Abstract
Round cell sarcomas harboring CIC-DUX4 fusions have recently been described as highly aggressive soft tissue tumors of children and young adults. Due to partial morphologic and immunohistochemical overlap with Ewing sarcoma (ES), CIC-DUX4-positive tumors have generally been classified as Ewing sarcoma-like and managed similarly, however, a systematic comparison at the molecular and immunohistochemical levels between these two groups has not yet been conducted. Based on an initial observation that CIC-DUX4-positive tumors show nuclear immunoreactivity for WT1 and ETS transcription factors, FLI1 and ERG, we performed a detailed immunohistochemical and molecular analysis including these markers, to further investigate the relationship between CIC-DUX4 tumors and ES. The study group included 21 CIC-DUX4-positive sarcomas and 20 EWSR1-rearranged ES. Immunohistochemically, CIC-DUX4 sarcomas showed membranous CD99 positivity in 18 (86%) cases, but only 5 (24%) with a diffuse pattern, while WT1 and FLI1 were strongly positive in all cases. ERG was positive in 18% of cases. All ES expressed CD99 and FLI1, while ERG positivity was only seen in EWSR1-ERG fusion positive ES. WT1 was negative in all ES. Expression profiling validated by q-PCR revealed a distinct gene signature associated with CIC-DUX4 fusion, with upregulation of ETS transcription factors (ETV4, ETV1 and ETV5) and WT1, among top overexpressed genes compared to ES, other sarcomas and normal tissue. In conclusion, the distinct gene signature and immunoprofile of CIC-DUX4 sarcomas suggest a distinct pathogenesis from ES. The consistent WT1 expression may provide a useful clue in the diagnosis in the context of round cell sarcomas negative for EWSR1-rearrangement.
Keywords: small blue round cell sarcoma, CIC-DUX4, WT1, Ewing Sarcoma
INTRODUCTION
According to the current WHO classification (Fletcher et al., 2013), round cell undifferentiated soft tissue sarcomas (round cell USTS) lack consistent genetic abnormalities and are characterized by relatively monotonous round to ovoid cytomorphology, with a high nuclear to cytoplasmic ratio and no distinct line of differentiation. As they most often resemble Ewing sarcoma (ES), for practical and treatment purposes, round cell USTS have been classified as ‘Ewing sarcoma-like’ or lumped within the Ewing sarcoma family of tumors. However, in contrast to classic ES, round cell USTS lack the pathognomonic translocations involving the EWSR1 gene on chromosome 22 fused to a member of the ETS transcription factor family, namely FLI1 (Delattre et al., 1992), ERG (Zucman et al., 1993; Sorensen et al., 1994), ETV1 (Jeon et al., 1995), ETV4 (Urano et al., 1996) or FEV (Peter et al., 1997), or similar translocations involving the FUS gene (FUS-ERG or FUS-FEV) (Shing et al., 2003), (Ng et al., 2007).
We and others recently described a series of USTS with primitive round cell morphology harboring a novel gene fusion, CIC-DUX4, resulting from either t(4;19)(q35;q13) or a t(10;19)(q26;q13) (Kawamura-Saito et al., 2006; Yoshimoto et al., 2009; Italiano et al., 2012; Graham et al., 2012; Choi et al., 2013). The genes involved in the fusion are CIC, a transcriptional repressor in chromosome band 19q13.1 and DUX4, a double homeobox transcription factor, located in either 4q35 or 10q26.3. Due to its aggressive clinical course and potential therapeutic implications, recognition of this recently defined subgroup of round cell USTS is important. However, the lack of any lineage-specific markers can make diagnosis and classification difficult. Focal and weak expression of CD99 has been the only consistently reported immunohistochemical finding in these tumors (Kawamura-Saito et al., 2006; Italiano et al., 2012; Choi et al., 2013). Molecular confirmation by either FISH or RT-PCR currently represents the gold standard to establish a definitive diagnosis of CIC-DUX4 round cell sarcomas, yet these tests are presently offered only at few centers.
As no systematic study has so far attempted to investigate the pathogenetic relationship between CIC-DUX4-positive tumors and ES, we carried out a detailed comparative immunohistochemical and molecular analysis using global gene expression profiling and validated by quantitative RT-PCR techniques.
MATERIALS AND METHODS
The consultation files of the corresponding authors (CRA, CDF) and the Surgical Pathology files of Memorial Sloan Kettering Cancer Center, New York, NY (CRA) and the Technische Universität München, Germany (KS) were searched for diagnosis of small blue round cell tumors/sarcomas and Ewing sarcomas, with tissue available for further immunohistochemical and molecular analysis, between 2001–2012. A total of 33 round cell USTS lacking EWSR1 gene rearrangements (or other common sarcoma-associated translocations) were identified. Hematoxylin and eosin-stained slides and previously performed immunohistochemical stains were reviewed in all cases. Among the 33 round cell USTS, 21 (64%) cases harbored CIC gene rearrangements by FISH (Table 1), cases #1–6 being previously reported (Italiano et al., 2012). In addition, a comparison group of 20 Ewing sarcomas (ES) was used for immunohistochemical studies and validation q-PCR. The diagnosis of ES was confirmed by reverse transcriptase polymerase chain reaction (RT-PCR) and sequencing analysis in 11 cases, showing a EWSR1-FLI1 in 9 cases and a EWSR1-ERG fusion transcript in 3 cases. Additional 9 cases showed EWSR1 gene rearrangement by fluorescence in situ hybridization (FISH). One case was confirmed by both methodologies. All cases were handled in accordance with the ethical rules of the respective institutions.
Table 1.
