Abstract
The diversity and patchy phylogenetic distribution of genetic sex-determining mechanisms observed in some taxa is thought to have arisen by the addition, modification, or replacement of regulators at the upstream end of the sex-determining pathway. Here, I review the various evolutionary forces acting on upstream regulators of sexual development that can cause transitions between sex-determining systems. These include sex-ratio selection and pleiotropic benefits, as well as indirect selection mechanisms involving sex-linked sexually antagonistic loci or recessive deleterious mutations. Most of the current theory concentrates on the population–genetic aspects of sex-determination transitions, using models that do not reflect the developmental mechanisms involved in sex determination. However, the increasing availability of molecular data creates opportunities for the development of mechanistic models that can clarify how selection and developmental architecture interact to direct the evolution of sex-determination genes.
Sex-determining mechanisms are highly diverse across taxa. Several evolutionary forces (e.g., sexually antagonistic selection) act on regulators of sexual development to cause transitions between sex-determining systems.
Biparental sexual reproduction is a common mode of reproduction in higher organisms. It is found in gonochorous animals (Bull 1983; Barnes et al. 2001), heterothallic fungi (Heitman et al. 2013), and dioecious flowering plants and algae (Ainsworth 2000; Umen 2011). Species belonging to this diverse group of organisms have distinct sexes, and their development typically passes through a critical stage at which the zygote commits irreversibly to either the male or the female sexual fate (Valenzuela 2008) (except in sequential hermaphrodites, which change sex during their life). This ontogenetic process, known as sex determination, triggers the differentiation of specialized male or female reproductive organs and organizes many sex-specific differences in gene expression, physiology, morphology, and behavior (sex differentiation) (Badyaev 2002; Ellegren and Parsch 2007).
Despite the universal and simple dichotomous outcome of sex determination, sex-determining mechanisms are highly diverse across taxa. Some species use a specific environmental cue (e.g., temperature, photoperiod, or population density) as the primary sex-determining signal (environmental sex determination; ESD), whereas others rely on various types of genetic sex determination (GSD), including male or female heterogamety, haplodiploidy or multilocus sex-determining mechanisms (Bull 1983; Marshall Graves 2008; Janousek and Mrackova 2010). In addition, sex determination can depend on epigenetic factors such as imprinting or maternal gene products deposited in the egg (Verhulst et al. 2010a). The apparent variability of sex determination is even more puzzling given that other processes acting in mid-development are evolutionarily conserved, presumably as a result of strong ontogenetic constraints (Marín and Baker 1998; Kalinka and Tomancak 2012). Considerable effort has therefore been directed at explaining the function and evolutionary origin of diversity in the mechanisms determining sex.
Here I review this literature, concentrating on transitions between genetic sex determination systems (for other recent reviews, see Beukeboom and Perrin 2014; van Doorn 2014). An important ultimate cause of such transitions is sexually antagonistic selection, which interacts with various other evolutionary forces shaping the sex-determining system. To disentangle these factors, I will first discuss the current paradigm for how sex-determination pathways have been modified, before reviewing the various mechanisms thought to be responsible for transitions in sex determination. The final part of this review will straddle the proximate/ultimate divide by exploring how adaptive mechanisms interact with the developmental architecture of sex determination.
PATTERNS OF EVOLUTION IN SEX-DETERMINATION CASCADES
Genetic and developmental analyses have revealed that the gene-regulatory networks responsible for sex determination in various model organisms have a clear hierarchical structure. For example, in the round worm Caenorhabditis elegans, the primary sex-determination signal is relayed by a cascade of inhibitory regulatory interactions involving the genes xol-1, sdc-1,2,3, her-1, tra-2,3, fem-1,2,3, and tra-1 (Stothard and Pilgrim 2003). The final gene in the cascade inhibits egl-1, mab-3 (male abnormal 3), and other genes responsible for male development. In the fruit fly Drosophila melanogaster, sex determination involves a cascade of regulatory genes whose expression is controlled by sex-specific alternative splicing. The topmost gene, Sxl (Sex-lethal), responds to the primary sex-determination system early in development and then maintains the female state by means of an auto-regulatory feedback loop (Schütt and Nöthiger 2000). The gene dsx (doublesex), at the bottom of the cascade, shares a characteristic cysteine rich DNA-binding domain (Zhu et al. 2000) with the mab-3 gene of C. elegans. This finding provided a first clue that sex-determination pathways in distantly related organisms derive from a common ancestral pathway. Consistent with their sequences being similar, dsx and mab-3 share biological functions important for male development and the protein product of the male-specific splice form of dsx can partly restore the wild-type phenotype in male worms mutant for mab-3 (Raymond et al. 1998). Other dsx-mab-3 (DM)-family genes with a role in sex determination or differentiation have later been discovered in crustaceans, vertebrates, and cnidarians, supporting a single origin of the pathways controlling sexual development in animals (Hodgkin 2002; Haag and Doty 2005; Shukla and Nagaraju 2010).
A striking pattern that is shared by the well- studied sex-determination pathways of insects (Nöthiger and Steinmann-Zwicky 1985; Gempe and Beye 2011), nematodes (Zarkower 2001; Stothard and Pilgrim 2003) and vertebrates (Marshall Graves 2008; Mawaribuchi et al. 2012), is that the downstream genes are more conserved than upstream ones (Zarkower 2001). For example, Sxl, the first gene in the D. melanogaster cascade, has no known function in sex determination outside the Drosophilidae (Sánchez 2008). However, its downstream target, tra (transformer), is found in several orders of the holometabolous insects (Verhulst et al. 2010b; Geuverink and Beukeboom 2014). Functional orthologs of tra have been identified, for instance, in other Dipterans (Ceratitis capitata, Musca domestica, Anastrepha sp., Bactrocera oleae, Lucilia cuprina) (Lagos et al. 2007; Ruiz et al. 2007; Concha and Scott 2009; Hediger et al. 2010), Coleoptera (Tribolium castaneum) (Shukla and Palli 2012) and in the Hymenoptera (Apis mellifera and Nasonia vitripennis) (Hasselmann et al. 2008; Verhulst et al. 2010a), the most basal order of the Holometabola. Thus far, no transformer homologs have been found in the Lepidoptera or outside the insects (Geuverink and Beukeboom 2014). The regulation of tra by upstream factors is highly diverse (Verhulst et al. 2010b). In the diploid Mediterranean fruitfly C. capitata, for example, the production of functional Tra protein is repressed by a dominant masculinizing factor; in the haplodiploid species A. mellifera, it is under control of the complementary sex-determiner gene csd, and in N. vitripennis (another haplodiploid species), its expression is regulated by imprinting of the maternal copy of tra (Verhulst et al. 2010a,b; Gempe and Beye 2011). However, in all cases, tra acts as a splice regulator of dsx, a putative ancient sex-determination gene that has been conserved across phyla.
The observation that downstream effectors (such as the DM-family genes in animals) are conserved, whereas the top most genes of sex-determination cascades are highly variable, supports the hypothesis that sex-determination pathways have evolved from the bottom up by the successive addition of novel upstream regulators (Nöthiger and Steinmann-Zwicky 1985; Wilkins 1995; Marín and Baker 1998). Theoretical studies on sex determination therefore tend to concentrate on the evolutionary forces responsible for the recruitment of upstream modifiers (implicitly), assuming that the downstream parts of the pathway remain fixed. A clear example of this approach is provided by Pomiankowski et al. (2004), who give a detailed step-by-step reconstruction of the recruitment of first tra and then Sxl to the sex-determining pathway of D. melanogaster.
