in TAR.
CONCLUDING REMARKS
Despite many years of intensive research on TAR using a large battery of different systems, further genetic and molecular analyses are needed to identify the intermediates involved. For example, a detailed and comparative analysis of TAR using different substrates leading to potentially different recombination events would be important to evaluate the prevalence of the kind of initiation event (DSB or ssDNA gap), recombination event (reciprocal or nonreciprocal), and involved pathways (break-induced replication, single-strand annealing, synthesis-dependent strand annealing, or DSB repair) in each case. It is unclear whether spontaneous TAR observed in wild-type cells is similar to that observed in mRNP processing mutants or mediated by R-loops. Although a genome-wide analysis in yeast has shown that the sites of major accumulation of the Rrm3 helicase are the open reading frames of actively transcribed genes, implying that replication has difficulties in traversing such DNA regions (Azvolinsky et al. 2009), a similar study on the distribution of γ-H2AX signals, as a marker of DSBs, has shown that they accumulate in other regions including tRNAs and promoters of protein-encoding genes (Szilard et al. 2010). An explanation for this apparent discrepancy could be that transcription does not necessarily induce DSBs, which would be consistent with recent work performed in mammalian cells (Gottipati et al. 2008) or the proposal that most TAR events induced in yeast THO mutants, although being replication-dependent, would not occur via DSBs (Gomez-Gonzalez et al. 2009). Indeed, hpr1 mutants show a strong transcription-dependent hyper-recombination leading to deletions between direct repeats but a mild increase in other types of events. It has been proposed that a DNA polymerase stalled in front of an R-loop may switch to another homologous region and use it as a template to resume replication, possibly explaining why direct-repeat recombination is the most favored type of event in R-loop accumulating mutants (Fig. 3B) (Gomez-Gonzalez et al. 2009).
It seems, therefore, important to identify the molecular event initiating recombination at transcribed DNA that could explain the different TAR events studied so far—by analogy with CFSs, specific chromosomal sites showing breaks or constrictions in metaphase chromosomes (Durkin and Glover 2007), which have recently been shown to occur in very long genes located in low-replication initiation regions (Helmrich et al. 2011; Le Tallec et al. 2011; Letessier et al. 2011). An interesting and open question is the mechanism by which TAR could be induced in terminally differentiated cells. It could be that cotranscriptionally formed R-loops interfere with DNA-repair activities or transcriptional RNA-binding proteins might have a role in DNA repair itself. However, the emerging field involving small noncoding RNAs in the DNA damage response to DSBs (Wei et al. 2012; d’Adda di Fagagna 2013) points to novel connections between RNA and the maintenance of genome integrity that would need to be further investigated in relation to TAR. Up to which point the nature of the transcriptional intermediates determine the outcome of recombination remains to be seen, but, no doubt, the more we know about the type of event involved, the more we will learn about the molecular nature of the cross talk between transcription and recombination and their connection to replication. This is of major interest in modern genetics and molecular biology not only for the increasing relevance of transcription in genome dynamics and, as recently proposed, accelerated evolution (Paul et al. 2013), but also because of the importance of genome integrity maintenance as a mechanism to prevent cancer and other genetic diseases.
ACKNOWLEDGMENTS
We thank R. Wellinger for helpful discussions and D. Haun for style supervision. Research in A.A.’s laboratory is funded by the Spanish Ministry of Economy and Competitiveness, the Junta de Andalucía, and the European Union (FEDER). We apologize to those whose work could not be cited because of space constraints.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
- Aguilera A 2002. The connection between transcription and genomic instability. EMBO J 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T 2012. R loops: From transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T 2013. Causes of genome instability. Annu Rev Genet 47: 1–32 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro D, Lisby M, Rothstein R 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet 3: e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. 2012. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151: 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Svejstrup J, Liu Y 2008. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci 105: 8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ 2009. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem 284: 23197–23203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA 2006. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bah A, Azzalin CM 2012. The telomeric transcriptome: From fission yeast to mammals. Int J Biochem Cell Biol 44: 1055–1059 [DOI] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B 2013. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Bausch C, Noone S, Henry JM, Gaudenz K, Sanderson B, Seidel C, Gerton JL 2007. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol 27: 8522–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P, Hochstrasser M, Jongeneel CV, Alberts BM 1983. Properties of the T4 bacteriophage DNA replication apparatus: The T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell 34: 115–123 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, et al. 2009. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell 138: 870–884 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M 2012. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol Cell 45: 710–718 [DOI] [PubMed] [Google Scholar]
- Boubakri H, de Septenville AL, Viguera E, Michel B 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 [DOI] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45: 977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Garcia-Muse T, Aguilera A 2012. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep 13: 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 52: 583–590 [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR 1988. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55: 413–425 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F 2013. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol 24: 171–178 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ 2001. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15: 2060–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels GA, Lieber MR 1995. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: Implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res 23: 5006–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O 2012. Common fragile sites: Mechanisms of instability revisited. Trends Genet 28: 22–32 [DOI] [PubMed] [Google Scholar]
- de la Loza MC, Wellinger RE, Aguilera A 2009. Stimulation of direct-repeat recombination by RNA polymerase III transcription. DNA Repair 8: 620–626 [DOI] [PubMed] [Google Scholar]