Clinical and Immunohistochemical Findings of CIC-rearranged Round Cell Sarcoma
| Case# | Age/ Gender | Location | FISH for DUX4 on 4q35/10q26 | CD99 | Fli1 | ERG | WT1 |
|---|---|---|---|---|---|---|---|
| 1 | 45/M | Back | t(10;19) CIC-DUX4 | 1+ | 4+ | Neg | 4+(N+C) |
| 2 | 49/M | Arm | t(10;19) CIC-DUX4 | 3+ | 4+ | Neg | 4+(N+C) |
| 3 | 30/F | Mandible | t(4;19) CIC-DUX4 | 3+ | 4+ | Neg | 4+(N+C) |
| 4 | 29/F | Arm | t(10;19) CIC-DUX4 | 1+ | ND | ND | 2+(C) |
| 5 | 28/M | Thigh | t(4;19) CIC-DUX4 | 3+ | ND | ND | 4+(N+C) |
| 6 | 26/M | Arm | t(10;19) CIC-DUX4 | 3+ | ND | ND | ND |
| 7 | 36/F | Paraspinal | t(4;19) CIC-DUX4 | 1+ | 3+ | Neg | 2+(N) |
| 8 | 19/F | Stomach | t(4;19) CIC-DUX4 | 3+ | 4+ | Neg | 4+(N) |
| 9 | 25/M | Neck | t(4;19) CIC-DUX4 | 1+ | ND | ND | 4+(N+C) |
| 10 | 51/F | Ankle | Neg | 1+ | ND | 3+ | 4+(N+C) |
| 11 | 14/F | Thigh | Neg | Neg | 4+ | Neg | 4+(N+C) |
| 12 | 27/F | Brain mets | Neg | 1+ | 4+ | Neg | 4+(N+C) |
| 13 | 50/F | Peritoneum | t(10;19) CIC-DUX4 | 1+ | ND | ND | 4+(N+C) |
| 14 | 33/F | Knee | t(10;19) CIC-DUX4 | Neg | ND | ND | 4+(N+C) |
| 15 | 36/F | Shoulder | t(4;19) CIC-DUX4 | 1+ | ND | ND | 4+(N+C) |
| 16 | 13/F | Paravertebral | Neg | 1+ | 4+ | 1+ | 4+(N+C) |
| 17 | 25/F | Paravertebral | t(4;19) CIC-DUX4 | 1+ | 4+ | Neg | 4+(N+C) |
| 18 | 47/M | Clavicular | t(4;19) CIC-DUX4 | 1+ | ND | ND | 4+(N+C) |
| 19 | 37/M | Peri-prostatic | t(4;19) CIC-DUX4 | 1+ | ND | Neg | 4+(N+C) |
| 20 | 6/M | Buttock | Neg* | 1+ | ND | ND | 3+(N) |
| 21 | 6/M | Scalp | Neg | Neg | ND | ND | 3+(N) |
M, male; F, female; N, nuclear, C, cytoplasmic; ND, not done, Neg, negative. Cases previously reported in ((Italiano et al., 2012): their case # 2,6,8,9,11,12); cases 1–3 and 6,7 were studied on U133A chip;
case showed a karyotype of t(2;10;19)(q35;p14;q13)
Interphase FISH
FISH analysis was performed on interphase nuclei from paraffin-embedded 4 μm sections using bacterial artificial chromosomes (BAC clones), flanking CIC in 19q13 and DUX4 in 4q35 and 10q26.3 as previously described (Italiano et al., 2012). Two hundred tumor nuclei were evaluated using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). A cut-off of >20% nuclei showing a break-apart signal was considered to be positive for rearrangement. Nuclei with incomplete set of signals were omitted from the score.
Immunohistochemistry
Immunohistochemistry was performed on whole tissue sections of the 21 CIC-DUX4 – positive round cell sarcoma cases and in the 20 cases of Ewing sarcoma using the following antibodies: an anti-CD99 mouse monoclonal antibody (1:150; clone O13, Covance), an anti-FLI1 mouse monoclonal antibody raised against a bacterially expressed FLI1 Ets domain fusion protein (1:100; G146-222, BD Bioscience), an anti-ERG rabbit monoclonal antibody raised against a synthetic peptide corresponding to the carboxy-terminal end of ERG (1:2000; EPR3864(2), Epitomics), and an anti-WT-1 mouse monoclonal antibody (1:50; 6F-H2, DAKO; which is directed at the N-terminal and reacts with all isoforms of the full-length WT1). Immunodetection was performed using the Envision Plus detection system (Dako). The extent of immunoreactivity was graded according to the percentage of positive tumor cells (0 <5% or no staining; 1+ 5– 25%; 2+ 26–50%; 3+ 51–75%, 4+ 76%–100%). The intensity of staining was graded as weak, moderate or strong.
Gene Expression Profiling
Total RNA from fresh-frozen tissue extracted from 5 CIC-DUX4 positive tumors, two with t(4;19)(q35;q13) and three with t(10;19)(q26;q13) translocation, was labeled and hybridized onto an Affymetrix U133A chip (22,000 transcripts), as previously described (Antonescu et al., 2004). Their expression was compared to a previously published, well-characterized dataset of 29 soft tissue sarcomas (Segal et al., 2003; Hadju et al., 2010) and 8 normal tissues.
In parallel, we investigated 5 EWSR1-FLI1 fusion positive ES tumors and 21 normal tissues using the Affymetrix Human Gene 1.0 ST arrays (32,000 transcripts); microarray data been previously deposited at the gene expression omnibus (GSE45544). The transcriptional signature was further compared to a comprehensive gene expression meta-analysis of ES tumors (Hancock and Lessnick, 2008). The overlapping upregulated and downregulated 95 genes from the two analyses were selected and subsequently compared to the CIC-DUX4 signature (Fig. 3).
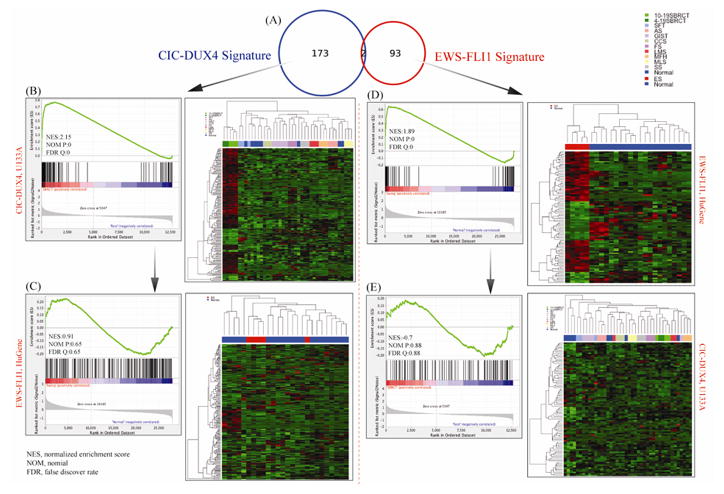
Fig. 3. Distinct Transcriptional Signature of CIC-DUX4-positive tumors compared to EWSR1-FLi1 Ewing sarcoma (ES) cases.
A. Venn diagram showing minimal genomic overlap between the two groups investigated (n=2 genes). The 175 gene list was obtained by comparing 5 CIC-DUX4 tumors with 29 soft tissue sarcomas and 8 normal tissues on Affymetrix U133A chip (1.4 FC; FDR 0.05 p-value). The ES 95 gene-signature was obtained by overlapping the 854 differentially expressed genes in the 5 ES tumors (Affymetrix Hu-Gene; 1.3FC; FDR 0.1 p-value) with a published meta-analysis of ES. B. The 175 gene-signature was applied for hierarchical clustering showing a distinct genomic group of CIC-DUX4-tumors from all the other control samples. The GSEA confirms highly ranked genes, with high normalized enrichment score (NES) values. C. In contrast, applying the 175 CIC-DUX4 gene-signature to the ES and controls resulted in poor clustering expression patterns and low GSEA scores. D. Hierarchical clustering using the robust 95 ES gene-list shows a well-defined ES genomic cluster from all the normal controls, with high NES scores on GSEA. E. In contrast, using the same 95 gene-signature on the CIC-DUX4 tumors and controls shows an ambiguous expression pattern on clustering and low NEM score.