The original motivation of the “bottom-up” hypothesis has been the idea that an ongoing evolutionary conflict between feminizing and masculinizing genetic factors may have driven the sequential invasion of novel master-sex determining switches, each one reversing the action of the previous master sex-determining gene (Wilkins 1995). The result would be a cascade of inhibitory interactions, as observed in the C. elegans pathway. Although the existence of a tug-of-war between the sexes is plausible, it is worth noting that a cascade of inhibitory interactions could also be explained simply by the fact that the invasion of a novel sex-determining gene is more likely to be successful if it is epistatically dominant over the ancestral one (van Doorn and Kirkpatrick 2010). Epistatically dominant modifiers can either replace the ancestral upstream regulator or lead to its fixation (Bull and Charnov 1977), causing the length of the pathway to remain equal or to grow by one step, respectively. Fixation of the ancestral gene occurs only when the novel and the ancestral gene have opposite effects, such as when a feminizing allele invades in a population with a masculinizing Y chromosome (XX♀/XY♂ heterogamety). So, any mechanism favoring the repeated invasion of epistatically dominant upstream regulators would result in a cascade of inhibitory interactions, irrespective of which selective forces are operating or the order in which feminizing and masculinizing alleles appear. Data on evolutionary patterns must therefore be complemented with mechanistic models to clarify how sex-determination pathways evolve.
NEUTRAL EVOLUTION
The first theoretical studies attempting to explain evolutionary transitions between different modes of sex determination were published by Bull and Charnov (1977) and Bull (1981), well before the molecular structure of sex-determination pathways was elucidated. As a consequence, these classical studies, as well as many later theoretical models building on them, are not explicit about the developmental mechanisms involved in sex determination, nor are they framed in terms of a specific hypothesis for the mode of pathway evolution (Pomiankowski et al. 2004; Uller and Helanterä 2011). Nevertheless, interpreted in the light of Wilkins’ bottom-up model, the early population-genetic models help clarify what different evolutionary forces act on upstream sex-determining genes when they first appear in a population.
A crucial insight offered by the early theoretical studies is that evolutionary transitions between sex-determining mechanisms can occur via selectively neutral intermediate states in which multiple polymorphic sex-determination loci determine sex (Scudo 1967). For example, one of the scenarios analyzed by Bull and Charnov (1977) concerns a population with XX♀/XY♂ GSD (male heterogamety) as the ancestral state, in which some individuals carry an epistatically dominant feminizing mutation W on one of the autosome pairs. When this mutant allele spreads in the population, it can establish a new ZZ♂/ZW♀ GSD system. Bull and Charnov (1977) showed that the new state of female heterogamety could be reached gradually through a connected set of intermediate states with multifactorial sex determination. They also showed that there is no tendency for selection to favor the ancestral sex-determining system, the novel one, or any one of the intermediate states once the sex ratio has equilibrated, unless some sex-determination genotypes are assumed to be intrinsically more fit than others. In the absence of other selective forces, it is thus conceivable that demographic stochasticity or other sources of random variation induce a transition from male to female heterogamety by causing the population to drift through genotype space along a line of equilibria connecting the alternative modes of sex determination (Fig. 1A). A similar continuous path of neutral equilibria between different modes of sex determination has been observed in population genetic analyzes of multifactorial GSD (Scudo 1967), one-locus multiallele sex determination (Karlin and Lessard 1984), and in models of transitions between genetic and environmental sex determination (Bull 1981).
Figure 1.
Neutral and adaptive models of a transition from male to female heterogamety. The two panels illustrate the change of the gamete frequencies of the ancestral (horizontal axes) and novel (vertical axes) sex-determining alleles during a transition from XX♀/XY♂ GSD (male heterogamety) to ZZ♂/ZW♀ GSD (female heterogamety). For each panel, genetic assumptions are illustrated by means of schematic representations of the ancestral sex chromosome pair (gray) with the sex-determining locus, and a pair of autosomes (white) carrying a sex-determination mutation (A,B) and a sexually antagonistic locus (B only). (A) Invasion of an autosomal, epistatically dominant feminizing mutation W. In the absence of intrinsic fitness differences between sex-determination genotypes, the allele W can drift to fixation as the population moves stochastically along a line of equilibria (thick gray line) (Bull and Charnov 1977), away from the ancestral state in the lower left corner of the diagram. By the time variation at the ancestral sex-determining locus is lost and the allele W reaches fixation (in the upper right corner of genotype space), the former X chromosome has disappeared from the population. Populations initialized with arbitrary combinations of allele frequencies quickly evolve toward the line of equilibria (thin gray trajectories), under the influence of selection for a balanced sex ratio. The process of drift along the line of equilibria is illustrated by the results from a stochastic, individual-based implementation of the population-genetic model (black trajectory with open circles; gamete frequencies are plotted every 50 generations for a population of 1000 individuals). (B) A sexually antagonistic gene located on the same autosome pair as the feminizing mutation causes the W allele to spread as a result of indirect selection, supported by the development of linkage disequilibrium between W and the female beneficial allele B (van Doorn and Kirkpatrick 2010). A simulation of a large population of 1 · 106 individuals illustrates the slow deterministic movement of the allele frequencies along a nearly neutral path close to the former line of equilibria (thick grey line). Small insets in A and B present a close-up view of evolutionary trajectories in the vicinity of the line of equilibria, confirming that movement along the line is no longer neutral in B.
The existence of a line of equilibria in models without intrinsic fitness differences implies that generic sex-determining mechanisms, like male and female heterogamety or ESD, do not necessarily represent different peaks in the adaptive landscape. So, populations in transition from one mechanism to another do not necessarily have to pass through a fitness valley (cf. Gavrilets 1997; Valenzuela 2008). Moreover, only small fitness differences between genotypes suffice to induce a consistent movement of the population along the line of equilibria (Fig. 1B), toward one of the alternative single-factor sex-determining mechanisms (Bull and Charnov 1977; Bull 1981). Sex-determination transitions may therefore be driven by subtle sources of selection, such as those acting indirectly via linkage disequilibria with other loci (van Doorn and Kirkpatrick 2007), or by a bias generated by the interaction of sex-ratio selection and demographic stochasticity around the line of equilibria (Vuilleumier et al. 2007).
Another implication is that neutral transitions serve mainly as a null model for the evolutionary turnover of sex-determination systems. In fact, any source of additional selection, however weak, acting on the sex-determining genes transforms the line of equilibria into a nearly neutral path along which populations will evolve deterministically in one direction or the other (Fig. 1B). This may be one reason why multifactorial sex determination is not often found in nature (Bull 1983) (even though its actual frequency is hard to estimate, given that the presence of a multifactorial system is only revealed by elaborate crossing experiments). The best-studied current examples among species with genetic sex determination are the housefly Musca domestica (Kozielska et al. 2006) and the platyfish Xiphophorus maculatus (Orzack et al. 1980). In both cases, rather than representing a temporary transitional state, multifactorial sex determination appears to be maintained as a consequence of specific relative fitness differences between genotypes (Orzack et al. 1980; Feldmeyer et al. 2008). Therefore, both the rarity of multifactorial sex determination and its apparent maintenance by selection in species with unusual sex determination provide an argument for exploring the role of adaptive mechanisms in sex determination transitions.
SOURCES OF DIRECT SELECTION ON NOVEL SEX-DETERMINING GENES
Both sex-ratio selection and pleiotropic fitness benefits can drive the spread of a novel sex-determining mutation by means of direct selection. This type of selection results from fitness differences between alternative alleles at the sex-determining locus that exist independently of alleles carried at other loci. In contrast, indirect selection mechanisms (which are reviewed after this section) rely on a nonrandom statistical association (linkage disequilibrium) between sex-determination alleles and loci experiencing direct selection.