- de Lange T 2009. How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM 2009. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, et al. 2006. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8: 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7: e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Strom L 2012. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol 22: R240–R250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli ΔtopA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci 92: 3526–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul JL, Drexler H 1988. Transcription stimulates recombination: II. Generalized transduction of Escherichia coli by phages T1 and T4. Virology 162: 471–477 [DOI] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18: 1618–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW 2007. Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D 2010. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias-Arnanz M, Salas M 1999. Resolution of head-on collisions between the transcription machinery and bacteriophage Φ29 DNA polymerase is dependent on RNA polymerase translocation. EMBO J 18: 5675–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M, Nomura M 1982. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J Bacteriol 149: 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Cheng KK, Klein HL 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics 142: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Merker RJ, Klein HL 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol Cell Biol 21: 5459–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J 2010. TERRA biogenesis, turnover and implications for function. FEBS Lett 584: 3812–3818 [DOI] [PubMed] [Google Scholar]
- French S 1992. Consequences of replication fork movement through transcription units in vivo. Science 258: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Gaillard H, Herrera-Moyano E, Aguilera A 2013. Transcription-associated genome instability. Chem Rev 113: 8638–8661 [DOI] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25: 2041–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley AR, Hayashi K, Horiuchi T, Kobayashi T 2005. Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc Natl Acad Sci 102: 11787–11792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio ML, Aguilera A 2012. Topological constraints impair RNA polymerase II transcription and causes instability of plasmid-borne convergent genes. Nucleic Acids Res 40: 1050–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A 2003. Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics 165: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavalda S, Gallardo M, Luna R, Aguilera A 2013. R-loop mediated transcription-associated recombination in trf4Δ mutants reveals new links between RNA surveillance and genome integrity. PLoS One 8: e65541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chedin F 2013. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23: 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Aguilera A 2007. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci 104: 8409–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A 2009. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29: 5203–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Tous C, Gomez-Gonzalez B, Huertas P, Luna R, Aguilera A 2008. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 19: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Barrera S, Garcia-Rubio M, Aguilera A 2002. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics 162: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati P, Cassel TN, Savolainen L, Helleday T 2008. Transcription-associated recombination is dependent on replication in mammalian cells. Mol Cell Biol 28: 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar J, Harinarayanan R 2004. Why is transcription coupled to translation in bacteria? Mol Microbiol 54: 598–603 [DOI] [PubMed] [Google Scholar]
- Grabczyk E, Mancuso M, Sammarco MC 2007. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res 35: 5351–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Schaer P, Munz P, Kohli J 1991. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol 11: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolimund L, Aeby E, Hamelin R, Armand F, Chiappe D, Moniatte M, Lingner J 2013. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun 4: 2848. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44: 966–977 [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44: 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM, Marians KJ 1990. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci 87: 2481–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y 1995. Recombinational rescue of the stalled DNA replication fork: A model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol 177: 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G 2005. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol 25: 3431–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P 2010. DNA resection in eukaryotes: Deciding how to fix the break. Nat Struct Mol Biol 17: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Huertas P, Garcia-Rubio ML, Wellinger RE, Luna R, Aguilera A 2006. An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol Cell Biol 26: 7451–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696 [DOI] [PubMed] [Google Scholar]
- Ikeda H, Kobayashi I 1977. Involvement of DNA-dependent RNA polymerase in a recA-independent pathway of genetic recombination in Escherichia coli. Proc Natl Acad Sci 74: 3932–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Matsumoto T 1979. Transcription promotes recA-independent recombination mediated by DNA-dependent RNA polymerase in Escherichia coli. Proc Natl Acad Sci 76: 4571–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Fox D III, Guo R, Enomoto T, Wang W 2010. RecQL5 promotes genome stabilization through two parallel mechanisms—Interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol 30: 2460–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome A, Fernandez-Capetillo O 2011. Lac operator repeats generate a traceable fragile site in mammalian cells. EMBO Rep 12: 1032–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassube SA, Jinek M, Fang J, Tsutakawa S, Nogales E 2013. Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5. Nat Struct Mol Biol 20: 892–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil RL, Roeder GS 1984. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386 [DOI] [PubMed] [Google Scholar]
- Khatri GS, MacAllister T, Sista PR, Bastia D 1989. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell 59: 667–674 [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S 2012. Transcription as a source of genome instability. Nat Rev Genet 13: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC 1989. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57: 975–985 [DOI] [PubMed] [Google Scholar]
- Kim HD, Choe J, Seo YS 1999. The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38: 14697–14710 [DOI] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S 2011. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332: 1561–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453 [DOI] [PubMed] [Google Scholar]