For the data analysis, RMA-normalization was performed, including background correlation, quantile normalization, and median polish summary method (Richter et al., 2009). Subsequent analysis was carried out with signal intensities that were log2-transformed to remove biases based on signal expression values (Hauer et al., 2013). Statistical t-test and FDR were performed to identify differentially expressed gene list, and subsequent hierarchical clustering was accomplished by heatmap function in R and bioconductor (Antonescu et al., 2009). Annotation files for Affymetrix U133A and Human Gene arrays were obtained from Affymetrix website (http://www.affymetrix.com/support/technical/annotationfilesmain.affx) and analyzed using PERL script in order to match the probe IDs. Subsequently, gene set enrichment analysis (GSEA) was performed for investigating statistical associations between variable gene sets and phenotype of interest (Wang and Cairns, 2013). The algorithm in GSEA calculates the enrichment score, with corresponding significance level based on permutation tests (empirical p-values and FDRs controlling global false positives). Each sample group was permutated 1,000 times to yield statistical significances.
Real-Time Quantitative RT-PCR (qPCR)
Total RNA was extracted from macrodissected formalin-fixed, paraffin-embedded tumor tissue using the High-Pure RNA Paraffin Kit reagent according to the manufacturer’s instructions (Roche Diagnostics, Penzberg, Germany). One microgram of RNA was reverse transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen) and 250 ng of random hexamers (Roche Diagnostics) in a final volume of 20 μl. Gene expression was assessed using RealTime ready single assays (Roche Diagnostics) for the target genes ETV1 (ID 140599), ETV4 (ID 137042), ETV5 (ID 127143) and the housekeeping gene ACTB (ID 101125). Quantitative RT-PCR was performed in duplicate with the LightCycler 480 Instrument using LightCycler 480 Probes Master (Roche Diagnostics) and 10 ng of cDNA per well. Relative mRNA expression was calculated by the ΔΔCt method using the LightCycler 480 Software with normalization to ACTB as reference gene.
RESULTS
Clinicopathologic and Molecular Characteristics of CIC-DUX4-Fusion-Positive Round Cell Sarcomas
Among the 33 round cell USTS negative for EWSR1-associated gene fusions, a total of 21 tumors were positive for CIC gene rearrangements by FISH analysis (Table 1). Subsequent FISH analysis confirmed the fusion of CIC with the DUX4 gene in either chromosome band 4q35 in 9 cases or in 10q26.3 in 6 cases. In 6 cases there was no abnormality identified in the two DUX4 genes, suggesting alternative fusions events. In one DUX4 negative tumor, the outside karyotype found a three-way translocation, t(2;10;19)(q35;p14;q13); the break on 2q35 being outside the PAX3 locus by FISH (not shown). The cases affected 9 males and 12 females, with a mean and median age at diagnosis of 30 and 29, respectively (range 6–51 years). Five patients (24%) were younger than 20 years. Nineteen cases (90%) arose in somatic soft tissue, one tumor originated in the stomach and one patient presented with brain metastases, the primary tumor site being unknown. The anatomic distribution included upper extremity (3), lower extremity (5), trunk (9), head and neck (2) and stomach (1).
Microscopic Features
On microscopic examination, the CIC-DUX4-positive tumors were characterized by a diffuse or vaguely nodular growth of undifferentiated cells, arranged in solid sheets separated by thin fibrous septa or areas of necrosis (Fig. 1A, B). The majority of tumors exhibited infiltrative growth into adjacent anatomic structures, such as skeletal muscle or adipose tissue. Geographic tumor necrosis was common and prominent in the majority of cases, (15/21), with frequent apoptosis and individual tumor cell necrosis with a ‘starry-sky’ appearance or foci of pyknotic, ‘dark’ cells (Figs. 1A–C). The tumor cells contained small to medium-sized round to oval nuclei, with minor variability in size and shape, surrounded by variable amounts of amphophilic, clear or pale eosinophilic cytoplasm, typically more abundant than in ES (Fig. 1D). In contrast to classic ES, nuclei showed vesicular chromatin, with distinct and occasional prominent nucleoli (Figs. 1D, E). Mitotic counts ranged from 6–92 per 10 high-power fields (median count of 40). Some cases showed focally more prominent pleomorphism (Figs. 1D, E) and spindle cell areas (Fig. 1F). Patchy myxoid or edematous stromal change was frequently seen, sometimes with formation of microcystic spaces. No rosette formation was observed.
Figure 1. Pathologic features of CIC-DUX4-positive sarcomas.
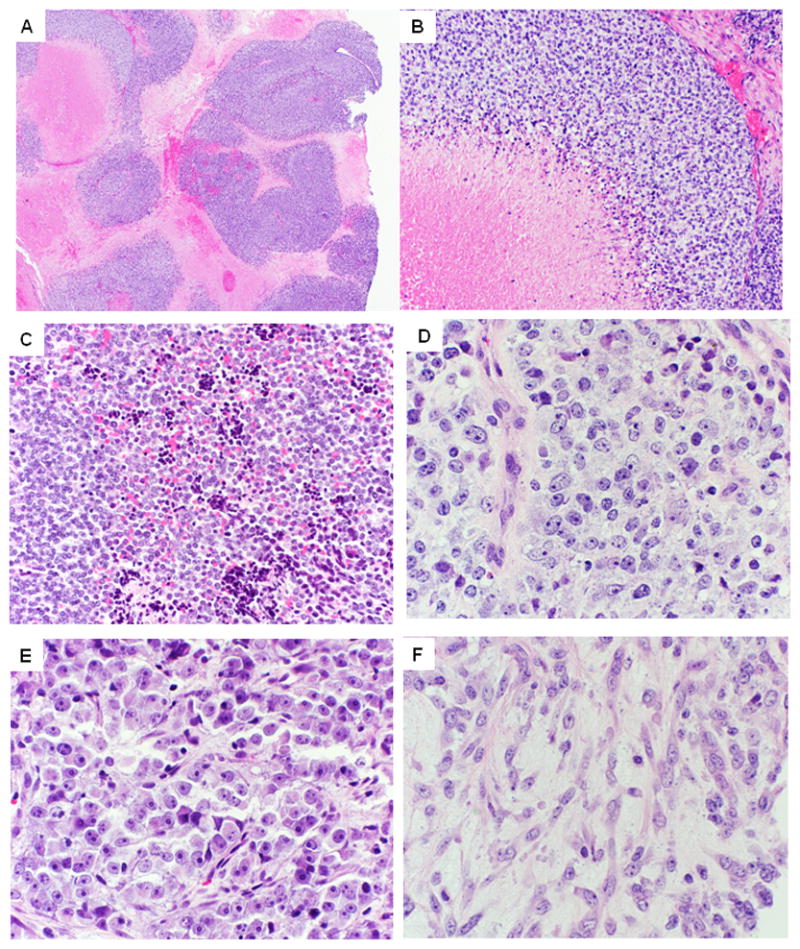
A. At low-power, tumors show a vague nodular growth, often outlined by confluent geographic areas of necrosis (case 10)
B. Higher power show tumor cells arranged in solid sheets, surrounding areas of necrosis (case 18).
C. Admixed with viable cells are often pyknotic, degenerating cells (case 20; x100).
D. Higher power showing ill-defined cell borders with vesicular chromatin and distinct nucleoli. Of note the nuclei show a round, oval to more angulated appearance, with subtle but increased pleomorphism than typical ES (case 16; x200).