Sex-Ratio Selection
Because of the immediate connection between sex determination and the sex ratio, any deviation of the sex ratio from its optimum generates selection on sex-determination genes. A sex-determination transition is a likely response to sex-ratio selection if there is genetic variation for an alternative sex-determining mechanism capable of producing a sex ratio closer to the optimum. Bulmer and Bull (1982) were the first to point out that this mechanism could lead to transitions from ESD to GSD. In particular, the accumulation of genetic variation at loci affecting the reaction norm for ESD would help to dampen maladaptive fluctuations in the population sex ratio induced by spatial or temporal variation in environmental conditions. If the cost of producing a suboptimal sex ratio outweighs the fitness benefit of using ESD (i.e., being able to adaptively match offspring sex to the state of the environment) (Charnov and Bull 1977), pure ESD, using a switch-like threshold reaction norm, is no longer evolutionarily stable. Selection then favors the establishment of genetic polymorphism at loci that control the location of the threshold of the ESD reaction norm. Eventually, a gene with major effect on sex determination may invade, marking the transition to GSD (Bulmer and Bull 1982; van Dooren and Leimar 2003).
Although species with ESD are more prone to exhibit suboptimal sex ratio fluctuations, they might enjoy a fitness advantage in situations in which selection favors a biased sex ratio, for example, because of local mate competition or different energetic costs to the parents for the production of sons versus daughters (Charnov 1982). According to this argument, ESD is more flexible and permits faster evolution toward an optimal biased sex ratio than common GSD mechanisms, which are constrained by the segregation ratio in the heterogametic sex (Bull 1983; Charnov and Bull 1989; Freedberg and Wade 2001). Empirical studies in species with ESD offer no support for the hypothesis that selection for a biased sex ratio has been a key driving factor in the origin or maintenance of ESD (Shine 1999; Janzen and Phillips 2006). Several lines of evidence do suggest, however, that male or female heterogamety in species with GSD cannot easily be modified into a flexible mechanism capable of producing a biased sex ratio (Uller et al. 2007). For example, Kozielska et al. (2006) calculated the response of the multifactorial sex-determining mechanism of the housefly M. domestica to sex-ratio selection and found that the sex-ratio bias produced by the system was much smaller than predicted by sex-allocation theory owing to population-genetic constraints. Similarly, Werren et al. (2002) showed that even a flexible GSD mechanism could evolve to lose its capacity to produce a biased sex ratio, as a result of an arms race between maternally and zygotically expressed genes induced by selection for a skewed sex ratio. Consistent with these findings, there is overall little evidence of modifications to the sex-determining mechanism in response to sex-ratio selection among diplo-diploid species with sex chromosomes (Krackow 2002). The same species also show almost no heritable genetic variation for sex ratio (Bulmer and Bull 1982; Bull and Charnov 1988). Nevertheless, it is unlikely that the lack of adaptive control over sex determination poses more than a mild evolutionary constraint for species with GSD, given that several other mechanisms to accommodate a biased sex ratio (e.g., sperm selection or selective resorption of embryos) have evolved (Krackow 2002).
When selection favors a balanced sex ratio and the sex-determining mechanism is capable of producing equal number of males and females, population sex ratios may still deviate markedly from 1:1, owing to the manipulation of sex determination by selfish genetic elements. The resulting distortion of the sex ratio generates sustained sex-ratio selection that may evoke counteradaptations of the sex-determining system (Werren and Beukeboom 1998). For example, in the Spanish mole, Talpa occidentalis (McVean and Hurst 1996), the creeping vole, Microtus oregoni (Charlesworth and Dempsey 2001), and several other rodent species, genetic conflict between sex-linked segregation distorters and their modifiers seems to have driven the evolution of unusual sex chromosome systems. Genetic conflicts can also drive transitions between male and female heterogamety, as shown by various models. For example, under sex-chromosome meiotic drive (Jaenike 2001), sex-ratio selection favors the invasion of a novel sex-determining allele that produces individuals of the underrepresented sex (Kozielska et al. 2010). As the new sex-determining locus spreads, polymorphism at the ancestral locus is lost, eventually leading to the demise of the sex-ratio distorter. Similarly, the repression of cytoplasmic sex-ratio distorters by host masculinizing alleles can induce a switch from female heterogamety to a multifactorial system dominated by male heterogamety (Caubet et al. 2000).
Pleiotropic Benefits
If a novel sex-determining mutation confers a higher viability or other intrinsic fitness advantage to its carrier, the mutation can spread as the result of simple natural selection. Bull and Charnov (1977) observed that models incorporating such direct selection behaved similar to their equal fitness counterparts in that the genotype frequencies initially approached a region near the line of equilibria of the corresponding equal-fitness model. In that region of genotype space, the invading sex-determining mutant is unaffected by sex-ratio selection, but still experiences positive natural selection. The frequency of the mutant therefore increases, whereas the genotype frequencies continue to follow approximately the neutral line of equilibria, until genetic polymorphism at the ancestral sex-determining locus is lost.
A serious problem for the pleiotropic-benefit hypothesis of sex-determination transitions, already noted by Bull and Charnov (1977), is that there are few obvious biological reasons why a novel sex-determining allele would have an intrinsic fitness advantage over the ancestral one. One realistic possibility, that is nearly indistinguishable from pleiotropy in practice, is that the invading sex determiner is in strong positive linkage disequilibrium with an allele that is favored by selection (Bull 1983). For example, some sex-determination variants in the housefly appear to be linked to pesticide-resistance alleles (Werren and Beukeboom 1998). As the beneficial allele sweeps to fixation, it drags along the genetically associated sex-determining allele, giving it the appearance of being favored by selection itself (Barton 2000). However, such “genetic hitchhiking” proceeds for as long as there is genetic variation at the locus under selection and recombination has not broken down the linkage equilibrium. Accordingly, hitchhiking can only facilitate a sex-determination transition if the sex determiner is tightly physically linked to a polymorphic locus under strong positive selection. This is quite unlikely, except perhaps when both the sex-determining mutation and the novel beneficial allele arise on a chromosomal inversion (Kirkpatrick 2010).
The alternative possibility that the novel sex determiner itself is favored by direct selection has sometimes been considered in theoretical studies, but is then usually discarded on the basis of parsimony, because it requires a fortuitous type of pleiotropy (e.g., Rice 1986). A notable exception is provided by Kraak and De Looze (1993), who proposed a verbal model for the evolution of sex determination in vertebrates, in which a gene under selection gains control over sex determination. Their hypothesis builds on the observation that the growth rate of the undifferentiated gonads in the developing embryo acts as a proximate cue for sex determination in several species with ESD. Therefore, a gene causing the undifferentiated gonads to develop more rapidly would be able to substitute for the role of the environment and act as a trigger for sex determination. Individual-based computer simulations show that the proposed transition from ESD to GSD is feasible if genetic variation for growth rate is concentrated at a single locus (Kraak et al. 2000). However, when genetic variation can accumulate at multiple loci, growth-rate accelerating alleles become genetically associated with each other only when size benefits differ maximally between males and females and several additional restrictive conditions are met (Kraak and Pen 2002).
Without empirical evidence for major-effect genes that modify sex determination and that also differentially affect the fitness of males and females by their impact on embryonic growth rate (or any other fitness-relevant process involved in sex determination), the role for pleiotropic benefits in triggering sex-determination transitions appears rather limited. Still, this conclusion does not rule out the possibility that pleiotropic fitness effects are important to stabilize an established sex-determination system. For example, the Sxl gene in Drosophila regulates both tra, its downstream gene in the sex-determination cascade, and msl-2, a gene involved in dosage compensation. Because of the latter interaction, which became critical after the establishment of Sxl and the differentiation of its sex chromosome (Pomiankowski et al. 2004), replacement of Sxl by another sex-determining switch is likely to have severe detrimental effects.