- Kogoma T 1997. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61: 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM 1998. Transcription through a simple DNA repeat blocks replication elongation. EMBO J 17: 5095–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M 2011. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol 18: 1421–1423 [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Li M, Xu X, Liu Y 2011. The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol Cell Biol 31: 2090–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M 2010. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci 107: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O’Shea SH, Jinks-Robertson S 2011. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci 108: 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Alberts BM 1995. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC 1987. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci 84: 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wong ML, Tinker RL, Geiduschek EP, Alberts BM 1993. The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature 366: 33–39 [DOI] [PubMed] [Google Scholar]
- Lu W, Zhang Y, Liu D, Songyang Z, Wan M 2013. Telomeres-structure, function, and regulation. Exp Cell Res 319: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell 18: 711–722 [DOI] [PubMed] [Google Scholar]
- Marinello J, Chillemi G, Bueno S, Manzo SG, Capranico G 2013. Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res 41: 10110–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y 2012. DNA–RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17: 218–233 [DOI] [PubMed] [Google Scholar]
- Nasmyth K 2011. Cohesin: A catenase with separate entry and exit gates? Nat Cell Biol 13: 1170–1177 [DOI] [PubMed] [Google Scholar]
- Nickoloff JA, Reynolds RJ 1990. Transcription stimulates homologous recombination in mammalian cells. Mol Cell Biol 10: 4837–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olavarrieta L, Hernandez P, Krimer DB, Schvartzman JB 2002. DNA knotting caused by head-on collision of transcription and replication. J Mol Biol 322: 1–6 [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J 2010. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11: 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H 2013. Accelerated gene evolution through replication-transcription conflicts. Nature 495: 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J 2013. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J 32: 2861–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT 2010. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J 277: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A 1996. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics 143: 1533–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 17: 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M 2008. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M 2010. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 327: 590–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem 276: 2790–2796 [DOI] [PubMed] [Google Scholar]
- Powell WT, Coulson RL, Gonzales ML, Crary FK, Wong SS, Adams S, Ach RA, Tsang P, Yamada NA, Yasui DH, et al. 2013. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci 110: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118: 431–438 [DOI] [PubMed] [Google Scholar]
- Reaban ME, Griffin JA 1990. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348: 342–344 [DOI] [PubMed] [Google Scholar]
- Reaban ME, Lebowitz J, Griffin JA 1994. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin α switch region. J Biol Chem 269: 21850–21857 [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135: 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Gomez-Gonzalez B, Aguilera A 2011. AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet 7: e1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM, Tornow J, McLaughlin CS, Moldave K 1988. Properties of promoters cloned randomly from the Saccharomyces cerevisiae genome. Mol Cell Biol 8: 4217–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Herrero AB, Garcia-Rubio ML, Marin A, Moreno S, Aguilera A 2013. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev 27: 2445–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Aguilera A 1995. Isolation and genetic analysis of extragenic suppressors of the hyper-deletion phenotype of the Saccharomyces cerevisiae hpr1 δ mutation. Genetics 139: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen L, Helleday T 2009. Transcription-associated recombination is independent of XRCC2 and mechanistically separate from homology-directed DNA double-strand break repair. Nucleic Acids Res 37: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen L, Cassel T, Helleday T 2010. The XPD subunit of TFIIH is required for transcription-associated but not DNA double-strand break-induced recombination in mammalian cells. Mutagenesis 25: 623–629 [DOI] [PubMed] [Google Scholar]
- Saxe D, Datta A, Jinks-Robertson S 2000. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol Cell Biol 20: 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, Roeder GS 1989. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol 9: 3464–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Jacques PE, Laramee L, Cheng B, Galicia S, Bataille AR, Yeung M, Mendez M, Bergeron M, Robert F, et al. 2010. Systematic identification of fragile sites via genome-wide location analysis of γ-H2AX. Nat Struct Mol Biol 17: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghian DG, Nickoloff JA 1997. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol 17: 6386–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S 2011. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci 108: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S 2014. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol 21: 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Tsang E, Carr AM 2008. Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair 7: 1613–1623 [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D, Uzest M, Ehrlich SD, Michel B 1992. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J 11: 3629–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 44: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D 2013. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife 2: e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y 2012. A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112 [DOI] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A 2006. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YS, Xing D, Clikeman JA, Nickoloff JA 2000. Transcriptional effects on double-strand break-induced gene conversion tracts. Mutat Res 461: 119–132 [DOI] [PubMed] [Google Scholar]
- Xu Y, Suzuki Y, Ito K, Komiyama M 2010. Telomeric repeat-containing RNA structure in living cells. Proc Natl Acad Sci 107: 14579–14584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ishizuka T, Yang J, Ito K, Katada H, Komiyama M, Hayashi T 2012a. Oligonucleotide models of telomeric DNA and RNA form a hybrid G-quadruplex structure as a potential component of telomeres. J Biol Chem 287: 41787–41796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pone EJ, Mai T, Casali P 2012b. Immunoglobulin class-switch DNA recombination: Induction, targeting and beyond. Nat Rev Immunol 12: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4: 442–451 [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bahler J, Arcangioli B, Martienssen RA 2011. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature 469: 112–115 [DOI] [PMC free article] [PubMed] [Google Scholar]