E. Case 10 displaying greater variability in nuclear size, prominent nucleoli and more abundant cytoplasm, pushing the nuclei at the periphery.
F. Areas with spindle cell morphology are typically rare and present as a focal phenomenon. However, not uncommon is the presence of a edematous, myxoid stroma (case 16; 200x).
Immunohistochemical Findings
The results of the immunohistochemical studies are summarized in Table 1. Membranous CD99 staining was seen in 18/21 (86%) CIC-DUX4-positive cases (Fig. 2A). Staining was mostly focal and patchy (1+, <25% of the cells) with weak to moderate intensity, as seen in 13 (62%) cases. Three cases (14%) were completely negative for CD99. All cases tested were uniformly and strongly positive for Fli1 (9/9; Fig. 2B). ERG positivity was seen in 2 cases (2/11; 18%), with either focal (1+) and weak or multifocal (3+) and moderate intensity (Fig. 2C). WT-1 showed positivity in all cases tested (20/20), displaying mostly a combined nuclear and cytoplasmic pattern of staining, with moderate to strong intensity in 15/20 cases tested (Fig. 2D); 4 showed only nuclear staining, while one tumor had only cytoplasmic reactivity. All neural and neuroendocrine markers including S-100, GFAP, synaptophysin, chromogranin, as well as all myogenic markers and lymphoid markers tested were negative. Similarly, all epithelial markers were negative, except for one tumor displaying focal Cam5.2 positivity.
Figure 2. Immunohistochemical findings in CIC-DUX4 sarcoma.
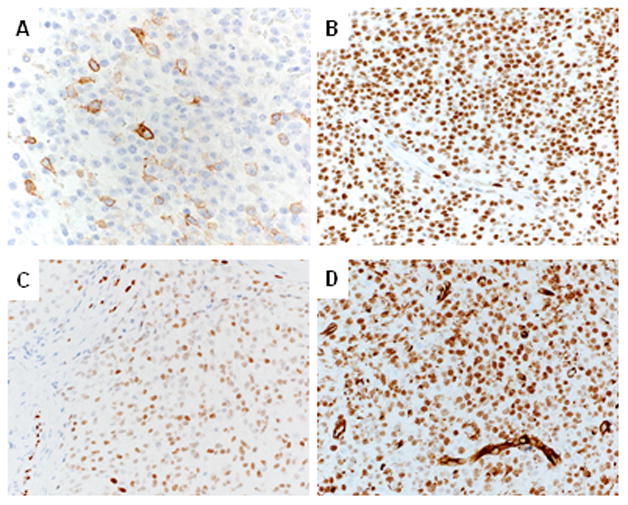
A. CD99 expression is typically focal (1+) and of moderate intensity (case 16).
B. FLI1 labeling is uniformly strongly positive in the majority of tumor cells (case 16).
C. ERG expression is variable, with multifocal (3+) and moderate staining intensity (case 10).
D. WT-1 staining is seen in all CIC-DUX4 – positive sarcomas, with most cases displaying both cytoplasmic and nuclear staining (case 10).
In the ES group, as expected, all tumors showed strong and diffuse membranous CD99 expression (20/20). Strong and diffuse Fli1 nuclear reactivity was equally seen in all EWSR1-FLI1 (n=13) and EWSR1-ERG (n=2) positive ES. ERG positivity was only seen in the 2 EWSR1-ERG but not in EWSR1-FLI1-positive Ewing sarcomas (2/17; 17%). All Ewing tumors assessed were negative for WT1 (0/17).
Gene Expression Signature
We further explored the pathogenetic relationship between CIC-DUX4-positive sarcomas and ES by comparing their transcriptional profiles. The CIC-DUX4 associated gene expression signature of two t(4;19) and three t(10;19) tumors was obtained by comparison with a wide spectrum of soft tissue tumors and normal tissues. By unsupervised clustering the CIC-DUX4-positive sarcomas grouped in a tight genomic cluster from all other sarcomas and normal samples, regardless of the fusion type (Fig. 3A). A 175 gene-list of differentially expressed genes were identified (fold change, FC, >1.4, FDR p<0.05); most of them 144/175 (82%) being over-expressed (Table 2). Interestingly, among the highest ranked upregulated genes were CRF, corticotropin releasing hormone and VGF, VGF nerve growth factor inducible. Other top up-regulated genes were PEA family members of ets transcription factors, ETV4, ETV1 and ETV5, as well as other notable transcription factors with function in sarcomagenesis: ZIC1, HEY1, and WT1 (Table 2, Fig. 3). Additional overexpressed genes were involved either in neuronal (neuronal pentraxin I/II) or skeletal muscle (DLK1) function. Among the tyrosine kinase receptors, TIE1 (tyrosine kinase with immunoglobulin-like and EGF-like domains 1) was significantly upregulated.
Table 2.
CIC-DUX4 Associated transcriptional signatureΩ
| Gene Symbol | U133A Probe ID | Chromosomal Location | Gene Ontology Molecular Function | Gene Title | Log FC | p-value |
|---|---|---|---|---|---|---|
| CRH | 205629_s_at | chr8q13 | hormone activity | corticotropin releasing hormone | 4.8 | 0.006 |
| NPTX2 | 213479_at | chr7q21.3-q22.1 | metal ion binding | neuronal pentraxin II | 4.2 | 0.005 |
| ETV4α | 211603_s_at | chr17q21 | sequence-specific DNA binding transcription factor activity | ets variant 4 | 4.0 | 4.75E-05 |
| HMGA2 | 208025_s_at | chr12q15 | nucleic acid binding transcription factor activity | high mobility group AT-hook 2 | 4.0 | 2.25E-05 |
| DUSP4 | 204014_at | chr8p12-p11 | protein tyrosine phosphatase activity | dual specificity phosphatase 4 | 3.9 | 5.07E-06 |
| VGF | 205586_x_at | chr7q22.1 | neuropeptide hormone activity | VGF nerve growth factor inducible | 3.7 | 0.0005 |
| CDH4 | 206866_at | chr20q13.3 | calcium ion binding | cadherin 4, type 1, R-cadherin (retinal) | 3.3 | 0.008 |
| NPTX1 | 204684_at | chr17q25.3 | metal ion binding | neuronal pentraxin I | 3.2 | 0.0009 |
| DLK1 | 209560_s_at | chr14q32 | calcium ion binding | delta-like 1 homolog (Drosophila) | 3.1 | 0.02 |
| ZIC1 | 206373_at | chr3q24 | sequence-specific DNA binding transcription factor activity | Zic family member 1 | 2.9 | 0.004 |
| CCND2 | 200951_s_at | chr12p13 | protein kinase binding | cyclin D2 | 2.7 | 0.0009 |
| CCNE1 | 213523_at | chr19q12 | transcription co-activator activity /kinase activity | cyclin E1 | 2.7 | 0.016 |
| TIE1 | 204468_s_at | chr1p34-p33 | transmembrane receptor protein tyrosine kinase activity | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | 2.7 | 5.32E-06 |
| ETV5α | 203348_s_at | chr3q28 | sequence-specific DNA binding transcription factor activity | ets variant 5 | 2.5 | 0.016 |
| HEY1 | 218839_at | chr8q21 | sequence-specific DNA binding transcription factor activity | hairy/enhancer-of-split related with YRPW motif 1 | 2.4 | 0.008 |
| ETV1α | 206501_x_at | chr7p21.3 | sequence-specific DNA binding transcription factor activity | ets variant 1 | 2.1 | 0.004 |
| WT1 | 206067_s_at | chr11p13 | sequence-specific DNA binding transcription factor activity | Wilms tumor 1 | 2.1 | 0.10 |
| HOXA5 | 213844_at | chr7p15.2 | sequence-specific DNA binding transcription factor activity | homeobox A5 | −2.1 | 3.67E-05 |
| HOXC6 | 206858_s_at | chr12q13.3 | sequence-specific DNA binding transcription factor activity | homeobox C6 | −2.11 | 5.32E-06 |
| SFRP1 | 202035_s_at | chr8p11.21 | Wnt-protein binding | secreted frizzled-related protein 1 | −2.31 | 0.0003 |
selected from a 175 gene list;
genes up-regulated by overexpression of CIC-DUX4 (Kawamura-Saito et al., 2006);
genes validated by qPCR.