SOURCES OF INDIRECT SELECTION ON NOVEL SEX-DETERMINING GENES
Novel sex-determination factors can enjoy an indirect fitness advantage because of their association with sexually antagonistic genes or sex-linked deleterious mutations. In general, such indirect selection is much weaker than direct selection, because the linkage disequilibria between loci are often only modest (Kirkpatrick et al. 2002). However, many sex-determination genes occur in genomic regions of reduced recombination, which facilitate the maintenance of linkage disequilibrium. As a result, indirect selection is an important factor in the evolution of GSD systems.
Intralocus Sexual Conflict
Many traits that are expressed in the context of sexual selection and reproduction have sex-specific optima, owing to the functional divergence of male and female gender roles (Parker 1979). Because males and females share a common gene pool, the genetic conflict between the sexes over the expression of these traits (intralocus sexual conflict) cannot always be resolved by sex-specific gene regulation or other means of evolving sexual dimorphism (van Doorn 2009; Stewart et al. 2010; Pennell and Morrow 2013). As a result, sexually antagonistic variation in fitness is maintained in populations (Brommer et al. 2007; Foerster et al. 2007), with some individuals carrying alleles that are beneficial in males but deleterious in females, and others carrying alleles with the opposite fitness effects.
Intralocus sexual conflict has major consequences for the evolution of the sex chromosomes. In particular, selection favors the accumulation of sexually antagonistic genes on the sex chromosomes (Charlesworth and Charlesworth 1980; Rice 1984), as well as the evolution of reduced recombination between the sex-determining gene and sexually antagonistic loci in its vicinity (Bull 1983; Rice 1987; Otto et al. 2011). Both processes inhibit the breakdown of adaptive linkage disequilibria, which partially resolve the sexual conflict. These linkage disequilibria arise because alleles at a sex-determination locus automatically become statistically associated with sexually antagonistic alleles segregating at loci nearby on the same chromosome, in such a way that the transmission of each sexually antagonistic allele is biased toward the sex in which it is beneficial. For example, a young (recombining) sex chromosome carrying a dominant feminizing allele (e.g., a proto-W chromosome) is predicted to become enriched for female-beneficial alleles, particularly in the region surrounding the sex-determination locus, which is transmitted almost exclusively from mother to daughter (Clark 1988; Jordan and Charlesworth 2011). The same region on the homologous proto-Z chromosome spends up to two-thirds of its time in males, and is therefore expected to become enriched for male-beneficial alleles.
The genetic association between sex-determination genes and sex-linked sexually antagonistic loci enables evolution to implement a conditional sex-determination strategy that adaptively matches the sex of a zygote to the sexually antagonistic alleles it carries. Hence, in the presence of sexually antagonistic variation, GSD enjoys a clear selective advantage over alternative sex-determination mechanisms that assign sex randomly with respect to genetic background. To see this, consider once more the example of a dominant feminizing allele that is linked to a sexually antagonistic locus. If the recombination between the loci is low, selection can maintain strong linkage disequilibrium between them. Zygotes carrying a female-beneficial allele are then highly likely to carry the feminizing allele as well, in which case they will develop as females. Conversely, most of the zygotes carrying two male-beneficial alleles will develop as males, because they are unlikely to have inherited the feminizing allele. GSD thus results in offspring with, on average, a higher fitness than when the sex antagonistic alleles would be transmitted randomly to male and female offspring.
In line with this argument, Rice (1986) showed that a sexually antagonistic gene, tightly linked to a dominant masculinizing mutation, facilitated the spread of the mutant sex-determining allele in a population with polygenic sex determination as ancestral state. As expected, the transition from polygenic to single-locus GSD was critically supported by the genetic association between the masculinizing mutation and the sexually antagonistic gene. Individuals that inherited the mutation were more likely to have inherited the male-beneficial allele as well, which gave them a selective advantage over other types of males. This indirect benefit allowed the major sex-determining gene to spread, eventually resulting in the loss of polygenic sex determination.
The same mechanism can cause transitions between different single-locus genetic sex-determining systems, resulting in the establishment of a new sex-determining locus, the evolution of a new sex chromosome pair, or a transition between male and female heterogamety. The evolutionary dynamic of such GSD transitions is complicated by the fact that also the ancestral sex-determining locus might be linked to sexually antagonistic genes. van Doorn and Kirkpatrick (2007) therefore analyzed the relatively simple scenario of a novel masculinizing mutation arising on an autosome, in a population with male heterogamety. In their model, both sex-determining alleles were assumed to be linked to a sexually antagonistic gene. The autosomal sexually antagonistic gene favored the invasion of the masculinizing mutation, in the same way as discussed by Rice (1986). However, sexually antagonistic variation segregating on the ancestral sex chromosomes inhibited the spread of the mutant, because males carrying the novel masculinizing allele also carried two X chromosomes enriched for female-beneficial alleles. Mutant males were therefore more likely to inherit the autosomal male-beneficial allele, but also more likely to inherit the female-beneficial allele on the ancestral sex chromosomes than males carrying an ancestral Y chromosome.
Depending on the net fitness effect of these two indirect selection forces, van Doorn and Kirkpatrick (2007) identified two generic evolutionary outcomes. Either the masculinizing mutation was lost or it spread and replaced the ancestral sex-determining gene, leading to the loss of the ancestral Y chromosome and the establishment of a new sex chromosome pair. In some cases, selection could maintain a stable sex-chromosome polymorphism, causing a transition from single-locus GSD to a multifactorial sex determining system. However, this outcome required rather specific combinations of sexually antagonistic selection coefficients, as well as tight linkage. The same conclusions were later shown to apply to more complicated transitions between genetic sex-determining mechanisms (van Doorn and Kirkpatrick 2010), including the invasion of a novel sex-determining allele at the ancestral sex-determining locus, the establishment of a novel sex-determining gene located on the ancestral sex chromosomes, and heterogamety transitions involving an arbitrary number of sexually antagonistic loci or recessive deleterious alleles on the ancestral Y chromosome (Fig. 2). In all cases, fixation of the mutant gene occurred when the sexually antagonistic loci associated with the sex-determination modifier harbored more genetic variation, when their alleles had stronger sexually antagonistic fitness effects, or when they were more tightly linked to their nearby sex-determining gene than the sexually antagonistic genes on the ancestral sex chromosome.
Figure 2.
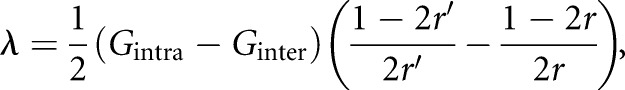 |
(1) |
One way to interpret these results is that evolution favors whichever GSD mechanism responds most effectively to predictors of sex-specific fitness at the time of sex determination (Leimar 2005). In the case of sex-chromosome turnovers, these predictors are represented by (groups of) sexually antagonistic loci segregating at different linkage groups. However, in a broader context, they may also include environmental factors (Charnov and Bull 1977) or variation in parental condition with sex-differential fitness effects (Trivers and Willard 1973; Langkilde and Shine 2005). In all of these cases, similar principles underlie the evolution of the sex-determining mechanism (van Doorn 2014). When individuals differ in their genetic background or experience different environmental conditions during development, and not all components of such variation affect males and females equally (e.g., Chippindale et al. 2001; Warner and Shine 2008), selection favors a conditional sex-determination strategy. Moreover, the optimal conditional strategy adaptively switches development into the male or female trajectory contingent on whichever genetic or environmental cue proves to be the most reliable predictor of sex-specific fitness (Charnov and Bull 1977; Rice 1986; van Dooren and Leimar 2003; Pen et al. 2010).
Simulations of sex-chromosome transitions with a large number of sexually antagonistic loci suggest that the few sexually antagonistic genes that are most tightly linked to a sex-determining gene have a decisive influence on the direction of sex-determination transitions. This would imply that the sexually antagonistic variation effectively responsible for sex-determination transitions segregates at a small subset of the sexually antagonistic loci. Therefore, even a small inversion that captures both a sex-determining mutation and a single fitness-relevant sexually antagonistic locus on an autosome, could conceivably hijack sex-determination from the ancestral sex chromosomes. Consistent with this suggestion, the sex-determining region on the recently evolved Y chromosome in the three-spined stickleback has experienced multiple inversions (Ross and Peichel 2008; Kirkpatrick 2010), and these chromosomal rearrangements appear to have occurred differently between the Japan Sea lineage (which has a novel sex-chromosome system) (Kitano et al. 2009) and its Pacific Ocean ancestor (Natri et al. 2013). However, the molecular data allow for multiple interpretations (Natri et al. 2013), and it remains unclear whether inversions have triggered the sex-chromosome shifts that have occurred in sticklebacks, or occurred after the sex-determining locus was established (as suggested by the traditional theory for sex-chromosome evolution) (Rice 1987; Charlesworth et al. 2005). This can, in principle, be tested by analyzing the patterns of neutral variation on the inversion and the neo-sex chromosome (Kirkpatrick et al. 2010; Guerrero et al. 2012a). Further empirical support for a role of sexual antagonism in sex-determination transitions comes from a growing number of cases of recently derived sex chromosomes that carry sexually selected loci (Kallman 1970; Wada et al. 1998; Lindholm and Breden 2002; Fernandez and Morris 2008; Kitano et al. 2009). A particularly interesting example is provided by a mutation in the Pax7 gene in lake Malawi cichlid fish, which causes an orange-blotched phenotype subject to sexually antagonistic selection. This mutation originated once in lake Malawi cichlids, and appears to have spread in close association with a feminizing allele that is epistatically dominant over the ancestral XX♀/XY♂ sex-determining system (Roberts et al. 2009).
Deleterious Mutations Accumulating on the Heterogametic Sex Chromosome
The theory reviewed in this section so far considers evolutionarily young sex chromosomes that have not yet stopped recombining. Older, heteromorphic sex chromosomes typically contain large regions in which recombination is suppressed (Rice 1987; Otto et al. 2011). The nonrecombining portion of the heterogametic sex chromosome tends to accumulate deleterious mutations, which can eventually lead to a complete degeneration of the chromosome (Charlesworth et al. 2005). Similar to sexually antagonistic selection, sex-chromosome degeneration does not affect both sexes equally, and it therefore generates qualitatively similar indirect selection pressures on the sex-determination system (Fig. 2B). A potential evolutionary response to sex-chromosome degeneration is that the old sex chromosome is replaced by a new one after a transposition of the sex-determining gene to an autosome pair, or the origin of a new autosomal sex-determining gene. Blaser et al. (2013) recently showed that such transitions are favored when the deleterious mutation load outweighs the disadvantage of losing beneficial sexually antagonistic alleles fixed on the decaying chromosome. This cost/benefit argument ignores the disruptive effect the transition might have on the expression of genes on the ancestral sex chromosomes, which would occur if specific sex-linked gene regulation (e.g., dosage compensation) had evolved. As these effects might often be substantial, sex-chromosome transitions involving heteromorphic sex chromosomes are expected to be rare. Besides, if such transitions occur, they are predicted to conserve the ancestral pattern of heterogamety (consistent with the pattern observed in some clades of vertebrates) (Woram et al. 2003; Miura 2007), because turnovers leading to the fixation of a degenerated sex chromosome are particularly likely to be associated with severe fitness costs.
The accumulation of sexually antagonistic variation on the sex chromosomes, the subsequent arrest of recombination and the resulting accumulation of deleterious mutations on the heterogametic sex chromosome can combine, over extended periods of time, to generate repeated sex-chromosome turnovers (Blaser et al. 2014). Several studies indicate that some autosomes are more likely than others to be co-opted as sex chromosomes in this process (Miura 2007; Graves and Peichel 2010; O’Meally et al. 2012; Brelsford et al. 2013), suggesting that the involvement of a chromosome pair in sex determination leaves a long-lasting signature. In particular, former sex chromosomes could be enriched for modifiers of recombination and potential targets of mutations with sexually antagonistic effects or effects on sex determination (Blaser et al. 2014; Beukeboom and Perrin 2014), all of which would increase the probability of recapturing the role of sex chromosome in a turnover event. A possible example of a chromosome that carries signatures of a previous role in sex determination is the dot chromosome of Drosophila, which has lost its ancestral function as an X chromosome after a sex-chromosome turnover in early drosophilid evolution, but which still has a minor feminizing role in sex determination and is subject to a specific mechanism of gene regulation similar to that of sex-linked genes (Vicoso and Bachtrog 2013).
GENETIC AND DEVELOPMENTAL ARCHITECTURE
As the structure of sex-determining pathways is being elucidated in a growing number of species, it becomes increasingly clear that the genetic and developmental architecture of a species shapes the selection pressures acting on its sex-determining genes. Understanding the interplay between adaptation and developmental mechanisms is a key challenge for future theoretical and empirical work on sex determination.
One obvious question to ask in this context is why sex determination appears to be labile in some taxa, but stable in others. For example, the evolutionary stability of sex determination in birds and mammals contrasts sharply with the high rate of transitions observed in some clades of cold-blooded vertebrates (Marín and Baker 1998; Kraak and Pen 2002; Ezaz et al. 2006). Part of this difference arises because the evolution of heteromorphic sex chromosomes constrains future sex-chromosome transitions, owing to the accumulation of sexually antagonistic genes on the sex chromosomes, the reduction of recombination rates in the vicinity of the master sex-determining gene, the loss of functional genes from the Y chromosome, the evolution of dosage compensation, and the translocation of genes essential for male fitness to the Y chromosome (Bull 1983; Rice 1987; Charlesworth et al. 2005). Once the sex chromosomes have become heteromorphic, XX males, XY females, or YY individuals of either sex, which arise when sex determination is hijacked by another chromosome pair, are likely to suffer from reduced fertility and/or viability, inhibiting the spread of new sex-determining genes (Bull and Charnov 1985; van Doorn and Kirkpatrick 2007; Mawaribuchi et al. 2012).
Sex-chromosome heteromorphism can thus explain why sex determination appears to be locked into its current state in birds and mammals. Yet, so far, the argument leaves unaddressed why sex determination should continue to be labile in taxa lacking heteromorphic sex chromosomes. The lower vertebrates, which generally have undifferentiated sex chromosomes, show evidence of frequent sex-chromosome transitions. For example, a phylogenetic analysis of genetic sex determination in teleost fishes found that eight of 26 families include both species with XY and species with ZW sex determination (Mank et al. 2006), and within the subfamily Salmoninae (including char, trout, and salmon), at least four different chromosomes determine sex in different species (Woram et al. 2003). A further example of frequent turnovers between male and female heterogamety is provided by a survey in amphibians, which identified eight transitions in a phylogenetically diverse sample of 63 species (Hillis and Green 1990). However, the same data also show that many species of fish and amphibians have not undergone a change in sex determination since more than several mya, and yet, almost none of these species show evidence of sex-chromosome differentiation (Stöck et al. 2011).