By comparing 5 EWSR1-FLI1 fusion-positive tumors with a large spectrum of normal tissues (FC >1.3 or FC<−1.3 and adjusted p-value (FDR) < 0.1) 854 differentially expressed genes were identified. In parallel, we obtained the transcriptional signature of ES from a previously published comprehensive meta-analysis of ES tumors and cell lines (Hancock and Lessnick, 2008). Subsequently, we cross-referenced the gene lists from these two analyses and obtained 95 overlapping genes, representative of ES signature (Supplementary Table 1), which was then validated by hierarchical clustering of the ES and normal samples (Fig. 3D). The robust 95 gene-list of ES was then cross-referenced to the 175 genes differentially expressed in CIC-DUX4 sarcomas and there were only 2 common genes identified (Fig. 3A).
Using GSEA, the 175 CIC-DUX4 gene-list and the 95 EWSR1-FLI1 gene-signature to the CIC-DUX4-positive cohort and ES group, respectively, high normalized enrichment scores were obtained, of 2.15 and 1.89, respectively, as well as clustered as distinct genomic groups due to their unique signature list (Figs. 3B, D). In contrast, applying CIC-DUX4 and EWSR1-FLI1 signature to ES and SBRCT samples, respectively, the enrichment scores obtained were low, of −0.7 and 0.91, respectively (Figs. 3C, E); suggesting that these signatures do not correlate with the opposite phenotype.
Q-PCR Validation of Top Upregulated Genes
To validate the gene array results as well as screen a larger cohort of CIC-DUX4–positive tumors, real-time quantitative RT-PCR analysis of selected ETV1, ETV4, ETV5 was performed in 8 CIC-DUX4 sarcomas and in 11 ES tumors. Median mRNA expression levels (ratio of target gene/housekeeping gene) were ETV1: 0.32 (range 0.14–0.38), ETV4: 3.42 (range 1.22–7.15), and ETV5 1.54 (range 0.8–2.56) in CIC-DUX-positive cases and ETV1: 0.013 (range 0.001–0.04), ETV4: 0.002 (range 0.0005–0.01), and ETV5 0.051 (range 0.02–0.54) in ES, with no significant difference in EWSR1-FLI1 and EWSR1-ERG-fusion positive cases (Fig. 4). Gene expression level of ETV6, another member of ETS family of transcription members, performed as a control, was not significantly elevated in CIC-DUX4 tumors as compared to ES (data not shown).
Fig. 4.
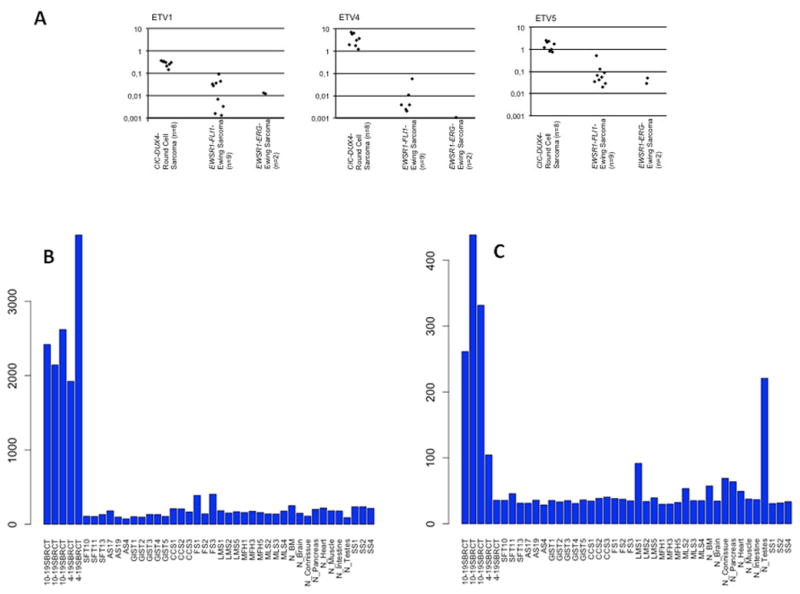
A. Quantitative RT-PCR reveals upregulation of ETV1, ETV4 and ETV5 in CIC-DUX4 sarcomas (n=8) as compared to EWSR1-FLI1 (n=9) or EWSR1-ERG (n=2) rearranged Ewing sarcomas (median ETV1: 24xfold; median ETV4: 1720x-fold; median ETV5: 30x-fold upregulation). B, C. Bar chart of mRNA overexpression of ETV4 (B) and WT1 (C) in CIC-DUX4-postiive tumors compared to other sarcoma types and normal tissue. WT1 shows overexpression mainly in the t(10;19) tumors.
DISCUSSION
Round cell sarcomas harboring a t(4;19)(q35;q13) or a t(10;19)(q26;q13) with CIC-DUX4 fusion are recently described aggressive tumors arising in soft tissues of children and young adults. Due to partial morphologic overlap with Ewing sarcoma (ES) and weak/patchy CD99 expression as the only consistent immunoprofile, they have been referred to as ‘Ewing sarcoma-like’, although they lack the pathognomonic canonical EWSR1-ETS translocation of ES. Whether the group of CIC-DUX4-positive round cell sarcomas represents a stand-alone category of tumors or a subgroup of the Ewing sarcoma family of tumors is still a matter of debate. In the present study we provide novel immunohistochemical and molecular genetic evidence that CIC-DUX4-positive sarcomas represent a pathologic entity distinct from ES.