A plausible hypothesis for the maintenance of homomorphic sex chromosomes in lower vertebrates is that the sex chromosomes of cold-blooded vertebrates are prevented from degenerating because of rare recombination events prompted by a low rate of sex reversal (Perrin 2002). Because of the general effect of temperature on the development of ectotherms, such sex reversal events occur once in a while under natural conditions. Consistent with this idea, a recent analysis of neutral genetic variation on the sex chromosomes provides statistical support for a low rate of X-Y recombination in European tree frogs (Hyla spp.) (Guerrero et al. 2012b), a group of species with cryptic sex chromosomes and stable sex determination (Stöck et al. 2011). Furthermore, a theoretical study of mutation accumulation on the sex chromosomes shows that occasional sex reversal events provide sufficient opportunity for recombination to purge the Y chromosome from its deleterious mutation load (Grossen et al. 2012). The evolution of sex chromosomes (or the lack thereof in species with ESD) has several other consequences for the genetic architecture that can interact with the evolution of sex determination. Some of these are discussed by Ewert and Nelson (1991) (inbreeding), Reeve and Pfennig (2003) (sexual selection), and Kitano and Peichel (2012) (speciation).
A central challenge for future work is to integrate our understanding of the population genetics of sex-determination transitions with molecular data (Pomiankowski et al. 2004). Models that incorporate the available knowledge of sex-determination pathways are essential to complement the black-box approach of population-genetic analysis, to validate its implicit assumptions and to generate testable predictions. For example, combining evolutionary and systems biology modeling approaches can help clarify why it is so rare to observe “leaky” sex determination. Is it because selection generally disfavors (partially) random sex determination, or is a canalized all-or-nothing switch easier to implement, considering that the developmental decision triggered by the initial sex-determination signal must be stabilized and retained during development (Valenzuela 2008)? This issue is relevant for some sex-ratio selection models of sex-determination transitions, as these assume that developmental constraints prevent optimization of the sex ratio in species with GSD. A second possibility is to look into how sex determination interacts with downstream developmental processes, and how these interactions generate potential pleiotropic fitness effects. The results of such an analysis may validate the common assumption that sex determination can be studied in isolation from other developmental processes (which means that sex-determination modifiers can safely be assumed to have no other fitness effects). Alternatively, they could support the pleiotropic benefits hypothesis for the evolution of sex-determining genes.
Finally, realistic models of evolving sex-determining pathways are needed to clarify how selection and molecular mechanisms interact to generate the variation of evolutionary rates between genes at different positions in the sex-determining cascade (MacCarthy et al. 2010). Despite that population genetic models tend to ignore the complexity of development, they do offer suggestions for why the flexibility of sex determination may predominantly rely on the recruitment of novel upstream regulators to the pathway. In particular, it seems reasonable to suppose that a novel upstream regulator that interacts directly with the original master switch (for example, by mimicking or blocking the primary sex-determination signal) can reverse the outcome of the original switch for a certain set of environmental or genetic conditions and, in this way, act as an epistatically dominant modifier of sex determination. In contrast, regulators interacting with downstream sex-determination genes would perhaps interfere with how the sex-determination signal is processed, which would be likely to have negative pleiotropic effects or result in a breakdown of canalization (Uller and Helanterä 2011). Selection against leaky sex determination may therefore be one of the factors preventing the spread of downstream sex-determination modifiers. A thorough test of this hypothesis requires investigating how genes in the sex-determination pathway coevolve. There is growing evidence that such coevolution is occurring (Haag and Ackerman 2005; Stothard and Pilgrim 2006), and that it is associated with rapid evolution of sex-determining genes, often without sex-determination transitions or structural pathway rearrangements (Ferris et al. 1997; O’Neill and O’Neill 1999; Chandler et al. 2012). The evolutionary flexibility of sex determination may therefore be even higher than is currently appreciated.
ACKNOWLEDGMENTS
The author thanks Michael Wade, Sergey Gavrilets, and an anonymous referee for their comments on this manuscript, and gratefully acknowledges financial support from the European Research Council (ERC Starting grant 30955) and the Netherlands Organization for Scientific Research (NWO VIDI grant 864.11.012).
Footnotes
Editors: William R. Rice and Sergey Gavrilets
Additional Perspectives on The Genetics and Biology of Sexual Conflict available at www.cshperspectives.org
REFERENCES
- Ainsworth C 2000. Boys and girls come out to play: The molecular biology of dioecious plants. Ann Bot 86: 211–221 [Google Scholar]
- Badyaev AV 2002. Growing apart: An ontogenetic perspective on the evolution of sexual size dimorphism. Trends Ecol Evol 17: 369–378 [Google Scholar]
- Barnes RSK, Calow P, Olive PJW, Golding DW, Spicer JI 2001. The Invertebrates: A Synthesis. Blackwell Science, Oxford [Google Scholar]
- Barton NH 2000. Genetic hitchhiking. Phil Trans R Soc B 355: 1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom LW, Perrin N 2014. Transitions among sex-determination systems. In Evolution of sex determination (ed. Beukeboom LW, Perrin N). Oxford University Press, Oxford [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N 2013. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67: 635–645 [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, Perrin N 2014. Sex-chromosome turnovers: The hot-potato model. Am Nat 183: 140–146 [DOI] [PubMed] [Google Scholar]
- Brelsford A, Stöck M, Betoo-Colliard C, Dubey S, Dufresnes C, Jourdan-Pineau H, Rodrigues N, Savary R, Sermier R, Perrin N 2013. Homologous sex chromosomes in three deeply divergent anuran species. Evolution 67: 2434–2440 [DOI] [PubMed] [Google Scholar]
- Brommer JE, Kirkpatrick M, Qvarnström A, Gustafsson L 2007. The intersexual genetic correlation for lifetime fitness in the wild and its implications for sexual selection. PLoS ONE 2: e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ 1981. Evolution of environmental sex determination from genotypic sex determination. Heredity 47: 173–184 [Google Scholar]
- Bull JJ 1983. Evolution of sex determining mechanisms. Benjamin Cummings, Menlo Park, CA [Google Scholar]
- Bull JJ, Charnov EL 1977. Changes in the heterogametic mechanism of sex determination. Heredity 39: 1–14 [DOI] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL 1985. On irreversible evolution. Evolution 39: 1149–1155 [DOI] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL 1988. How fundamental are Fisherian sex ratios? Oxford Surv Evol Biol 5: 98–135 [Google Scholar]
- Bulmer MG, Bull JJ 1982. Models of polygenic sex determination and sex ratio control. Evolution 36: 13–26 [DOI] [PubMed] [Google Scholar]
- Caubet Y, Hatcher MJ, Mocquard J-P, Rigaud T 2000. Genetic conflict and changes in heterogametic mechanisms of sex determination. J Evol Biol 13: 766–777 [Google Scholar]
- Chandler CH, Chadderdon GE, Phillips PC, Dworkin I, Janzen FJ 2012. Experimental evolution of the Caenorhabditis elegans sex determination pathway. Evolution 66: 82–93 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res Camb 35: 205–214 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Dempsey ND 2001. A model of the evolution of the unusual sex chromosome system of Microtus oregoni. Heredity 86: 387–394 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128 [DOI] [PubMed] [Google Scholar]
- Charnov EL 1982. The theory of sex allocation. Monogr Popul Biol 18: 1–355 [PubMed] [Google Scholar]
- Charnov EL, Bull J 1977. When is sex environmentally determined? Nature 266: 828–830 [DOI] [PubMed] [Google Scholar]
- Charnov EL, Bull JJ 1989. The primary sex ratio under environmental sex determination. J Theor Biol 139: 431–436 [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Gibson JR, Rice WR 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci 98: 1671–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG 1988. The evolution of the Y chromosome with X-Y recombination. Genetics 119: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha C, Scott MJ 2009. Sexual development in Lucilia cuprina (Diptera, Calliphoridae) is controlled by the transformer gene. Genetics 182: 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet 8: 689–698 [DOI] [PubMed] [Google Scholar]
- Ewert MA, Nelson CE 1991. Sex determination in turtles: Diverse patterns and some possible adaptive values. Copeia 1991: 50–69 [Google Scholar]
- Ezaz T, Stiglec R, Veyrunes F, Marshall Graves JA 2006. Relationships between vertebrate ZW and XY sex chromosome systems. Curr Biol 16: R736–R743 [DOI] [PubMed] [Google Scholar]
- Feldmeyer B, Kozielska M, Kuijper B, Weissing FJ, Beukeboom LW, Pen I 2008. Climatic variation and the geographical distribution of sex-determining mechanisms in the housefly. Evol Ecol Res 10: 797–809 [Google Scholar]
- Fernandez AA, Morris MR 2008. Mate choice for more melanin as a mechanism to maintain a functional oncogene. Proc Natl Acad Sci 105: 13503–13507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Pavlovic C, Fabry S, Goodenough UW 1997. Rapid evolution of sex-related genes in Clamydomonas. Proc Natl Acad Sci 94: 8634–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster K, Coulson T, Sheldon BC, Pemberton JM, Clutton-Brock TH, Kruuk LEB 2007. Sexually antagonistic genetic variation for fitness in red deer. Nature 447: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Freedberg S, Wade MJ 2001. Cultural inheritance as a mechanism for population sex-ratio bias in reptiles. Evolution 55: 1049–1055 [DOI] [PubMed] [Google Scholar]
- Gavrilets S 1997. Evolution and speciation on holey adaptive landscapes. Trends Ecol Evol 12: 307–312 [DOI] [PubMed] [Google Scholar]
- Gempe T, Beye M 2011. Function and evolution of sex-determination mechanisms, genes and pathways in insects. Bioessays 33: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuverink E, Beukeboom LW 2014. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex Dev 8: 38–49 [DOI] [PubMed] [Google Scholar]
- Graves JAM, Peichel CL 2010. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol 11: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossen C, Neuenschwander S, Perrin N 2012. The evolution of XY recombination: Sexually antagonistic selection versus deleterious mutation load. Evolution 66: 3155–3166 [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Rousset F, Kirkpatrick M 2012a. Coalescent patterns for chromosomal inversions in divergent populations. Phil Trans R Soc B 367: 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Kirkpatrick M, Perrin N 2012b. Cryptic recombination in the ever-young sex chromosomes of Hylid frogs. J Evol Biol 25: 1947–1954 [DOI] [PubMed] [Google Scholar]
- Haag ES, Ackerman AD 2005. Intraspecific variation in fem-3 and tra-2, two rapidly coevolving nematode sex-determining genes. Gene 349: 35–42 [DOI] [PubMed] [Google Scholar]
- Haag ES, Doty AV 2005. Sex determination across evolution: Connecting the dots. PLoS Biol 3: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann M, Gempe T, Schiott M, Nunes-Silva CG, Otte M, Beye M 2008. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454: 519–522 [DOI] [PubMed] [Google Scholar]
- Hediger M, Henggeler C, Meier N, Perez R, Saccone G, Bopp D 2010. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 184: 155–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Sun S, James TY 2013. Evolution of fungal sexual reproduction. Mycologia 105: 1–27 [DOI] [PubMed] [Google Scholar]
- Hillis DM, Green DM 1990. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J Evol Biol 3: 49–64 [Google Scholar]
- Hodgkin J 2002. The remarkable ubiquity of DM domain factors as regulators of sexual phenotype: Ancestry or aptitude? Genes Dev 16: 2322–2326 [DOI] [PubMed] [Google Scholar]
- Jaenike J 2001. Sex chromosome meiotic drive. Annu Rev Ecol Syst 32: 25–49 [Google Scholar]
- Janousek B, Mrackova M 2010. Sex chromosomes and sex-determination pathway dynamics in plant and animal models. Biol J Linn Soc 100: 737–752 [Google Scholar]
- Janzen FJ, Phillips PC 2006. Exploring the evolution of environmental sex determination, especially in reptiles. J Evol Biol 19: 1775–1784 [DOI] [PubMed] [Google Scholar]
- Jordan CY, Charlesworth D 2011. The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66: 505–516 [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Tomancak P 2012. The evolution of early animal embryos: Conservation or divergence? Trends Ecol Evol 27: 385–393 [DOI] [PubMed] [Google Scholar]
- Kallman KD 1970. Different genetic basis of identical pigment patterns in two populations of platyfish, Xiphophorus maculatus. Copeia 1970: 472–487 [Google Scholar]
- Karlin S, Lessard S 1984. On the optimal sex-ratio: A stability analysis based on a characterization for one-locus multiallele viability models. J Math Biol 20: 15–38 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M 2010. How and why chromosome inversions evolve. PLoS Biol 8: e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Johnson T, Barton N 2002. General models of multilocus evolution. Genetics 161: 1727–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Guerrero RF, Scarpino SV 2010. Patterns of neutral genetic variation on recombining sex chromosomes. Genetics 184: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Peichel CL 2012. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fish 94: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielska M, Pen I, Beukeboom LW, Weissing FJ 2006. Sex ratio selection and multi-factorial sex determination in the housefly: A dynamic model. J Evol Biol 19: 879–888 [DOI] [PubMed] [Google Scholar]
- Kozielska M, Weissing FJ, Beukeboom LW, Pen I 2010. Segregation distortion and the evolution of sex-determining mechanisms. Heredity 104: 100–112 [DOI] [PubMed] [Google Scholar]
- Kraak SBM, de Looze EMA 1993. A new hypothesis on the evolution of sex determination in vertebrates: Big females ZW, big males XY. Neth J Zool 43: 260–273 [Google Scholar]
- Kraak SBM, Pen I 2002. Sex-determining mechanisms in vertebrates. In Sex ratios: Concepts and research methods (ed. Hardy ICW), pp. 158–177 Cambridge University Press, Cambridge [Google Scholar]
- Kraak SBM, Pen I, Weissing FJ 2000. “Joint evolution of environmental and genetic sex determination, in Pen I: Sex allocation in a life history context.” PhD thesis, University of Groningen, Groningen, Netherlands, pp. 149–160 [Google Scholar]
- Krackow S 2002. Why parental sex ratio manipulation is rare in higher vertebrates. Ethology 108: 1041–1056 [Google Scholar]
- Lagos D, Koukidou M, Savakis C, Komitopoulou K 2007. The transformer gene in Bactrocera oleae: The genetic switch that determines its sex fate. Insect Mol Biol 16: 221–230 [DOI] [PubMed] [Google Scholar]
- Langkilde T, Shine R 2005. Different optimal offspring sizes for sons versus daughters may favor the evolution of temperature-dependent sex determination in viviparous lizards. Evolution 59: 2275–2280 [PubMed] [Google Scholar]
- Leimar O 2005. The evolution of phenotypic polymorphism: Randomized strategies versus evolutionary branching. Am Nat 165: 669–681 [DOI] [PubMed] [Google Scholar]
- Lindholm A, Breden F 2002. Sex chromosomes and sexual selection in poeciliid fishes. Am Nat 160: S214–S224 [DOI] [PubMed] [Google Scholar]
- MacCarthy T, Seymour RM, Pomiankowski A 2010. Differential regulation drives plasticity in sex determination gene networks. BMC Evol Biol 10: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Promislow DEL, Avise JC 2006. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol J Linn Soc 87: 83–93 [Google Scholar]