The first case of sarcoma with a t(4;19)(q35;q13.1) as the sole cytogenetic abnormality was reported by Richkind et al (Richkind et al., 1996) in a 12 year-old boy, who presented with an ankle soft tissue mass and synchronous lung metastases and died of disease within 10 months. Ten years later, Kawamura-Saito et al. identified the genes CIC in chromosome band 19q13 and DUX4 in 4q35 resulting from the t(4;19)(q35;q13) in two round cell sarcoma cases of adults (Kawamura-Saito et al., 2006). More recently, it has been shown that a CIC-DUX4 fusion can result not only from a t(4;19)(q35;q13), but also from a t(10;19)(q26;q13); the DUX4 retrogene being located within a D4Z4 repeat array in the subtelomeric regions of chromosome bands 4q35 and 10q26.3 (Italiano et al., 2012).
To our knowledge, 25 cases with a t(4;19) or a t(10;19) with CIC-DUX4 fusion have been reported in the literature so far (Richkind et al., 1996; Somers et al., 2004; Kawamura-Saito et al., 2006; Yoshimoto et al., 2009; Graham et al., 2012; Italiano et al., 2012; Choi et al., 2013). Together with the 15 new cases described in the current series, the total number of patients with CIC-DUX4-positive sarcomas reported is expanded to 40. Combined clinicopathologic data indicate an almost equal gender distribution (male: female ratio = 1.08), a median age at diagnosis of 26 years (range 6–62 years), a tumor location within soft tissue (>95%), and a high rate of metastatic relapse (75% cases with available follow-up), with lung as the most frequent site of metastasis (65%).
Although CIC-DUX4 round cell sarcomas have been classified under the umbrella of Ewing sarcoma family of tumors, emerging clinicopathologic immunohistochemical and genetic evidence suggest important differences. First, the anatomic distribution is different, with the majority of CIC-DUX4-sarcomas reported so far, including our series, occurring in the deep soft tissue of extremities and trunk, whereas typical ES presents much more common as a bone tumor. Second, the CIC-DUX4-positive tumors mainly affect young adults with a peak incidence in the third decade, with equal gender distribution. In contrast, patients with ES have a mean age at diagnosis of 15 years (Sankar and Lessnick, 2011), with a slight male predominance. Although both tumor entities share an undifferentiated and mostly monotonous cytomorphology, there are subtle but distinctive features associated with CIC-DUX4-positive tumors, such as increased nuclear size and shape variability, vesicular chromatin with focally prominent nucleoli, in addition to more abundant cytoplasm. Myxoid matrix and prominent stromal edema, often present in CIC-DUX4-associated tumors, are usually not seen in ES. In contrast, neural differentiation, i.e. Homer-Wright rosettes, a distinctive if infrequent morphologic feature of primitive neuroectodermal tumor (PNET), are not observed in CIC-DUX4-positive sarcomas.
The immunophenotype of CIC-DUX4-associated tumors is also not identical with ES. Based on our immunohistochemical data, most CIC-DUX4-positive tumors show weak and patchy staining with CD99 (Somers et al., 2004; Kawamura-Saito et al., 2006; Rakheja et al., 2008; Yoshimoto et al., 2009; Italiano et al., 2012; Graham et al., 2012). In contrast, ES shows diffuse and strong membranous reactivity with CD99. The MIC2 membrane-associated glycoprotein of ES cells is recognized by a number of monoclonal antibodies including 12E7, HBA71, O13, and HO36-1.1. Although CD99 immunopositivity has been documented in a significant subset of small blue round cell tumors (but mostly cytoplasmic, weak), including lymphoblastic leukemia and lymphoma (Riopel et al., 1994), rhabdomyosarcoma (Stevenson et al., 1994), poorly differentiated synovial sarcoma (Dei Tos et al., 1995), desmoplastic round cell tumor (Ordonez, 1998) and small cell osteosarcoma (Devaney et al., 1993), it continues to be an important and helpful marker in the diagnosis of ES.
WT1 staining was observed in all CIC-DUX4-positive tumors examined, with a diffuse (3+ or 4+) and moderate to strong nuclear or nuclear and cytoplasmic pattern. Only one case showed exclusively diffuse cytoplasmic staining (3+). In contrast, WT1 immunohistochemistry was negative in all ES tested (0/17), confirming previous reports of absence of WT1 staining in Ewing family of tumors (Barnoud et al., 2000). WT1 is a diagnostic immunomarker best known for its utility in the diagnosis of Wilms tumor, desmoplastic small round cell tumor and mesothelioma (Grubb et al., 1994; Amin et al., 1995; Charles et al., 1997). Due to its absence in ES, the use of WT1 antibody can help in the differential diagnosis with other small round cell tumors in the appropriate clinical, morphological and immunohistochemical context. Our finding of consistent WT1 immunoreactivity in CIC-DUX4-positive sarcomas may serve as a useful marker in the differential diagnosis of ES, when molecular confirmation is not available. WT1 protein overexpression is most likely due to its transcriptional upregulation (see below) and not secondary to genetic abnormalities or rearrangements of the WT1 gene locus, excluded by WT1 FISH in 8 CIC-rearranged tumors, including 6 of the 7 tumors lacking DUX4 gene abnormalities (data not shown).
Based on our initial observation of FLI1 and ERG immunoreactivity in cases of CIC-DUX4 sarcomas, we systematically examined the expression of these two markers in this series. Anti-FLI1 antibody labeled all eight CIC-DUX4 cases with a diffuse and strong labeling pattern (8/8). Similarly, diffuse and strong nuclear FLI1 expression was present in the 15 ES investigated, regardless of fusion type. The latter findings are similar to those of Folpe et al. (Folpe et al., 2000) and Wang et al. (Wang et al., 2012) who also found strong FLI1 expression in ES, regardless of its EWSR1 fusion partner. As noted previously (Wang et al., 2012), one possible explanation for this phenomenon is the cross-reactivity of FLI1 antibody with the highly conserved and homologous Ets DNA binding domain present in the C-terminus of both FLI and ERG. Whether strong FLI1 expression observed in CIC-DUX4 sarcoma reflects ‘true’ FLI1 protein overexpression or activation of related Ets proteins remains debatable; our gene expression data from CIC-DUX4-sarcomas, however, favors the latter possibility. Using a specific anti-ERG monoclonal antibody, ERG positivity was demonstrated in 2/11 CIC-DUX4 sarcoma cases (18%), with either focal (1+) and weak, or multifocal (3+) with moderate intensity. In comparison, strong ERG staining was only seen in EWSR1-ERG-positive Ewing sarcoma (2/2; 100%), but not in EWSR1-FLI1-positive Ewing sarcoma (0/15).
Finally, the molecular genetic abnormalities of CIC-DUX4-positive sarcomas are different from Ewing sarcoma family tumors. ES tumors harbor the canonical fusion between EWSR1 and ETS family members; the resulting chimeric genes encode aberrant transcription factors that play a crucial role in their pathogenesis (Sankar and Lessnick, 2011). In contrast, CIC-DUX4 fusion appears functionally unrelated to EWSR1-ETS. CIC is the human homologue of Drosophila capicua, a gene identified in a screen for mutations affecting the anterior-posterior pattern of Drosophila embryos (Jimenez et al., 2000). It encodes a transcriptional repressor with a high-mobility group (HMG)-box containing DNA binding domain. CIC gene abnormalities have been implicated in neoplasia - for example, CIC loss of function mutations are identified in 83% of oligodendrogliomas (Bettegowda et al., 2011, Sahm et al., 2012). The DUX4 gene is normally expressed in germ cells and is epigenetically silenced in somatic differentiated tissues. Aberrant expression of DUX4 has been implicated in the development of facioscapulohumeral muscular dystrophy (van der Maarel et al., 2011).
Despite different and functionally unrelated genes involved in the translocation, a possible pathogenetic link between CIC-DUX4 sarcoma and Ewing family of tumors is suggested by our gene expression results. Although the transcriptional profile of CIC-DUX4-positive sarcomas is distinct from that of ES and other sarcoma subtypes, in keeping with them being separate tumor entities, CIC-DUX4 fusion overexpresses three ETS transcription factors (ETV4, ETV1 and ETV5) from the PEA3 (polyoma enhancer activator 3) subfamily (Sankar and Lessnick, 2011). Thus, up-regulation of the PEA3 family genes by CIC-DUX4 could serve as an equivalent molecular change to the EWSR1-ETS fusion. Interestingly, ETV1 and ETV4 can serve as fusion partners to EWSR1 in rare cases of ES (Sankar and Lessnick, 2011). In analogy to EWS/ETS fusion-positive Ewing sarcoma, one might speculate that CIC-DUX4 fusion induces an aberrant transcriptional program with deregulation of overlapping/converging or functionally similar key oncogenic target genes, thereby sharing a basic pathogenesis with Ewing sarcoma. It is of interest in this context that 92% of the reported ES rare variant translocations (EWSR1-ETV1, EWSR1-ETV4 or EWSR1-FEV) occurred at extraskeletal sites (Wang et al., 2007). Whether this is a reporting bias or represents a specific feature of these rare translocation variants remains undetermined at this time. Our study confirms and extends the previous report of upregulation of ETV5 and ETV1 (but not of ETV4) genes by CIC-DUX4 fusion (Kawamura-Saito et al., 2006). Using an experimental cell line model system, Kawamura-Saito et al. demonstrated binding of the HMG-box of CIC to a DNA sequence within the promoter of PEA genes ETV1 and ETV5. Their results further revealed that fusion of DUX4 to CIC sequence provides strong transcriptional activity, resulting in mostly upregulated gene expression, with minimal down-regulated genes, in keeping with our own results.
Apart from ETS family members, other genes upregulated in CIC-DUX4-positive sarcomas include transcription factors such as HMGA2, ZIC1, HEY1 and WT1, previously shown to play a pathogenetic role in other sarcoma types. There was no transcriptional overlap between the CIC-DUX4 and ES gene expression signature.
In summary, CIC-DUX4-positive round cell sarcomas have a unique clinical presentation, morphology, immunoprofile and genetic signature that are different from ES. CIC-DUX4-positive sarcomas occur almost exclusively in deep soft tissue of young adults. The immunoprofile comprises weak and focal CD99 reactivity and diffuse nuclear WT1. Frequent FLI1 and occasional ERG immunoreactivity observed in CIC-DUX4 sarcoma are similarly seen in ES. The gene signature of CIC-DUX4-positive sarcomas separates them from classic ES and justifies designation as a “stand alone” category.
Supplementary Material
Acknowledgments
Supported in part by: Wilhelm-Sander-Stiftung (KS), TransSaRNet; 01GM1104B of the BMBF, Germany (GHSR), P01CA47179 (CRA), P50 CA 140146-01 (CRA), Kristen Ann Carr Fund (CRA).
We thank Brigit Geist for expert technical assistance and Milagros Soto for excellent editorial assistance.
References
- Amin KM, Litzky LA, Smythe WR, Mooney AM, Morris JM, Mews DJ, Pass HI, Kari C, Rodeck U, Rauscher FJ, Kaiser LR, Albelda SM. Wilms’ tumor 1 susceptibility (WT1) gene products are selectively expressed in malignant mesothelioma. Am J Pathol. 1995;146:344–356. [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Viale A, Sarran L, Tschernyavsky SJ, Gonen M, Segal NH, Maki RG, Socci ND, DeMatteo RP, Besmer P. Gene expression in gastrointestinal stromal tumors is distinguished by KIT genotype and anatomic site. Clin Cancer Res. 2004;10:3282–3290. doi: 10.1158/1078-0432.CCR-03-0715. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Yoshida A, Guo T, Chang NE, Zhang L, Agaram NP, Qin LX, Brennan MF, Singer S, Maki RG. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009;69:7175–7179. doi: 10.1158/0008-5472.CAN-09-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnoud R, Sabourin JC, Pasquier D, Ranchere D, Bailly C, Terrier-Lacombe MJ, Pasquier B. Immunohistochemical expression of WT1 by desmoplastic small round cell tumor: a comparative study with other small round cell tumors. Am J Surg Pathol. 2000;24:830–836. doi: 10.1097/00000478-200006000-00008. [DOI] [PubMed] [Google Scholar]
- Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, Velculescu VE, Oba-Shinjo SM, Marie SK, Vogelstein B, Bigner D, Yan H, Papadopoulos N, Kinzler KW. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles AK, Moore IE, Berry PJ. Immunohistochemical detection of the Wilms’ tumour gene WT1 in desmoplastic small round cell tumour. Histopathology. 1997;30:312–314. doi: 10.1046/j.1365-2559.1997.d01-620.x. [DOI] [PubMed] [Google Scholar]
- Choi EY, Thomas DG, McHugh JB, Patel RM, Roulston D, Schuetze SM, Chugh R, Biermann JS, Lucas DR. Undifferentiated Small Round Cell Sarcoma With t(4;19)(q35;q13.1) CIC-DUX4 Fusion: A Novel Highly Aggressive Soft Tissue Tumor With Distinctive Histopathology. Am J Surg Pathol. 2013;37:1379–1386. doi: 10.1097/PAS.0b013e318297a57d. [DOI] [PubMed] [Google Scholar]
- Dei Tos AP, Wadden C, Calonje E, Sciot R, Pauwels P, Knight JC, Dal Cin P, CDF Immunohistochemical demonstration of gylcoprotein p30/32(MIC2) (CD99) in synovial sarcoma: a potential cause of diagnostic confusion. Applied Immunohistochemistry. 1995;3:168–173. [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Devaney K, Vinh TN, Sweet DE. Small cell osteosarcoma of bone: an immunohistochemical study with differential diagnostic considerations. Hum Pathol. 1993;24:1211–1225. doi: 10.1016/0046-8177(93)90218-6. [DOI] [PubMed] [Google Scholar]