- Marín I, Baker BS 1998. The evolutionary dynamics of sex determination. Science 281: 1990–1994 [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet 42: 565–586 [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S, Yoshimoto S, Ohashi S, Takamatsu N, Ito M 2012. Molecular evolution of vertebrate sex-determining genes. Chromosome Res 20: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Hurst LD 1996. Genetic conflicts and the paradox of sex determination: Three paths to the evolution of female intersexuality in a mammal. J Theor Biol 179: 199–211 [DOI] [PubMed] [Google Scholar]
- Miura I 2007. An evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex Dev 1: 323–331 [DOI] [PubMed] [Google Scholar]
- Natri HM, Shikano T, Merilä J 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol 30: 1131–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthiger R, Steinmann-Zwicky M 1985. A single principle for sex determination in insects. Cold Spring Harb Symp Quant Biol 50: 615–621 [DOI] [PubMed] [Google Scholar]
- O’Meally D, Ezaz T, Georges A, Sarre SD, Graves JAM 2012. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res 20: 7–19 [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, O’Neill RJW 1999. Whatever happened to SRY? Cell Mol Life Sci 56: 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzack SH, Sohn JJ, Kallman KD, Levin SA, Johnston R 1980. Maintenance of the three sex chromosome polymorphism in the platyfish Xiphophorus maculatus. Evolution 34: 663–672 [DOI] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF 2011. About PAR: The distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet 27: 358–367 [DOI] [PubMed] [Google Scholar]
- Parker GA 1979. Sexual selection and sexual conflict. In Sexual selection and reproductive competition in insects (ed. Blum MS, Blum NA), pp. 123–166 Academic, London [Google Scholar]
- Pen I, Uller T, Feldmeyer B, Harts A, While GM, Wapstra E 2010. Climate-driven population divergence in sex-determining systems. Nature 468: 436–438 [DOI] [PubMed] [Google Scholar]
- Pennell TM, Morrow EH 2013. Two sexes, one genome: The evolutionary dynamics of intralocus sexual conflict. Ecol Evol 3: 1819–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin N 2009. Sex reversal: A fountain of youth for sex chromosomes? Evolution 63: 3043–3049 [DOI] [PubMed] [Google Scholar]
- Pomiankowski A, Nöthiger R, Wilkins A 2004. The evolution of the Drosophila sex-determination pathway. Genetics 166: 1761–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391: 691–695 [DOI] [PubMed] [Google Scholar]
- Reeve HK, Pfennig DW 2003. Genetic basis for showy males: Are some genetic systems especially conducive to sexual selection? Proc Natl Acad Sci 100: 1089–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742 [DOI] [PubMed] [Google Scholar]
- Rice WR 1986. On the instability of polygenic sex determination: The effect of sex-specific selection. Evolution 40: 633–639 [DOI] [PubMed] [Google Scholar]
- Rice WR 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914 [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD 2009. Sexual conflict resolved by invasion of a novel sex determiner in lake Malawi cichlid fishes. Science 326: 998–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Peichel CL 2008. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics 179: 2173–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MF, Milano A, Salvemini M, Eirín-López JM, Perondini ALP, Selivon D, Polito C, Saccone G, Sánchez L 2007. The gene transformer of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. PLoS ONE 2: e1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L 2008. Sex-determining mechanisms in insects. Int J Dev Biol 52: 837–856 [DOI] [PubMed] [Google Scholar]
- Schütt C, Nöthiger R 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127: 667–677 [DOI] [PubMed] [Google Scholar]
- Scudo FM 1967. Criteria for the analysis of multifactorial sex determination. Monitore Zool Ital 1: 1–21 [Google Scholar]
- Shine R 1999. Why is sex determined by nest temperature in many reptiles? Trends Ecol Evol 14: 186–189 [DOI] [PubMed] [Google Scholar]
- Shukla JN, Nagaraju J 2010. Doublesex: A conserved downstream gene controlled by diverse upstream regulators. J Genet 89: 341–356 [DOI] [PubMed] [Google Scholar]
- Shukla JN, Palli SR 2012. Sex determination in beetles: Production of all male progeny by parental RNAi knockdown of transformer. Sci Rep 2: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AD, Pischedda A, Rice WR 2010. Resolving intralocus sexual conflict: Genetic mechanisms and time frame. J Hered 101: S94–S99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck M, Horn A, Grossen C, Lindtke D, Sermier R, Betto-Colliard C, Dufresnes C, Bonjour E, Dumas Z, Luquet E, et al. 2011. Ever-young sex chromosomes in European tree frogs. PLoS Biol 9: e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P, Pilgrim D 2003. Sex-determination gene and pathway evolution in nematodes. Bioessays 25: 221–231 [DOI] [PubMed] [Google Scholar]
- Stothard P, Pilgrim D 2006. Conspecific and interspecific interactions between the FEM-2 and the FEM-3 sex-determining proteins despite rapid sequence divergence. J Mol Evol 62: 281–291 [DOI] [PubMed] [Google Scholar]
- Trivers RL, Willard DE 1973. Natural selection of parental ability to vary the sex ratio of offspring. Science 179: 90–92 [DOI] [PubMed] [Google Scholar]
- Uller T, Helanterä H 2011. From the origin of sex-determining factors to the evolution of sex-determining systems. Q Rev Biol 86: 163–180 [DOI] [PubMed] [Google Scholar]
- Uller T, Pen I, Wapstra E, Beukeboom LW, Komdeur J 2007. The evolution of sex ratios and sex-determining systems. Trends Ecol Evol 22: 292–297 [DOI] [PubMed] [Google Scholar]
- Umen JG 2011. Evolution of sex and mating loci: An expanded view from Volvocine algae. Curr Opin Microbiol 14: 634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela N 2008. Sexual development and the evolution of sex determination. Sex Dev 2: 64–72 [DOI] [PubMed] [Google Scholar]
- van Dooren TJM, Leimar O 2003. The evolution of environmental and genetic sex determination in fluctuating environments. Evolution 57: 2667–2677 [PubMed] [Google Scholar]
- van Doorn GS 2009. Intralocus sexual conflict. Ann NY Acad Sci 1168: 52–71 [DOI] [PubMed] [Google Scholar]
- van Doorn GS 2014. Evolutionary transitions between sex-determining mechanisms: A review of theory. Sex Dev 8: 7–19 [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912 [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M 2010. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186: 629–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst EC, Beukeboom LW, van de Zande L 2010a. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328: 620–623 [DOI] [PubMed] [Google Scholar]
- Verhulst EC, van de Zande L, Beukeboom LW 2010b. Insect sex determination: It all evolves around transformer. Curr Opin Genet Dev 20: 376–383 [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499: 332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier S, Lande R, van Alphen JJM, Seehausen O 2007. Invasion and fixation of sex-reversal genes. J Evol Biol 20: 913–920 [DOI] [PubMed] [Google Scholar]
- Wada H, Shimada A, Fukamachi S, Naruse K, Shima A 1998. Sex-linked inheritance of the lf locus in the medaka fish (Oryzias latipes). Zool Sci 15: 123–126 [DOI] [PubMed] [Google Scholar]
- Warner DA, Shine R 2008. The adaptive significance of temperature-dependent sex determination in a reptile. Nature 451: 566–568 [DOI] [PubMed] [Google Scholar]
- Werren JH, Beukeboom LW 1998. Sex determination, sex ratios, and genetic conflict. Annu Rev Ecol Syst 29: 233–261 [Google Scholar]
- Werren JH, Hatcher MJ, Godfray HCJ 2002. Maternal-offspring conflict leads to the evolution of dominant zygotic sex determination. Heredity 88: 102–111 [DOI] [PubMed] [Google Scholar]
- Wilkins AS 1995. Moving up the hierarchy: A hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17: 71–77 [DOI] [PubMed] [Google Scholar]
- Woram RA, Gharbi K, Sakamoto T, Hoyheim B, Holm LE, Naish K, McGowan C, Ferguson MM, Phillips RB, Stein J, et al. 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res 13: 272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D 2001. Establishing sexual dimorphism: Conservation amidst diversity? Nat Rev Genet 2: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, Kent SB, Weiss MA 2000. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev 14: 1750–1764 [PMC free article] [PubMed] [Google Scholar]