- Fletcher CDM World Health Organization., International Agency for Research on Cancer. WHO classification of tumours of soft tissue and bone. 4. Lyon: IARC Press; 2013. p. 468. [Google Scholar]
- Folpe AL, Hill CE, Parham DM, O’Shea PA, Weiss SW. Immunohistochemical detection of FLI-1 protein expression: a study of 132 round cell tumors with emphasis on CD99-positive mimics of Ewing’s sarcoma/primitive neuroectodermal tumor. Am J Surg Pathol. 2000;24:1657–1662. doi: 10.1097/00000478-200012000-00010. [DOI] [PubMed] [Google Scholar]
- Graham C, Chilton-MacNeill S, Zielenska M, Somers GR. The CIC-DUX4 fusion transcript is present in a subgroup of pediatric primitive round cell sarcomas. Hum Pathol. 2012;43:180–189. doi: 10.1016/j.humpath.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Grubb GR, Yun K, Williams BR, Eccles MR, Reeve AE. Expression of WT1 protein in fetal kidneys and Wilms tumors. Lab Invest. 1994;71:472–479. [PubMed] [Google Scholar]
- Hadju M, Singer S, Maki RG, Schwartz G, Keohan M, Antonescu CR. IGF2 over-expression in solitary fibrous tumours is independent of anatomical location and is related to loss of imprinting. J Pathol. 2010;221:300–307. doi: 10.1002/path.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7:250–256. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- Hauer K, Calzada-Wack J, Steiger K, Grunewald TG, Baumhoer D, Plehm S, Buch T, Prazeres da Costa O, Esposito I, Burdach S, Richter GH. DKK2 mediates osteolysis, invasiveness, and metastatic spread in Ewing sarcoma. Cancer Res. 2013;73:967–977. doi: 10.1158/0008-5472.CAN-12-1492. [DOI] [PubMed] [Google Scholar]
- Italiano A, Sung YS, Zhang L, Singer S, Maki RG, Coindre JM, Antonescu CR. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51:207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon IS, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- Jimenez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, Takizawa T, Nakamura T. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- Ng TL, O’Sullivan MJ, Pallen CJ, Hayes M, Clarkson PW, Winstanley M, Sorensen PH, Nielsen TO, Horsman DE. Ewing sarcoma with novel translocation t(2;16) producing an in-frame fusion of FUS and FEV. J Mol Diagn. 2007;9:459–463. doi: 10.2353/jmoldx.2007.070009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez NG. Desmoplastic small round cell tumor: II: an ultrastructural and immunohistochemical study with emphasis on new immunohistochemical markers. Am J Surg Pathol. 1998;22:1314–1327. doi: 10.1097/00000478-199811000-00002. [DOI] [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- Rakheja D, Goldman S, Wilson KS, Lenarsky C, Weinthal J, Schultz RA. Translocation (4;19)(q35;q13.1)-associated primitive round cell sarcoma: report of a case and review of the literature. Pediatr Dev Pathol. 2008;11:239–244. doi: 10.2350/07-06-0296.1. [DOI] [PubMed] [Google Scholar]
- Richkind KE, Romansky SG, Finklestein JZ. t(4;19)(q35;q13.1): a recurrent change in primitive mesenchymal tumors? Cancer Genet Cytogenet. 1996;87:71–74. doi: 10.1016/0165-4608(95)00240-5. [DOI] [PubMed] [Google Scholar]
- Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, Hotfilder M, Lowel D, von Luettichau I, Mossbrugger I, Quintanilla-Martinez L, Kovar H, Staege MS, Muller-Tidow C, Burdach S. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riopel M, Dickman PS, Link MP, Perlman EJ. MIC2 analysis in pediatric lymphomas and leukemias. Hum Pathol. 1994;25:396–399. doi: 10.1016/0046-8177(94)90149-x. [DOI] [PubMed] [Google Scholar]
- Sahm F, Koelsche C, Meyer J, Pusch S, Lindenberg K, Mueller W, Herold-Mende C, von Deimling A, Hartmann C. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol. 2012;123:853–860. doi: 10.1007/s00401-012-0993-5. [DOI] [PubMed] [Google Scholar]
- Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204:351–365. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal NH, Pavlidis P, Antonescu CR, Maki RG, Noble WS, DeSantis D, Woodruff JM, Lewis JJ, Brennan MF, Houghton AN, Cordon-Cardo C. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing DC, McMullan DJ, Roberts P, Smith K, Chin SF, Nicholson J, Tillman RM, Ramani P, Cullinane C, Coleman N. FUS/ERG gene fusions in Ewing’s tumors. Cancer Res. 2003;63:4568–4576. [PubMed] [Google Scholar]
- Somers GR, Shago M, Zielenska M, Chan HS, Ngan BY. Primary subcutaneous primitive neuroectodermal tumor with aggressive behavior and an unusual karyotype: case report. Pediatr Dev Pathol. 2004;7:538–545. doi: 10.1007/s10024-004-2024-6. [DOI] [PubMed] [Google Scholar]
- Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- Stevenson AJ, Chatten J, Bertoni F, MM CD99 (p30/32MIC2) neuroectodermal/Ewing’s sarcoma antigen as an immunohistochemical marker: review of more than 600 tumors and the literature experience. Applied Immunohistochemistry. 1994;2:231–240. [Google Scholar]
- Urano F, Umezawa A, Hong W, Kikuchi H, Hata J. A novel chimera gene between EWS and E1A-F, encoding the adenovirus E1A enhancer-binding protein, in extraosseous Ewing’s sarcoma. Biochem Biophys Res Commun. 1996;219:608–612. doi: 10.1006/bbrc.1996.0281. [DOI] [PubMed] [Google Scholar]
- van der Maarel SM, Tawil R, Tapscott SJ. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bhargava R, Zheng T, Wexler L, Collins MH, Roulston D, Ladanyi M. Undifferentiated small round cell sarcomas with rare EWS gene fusions: identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. J Mol Diagn. 2007;9:498–509. doi: 10.2353/jmoldx.2007.070053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WL, Patel NR, Caragea M, Hogendoorn PC, Lopez-Terrada D, Hornick JL, Lazar AJ. Expression of ERG, an Ets family transcription factor, identifies ERG-rearranged Ewing sarcoma. Mod Pathol. 2012;25:1378–1383. doi: 10.1038/modpathol.2012.97. [DOI] [PubMed] [Google Scholar]
- Wang X, Cairns MJ. Gene set enrichment analysis of RNA-Seq data: integrating differential expression and splicing. BMC Bioinformatics. 2013;14(Suppl 5):S16. doi: 10.1186/1471-2105-14-S5-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Graham C, Chilton-MacNeill S, Lee E, Shago M, Squire J, Zielenska M, Somers GR. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195:1–11. doi: 10.1016/j.cancergencyto.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Triche TJ, Sheer D, Turc-Carel C, et al. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. Embo J. 1993;12:4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.